Abstract
Escherichia coli BL21(DE3) has long served as a model organism for scientific research, as well as a workhorse for biotechnology. Here we present the most current genome annotation of E. coli BL21(DE3) based on the transcriptome structure of the strain that was determined for the first time. The genome was annotated using multiple automated pipelines and compared to the current genome annotation of the closely related strain, E. coli K-12. High-resolution tiling array data of E. coli BL21(DE3) from several different stages of cell growth in rich and minimal media were analyzed to characterize the transcriptome structure and to provide supporting evidence for open reading frames. This new integrated analysis of the genomic and transcriptomic structure of E. coli BL21(DE3) has led to the correction of translation initiation sites for 88 coding DNA sequences and provided updated information for most genes. Additionally, 37 putative genes and 66 putative non-coding RNAs were also identified. The panoramic landscape of the genome and transcriptome of E. coli BL21(DE3) revealed here will allow us to better understand the fundamental biology of the strain and also advance biotechnological applications in industry.
INTRODUCTION
Genome sequence with an accurate annotation is now almost essential for traditional experimental and systems biological studies (1,2), and interpretation and analysis of multi-omics data depend heavily on the quality of annotation. Genome re-annotation aims to ensure that the descriptions and locations of all identifiable genes are as accurate and most up-to-date as possible (3). Deciphering the precise locations of open reading frames (ORFs) and their functions requires constant re-evaluation and correction as new data become available. The most common new data emerging nowadays include (i) translation initiation sites (TISs) revealed by high-throughput and high-resolution transcriptome and proteome analysis, (ii) experimental data and newly identified sequence-function information that can allocate specific functions to genes previously annotated as ‘(conserved) hypothetical proteins’, (iii) predicted genes without any transcription and/or protein expression evidences can be eliminated and (iv) the identification of genes and non-coding RNAs (ncRNAs) overlooked in the original annotation. Various automated genome annotation pipelines such as Integrated Microbial Genomes (IMG) (4) and Rapid Annotations using Subsystems Technology (RAST) (4) have been developed. However, the number, size and description of predicted ORFs can vary widely depending on the algorithm used (3,5). It is thus important to correct outdated genome annotation as complete as possible, particularly for vital model organisms like Escherichia coli BL21(DE3).
Escherichia coli BL21(DE3) has long been a mainstay of numerous biotechnological applications, most notably recombinant protein production. Several prominent features of the BL21(DE3) strain make it ideal for its role as an industrial host, including fast cell growth in minimal media, low acetate production when grown on high levels of glucose, low protease abundance and an amenability to high-density culture (6). E. coli K-12 strains have been widely used for genetic analysis in laboratory settings, and international efforts have been dedicated to updating the annotation of the K-12 genome (7–9). In 2009, the genome sequences of two E. coli B strains, BL21(DE3) and REL606, were the first to be completed and annotated (10,11), although we have made slight modification to the genome sequence by adding a new copy of insertion sequence. Hereafter, we refer to the current release of GenBank entry (CP001509.3) as the original annotation of E. coli BL21(DE3). This near complete genome annotation has enabled multifaceted holistic analyses of E. coli BL21(DE3) using genomics, transcriptomics, proteomics and phenomics (12–15). An up-to-date description of the BL21(DE3) genome is particularly important to the biotechnological community as this strain is one of the most commonly used host strains for various biotechnological applications in academia and industry.
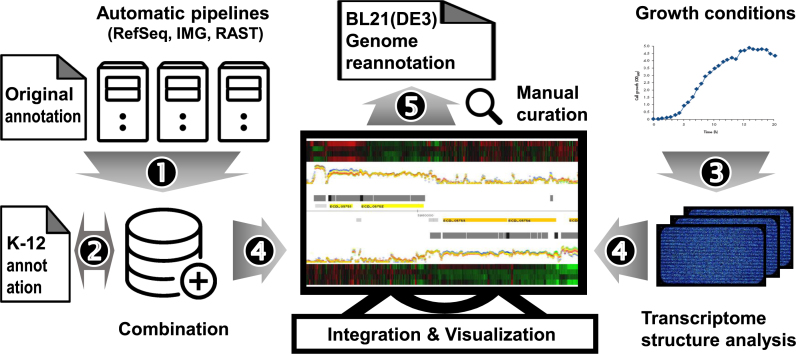
In this study, we report an updated, consolidated annotation for E. coli BL21(DE3) genome, performed through an integrated analysis of genome and transcriptome structure (Figure 1). We integrated de novo genome annotation from three different annotation pipelines with the original annotation. Gene boundaries and gene product descriptions were revised and putative novel ORFs and ncRNAs that were not annotated in the original analysis were identified. Transcriptome profiles were generated from different stages of growth in batch fermentations using rich and minimal media. Through high-resolution analysis of the transcriptome structure, transcription units (TUs) of mono- and poly-cistronic mRNAs, including ncRNAs and transcripts not described in the original genome annotation, were identified. All of these data were used to re-annotate the E. coli BL21(DE3) genome, which will be invaluable for biological and biotechnological studies on this strain of scientific and industrial importance.
Figure 1.
Overview of the genome re-annotation of Escherichia coli BL21(DE3) based on sequence homology and experimental evidence. Results from three automatic genome annotation pipelines (RefSeq, IMG and RAST) were combined (1) and compared to the high-quality genome annotation of the closely related E. coli strain K-12 MG1655 (2). Total RNAs from various growth stages and culture conditions were directly labeled with fluorescence and hybridized to high-resolution tiling arrays (3). Known and predicted ORFs were mapped to genome coordinates, along with the strand-specific transcriptome data (4) and then manually curated (5).
MATERIALS AND METHODS
Genome annotation update
The annotation results from three annotation pipelines were integrated with the original annotation (GenBank: CP001509.3) that we had previously generated (10). The genome sequence of E. coli BL21(DE3) was queried with the IMG Expert Review (IMG/ER) (4) and RAST servers (16). These two annotation results and the current release of RefSeq annotation re-annotated by the NCBI Prokaryotic Genome Annotation Pipeline (NC_012971.2) (17) were downloaded as GenBank flat files. These were parsed using custom Perl scripts to integrate features.
Bacterial strain and growth conditions
Escherichia coli strain BL21(DE3) was provided by F William Studier, Brookhaven National Laboratory (18). Cells were grown in LB medium or modified R (MR) medium. The MR medium (pH 7.0) contained 10 g/l glucose, 4 g/l (NH4)2HPO4, 6.67 g/l KH2PO4, 0.8 g/l citric acid, 0.8 g/l MgSO4·7H2O and 5 ml/l trace metal solution (19). The trace metal solution contained 0.5 M HCl, 10 g/l FeSO4·7H2O, 2.2 g/l ZnSO4·7H2O, 1 g/l CuSO4·5H2O, 0.5 g/l MnSO4·4H2O, 0.02 g/l Na2B4O7·10H2O, 2 g/l CaCl2 and 0.1 g/l (NH4)6MO7O24·4H2O.
Seed cultures were prepared by growing cells in 125 ml flasks containing 25 ml of medium at 37°C and 200 rpm for 12 h. Next, 10 ml of seed culture was transferred to a 2.5 l bioreactor (BioFlo 310, New Brunswick Scientific, Edison, NJ, USA) containing 1 l of medium. pH was maintained at 7.0 by the automatic feeding of 25% (v/v) NH4OH. The dissolved oxygen concentration was kept above 40% air saturation by supplying air (1.5 l/min) and automatically varying agitation speed between 300 and 800 rpm. Cell growth was monitored by measuring absorbance at 600 nm (OD600). The concentrations of glucose, acetate, lactate and other organic acids in the culture supernatants were measured using an Agilent 1260 Infinity HPLC (Agilent Technologies, Santa Clara, CA, USA) equipped with an ion exchange column (Aminex HPX-87H, 300 × 7.8 mm, Bio-Rad Laboratories, Hercules, CA, USA).
Construction of a high-resolution tiling microarray, RNA hybridization and image analysis
A whole-genome high-resolution tiling array was designed, containing 957 515 60-mer probes with strand-specific sequences, in addition to the manufacturer's included controls (Agilent custom GE microarray 1 × 1M). Probes were tiled every 10 bp (i.e. 50 bp overlap between adjacent probes) for E. coli BL21(DE3) (10). To accurately analyze regions upstream of each ORF, probes were added with a tiling resolution of 4 nt. As a control for non-specific background hybridization, 10 000 negative control probes (NCPs) were added. These NCPs were designed by random concatenation of 12-mers absent from the genome, followed by random concatenation of five such 12-mers. This ensured that each NCP had at least five mismatches relative to any 60 nt stretch of the BL21(DE3) genome (20).
Culture broth (4.0 × 108 cells) was immediately mixed with two volumes of RNAprotect Bacteria Reagent (Qiagen, Düsseldorf, Germany). Bacterial cells were lysed with lysozyme for 5 min at 25°C. Total RNA was prepared using a mirVana miRNA Isolation Kit (Thermo Fisher Scientific Inc., Waltham, MA, USA), per the manufacturer's instruction. Any contaminating DNA was digested with Turbo DNase (Applied Biosystems, Austin, TX, USA) for 30 min at 37°C and then removed using the filter cartridges supplied with the mirVana miRNA Isolation Kit. The quantity and purity of the purified RNA was determined by bioanalyzer (Agilent Technologies, Palo Alto, CA, USA), and high quality RNAs (those with an RNA integrity number (RIN) > 7.5) were used in subsequent array tests.
The resulting RNA was directly labeled with Cy3 using a Label IT μArray Labeling Kit (Mirus, Madison, WI, USA), per the manufacturer's instruction. Cy3-labeled RNA was purified using Quick Spin columns (Roche, Basel, Switzerland) and then fragmented through incubation with blocking agent and fragmentation buffer for 30 min at 60°C. The fragmented and labeled RNA was hybridized with the tiling array for 17 h at 65°C. After the hybridization and wash steps described in the array manufacturer's instructions, arrays were scanned using an Agilent DNA microarray Scanner (Agilent Technologies). Signal intensity and local background were determined using Feature Extraction software (Agilent Technologies).
Identification of transcription units
Probe intensities from the tiling array tests were quantile-normalized to ensure all the arrays had an equivalent intensity distribution. The 10 000 NCPs were used to evaluate the statistical significance of expression of the individual probes. A TranscriptionDetector algorithm (20) was used to determine probes that were expressed above the background level with a false discovery rate (FDR) of 0.01. To assess statistical significance of expression for each locus, a P-value estimating the likelihood of over-representation of the expressed probes for each locus was calculated based on the cumulative hypergeometric distribution. These P-values were corrected for multiple comparison testing by the Bonferroni method. Loci with a P-value < 0.01 were considered expressed.
Adjacently expressed probes were concatenated to detect transcriptionally active genomic regions. To reduce false positives arising from use of the TranscriptionDetector algorithm, short areas below 100 bp in length were removed. If there were densely expressed regions in the opposite strand, orphan calls of the main strand were also removed (21). Array data and computational calculations were plotted against coordinates on the genome, and TUs were manually inspected and curated through an interactive exploration process using the Gaggle Genome Browser (GGB) (22).
Gene expression analysis
Log2-transformed ratios were calculated for each probe (stationary phase/exponential phase in LB or MR media), and a median value of the expression ratios of probes was assigned to each loci. Likewise, a median value of the intensities of probes was assigned to each loci at each of the growth conditions. For functional enrichment analysis, amino acid sequences of coding DNA sequences (CDSs) were assigned to the latest version of clusters of orthologous groups (COGs) (23) using COG software (24). When multiple COGs were assigned to a single query protein, the best was chosen on the basis of cognitor score.
RESULTS
Genome re-annotation by integration of multiple sources
The genome annotations from four different sources were consolidated for further analyses in this study (Supplementary Table S1). The ORFs identified redundantly in the four sources were annotated based on the genomic location of the stop codon. All the identified ORFs were classified according to co-occurrence across each of the annotation sources (Supplementary Figure S1). Three annotation pipelines and original annotation reported 3949 genes in common and 3243 of them (82.1%) were congruent in terms of size. As for each of the remaining incongruent genes, the TIS which was the farthest site from the stop codon was chosen. All the TISs were further verified and determined by comparison with those of E. coli K-12 genes, followed by inspection of tiling array data (see below). List of tRNA and rRNA was almost identical between the four annotation sources, and remained the same as the original annotation. The IMG/ER server also predicted 120 miscellaneous ‘other’ RNAs (termed misc_RNA). Newly characterized pseudogenes identified using RefSeq (NC_012971.2) were added to those previously described. Our newly integrated annotation led to the identification of 4872 feature loci, including 4559 CDSs, 22 rRNAs, 85 tRNAs, 86 pseudogenes and 120 miscellaneous other RNA (misc_RNAs) (Supplementary Table S1). Also, 536 putative novel loci were predicted (400 CDSs, 16 pseudogenes and 120 misc_RNAs).
The genome annotation of the closely related E. coli K-12 MG1655 (7,8), was used as a gold standard for the curation of the BL21(DE3) genome re-annotation. The genomes of E. coli BL21(DE3) and K-12 MG1655 are similar in terms of overall genetic organization and sequence identity, although there are several notable differences due to IS-element activity, the decay of cryptic prophages and horizontal acquisition of genetic islands (10,25). Each of the identified loci were compared to those in the latest genome annotation of K-12 MG1655 (GenBank™ U00096.3) in terms of their size, name and genomic location. Among the 4159 CDSs in the original annotation, 660 gene names and most gene products required updating (Supplementary Table S2). We inspected the inconsistent annotations manually, and found that K-12 annotation was more elaborate than the original annotation of E. coli BL21(DE3) without exception.
Determination of the transcriptome structure
Using statistical analysis of the expression of probes complementary to genomic sites, the TUs for the majority of genes and ncRNAs in E. coli BL21(DE3) were determined (Table 1). For the analysis, five samples were taken from batch fermentations in complex LB medium (two exponential phases and one stationary phase) and minimal MR medium (one exponential phase and one stationary phase) (Supplementary Figure S2). Samples representing the key phases of the E. coli growth curve were selected. For example, the two samples taken during the exponential phase in LB medium (cell densities of 0.24 and 1.54 in OD600) are distinct in terms of their carbon source usage (26).
Table 1. Summary of the genome and transcriptome structures of E. coli BL21(DE3).
| Genome structure | |
|---|---|
| Genome size | 4 558 953 bp |
| No. previous ORFsa | 4336 ea |
| No. added genes | 37 ea |
| No. added ncRNAs | 66 ea |
| No. modified TISs | (88 ea) |
| No. total features | 4439 ea |
| Transcriptome structure | |
| TU coverage (total)b | 3 461 841 bp (75.9%) |
| TU coverage (forward strand) | 1 766 138 bp (38.7%) |
| TU coverage (reverse strand) | 1 879 144 bp (41.2%) |
| No. TUs | 1609 ea |
aThe number is the sum of 4159 CDSs, 70 pseudogenes, 22 tRNAs and 85 rRNAs which were reported from the original annotation.
bPercentage of transcription unit (TU) coverage is the total length of TUs divided by the genome size.
Total RNA was labeled directly with fluorescence to avoid the synthesis of cDNA artefacts that can be introduced by reverse transcription and amplification in conventional transcriptome assays (27–29). In addition, one-color gene expression platform was used for genome-wide detection of transcript levels based on the absolute intensities of the expressed probe, rather than the traditional two-color, ratio-based microarray platform (30). TUs were identified based on the proximity of expressed probes (FDR < 0.01) and short regions of <100 bp were removed. Each of the annotated features, tiling array data and computational results were plotted against genome coordinates using the GBB (22). The computationally predicted 2925 TUs were manually inspected and curated (Figure 2). As the probes were tiled every 10 bp, and additional probes in the upstream regions of each gene were added with a tiling resolution of 4 bp, the resolution of the TU boundaries is ∼10 bp.
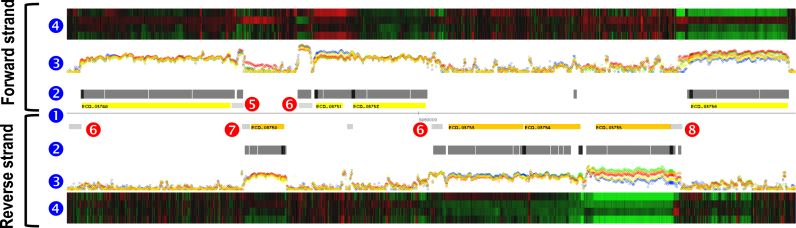
Figure 2.
Examples of integrating the genome re-annotation and transcriptome data of Escherichia coli BL21(DE3). Annotated features and tiling array data were plotted against genome coordinates (13.6 kb genomic region from 3 953 400 to 3 967 000) using a genome browser. This example showcases the value of strand-specific transcriptome analysis. Genes in the forward and reverse strands (1) are denoted in yellow and orange, respectively. Corresponding transcriptome data are aligned above the forward strand and below the reverse strand. The black bars (2) denote significantly expressed probes (FDR < 0.01). Dots (3) represent the normalized probe intensities on a log2 scale at the corresponding genomic location for samples from LB media (cell density of 0.24 in OD600 is colored red, 1.54 orange, 5 yellow) and MR media (1.5 is colored green, 14 blue). The heat map (4) shows transcript level changes (log2 scale) of four samples against a reference sample from MR media (1.5 in OD600) (red is upregulated; green downregulated). In the gene panel, newly identified features are shown in light gray. These included non-coding RNAs (5), ORFs (6), repetitive extra-genic palindrome elements (7) and 5΄ untranslated regions (8).
This analysis identified 1609 transcriptional units covering 76% of the genome, with an average length of 2.3 kb (Supplementary Table S3). Among them, 781 were poly-cistronic transcripts and 828 were mono-cistronic. The strand-specific transcriptome profiles for the five different culture conditions revealed dynamic changes in genome-wide transcription patterns for both coding and non-coding regions (Figure 2).
Discovery of putative novel genes and ncRNAs
The genome re-annotation project generated 536 features within intergenic and antisense regions that had not been previously notified. These features were queried using BLASTX against the nr database. Most features matched hypothetical or conserved proteins. Therefore, through manual inspection using GBB and EcoCyc databases (9), we examined equivalent K-12 MG1655 genome coordinates to identify any features located at the corresponding positions of the putative ORFs or ncRNAs predicted in BL21(DE3). This effort led to the identification of 37 genes and 66 non-coding RNAs. Remaining loci were removed as they were identified as repetitive extragenic palindromic elements (short base-pair sequences capable of producing stem-loop structures, 51 ea) or other repetitive sequences (9). Loci were also removed if they overlapped with ORFs reported in the original annotation (59), were adjacent to the reported CDSs and thus likely to be 5΄ UTR or 3΄ UTR (234) or were not expressed in any culture conditions (80). All pseudogenes newly reported from RefSeq were removed as they were related to hypothetical proteins or phage proteins.
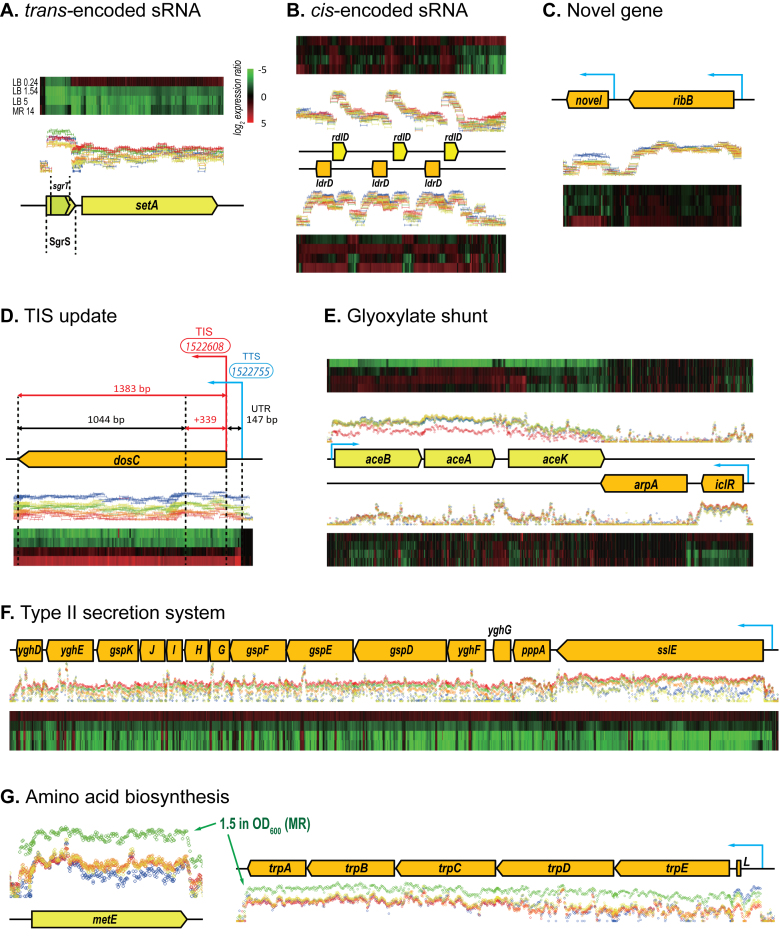
Among the 103 loci newly identified in BL21(DE3) genome, 30 genes and 63 ncRNAs matched those in the K-12 genome in terms of sequence, size and genomic location, and their gene names and products were assigned as per K-12 annotation (U00096.3) (Supplementary Table S4). The 70 loci having the K-12 homologs were found to be in at least one culture condition (P-value < 0.01). For example, a transcript (227 bp) located upstream of setA was expressed in BL21(DE3) under all culture conditions (Figure 3A). Its sequence and location were similar to that of the small regulatory RNA (sRNA) sgrS in K-12 MG1655, which also contains a polypeptide sgrT. The small RNA SgrS induces the degradation of the major glucose transporter ptsG transcript and hence regulates glucose transport (31). Interestingly, sgrS was reported to be differentially expressed in E. coli BL21 and K-12 strains when grown at high glucose concentration (32). Another example of sRNA annotation is the location of RdlD antisense RNAs (Figure 3B). While the K-12 genome has one toxin-antitoxin pair of ldrD/rdlD sequences (33), the BL21(DE3) genome has three pairs between the bcsG and yhjV genes.
Figure 3.
Examples of dynamic changes in the transcriptome structure of Escherichia coli BL21(DE3). Discovery of small regulatory RNAs (sRNAs) that are trans-encoded (A), cis-encoded (B) and a putative novel gene (C). Correction of a translation initiation site (D). Expression of gene clusters of the glyoxylate shunt (E), secondary type II secretion system (F) and amino acid biosynthesis (G). See Figure 2 for interpreting the notations.
In addition, we observed the expression of seven ORFs and three ncRNAs that were not reported in the K-12 genome annotation. These included a novel transcript (171 bp in length) that was highly expressed only in the stationary phase when grown in MR medium. This transcript encodes a protein homologous to a hypothetical E. coli protein (WP_049143758.1, E-value = 8 × 10−33) (Figure 3C).
Revision of ORF boundaries
The TISs of 88 CDSs from the original BL21(DE3) annotation were different when compared to the K-12 annotation, and were corrected accordingly (Supplementary Table S5). The positions of all stop sites were the same in both annotations. There was a wide distribution of absolute changes in length for the 88 CDSs, with an average change of 57 nt and a standard deviation of 71 nt. The updated gene boundaries were verified by transcriptome structure analysis, revealing numerous revisions required. For example, the first gene of the dosCP operon was annotated as yddV with a length of 1044 bp in the original genome annotation. However, the new re-annotation predicted that the TIS of dosC was actually 339 bp away from the previous site. This prediction was supported by the expression of a 1383 bp coding sequence for dosC with 147 bp of the 5΄ UTR (Figure 3D).
Possible changes in gene products due to the revised TISs were inspected in two ways. First, we searched conserved domains within nucleotide sequences unique to the revised or their original ORFs using NCBI's Conserved Domain Database (34). While only a few protein domains were detected within sequences exclusively present in the original ORFs (14%), lots of them were found within those sequences in the revised ORFs (72%) (Supplementary Table S5). However, each of those sequences had a small portion of the whole protein domain: one exception was 23S rRNA methyltransferase gene (rlmI) whose extended upstream region had most of acylphosphatase superfamily domain, without changing the product information. Second, we performed and compared BLASTP searches against UniProtKB/Swiss-Prot protein database (35) queried with each of the 88 ORFs having the revised or original TISs (Supplementary Table S6). The retrieved homologs were not different according to the varied ORF lengths. However, in terms of top hits having the same length with the query, 83 hits were retrieved for BLASTP searches when queried with TIS-revised ORFs, compared to five hits for those queried with original ORFs. Thus, revision of TISs does not seem to affect annotation of gene products. These analyses, taken together with the experimental evidence of gene expression, verify revision of the TISs in this study.
Dynamic changes in transcriptome structure
Escherichia coli BL21(DE3) has been widely used for the production of recombinant proteins as it possesses features desirable for high cell density culture, such as low acetate production and enhanced permeability (6,12). One of the major differences in central metabolism between E. coli B and K-12 strains is found in the expression patterns of the glyoxylate shunt pathway genes that are involved in acetate accumulation (12,36). The arpA gene of unknown function is located between the aceBAK and iclR genes, but is disrupted in B strains (12). We observed that transcript levels of the aceBAK operon and its negative regulator iclR depended on culture conditions, while arpA was not expressed at all (Figure 3E). Although the aceBAK operon genes are expressed from the same promoter, the absolute expression intensity of aceK was much lower than that of aceA (Supplementary Figure S3) and premature transcriptional termination was observed in aceK (Figure 3E). This unusual operon expression supports prior findings demonstrating that the cellular level ratios of products of aceB, aceA and aceK were ∼0.3:1:0.003 (37).
Compared to the E. coli K-12 genome, E. coli B genome possesses an additional gene cluster (gspDEFGHIJK located between the yghE and yghF genes) for type II secretion (T2S), indicating that B strains might have additional secretory potential that is absent in K-12 (10,12). Another gene cluster for T2S (gspAB and gspCDEFGHIJKLMO between the rpsJ and bfr genes) exists in both B and K-12 strains (10), although it was reported to be cryptic in K-12 under standard laboratory conditions (38). As expected, in all the culture conditions tested in this study, standard T2S genes were not expressed in BL21(DE3). However, additional T2S genes were found to be expressed and their transcript levels were regulated depending on growth conditions (Figure 3F). Their expression levels were higher during the exponential phase and decreased in the stationary phase. Interestingly, four genes (yghF, yghG, pppA and sslE) that are located upstream of the additional T2S operon were co-transcribed with the T2S operon genes. Although their functional role in T2S is not fully understood (39), they appear to be representatives of T2S in BL21(DE3). This example underlines the value of direct single-color labeling of total RNAs to measure absolute intensities of probes expressed in a strand-specific manner.
Differentially expressed genes (DEGs) according to the growth stage and culture media
Different stage of cell growth and usage of different culture media cause drastic changes in genome-wide gene expression and have been broadly used as culture conditions for determining transcriptome structures of diverse micro-organisms (29,40,41). To understand distinct effect of the growth stage and culture media on the transcriptome structure, we identified DEGs at each growth stage by pairwise comparison of transcriptomes at the stationary and exponential phases in each culture media (5.0 versus 1.54 in OD600 in LB media; 14 versus 1.5 in MR media). ORFs expressed in at least one culture condition were subjected to analyses of differentially expressed genes (DEGs) and COGs (Supplementary Table S7). Genes showing growth stage-specific differences (stationary phase/exponential phase in LB or MR media) in expression levels of ≥2- or ≤0.5-fold were considered to be DEGs.
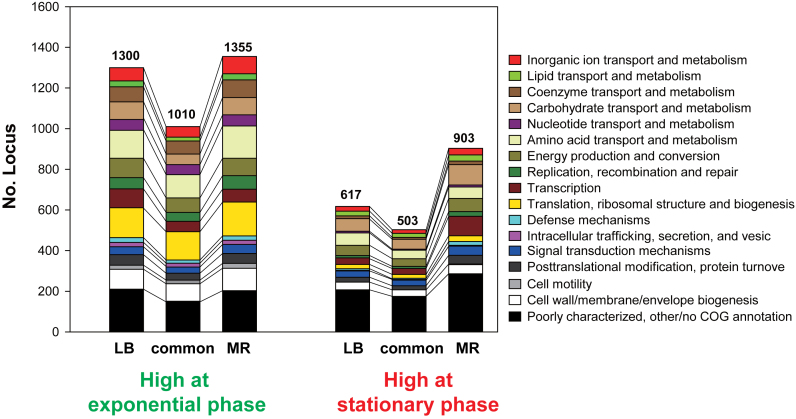
Distribution of the COG functional categories (23) were compared for genes expressed in complex LB media, minimal MR media and both media (Figure 4). Genes highly expressed at exponential phase outnumbered those at stationary phase. During the exponential growth phase, the most highly expressed genes were those involved in ribosomal biosynthesis and transport/metabolism of nucleotides and amino acids, while many of the highly expressed genes at stationary phase encoded stress-responsive proteins and membrane proteins (Supplementary Table S7). Interestingly, many more genes were identified as DEGs when cells were grown in MR media than in LB media. This is due to that correlation of transcript levels (in log2-transformed intensities) between exponential and stationary phases was lower in the minimal media (r = 0.60) than in the complex media (r = 0.78). Previous proteomic comparison of E. coli BL21 growing exponentially in complex and minimal media revealed that the most prominent proteomic differences occurred in enzymes for amino acid biosynthesis which constituted 6 and 20% of the proteome in complex and minimal medium, respectively (42). In this study, compared with growth in LB media, transcript levels of amino acid biosynthesis genes at the exponential stage in MR media were much higher and they decreased at the stationary phase to a greater extent (Figure 3G).
Figure 4.
Functional enrichment of differentially expressed genes at exponential and stationary phases in complex LB media and minimal MR media. Genes which were highly expressed in LB, MR and both media were categorized by clusters of orthologous groups (COGs). Left grouped bars are for genes highly expressed at exponential growth phase, and right for those at stationary phase.
DISCUSSION
Integration of the results from multiple annotation pipelines can be challenging due to different number of features and discrepancy of TIS assignment among the pipelines (3,43). To get around the difficulty, first, we collected all the features reported by the annotation sources without preference for a specific pipeline (Figure 1 and Supplementary Table S1). Then, we took full advantage of the well-annotated K-12 genome by comparing the complied features of BL21(DE3) genome with those of K-12. Lastly, the novel features and revised TISs were identified through computational analysis and manual inspection of gene expression data (Figures 2 and 3). This effort led to the identification of 103 novel features including 10 features only present in BL21(DE3) genome (Supplementary Table S4). Further, 88 revised TISs were validated by BLASTP and protein domain searches.
Escherichia coli B and its derivatives, along with E. coli K-12 and its derivatives, are the most widely studied laboratory strains of bacteria. The E. coli B derivative strain BL21(DE3) was engineered to harbor the T7 RNA polymerase gene for efficient expression of recombinant proteins (18) and has been a major workhorse for biotechnological applications (6). E. coli BL21(DE3) continues to be an important strain for both basic and applied research. Previously, we have reported important features of E. coli B strains making them great industrial host strains through comparative analysis of the E. coli B and K-12 genomes, transcriptomes, proteomes, phenomes and also metabolic models that capture features of the whole cellular system (12). However, much better understanding on the specific traits of E. coli BL21(DE3) has been needed.
While the K-12 MG1655 genome has been re-annotated through a continuing community effort (7,9,44), the BL21(DE3) genome re-annotation has received less attention; it had not been updated since the genome sequence was first released in 2009 (10). In the present study, we combined the results from multiple automated annotation pipelines with comparative genome analysis of BL21(DE3) and the closely related K-12 MG1655 strain to provide an accurate and contemporary re-annotation of the E. coli BL21(DE3) genome. Our study also determined for the first time the transcriptome structure of BL21(DE3), allowing high-resolution analysis of TUs at the genome-scale that provides experimental evidence of gene expression. Through integrated automated analysis and subsequent manual curation, we updated 660 gene names and the majority of gene product descriptions and corrected the TISs of 88 genes. Importantly, 37 novel ORFs and 66 non-coding RNAs were newly identified. The updated annotation was presented both in spreadsheet (Supplementary Tables S2 and 4) and a GenBank-format text file. The results of up-to-date genome re-annotation and transcription study on E. coli BL21(DE3) reported here will serve as an essential resource for systems modeling and functional genomic analysis, and will be useful for further improving the performance of this strain for industrial applications. Last but not least, the re-annotation strategy reported in this study can be applied generally to other sequenced genomes with well-annotated reference genomes.
ACCESSION NUMBERS
Tiling array data were deposited in Gene Expression Omnibus database under entry GSE93565. The latest annotation of E. coli BL21(DE3) genome was deposited in GenBank to update CP001509.3.
Supplementary Material
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Research Foundation of Korea through the Technology Development Program to Solve Climate Changes on Systems Metabolic Engineering for Biorefineries [2012M1A2A2026559 to S.H.Y., 2012M1A2A2026556 to S.Y.L.]; Ministry of Agriculture, Food, and Rural Affairs through the Strategic Initiative for Microbiomes in Agriculture and Food [916006-2 to S.H.Y.]. Funding for open access charge: National Research Foundation of Korea [2012M1A2A2026559].
Conflict of interest statement. None declared.
REFERENCES
- 1. Stein L. Genome annotation: from sequence to biology. Nat. Rev. Genet. 2001; 2:493–503. [DOI] [PubMed] [Google Scholar]
- 2. Reed J.L., Famili I., Thiele I., Palsson B.O.. Towards multidimensional genome annotation. Nat. Rev. Genet. 2006; 7:130–141. [DOI] [PubMed] [Google Scholar]
- 3. Siezen R.J., van Hijum S.A.. Genome (re-)annotation and open-source annotation pipelines. Microb. Biotechnol. 2010; 3:362–369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Markowitz V.M., Chen I.M., Palaniappan K., Chu K., Szeto E., Pillay M., Ratner A., Huang J., Woyke T., Huntemann M. et al. IMG 4 version of the integrated microbial genomes comparative analysis system. Nucleic Acids Res. 2014; 42:D560–D567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Poole F.L. 2nd, Gerwe B.A., Hopkins R.C., Schut G.J., Weinberg M.V., Jenney F.E. Jr, Adams M.W.. Defining genes in the genome of the hyperthermophilic archaeon Pyrococcus furiosus: implications for all microbial genomes. J. Bacteriol. 2005; 187:7325–7332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Yoon S.H., Jeong H., Kwon S.-K., Kim J.F.. Lee SY. Genomics, biological features, and biotechnological applications of Escherichia coli B: ‘Is B for better?!’. Systems Biology and Biotechnology of Escherichia coli. 2009; Berlin: Springer; 1–17. [Google Scholar]
- 7. Riley M., Abe T., Arnaud M.B., Berlyn M.K., Blattner F.R., Chaudhuri R.R., Glasner J.D., Horiuchi T., Keseler I.M., Kosuge T. et al. Escherichia coli K-12: a cooperatively developed annotation snapshot–2005. Nucleic Acids Res. 2006; 34:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Hayashi K., Morooka N., Yamamoto Y., Fujita K., Isono K., Choi S., Ohtsubo E., Baba T., Wanner B.L., Mori H. et al. Highly accurate genome sequences of Escherichia coli K-12 strains MG1655 and W3110. Mol. Syst. Biol. 2006; 2:doi:10.1038/msb4100049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Keseler I.M., Mackie A., Peralta-Gil M., Santos-Zavaleta A., Gama-Castro S., Bonavides-Martinez C., Fulcher C., Huerta A.M., Kothari A., Krummenacker M. et al. EcoCyc: fusing model organism databases with systems biology. Nucleic Acids Res. 2013; 41:D605–D612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Jeong H., Barbe V., Lee C.H., Vallenet D., Yu D.S., Choi S.H., Couloux A., Lee S.W., Yoon S.H., Cattolico L. et al. Genome sequences of Escherichia coli B strains REL606 and BL21(DE3). J. Mol. Biol. 2009; 394:644–652. [DOI] [PubMed] [Google Scholar]
- 11. Daegelen P., Studier F.W., Lenski R.E., Cure S., Kim J.F.. Tracing ancestors and relatives of Escherichia coli B, and the derivation of B strains REL606 and BL21(DE3). J. Mol. Biol. 2009; 394:634–643. [DOI] [PubMed] [Google Scholar]
- 12. Yoon S.H., Han M.J., Jeong H., Lee C.H., Xia X.X., Lee D.H., Shim J.H., Lee S.Y., Oh T.K., Kim J.F.. Comparative multi-omics systems analysis of Escherichia coli strains B and K-12. Genome Biol. 2012; 13:R37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Barrick J.E., Yu D.S., Yoon S.H., Jeong H., Oh T.K., Schneider D., Lenski R.E., Kim J.F.. Genome evolution and adaptation in a long-term experiment with Escherichia coli. Nature. 2009; 461:1243–1247. [DOI] [PubMed] [Google Scholar]
- 14. Kwon S.K., Kim S.K., Lee D.H., Kim J.F.. Comparative genomics and experimental evolution of Escherichia coli BL21(DE3) strains reveal the landscape of toxicity escape from membrane protein overproduction. Sci. Rep. 2015; 5:16076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Han M.J., Yun H., Lee J.W., Lee Y.H., Lee S.Y., Yoo J.S., Kim J.Y., Kim J.F., Hur C.G.. Genome-wide identification of the subcellular localization of the Escherichia coli B proteome using experimental and computational methods. Proteomics. 2011; 11:1213–1227. [DOI] [PubMed] [Google Scholar]
- 16. Aziz R.K., Bartels D., Best A.A., DeJongh M., Disz T., Edwards R.A., Formsma K., Gerdes S., Glass E.M., Kubal M. et al. The RAST Server: rapid annotations using subsystems technology. BMC Genomics. 2008; 9:75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Tatusova T., DiCuccio M., Badretdin A., Chetvernin V., Nawrocki E.P., Zaslavsky L., Lomsadze A., Pruitt K.D., Borodovsky M., Ostell J.. NCBI prokaryotic genome annotation pipeline. Nucleic Acids Res. 2016; 44:6614–6624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Studier F.W., Moffatt B.A.. Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. J. Mol. Biol. 1986; 189:113–130. [DOI] [PubMed] [Google Scholar]
- 19. Yoon S.H., Han M.J., Lee S.Y., Jeong K.J., Yoo J.S.. Combined transcriptome and proteome analysis of Escherichia coli during high cell density culture. Biotechnol. Bioeng. 2003; 81:753–767. [DOI] [PubMed] [Google Scholar]
- 20. Halasz G., van Batenburg M.F., Perusse J., Hua S., Lu X.J., White K.P., Bussemaker H.J.. Detecting transcriptionally active regions using genomic tiling arrays. Genome Biol. 2006; 7:R59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Cho B.K., Zengler K., Qiu Y., Park Y.S., Knight E.M., Barrett C.L., Gao Y., Palsson B.O.. The transcription unit architecture of the Escherichia coli genome. Nat. Biotechnol. 2009; 27:1043–1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Bare J.C., Koide T., Reiss D.J., Tenenbaum D., Baliga N.S.. Integration and visualization of systems biology data in context of the genome. BMC Bioinformatics. 2010; 11:382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Galperin M.Y., Makarova K.S., Wolf Y.I., Koonin E.V.. Expanded microbial genome coverage and improved protein family annotation in the COG database. Nucleic Acids Res. 2015; 43:D261–D269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kristensen D.M., Kannan L., Coleman M.K., Wolf Y.I., Sorokin A., Koonin E.V., Mushegian A.. A low-polynomial algorithm for assembling clusters of orthologous groups from intergenomic symmetric best matches. Bioinformatics. 2010; 26:1481–1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Studier F.W., Daegelen P., Lenski R.E., Maslov S., Kim J.F.. Understanding the differences between genome sequences of Escherichia coli B strains REL606 and BL21(DE3) and comparison of the E. coli B and K-12 genomes. J. Mol. Biol. 2009; 394:653–680. [DOI] [PubMed] [Google Scholar]
- 26. Sezonov G., Joseleau-Petit D., D’Ari R.. Escherichia coli physiology in Luria-Bertani broth. J. Bacteriol. 2007; 189:8746–8749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Cole K., Truong V., Barone D., McGall G.. Direct labeling of RNA with multiple biotins allows sensitive expression profiling of acute leukemia class predictor genes. Nucleic Acids Res. 2004; 32:e86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Yu W.H., Hovik H., Olsen I., Chen T.. Strand-specific transcriptome profiling with directly labeled RNA on genomic tiling microarrays. BMC Mol. Biol. 2011; 12:3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Yoon S.H., Reiss D.J., Bare J.C., Tenenbaum D., Pan M., Slagel J., Moritz R.L., Lim S., Hackett M., Menon A.L. et al. Parallel evolution of transcriptome architecture during genome reorganization. Genome Res. 2011; 21:1892–1904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Patterson T.A., Lobenhofer E.K., Fulmer-Smentek S.B., Collins P.J., Chu T.M., Bao W., Fang H., Kawasaki E.S., Hager J., Tikhonova I.R. et al. Performance comparison of one-color and two-color platforms within the MicroArray Quality Control (MAQC) project. Nat. Biotechnol. 2006; 24:1140–1150. [DOI] [PubMed] [Google Scholar]
- 31. Vanderpool C.K., Gottesman S.. Involvement of a novel transcriptional activator and small RNA in post-transcriptional regulation of the glucose phosphoenolpyruvate phosphotransferase system. Mol. Microbiol. 2004; 54:1076–1089. [DOI] [PubMed] [Google Scholar]
- 32. Negrete A., Ng W.I., Shiloach J.. Glucose uptake regulation in E. coli by the small RNA SgrS: comparative analysis of E. coli K-12 (JM109 and MG1655) and E. coli B (BL21). Microb. Cell Fact. 2010; 9:75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kawano M., Oshima T., Kasai H., Mori H.. Molecular characterization of long direct repeat (LDR) sequences expressing a stable mRNA encoding for a 35-amino-acid cell-killing peptide and a cis-encoded small antisense RNA in Escherichia coli. Mol. Microbiol. 2002; 45:333–349. [DOI] [PubMed] [Google Scholar]
- 34. Marchler-Bauer A., Bo Y., Han L., He J., Lanczycki C.J., Lu S., Chitsaz F., Derbyshire M.K., Geer R.C., Gonzales N.R. et al. CDD/SPARCLE: functional classification of proteins via subfamily domain architectures. Nucleic Acids Res. 2017; 45:D200–D203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. UniProt Consortium UniProt: a hub for protein information. Nucleic Acids Res. 2015; 43:D204–D212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Phue J.N., Noronha S.B., Hattacharyya R., Wolfe A.J., Shiloach J.. Glucose metabolism at high density growth of E. coli B and E. coli K: differences in metabolic pathways are responsible for efficient glucose utilization in E. coli B as determined by microarrays and Northern blot analyses. Biotechnol. Bioeng. 2005; 90:805–820. [DOI] [PubMed] [Google Scholar]
- 37. Chung T., Resnik E., Stueland C., LaPorte D.C.. Relative expression of the products of glyoxylate bypass operon: contributions of transcription and translation. J. Bacteriol. 1993; 175:4572–4575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Francetic O., Belin D., Badaut C., Pugsley A.P.. Expression of the endogenous type II secretion pathway in Escherichia coli leads to chitinase secretion. EMBO J. 2000; 19:6697–6703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Strozen T.G., Li G., Howard S.P.. YghG (GspSβ) is a novel pilot protein required for localization of the GspSβ type II secretion system secretin of enterotoxigenic Escherichia coli. Infect. Immun. 2012; 80:2608–2622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Rasmussen S., Nielsen H.B., Jarmer H.. The transcriptionally active regions in the genome of Bacillus subtilis. Mol. Microbiol. 2009; 73:1043–1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Tjaden B., Saxena R.M., Stolyar S., Haynor D.R., Kolker E., Rosenow C.. Transcriptome analysis of Escherichia coli using high-density oligonucleotide probe arrays. Nucleic Acids Res. 2002; 30:3732–3738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Li Z., Nimtz M., Rinas U.. The metabolic potential of Escherichia coli BL21 in defined and rich medium. Microb. Cell Fact. 2014; 13:45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Ederveen T.H., Overmars L., van Hijum S.A.. Reduce manual curation by combining gene predictions from multiple annotation engines, a case study of start codon prediction. PLoS One. 2013; 8:e63523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Karp P.D., Keseler I.M., Shearer A., Latendresse M., Krummenacker M., Paley S.M., Paulsen I., Collado-Vides J., Gama-Castro S., Peralta-Gil M. et al. Multidimensional annotation of the Escherichia coli K-12 genome. Nucleic Acids Res. 2007; 35:7577–7590. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.