Abstract
In plants, tasiRNAs form a class of endogenous secondary siRNAs produced through the action of RNA-DEPENDENT-RNA-POLYMERASE-6 (RDR6) upon microRNA-mediated cleavage of non-coding TAS RNAs. In Arabidopsis thaliana, TAS1, TAS2 and TAS4 tasiRNA production proceeds via a single cleavage event mediated by 22nt-long or/and asymmetric miRNAs in an ARGONAUTE-1 (AGO1)-dependent manner. By contrast, tasiRNA production from TAS3 seems to follow the so-called ‘two-hit’ process, where dual targeting of TAS3, specifically mediated by the 21nt-long, symmetric miR390, initiates AGO7-dependent tasiRNA production. Interestingly, features for TAS3 tasiRNA production differ in other plant species and we show here that such features also enable TAS3 tasiRNA biogenesis in Arabidopsis, and that a single miR390 targeting event is, in fact, sufficient for this process, suggesting that the ‘one-hit’ model underpins all the necessary rudiments of secondary siRNA biogenesis from plant TAS transcripts. Further results suggest that the two-hit configuration likely enhances the fidelity of tasiRNA production and, hence, the accuracy of downstream gene regulation. Finally, we show that a ‘non-cleavable one-hit’ process allows tasiRNA production from both TAS1 and TAS3 transcripts, indicating that RDR6 recruitment does not require miRNA cleavage, nor does the recruitment, as we further show, of SUPRRESSOR-OF-GENE-SILENCING-3, indispensable for tasiRNA generation.
INTRODUCTION
Small RNAs (sRNAs) are important regulatory molecules involved in the proper control of gene expression, genome stability and defense, via the process of RNA silencing. In plants, sRNAs originate from long double-stranded RNA (dsRNA) processed by DICER-LIKE (DCL) RNase III enzymes into 20–24-nt long molecules. In Arabidopsis thaliana sRNAs are then loaded into one of ten ARGONAUTE (AGO) effector proteins as part of RNA-induced silencing complexes (RISCs). RISCs may promote silencing at the post-transcriptional level via endonucleolytic cleavage (‘slicing’) or translational inhibition of target RNAs, or at the transcriptional level by guiding cytosine methylation and chromatin compaction. Depending on their origin, i.e. from perfectly or imperfectly complementary dsRNA, plant sRNAs can be classified into small interfering RNAs (siRNAs) or microRNAs (miRNAs), respectively (1).
An important feature of RNA silencing in plants is that it can be amplified, notably also in a selective manner: in some cases, RNAs targeted by sRNAs, in parallel to their degradation, become templates for RNA-DEPENDENT RNA POLYMERASES (RDRs), resulting in de novo synthesis of dsRNA. This dsRNA is in turn processed by DCLs into so-called ‘secondary siRNAs’, which amplify the original sRNA signal and allow silencing of RNA related in sequence to the primarily targeted RNA. Trans-acting siRNAs (tasiRNAs) form a highly conserved class of plant sRNA that typifies this type of amplification mechanism. While miRNAs typically promote degradation of target mRNAs, miRNA-mediated cleavage of TAS ncRNAs triggers the recruitment of SUPPRESSOR OF GENE SILENCING 3 (SGS3) and RDR6, resulting in the production, by DCL4, of 21-nt secondary siRNAs acting to silence complementary mRNAs in trans (2,3). Four well-defined TAS gene families are found in A. thaliana (TAS1–4). TAS1 and TAS2 are both targeted by miR173 (4,5) and TAS4 by miR828 (6). TAS3 distinguishes itself from the other three families because it displays two independent target sites for miR390, which is loaded specifically into AGO7, in contrast to most miRNAs, including miR173 and miR828 evoked above, which are loaded into AGO1 (7–9). A second specific feature of A. thaliana TAS3 is that the second miR390 target site displays central mismatches that prevent slicing. In all cases, however, the cleavable miRNA target site is believed to be used as the defining point for the production of tasiRNAs within a prevalent sequence register, resulting in a defined phase. tasiRNA nomenclature takes into account which fragment, upon miRNA-mediated cleavage of the originally targeted ncRNA, is used for siRNAs biogenesis. For instance, in TAS1, TAS2 and TAS4, tasiRNAs production takes place from the 3΄ cleavage fragment, and therefore they are referred to as tasi_3’D1, 3’D2, 3’D3, etc., with the final digit referring to their position relative to the miRNA slicing site. TAS3 on the contrary, spawn secondary siRNAs from the region located between the two miR390 sites and, consequently, from the 5΄ fragment formed upon miRNA cleavage. Thus, these siRNAs are referred to as 5’D1, 5’D2, 5’D3, etc. More recently, it was found that protein-coding transcripts, as opposed to TAS ncRNAs, can also produce tasi-like siRNAs. Similarly to tasiRNAs, these are phased with respect to the miRNA cleavage site and are thus often referred to as ‘phasiRNAs’ (2,3).
How TAS transcripts are routed to the RDR6 amplification pathway unlike most miRNA targets is still a matter of debate. Currently, two main hypotheses exist to explain this peculiar phenomenon. The first hypothesis, often referred to as the ‘two-hit’ model, is based on the observation that, in many plant species, TAS3 displays two miR390 target sites and that other secondary siRNA-producing loci, such as many pentatricopeptide repeat (PPR) genes, are also targeted twice by sRNAs (7). It has been hypothesized that this two ‘hits’ process might stimulate the recruitment of SGS3 and RDR6 through unspecified means. However, this scheme does not apply to all cases, as exemplified by the A. thaliana TAS1 ncRNA in which a single cleavage event mediated by miR173 suffices for tasiRNA production (10,11). Notably, miR173 and other single-hit miRNAs that trigger secondary siRNA production are 22-nt- instead of 21-nt-long, the cognate size of most miRNAs including miR390 (12,13), and/or form asymmetric miRNA/miRNA* duplexes (14). These unusual features, often conserved across plant species, have been put forward as key determinants supporting the ‘one-hit’ hypothesis of tasiRNA generation.
The best-characterized regulatory network orchestrated by tasiRNAs involves the down-regulation, by TAS3, of AUXIN RESPONSE FACTOR transcripts displaying target sites for a discrete number of phased tasiRNAs. Production of these so-called ‘tasi-ARFs’ is a conserved feature found in many plants including angiosperms, gymnosperms, ferns and even mosses. In A. thaliana, silencing of ARF2, ARF3/ETT and ARF4 by TAS3a involves the phased 5’D7 and 5’D8 tasiRNAs, which regulate leaf and lateral root development, organ polarity and juvenile-to-adult transition (2). Hence, Arabidopsis mutants defective in the TAS3 pathway display premature expression of adult vegetative traits, including the emergence of elongated and curled rosette leaves early in development (4,15–17). Understanding TAS3 tasiRNA biogenesis is therefore not only relevant from a mechanistic point of view, but also to decipher the tenets and outputs of widely conserved physiological processes important for plant growth and architecture.
As explained above, in A. thaliana, the TAS3 RNA is targeted twice by the miR390/AGO7 RISC; however, only the 3΄ proximal site is cleaved, resulting in the production of tasiRNAs that are phased in relation to this specific position. The 5΄ site, by contrast, contains mismatches that prevent miRNA cleavage (4,7). Montgomery and colleagues (8) have shown that, despite not being cleaved, the miR390 5΄ site is essential for tasiRNA biogenesis. Indeed, replacing this site by another miRNA-target sequence was sufficient to inactivate secondary siRNA production. On the other hand, different miRNA-target sites could functionally replace the miR390 3΄ proximal site: as long as cleavage was maintained, bona fide TAS3 tasiRNA production occurred. These results suggest that a stable and prolonged interaction of the miR390/AGO7 RISC at the 5΄ site is an important feature for TAS3 tasiRNA generation. Corroborating this view, it has been recently suggested that this stable interaction might have a negative impact on the translation of an ORF located just upstream of the miR390 5΄ site, potentially leading to ribosome stalling, which could affect the transcript stability and consequently, the production of tasiRNAs (18). This feature, however, is not apparent in TAS3 ncRNAs from some plant species. For instance, in Pinus taeda and Physcomitrella patens, in which the conserved miR390 5΄ site was first identified, both miRNA sites are cleavable (7). Likewise, in spruce (Picea abies), both configurations (cleavable/non-cleavable) can be found among the various TAS3 paralogs of this species (19). These differences prompt the questions as to why TAS3 tasiRNA biogenesis requirements differ among distinct plant species and, more specifically, why distinct mechanisms underpin the interaction of miR390 with the TAS3 5΄ proximal target site?
Here, we used a variety of transgenic sensor constructs to explore these differences in TAS3 tasiRNA production. We show that the miR390 5΄ site can indeed be cleaved to synthesize bona fide TAS3 tasiRNA in A. thaliana, providing a possible unifying framework for tasiRNA production across plant species. Moreover, our results suggest that the ‘one-hit’ process might represent a rudimentary or fundamental mode of tasiRNA biogenesis, while the ‘two-hit’ process is a possible adaptation to improve the fidelity of secondary siRNA production and accuracy of downstream gene regulation. Finally, our study reveals hitherto unappreciated aspects of RNA silencing amplification in plants by uncovering a slicing-independent mode of RDR6 recruitment on its RNA templates and additional role(s) for SGS3 in tasiRNA biogenesis.
MATERIALS AND METHODS
Plant material
A. thaliana ecotype Columbia (Col-0) and sgs3-14 (20) plants were grown in 16 h light/8 h dark condition at 21°C. Transgenic plants were selected with BASTA® (Bayer).
Reporter lines
The atasiSUL sRNA targeting the SUL homolog CH42 (At4g18480) was designed as described in Felippes and Weigel (11). To construct the TAS3-based reporter lines, overlapping PCR was used to insert the atasiSUL sequence in tandem either at positions 5D7/8 or positions 3D5/6 of TAS3a (At3g17185), replacing the original sequences located at those sites. For TAS1-based constructs, the same technique was employed to replace the sequence located at the position 3D10 of TAS1c (At2g39675) with a single atasiSUL. Overlapping PCR was also used for mutation of miRNA target sites in all reporter constructs. All constructs were initially cloned into a gateway entry vector (pDonnor221; Invitrogen) and subsequently recombined into the CaMV35S promoter containing-vector pB2GW7 (gateway.psb.ugent.be) using Gateway technology (Invitrogen). The binary vectors were introduced into Agrobacterium tumefaciens strain GV3101 used to transform Col-0 or sgs3-14 plants (21).
Phenotypic analyses
For segregation analysis, bulk of at least 50, two-week-old T1 plants were scored for the presence of bleaching. Rosette size was calculated from, at least 15, four-week-old seedlings by measuring the diameter of the smallest circle encompassing the whole plant. Chlorophyll a and b levels were quantified using 0.012 g of tissue from 10 individual four-week-old plants. Tissue was incubated with 500 ml of acetone for 30 min in dark, followed by spectrophotometer measurements at 645 and 663 nm.
Molecular analyses
Molecular characterization of the reporter lines was done using 4–5-week-old plants. For all experiments described in this work, we used total RNA extracted using Isol-RNA lysis Reagent (5 Prime).
sRNA northern blot
Pools of T1 plants were used for the characterization of the sRNA population by Northern blot. Five microgram of total RNA were separated in a 17% polyacrylamide gel under denaturing conditions (7M urea); transferred to a positively charged nylon membrane and hybridized with specific probes (see Supplementary Material). DNA oligonucleotide probes were labeled using γ -32P-ATP and T4 Polynucleotide Kinase (Thermo Scientific). Random-primed DNA probes were labeled by α-32P-dCTP incorporation using the Prime-a-gene kit (Promega).
RT-qPCR analyses
SUL expression analyses by RT-qPCR involved three independent T1 transformants as biological replicas. RNA was extracted as described above from plants that were frozen in liquid Nitrogen and kept at –80°C. Quality of the RNA was checked on agarose gels. One microgram of total RNA (measured with NanoDrop2000, Thermo Scientific) was treated with DNase I (Thermo Scientific) and subsequently used to synthetize cDNA using the RevertAid First Strand cDNA Synthesis kit (Thermo Scientific) and oligo-dTs. The reaction was performed in a LightCycler480 II apparatus (Roche) using the KAPA SYBR® FAST qPCR Kit (Labgene Scientific) and gene-specific primers (see Supplementary Material). For the reaction, 25 ng of cDNA and 10 pmol of each primer were used in a final volume of 10 μl. Thermocycling parameters were as followed: 40 cycles of 95°C for 10 s; 60°C for 20 seconds and 72°C for 5 seconds. ACTIN2 (ACT2, At3g18780) and GAPC (At3g04120) were used as housekeeping genes for relative quantification. qPCR validation and data analysis were done using the LightCycler® 480 Software (Roche). Error bars represent standard deviation from three technical replicates of three biological replicates. A t-test of two-sample equal variance, two-tailed distribution was used to calculate the indicated P-values.
5΄-RACE assay
To map possible miRNA cleavage sites in the reporter lines 5΄-RACE was performed. Five micro-grams of total RNA from pooled T1 plants were used in a reaction with 5΄-RNA adaptor and RNA ligase (Thermo Scientific). After precipitation, samples were treated with DNase I (Thermo Scientific) and cDNA was synthetized using RevertAid First Strand cDNA Synthesis kit (Thermo Scientific) and an oligonucleotide designed to specific amplify sequences containing the atasiSUL sRNA. First round and nested-PCR were performed using Dreamtaq (Thermo Scientific) and primers specific to the 5΄-RNA adaptor and the gene of interest. An aliquot of the PCR reaction was cloned into pJET1.2 (Thermo Scientific) for producing the RACE library. Individual clones were sequenced using either the M13 or pJet1 forward primers. In the case of constructs based on the TAS3 gene (excluding the sgs3-14 experiment), RACE was performed in a dcl2-1 dcl3-1 dcl4-2 background (dcl234) (22). Sequences for the adaptors and primers used for 5΄-RACE and sequencing are given in Supplementary Material.
Detection of transgene expression
To confirm the expression of the reporter constructs in transformed sgs3-14 transgenics, 1 μg of total RNA from pooled T1 plants was converted to cDNA using RevertAid First Strand cDNA Synthesis kit (Thermo Scientific), after DNase treatment. PCR using specific primers for the transgene was performed. ACT2 was used as control.
Small RNA sequencing
Total RNA (10 μl, 200–300 ng/μl) from 4 weeks-old rosettes was processed into sequencing libraries using adapted Illumina protocols and sequenced at Fasteris (http://www.fasteris.com, Switzerland) using the Illumina HiSeq sequencer. FASTQ file generation, demultiplexing and adapter removal was done by Fasteris. These deep sequencing files have been deposited to the NCBI Gene Expression Omnibus (GEO) (GSE89345).
Bioinformatic analysis
Small RNA reads were filtered to 15–35nt long reads. Reads with the same sequence were grouped using the processReads function from the ncPRO-seq pipeline (23) and then matched against both the A. thaliana genome (TAIR10) and the transgenes (see supplementary material) using Bowtie (24). Only reads with a maximum of one mismatch over their entire length were analyzed further.
Simple genomic position comparison was applied to retrieve sRNA read counts and positions corresponding to the selected loci. Small RNA profile representations were done using R after normalization to the number of mapped reads (reads per 10 millions reads). Colored phase profiles were produced as 21 colors histogram for the first 5 prime nucleotide of 21nt long read mapping the plus strand and the last three prime nucleotide of the minus strand. Colored clocks illustrate the modulo 21 of the absolute positions of the first five prime and last three prime nucleotides of 21nt long reads mapped in-between the two cleavage sites respectively for the plus and minus strands.
RESULTS
TAS3 tasiRNA production can be initiated from cleavable 5΄ and 3΄ proximal miR390 target sites in Arabidopsis
The apparent difference in tasiRNA biogenesis from TAS3 ncRNAs across distinct species prompted us to investigate in more detail the requirements for secondary siRNA production from this locus. To that aim, we set up a reporter system based on the silencing of CHLORINA42 (CH42) also known as SULPHUR (SUL), which results in an easy-to-score and semi-quantitative phenotype caused by the bleaching of green tissues due to chlorosis (25). Because tasiRNA biogenesis is phased with respect to the miRNA cleavage site, the sequence of siRNAs originating from TAS ncRNAs is usually highly predictable. This feature allowed us to design an artificial tasiRNA reporter system based on A. thaliana TAS3a (the atasiSUL system) in which the sequences in position 5’D7 and 5’D8 (respectively in the seventh and eighth phase-register 5΄ to the 3΄ proximal miR390 cleavage site, Figure 1A) were replaced for an siRNA designed to silence SUL (atasiSUL_5D7/8) (11). Conditions altering tasiRNA production in the appropriate phase would thus be readily detected due to reduced biogenesis of the cognate atasiSUL siRNA, leading to reduced SUL silencing. Different versions of the atasiSUL reporter were introduced into A. thaliana (ecotype Col-0) under the transcriptional control of the constitutive 35S promoter from Cauliflower mosaic virus (CaMV). To take into account the variations in transgene insertions and the sterility of strongly silenced atasiSUL lines (11), phenotypic analyses were conducted statistically on bulks of individual T1 transformants. Both northern blot and RACE analyses were performed with pools of plants, while quantitative reverse-transcription PCR (RT-qPCR) involved individual plants in several biological replicates.
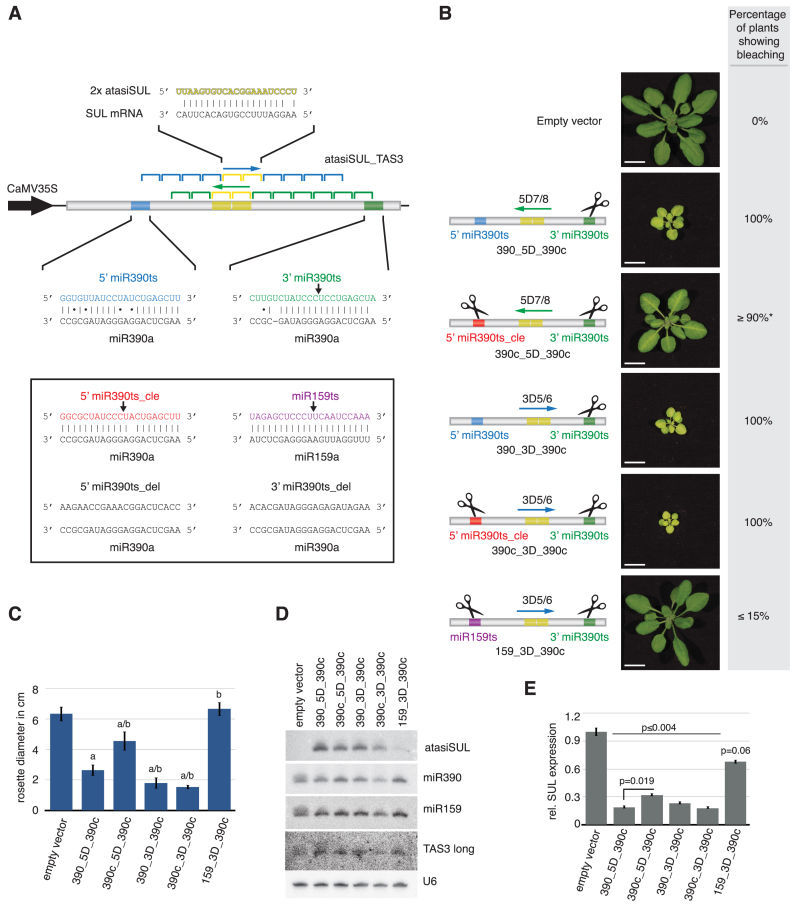
Figure 1.
Cleavage of the miR390 5΄ proximal target site allows cognate TAS3 tasiRNA production. (A) Schematic of the TAS3a-based atasiSUL reporter construct. The original target sequences, as well as mutations therein are presented alongside with the corresponding predicted miR390 base-pairing in the box. Black arrows indicate miR390 cleavage sites. The atasiSUL sequence and its predicted base-pairing with the SUL mRNA is shown on the top and the positions of tasiRNAs, in register to either the 5΄ or the 3΄ target site, are displayed below; arrows depict the predicted direction of phasing. Note that the color code is preserved throughout the panels of all figures. (B) Schematic of the different TAS3a-based constructs tested and the representative plant phenotypes generated upon transformation, assessed with a total of at least 50 individuals in each case. The respective percentage of non-silenced/silenced plants, assessed by bleaching intensity, is indicated in the right hand side panel. The asterisk (*) indicates consistent SUL silencing that is, however, weaker than that in other plants. (C) Quantification of the bleaching intensity by rosette size of 15 individual plants. (a) and (b) refers respectively, to a P-value ≤0.001 generated when a t-test (two-sample equal variance; two-tailed distribution) was performed between the different lines and the empty vector control; or between the reference reporter (390_5D_390c) and the remaining lines. (D) Northern analysis of atasiSUL, miR390 and control miR159 and U6 levels were conducted with complementary oligonucleotide probes. TAS3_long refers to a random-primed DNA probe corresponding to the tasiRNA-producing region of the TAS3a locus. (E) Steady state accumulation levels of the SUL mRNA in the various plants depicted in (B) relative to the empty vector control line was measured by RT-qPCR analyses involving each three biological replicates. Standard error bars are shown for each sample. P-values derived from a t-test (two-sample equal variance; two-tailed distribution) between the different lines and the empty vector control; or between two specific lines are indicated.
We first analysed transgenic plants expressing atasiSUL_5D7/8 in which the 5΄ proximal miR390 target site was mutated to allow cleavage (390c_5D_390c), mimicking the situation seen in P. patens. The corresponding plants consistently displayed a bleaching pattern centered on the central main vein of leaves (Figure 1B, Supplementary Figure S1), presumably as a result of AGO7-specific expression in this tissue (8). As an expected consequence of bleaching, 390c_5D_390c plants were smaller than plants from the control line carrying an empty vector (Figure 1C). Corroborating the observed phenotype, these plants produced the cognate atasiSUL accounting for the down-regulation of SUL as assessed by Northern blotting (Figure 1D) and RT-qPCR (Figure 1E), respectively. Nonetheless, the intensity and distribution of bleaching was much weaker in 390c_5D_390c plants than it was in transgenic control plants expressing a version of the atasiSUL_5D7/8 containing the wild-type target sites (390_5D_390c). These plants displayed intense chlorosis and severe stunting due to efficient silencing of SUL expression (Figure 1B and C, Supplementary Figure S1; Figure 1E). This observation corroborates the idea that the 5΄ proximal site plays an important function in the TAS3 tasiRNA pathway; however, it also indicates that cleavage at this position does not preclude tasiRNA production in A. thaliana.
A cleavable 5΄ proximal miR390 target site impairs the phasing, but not the overall production of atasiSUL
To ascertain the robustness of the atasiSUL system, we tested the effects of different mutations at the 5΄ and 3΄ proximal target sites of miR390 in the atasiSUL_5D7/8 reporter. Agreeing with previous work (8), production of the atasiSUL siRNA from the reporter required the specific targeting of miR390 at the 5΄ proximal site, while setting the proper phasing register could be achieved by different miRNA-cleavage events (Supplementary Figure S2).
One possible explanation for the decreased bleaching phenotype in plants expressing the construct with a cleavable 5΄-miR390 target site (Figure 1B and C, 390c_5D_390c) is that a stable and prolonged interaction of the miR390-loaded AGO7-RISC to this location is a key requirement for routing the TAS3 ncRNA into cognate secondary siRNA biosynthesis. In this scenario, 5΄-cleavage would be expected to strongly reduce or even possibly alleviate siRNA production. Indeed, AGO7 stable interaction to TAS3 transcript has been put forward as one possible model for tasiRNA biogenesis (8). A second possibility is that the abnormal occurrence of a 5΄ cleavage event generates an alternative 5΄ end in the resulting long ncRNA, which, upon amplification into dsRNA by RDR6, might be efficiently processed by DCL4. Since the 5΄- and the 3΄-miR390 target sites are not in phase relative to each other, the ensuing competition for Dicing from both the 5΄ and 3΄ ends of the cleaved and amplified long RNA would result in a fraction of the 21-nt long atasiSUL being produced out-of-phase and thus, less effective (Supplementary Figure S3). Of the two possibilities we favor the second one, because Northern analysis conducted with a labelled oligonucleotide complementary to atasiSUL showed reduced levels of this specific tasiRNA species in 390c_5D_390c compared to 390_5D_390c transformants; by contrast a random-primed DNA probe hybridizing the entire region producing tasiRNAs in TAS3a locus (and thus detecting the bulk of tasiRNAs rather than the specific atasiSUL species) gave a signal of near-equal intensity in both lines (Figure 1D).
In order to ascertain further the robustness of the second scenario, we re-engineered our reporter to now produce the atasiSUL in phase relative to the 5’ proximal miR390 target site. This was achieved by replacing the sequences 3’D5 and 3’D6 (fifth and sixth phase-register 3’ to the 5’ proximal miR390 cleavage site, Figure 1A) for the atasiSUL (atasiSUL_3D5/6). As expected, plants expressing the reporter containing the cleavable miR159 target site in the 5’ position (159_3D_390c) did not show any sign of bleaching. By contrast, expression of constructs bearing either the wild type (390_3D_390c) or the cleavable miR390 target site (390c_3D_390c) in the 5’ position promoted strong bleaching and stunting phenotypes (Figure 1B–C). These results indicate that tasiRNA biogenesis from A. thaliana TAS3 can be efficiently triggered by miR390-mediated cleavage of the 5’ upstream target site. 5’-cleavage, however, promotes production of out-of-phase tasiRNA species, but this can be remedied by artificially setting atasiSUL production in phase relative to the 5’ miR390 target site of the TAS3a ncRNA.
Bona fide TAS3 tasiRNA production can be triggered through a ‘one-hit’ process
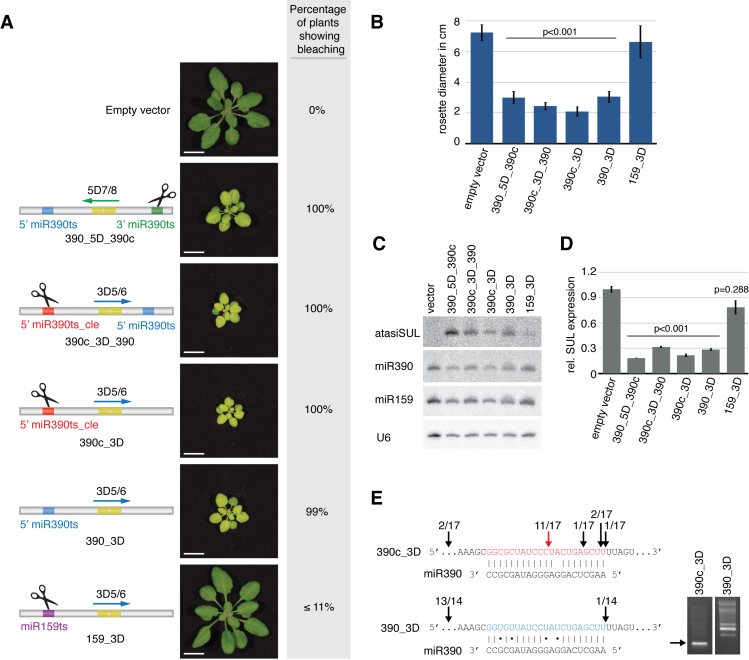
Among the four well-described TAS families found in A. thaliana, only TAS3 requires double targeting, following the ‘two-hit’ mode for initiation of tasiRNA production. Moreover, many miRNAs known to trigger secondary siRNA production from protein-coding genes apparently do so according to the ‘one-hit’ model (26,27). This observation, together with our findings, led us to investigate if tasiRNA biogenesis from TAS3 could also be triggered by a single-targeting event guided by miR390. Using the atasiSUL_3D5/6 reporter (in which tasiRNAs are now in phase relative to the 5’ target site for miR390), we first tested if an exchange/inversion of the two miR390 target sites (390c_3D_390) could still enable tasiRNA production. Transformants expressing this construct consistently displayed intense, uniform bleaching and stunted growth (Figure 2A–B); they also efficiently accumulated atasiSUL, as assessed by Northern blot analysis (Figure 2C).
Figure 2.
‘One-hit’ production of TAS3 tasiRNA. (A) Representative phenotypes of transgenic plants expressing the indicated versions of the atasiSUL reporter constructs, as compared to control plants engineered with an empty vector construct or with the original TAS3-based atasiSUL construct (the control plants depicted in each case were isolated from the same batch as that used in Figure 1). The percentage of plants displaying bleaching was calculated based on analysis of at least 50 individuals in each case, as in Figure 1. (B) Bleaching intensity was quantified by rosette size measurement. P-values derived from a t-test (two-sample equal variance; two-tailed distribution) between the different lines and the empty vector control is indicated. (C and D) Northern blot and RT-qPCR analyses of small RNAs and SUL mRNA steady state levels, respectively, were performed as described in Figure 1. (E) Schematic of the results of the 5΄-RACE analysis conducted for the indicated constructs. The region containing the 5΄-target site of miR390 is depicted (including 5-nt up- and down-stream of the target site). The mapped cDNA ends are indicated with black arrows alongside the number of corresponding clones obtained; the predicted miR390 cleavage site (construct 390c_3D) is indicated with a red arrow. Mapped cDNA ends located outside the presented area are depicted as single events. Agarose gel analysis of the corresponding PCR products for each library is displayed on the right hand side panel; the arrow indicates the expected size of the PCR product corresponding to mRNA cleavage predicted at the miR390 target site.
Next, we investigated if a single miR390-targeting event could trigger secondary siRNA production from the A. thaliana TAS3a. To that aim, we created a reporter construct with a degenerated 3’ proximal miR390 site and containing a cleavable miR390 site at the 5’ upstream position (390c_3D Figure 2A). As a control, the 5’ miR390 site in this reporter was replaced for a cleavable target site of the 21-nt miR159, which is unable to trigger secondary siRNA production on its own (159_3D, Figure 2A). Analysis of transformants expressing these constructs revealed that a single cleavage event mediated by miR390 is sufficient to trigger strong atasiSUL accumulation, extensive bleaching, stunted growth and reduction in SUL mRNA levels (Figure 2A–D and Supplementary Figure S1). By contrast, single 5’-cleavage mediated by miR159 did not result in increased atasiSUL production compared to vector-transformed control plants, with the corresponding plants remaining consistently unbleached.
Taken altogether, these data show that in A. thaliana the ‘one-hit’ configuration is sufficient to promote tasiRNA biogenesis from TAS3a, suggesting that the minimal requirements for RDR6/SGS3 recruitment are shared between the different TAS families.
miR390-mediated cleavage is not required for efficient TAS3 tasiRNA production
miRNA-mediated cleavage is considered to be an important and necessary aspect of tasiRNA biogenesis, not only for ensuring sRNA production in the proper register, but also to recruit RDR6 and SGS3 to the ncRNA transcript. Therefore, atasiSUL lines harbouring only the non-cleavable miR390 5’ target site (390_3D, Figure 2A) were generated as controls for the constructs used to test the ‘one-hit’ model for tasiRNA biogenesis Surprisingly, plants expressing the reporter 390_3D bearing the single, non-cleavable version of the miR390 5’ target site consistently showed strong bleaching, stunted growth and, accordingly, accumulated high levels of atasiSUL as well as reduced amount of SUL mRNA (Figure 2A–D and Supplementary Figure S1). The absence of cognate cleavage at the modified miR390 5’ target site was confirmed by RACE experiments (Figure 2E), corroborating the results of previous work (7,8).
High transcription levels driven by the CaMV 35S promoter are often associated with co-suppression, i.e. the silencing of the transgene due to secondary siRNAs, which, unlike tasiRNAs, are produced in a spontaneous manner. To ensure that the strong and uniform SUL silencing triggered in 390_3D plants was not caused by spontaneous co-suppression of the atasiSUL reporter, we designed an atasiSUL_3D5/6 construct in which both miR390 target sites were deleted (390del_3D, Supplementary Figure S4A). In contrast to the 390_3D T1 population, expression of this new construct led to a high incidence of transformants with a wild-type phenotype. Moreover, the bleaching phenotype of plants exhibiting SUL silencing was much more variable in the 390del_3D than in the 390_3D line (Supplementary Figure S4A and B). Furthermore, corroborating the differences in the phenotypes observed, northern blot analysis showed that the levels of atasiSUL are clearly more abundant in the lines carrying the 390_3D construct as they are in those expressing the 390del_3D reporter (Supplementary Figure S4C). Although we cannot formally exclude some incidence of co-suppression among the 390_3D T1 population, we rule out that this process accounts, alone, for the high frequency and uniformity of the bleaching phenotype displayed by the corresponding plants, indicating a clear effect of the non-cleavable miR390 target site on SUL silencing initiation.
These findings strongly suggest that miRNA-mediated cleavage is not essential to trigger the recruitment of RDR6/SGS3 for efficient dsRNA synthesis and downstream DCL4-dependent siRNA processing. Rather, a stable interaction of the miR390-loaded AGO7-RISC with the miR390 target sequence is likely sufficient for secondary siRNA synthesis.
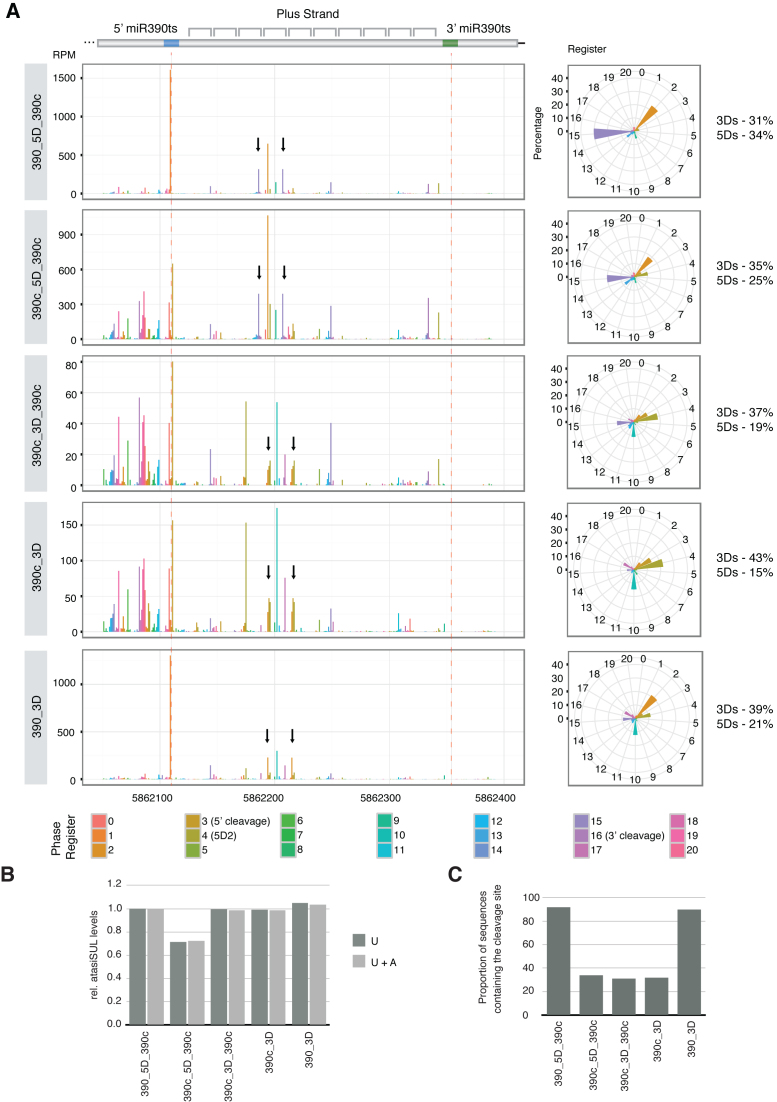
TAS3-tasiRNAs originate mainly from the reporter transgenes and display a conserved distribution profile among constructs
Based on the data presented so far, we propose that cleavage at the miR390 5’ target site does not impair the production of secondary sRNAs from TAS3, but instead generates an alternative entry point for DCL4 processing, resulting in phase competition and, consequently, reduction in the levels of cognate atasiSUL levels. To further test this hypothesis and clarify other aspects of our reporter system, we conducted deep sequencing of sRNAs from plants expressing our reporter constructs. Before proceeding with the analyses, we determined the respective contributions of the original atasiSUL reporter transgene and the endogenous TAS3 to the total amount of tasiRNA species found in the libraries. We calculated the amount of sequences that originated specifically from either the reporter or the endogenous loci (Supplementary Figure S5A-B). On average, transgene-specific sequences were four times more abundant than the ones derived from endogenous TAS3, indicating that variations in tasiRNA abundance/phasing associated with the strong expression of the intact or modified atasiSUL reporter constructs should be interpreted as resulting mostly from transgenic as opposed to endogenous tasiRNA production.
Next, we confirmed that sRNA profiles produced from most of the atasiSUL transgene constructs depicted in Figures 1B and 2A show overall little variation, agreeing with the nearly unchanged global production of atasiSUL in northern blot analysis (Figure 3A, Supplementary Figure S5C, Figures 1D and 2C). The only exception was a region located immediately upstream of the 5’ miR390 target site. In reporter lines where this site was designed to be uncleavable, very few siRNAs originated from this location. By contrast, a large amount of siRNAs mapped to this region in all cases where miR390-directed cleavage of the 5’ proximal site was expected. Because this pattern was conserved between lines showing distinctively different bleaching phenotypes, we conclude that this phenomenon is unlikely to be responsible for the differences observed. Therefore, our deep sequencing analyses focused on the region located between the two miR390 target sites, in which cognate tasiRNAs, including atasiSUL, are produced.
Figure 3.
Deep sequencing analysis of selected reporter lines. (A) Distribution of reads mapping the reporter transgene and their respective phase registers. Note that only the region containing both miR390 target sites and sRNAs mapping the plus strand are shown. The different phase registers are represented as a ‘clock’. The abundance of sRNAs being produced in phase regarding either the miR390 5΄- or 3΄-target site (3Ds or 5Ds, respectively) is given. For the abundance relative to the 3D and 5D positions, we have considered the predicted, plus 1nt up- and down-stream shifted sequences as the same register). The expected phase register set by cleavage of each of the miR390 target sites is indicated in the colour legend (5΄ cleavage and 3΄ cleavage). In addition, the expected register set by tasi-5D2(–) cleavage is also shown (5D2). Black arrows indicate the reads corresponding to the atasiSUL. (B) Levels of the atasiSUL relative to the reference line 390_5D_390c. Only the sequences potentially targeting SUL and starting with a 5΄ uracil (U) or both, uracil and adenine (U + A) were considered. (C) Proportion of sequences mapped around the miR390 5΄-target site that contain the miRNA cleavage site (positions 10–11).
miR390-mediated cleavage at the 5’ target site enhances production of phased siRNAs
As already discussed, one of the consequences of the phase competition created by cleavage at the miR390 5’ target site should be an increase in the levels of tasiRNAs being produced at positions 3Ds (from 3D1 to 3D11). This increase should be accompanied by a coincident decrease in siRNAs generated in the 5D register, especially in situations where the miR390 3’ target site is absent. To test this hypothesis, we analyzed the phase of all 21nt-long reads mapping to the various reporter constructs, taking into account that tasiRNAs targeting the SUL transcript can only originate from the top DNA strand. As expected, in the reference line (390_5D_390c) carrying wild-type miR390 target sites, a strong register compatible with phasing being generated by miRNA-directed cleavage of the 3’ site was observed (Figure 3A). However, this register was offset by 1nt relative to the predicted one. Such shift in phase register of tasiRNAs, which also comprised here the atasiSUL, is a common feature of TAS3 among many plant species, including A. thaliana (4). Interestingly, a second, strong register was also detected, and was linked to the accumulation of large amounts of siR1778, which, as suggested previously, is most likely produced by the re-attack, in cis, of tasiRNA-5D2(–) (4). Noteworthy, the phase register set by tasiRNA-5D2(–) cleavage is the same as the one defined by miR390-directed cut of the 5’ target site. Indeed, siR1778 production is predicted to occur from position 3D5 relative to the 5’ site, the same location of one of the atasiSUL in the 3D5/6 reporter lines. The implications of this phenomenon are discussed later on.
As expected, lines expressing the cleavable version of the miR390 5’ target site showed a clear decrease in tasiRNAs produced in the 5D register. While in our reference construct (390_5D_390c), reads generated from position 5D1 to 5D11 (including sequences offset by 1nt) account for approximately 34% of the siRNA species found in that region; this proportion fell to 25% and 19% when both target sites were cut (390c_5D_390c and 390c_3D_390c, respectively) and reached its lowest level (15%) when only the cleavable 5’ site was retained (390c_3D; Figure 3A). The level of tasiRNAs produced in the 5D register was slightly recovered (21%) when the original, not cleavable, miR390 5’ site was present on its own (390_3D). Accordingly, the opposite pattern was observed regarding the proportion of siRNAs produced from position 3D1 to 3D11 (including sequences offset by 1nt). The levels of these sRNAs increase from 31% in the reference line (390_5D_390c) to 35% and 37% when both cleavable miR390 target sites were present (390c_5D_390c and 390c_3D_390c, respectively), reaching 43% in the 390c_3D line. By contrast, in plants expressing the 390_3D construct, the levels of tasiRNAs in the 3D register was decreased to 39% (Figure 3A). Similarly, we could also detect a reduction in the proportions of tasiRNAs produced in the 5D register from the bottom DNA strand between the reference line and the other constructs (Supplementary Figure S6A). However, analysis of species generated in the 3D register from the minus strand was inconclusive, mainly due to the presence of a single, very abundant sequence with this same phase. In any case, the analysis of sRNA populations originating from the various atasiSUL reporters corroborates, overall, the initial hypothesis that the presence of two cleavable sites creates a competition between two phasing registers likely accounting for the different phenotypes observed.
The levels of atasiSUL are altered by conflicting phasing registers produced by double miR390-directed cleavage
According to our hypothesis, phasing competition created by the existence of two cleavable miR390 target sites would have a direct impact on the production of bona fide atasiSUL species. Based on the data shown so far, the weak phenotype displayed by 390c_5D_390c would result from a part of the atasiSUL being produced out of phase. Re-engineering the reporter construct so that the sRNA targeting SUL would now be in phase to the miR390 5’ site should thus enable normal levels of the predicted atasiSUL and, consequently, rescue the bleaching phenotype, especially in cases where the 5’ site is the main determinant of the phase register. In order to substantiate this scenario, we analysed directly the levels of atasiSUL in the different deep sequencing libraries. As discussed before, in lines where the sRNA targeting SUL was placed in positions 5D7/8 (390_5D_390c and 390c_5D_390c), the most abundant atasiSUL species displayed a 1nt shift upstream to the sequence predicted from the miR390 3’ cleavage site (Figure 3A, Supplementary Figure S6B). Like the predicted one, the alternative sequence also starts with a uracil, qualifying it for cognate loading into AGO1, and thus most likely accounts for the majority of the phenotype observed in transgenic plants. As explained, this 1nt shift seems to be a common feature of TAS3 and it was indeed suggested to represent the functional tasiARF (4). For the lines with the atasiSUL in position 3D5/6 (390c_3D_390c, 390c_3D and 390_3D), the corresponding siRNA seems to be produced as three main variants: the cognate, predicted sequence, and two additional ones displaying a 1nt shift, respectively up- and down-stream of the predicted one (Figure 3A, Supplementary Figure S6C). Given this observation, we identified all sRNAs that could potentially target the SUL transcript and compared their levels among the different lines (Figure 3B, Supplementary Figure S6B and C). For this comparison, we only considered sequences starting with a uracil or adenine, because they are the only ones expected to be efficiently loaded into AGO1 and AGO2, respectively, the main effectors of PTGS in plants. We excluded sRNAs beginning with a cytosine, since these are predicted to be effected by AGO5, which is not expressed in rosettes and therefore, unlikely to contribute to the phenotype (Supplementary Figure S6D) (8,9). As anticipated, the levels of sRNAs predicted to target SUL were reduced (29%) in 390c_5D_390c, but restored to levels comparable to the reference line if the atasiSUL was positioned within the same phase register set by the cleavage of the 5’ target site (Figure 3B). This result therefore agrees with the phenotypic variation observed among the different lines generated in this study (Figures 1B–C and 2A-B).
tasiRNA-5D2(–) has an important impact on the production of the atasiSUL
The analysis of sRNA populations from the selected lines also revealed important features regarding the way tasiRNAs are produced from the reporter constructs. In the case of atasiSUL_5D7/8 lines, the most abundant sequence in the region containing the atasiSUL matches the location giving rise naturally to siR1778, the phase register of which has been proposed to be set by re-attack, in cis, by tasiRNA-5D2(–) (4) (Figure 3A, Supplementary Figure S6B and C). Interestingly, the phase register set by the tasiRNA-5D2(–) is similar to the one defined when cleavage of the miR390 5’ target site is allowed. Moreover, siR1778 is predicted to be produced exactly at the position 3D5 regarding the miR390 5’ site. This has important implications on the interpretation of our results, as it helps clarifying unexpected aspects associated with some of the reporter lines, including the difference in phenotype displayed by lines 390c_5D_390c and 390c_3D_390c. Indeed, in both cases, cleavage occurs at both miR390 target sites, generating a competition between two distinct phase registers, yet only plants expressing 390c_5D_390c seem to display reduced bleaching (Figure 1B and C). Although we cannot exclude a preference of DCL4 for the dsRNA extremity generated by cleavage at the 5’ target site, a more likely explanation is that the re-attack by tasiRNA-5D2(–) contributes to the production of atasiSUL in the 390c_3D_390c lines, and thereby compensates for the competition between the two phase registers. The same rationale could be applied to explain intense bleaching observed in 390_3D_390c plants, which, despite having a single cleavable 3’ target site for miR390, still show the bleaching phenotype expected from phasing being set from the opposite, 5’ target position. Finally, re-attack by tasiRNA-5D2(-) would also explain why plants expressing the non-cleavable, one-hit reporter 390_3D display such a strong bleaching and high levels of atasiSUL (Figures 2A–C and 3B). In this case, the lack of a cleavable miR390 target site should theoretically result in out-of-phase tasiRNAs, decreased atasiSUL levels and less bleaching. It is thus likely that, in the absence of phasing set by the miR390-mediated cleavage, the influence of the re-attack by tasiRNA-5D2(–) is more prominent, and restores the coincidence of registers between this site and the miR390 5’ target site. Accordingly, such exchange of prominence between tasiRNA-5D2(–) and miR390 could be readily observed in the phasing pattern of the different lines (Figure 3A, phasing clock). Agreeing with these hypotheses, RACE products corresponding to cleavage mediated by the atasiRNA-5D2(–) were recovered in the corresponding transgenic lines (Supplementary Figure S6E). In addition, recent work using PARE data identified truncated TAS3 transcripts, which were likely generated by the atasiRNA-5D2(–)-mediated cleavage (18). Noteworthy, sRNAs generated in a phase register accountable for by tasiRNA-5D2(–)-directed cleavage are easily detected not only in Arabidopsis, but also other species (7,28).
tasiRNA production from TAS3 triggered via a ‘non-slicing, one-hit’ configuration is supported by sRNA deep sequencing analyses
We have shown that generation of tasiRNAs from TAS3 transcripts can be efficiently triggered in the absence of miRNA-mediated cleavage (Figure 2). Despite the strong indications, by RACE, of the absence of cleavage promoted by miR390, we decided to exploit the sRNA deep sequencing data generated to further substantiate this claim. We reasoned that the abundance of sRNA containing the predicted cleavage site would be strongly altered depending on the effective occurrence of miR390-mediated cleavage. Hence, in constructs where cleavage does not occur above a significant frequency, the amount of TAS3 transcript available, as template, for the biogenesis of sRNA containing the cleavage site would be higher, resulting in elevated levels of these species. Conversely, efficient cleavage would decrease steady-state accumulation of TAS3 transcripts containing the intact cleavage site, reflected in lower levels of sRNAs with such specificities. Therefore, we calculated the proportion of all possible sequences containing a predicted miR390 5’ cleavage site in the region surrounding the corresponding target site. As expected, the levels of these sRNAs dramatically decreased in all lines where the miR390 5’ target site was made cleavable compared to those where it remained mismatched (Figure 3C). In the cleavage-proficient lines, about 30% of the mapped sequences still contained the expected miRNA cleavage site, but they likely originated from inefficiently sliced transcripts and/or from the endogenous copy of TAS3. In addition to all the results presented so far, previously published data unambiguously support the notion that in A. thaliana there is no cleavage of the miR390 5΄ target site (7,8). Together, these observation strongly support our observation that tasiRNA biogenesis in TAS3 can be triggered independently of miRNA-mediated cleavage.
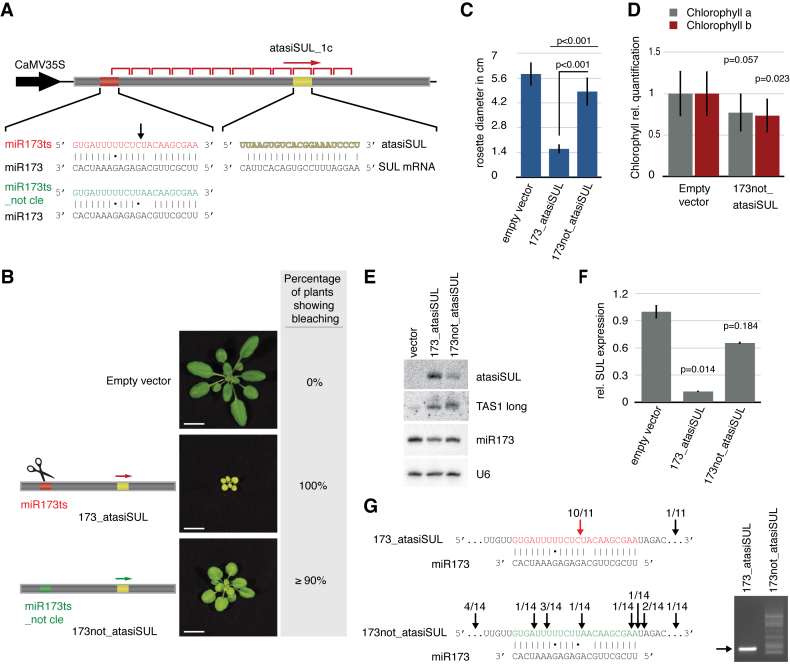
Efficient tasiRNA production from Arabidopsis TAS1 can be triggered in a cleavage-independent manner
The finding that miR390-mediated cleavage is not required for efficient production of tasiRNAs from TAS3a prompted us to investigate if the same can apply to TAS1, which is cleaved by the single, 22-nt long miR173 in an AGO1-dependent manner. We thus designed an atasiSUL system based on the TAS1c ncRNA backbone (atasiSUL-1c) in which the 21-nt long atasiSUL was inserted at position 3D10, downstream of the miR173 cleavage site (Figure 4A). Unlike in the TAS3a-based constructs used above, only one atasiSUL sequence was inserted into atasiSUL-1c, because, as previously documented (11,29), a single atasiSUL in TAS1c-based reporters is sufficient to trigger strong bleaching; whereas the existence of two sequences targeting SUL might result in plants displaying a phenotype too severe to be analyzed. As in the TAS3a-based experiments, the different versions of the atasiSUL-1c reporter were introduced into A. thaliana (ecotype Col-0) under the transcriptional control of the constitutive CaMV 35S promoter. Phenotypic analyses were conducted statistically on bulks of individual transformants and molecular analyses performed as described for the TAS3-based reporter.
Figure 4.
TAS1 tasiRNA production is slicing-independent. (A) Schematic of the TAS1-based atasiSUL reporter construct; the sequence targeting the SUL mRNA is located in position 3D10. The original miR173 target site and the mutations predicted to prevent cleavage are indicated. The black arrow indicates the miR173 cleavage site. (B) Plants expressing the non-cleavable form of the TAS1-based atasiSUL construct (173not_atasiSUL) show consistent bleaching throughout leaf tissue which is, however, weaker than the bleaching displayed by plants expressing the control, cleavable construct (173_atasiSUL). As in Figures 1 and 2, at least 50 individual plants were analysed in each case; representative phenotypes are depicted. (C) Bleaching intensity was measured by rosette size, taking in consideration at least 28 individual plants. P-values derived from a t-test (two-sample equal variance; two-tailed distribution) between the different lines and the empty vector control; or between two specific lines are indicated. (D) Relative quantification of chlorophyll contents of 10 individual T1 plants. P-values refer to t-test (two-sample equal variance; two-tailed distribution) performed between the reporter and the empty vector line. (E and F) Northern blot and RT-qPCR analyses of small RNAs and SUL mRNA steady state levels, respectively, were performed as described in Figure 1; TAS1_long refers to a random-primed DNA probe corresponding to the tasiRNA-producing region of the TAS1 locus. (G) 5΄-RACE analysis of reporter constructs. The mapped cDNA ends are indicated with black arrows alongside the number of corresponding clones obtained; the predicted miR173 cleavage site (construct 173_atasiSUL) is indicated with a red arrow. Agarose gel analysis of the corresponding PCR products for each library is displayed on the right hand side panel; the arrow indicates the expected size of the PCR product corresponding to mRNA cleavage predicted at the miR173 target site.
When expressing the atasiSUL-1c reporter containing the wild-type miR173 target site (173_atasiSUL), plants displayed a strong bleaching and stunted phenotype caused by silencing of SUL expression (Figure 4B, C and E, F). To test whether tasiRNA production from TAS1c could also be triggered via a slicing-independent mechanism, the miR173 target site in the atasiSUL-1c reporter construct was mutated to avoid cleavage (173not_atasiSUL). The corresponding transformants had a weak but consistent bleaching phenotype compared to empty vector controls (Figure 4B, C and Supplementary Figure S1). Accordingly, chlorophyll and the SUL mRNA levels were lower in 173not_atasiSUL than in empty vector-transformed plants. In addition, atasiSUL production, as assessed with a complementary oligonucleotide probe, could be detected by Northern blot analysis in the former, but not in the latter, albeit at significantly lower levels than in the 173_atasiSUL transformants (Figure 4D and F). The weak bleaching phenotype of 173not_atasiSUL plants was most likely caused by defects in phasing rather than processing of atasiSUL, since Northern analysis conducted with a random-primed DNA probe (TAS1 Long) hybridising the entire region producing tasiRNAs from the TAS1c locus (and thus detecting the bulk of tasiRNAs rather than the specific atasiSUL species) gave a signal of equal intensity in both 173_atasiSUL and 173not_atasiSUL transformants (Figure 4E).
5΄-RACE analyses conducted in 173_atasiSUL plants revealed that nearly all individual clones were indicative of cognate slicing between the 10th–11th nucleotide of the miR173–target RNA hybrid (Figure 4G). By contrast, only one out of 14 clones analysed for the 173not_atasiSUL reporter had a sequence potentially indicative of cognate miR173-mediated cleavage, a low-frequency unlikely to explain tasiRNA accumulation and SUL silencing observed among nearly all 173not_atasiSUL transformants. In order to investigate the possible effects of co-suppression in the phenotype of 173not_atasiSUL transformants, we generated a version of the TAS1-based reporter in which the miR173 target site was erased (173del_atasiSUL). Similar to the experiment involving TAS3a, T1 population analyses showed a much greater proportion of 173del_atasiSUL plants displaying a wild-type phenotype than that observed among the 173not_atasiSUL transformants (Supplementary Figure S4D and E). Furthermore, bleaching encompassed a range of variable phenotypes in silenced 173del_atasiSUL transformants not observed in silenced 173not_atasiSUL plants. As with the TAS3a experiments (Supplementary Figure S4C), northern blot analysis unambiguously shows that higher amounts of atasiSUL accumulate in lines expressing the 173not_atasiSUL reporter than in lines expressing the deleted version (173del_atasiSUL, Supplementary Figure S4F). These observations suggest that co-suppression alone cannot account for the uniformity and high frequency of SUL silencing in the 173not_atasiSUL T1 population, indicating a clear contribution of the non-cleavable miR173 target site. In line with the results obtained with the miR390-3D construct (Figure 2A), we conclude, therefore, that slicing is not necessary for TAS1 tasiRNA biogenesis, as recently shown by Arribas-Hernandez et al. (30).
TasiRNA biogenesis requires SGS3 even in non-slicing conditions
The tasiRNA pathway requires, among other factors, the action of SGS3 and RDR6 (20,31). RDR6 converts TAS ncRNAs into dsRNA molecules to be further processed by DCL4 into tasiRNAs. The function of SGS3, by contrast, is much less understood. Based on in vitro experiments, it is believed that SGS3 stabilizes and protects miRNA-cleaved RNA fragments against degradation (32), thereby possibly allowing them to act as templates for RDR6. Because of the apparent close relationship between miRNA-mediated cleavage and SGS3 function, we investigated if SGS3 was necessary when secondary siRNA biogenesis was initiated in vivo in a slicing-independent manner.
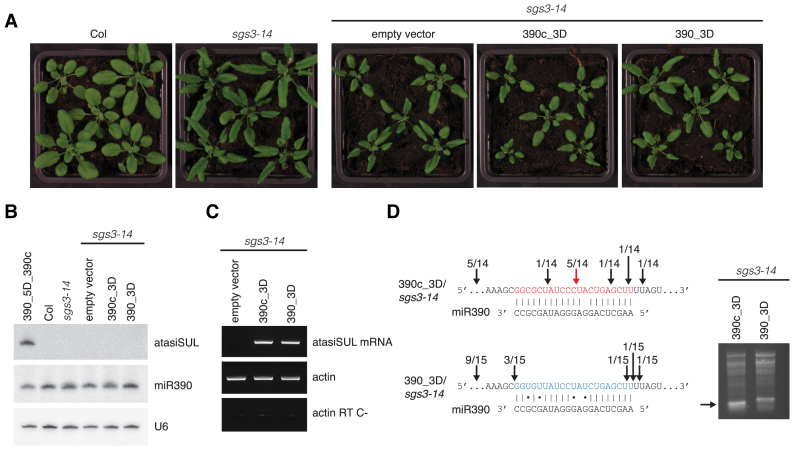
We transformed sgs3-14 mutant plants with both the cleavable and the non-cleavable ‘one-hit’ version of the TAS3-based atasiSUL reporter construct (390c_3D and 390_3D). A strong implication of SGS3 function in TAS3 RNA fragment stabilization/protection predicted that tasiRNA production would be compromised to a much greater extent in transformants expressing the reporter construct with a cleavable miR390 site. However, both reporter lines failed to accumulate atasiSUL and, accordingly, none exhibited a bleached and stunted phenotype (Figure 5A–B). We confirmed comparable transgene expression levels in the two types of transformants and the occurrence of cognate miR390-mediated cleavage for the 390c_3D, but not for the 390_3D construct, based on RACE analyses (Figure 5C and D). These results corroborate the notion that SGS3 is an essential component of tasiRNA biogenesis, but also suggests that the involvement of SGS3 in this process in vivo could go beyond stabilization/protection of cleaved RNA fragments as originally postulated. This would imply the existence of hitherto unknown functions for SGS3 in secondary siRNA production.
Figure 5.
SGS3 is required for TAS3 tasiRNA production triggered in a slicing-independent manner. (A) sgs3-14 mutant Arabidopsis was transformed with the cleavable (390c_3D) and non-cleavable (390_3D) versions of the TAS3a atasiSUL reporter as depicted in Figure 2A; control plants were transformed with an empty vector construct. Analyses were conducted on at least 50 individual plants in each case and representative phenotypes are depicted. (B) Northern analysis of the indicated small RNA was conducted as in Figure 2. (C) Top panel: transgene expression levels were analysed by semi-quantitative RT-PCR. Amplification of the Actin cDNA is depicted in the middle panel. To check for genomic contamination, DNase I treated samples before cDNA synthesis were checked for ACT2 amplification (Actin RT C-). (D) 5΄-RACE and cDNA library analyses as described in Figure 2D.
DISCUSSION
By experimentally testing previously-made inferences and uncovering hitherto unknown properties of TAS ncRNAs and their associated protein complexes, our study helps refining several current models for plant tasiRNA biogenesis and rationalizes the multiplicity of tasiRNA pathways found in various species. It also enables the elaboration of a possible scenario for the origin of single-hit TAS loci, widespread in plants, and suggests how A. thaliana TAS3-like loci might represent an embodiment of optimal tasiRNA biogenesis. Finally, our study sheds light on the mechanisms underlying amplification of post-transcriptional gene silencing in plants by identifying unanticipated modes of RDR6 recruitment, and SGS3 action, on TAS and perhaps other RNAs.
Production of most, if not all, tasiRNAs could be achieved via a ‘one hit’ process
Until now, two different models have been used to explain the biogenesis of tasiRNAs. Despite sharing some of the pathway components, the ‘one-hit’ and ‘two-hit’ models have been, so far, considered two distinct mechanisms to generate secondary siRNAs, each with their own particularities and requirements. Our finding that synthesis of tasiRNAs from TAS3, the main contender in the ‘two-hit’ model, can be triggered by a single miR390-targeting event sheds a significantly new light on our understanding of the biogenesis of this sRNA class. By unifying tasiRNA production, our results indeed suggest that the ‘one-hit’ process underlies the basic mechanism of secondary sRNA production. However, there is still some important differences regarding the way tasiRNAs are generated from TAS3 and other tasi- and phasiRNA loci. While in the former, secondary sRNA biogenesis relies on AGO7 loaded with miR390, other loci seem to require the action of AGO1 loaded with miRNAs displaying special qualities (8,12–14). Despite these differences, our results suggest that these two systems are more similar than previously suspected. We believe AGO1 association with such miRNAs leads to the re-configuration of RISC to promote transitivity and tasiRNA production. In contrast, AGO7 would have evolved to be constantly programed to trigger transitivity. This scenario together with the extreme specificity of AGO7 for miR390 (8), would explain why AGO7/TAS3 does not require 22nt long/asymmetric miRNAs to trigger tasiRNA biogenesis.
Secondary sRNA production can be effectively triggered via a non-cleavable miRNA interaction
A surprising result emerging from our analysis of ‘two-hit’ and ‘single-hit’ types of TAS ncRNAs in A. thaliana is that both experimental systems for atasiSUL production could be ultimately genetically streamlined to trigger consistent, albeit less severe, SUL silencing via a single non-cleavable miRNA target site (Figures 2A and 4B).
The results therefore suggest that, as long as the specifics of each system are maintained i.e. recruitment of the cognate 21-nt miR390-AGO7 RISC on a single site in TAS3 (Figure 2A) or the recruitment of the cognate 22-nt miR173-AGO1 RISC on a single site in TAS1 [as previously established: (10,11)], slicing is dispensable for RDR6-mediated dsRNA neo-synthesis from TAS ncRNAs. We note, nonetheless, that at least for 173not-atasiSUL, the extent of SUL silencing was consistently less pronounced than that triggered by the corresponding cleavable construct (173_atasiSUL, Figure 4B). Thus, while it unlikely alters the overall efficacy of tasiRNA production (Figures 1D and 4E), the absence of cleavage likely causes suboptimal phasing possibly due to ill-defined siRNA processing ends available for DLC4 action. Consistent with this interpretation, slicing defective AGO1 can guide efficient miR173-dependent tasiRNA production but the phasing of the resulting siRNAs is compromised (30). Collectively, these results strongly suggest that a single non-cleavable miRNA hit configuration, is a sufficient requirement for RDR6 recruitment and functional tasiRNA biogenesis in plants.
Slicing-independent recruitment of RDR6 and SGS3 in tasiRNA biogenesis
Although SGS3 has been identified early on as an essential component of tasiRNA production (20,31), its precise function(s) still remain(s) elusive. Using an in vitro approach SGS3 was shown to stabilize miRNA-cleaved RNA fragments to possibly facilitate RDR6 activity (32). Here, we have uncovered a key requirement for SGS3 when tasiRNA production is triggered independently of miRNA-mediated cleavage, suggesting additional roles for SGS3 in secondary siRNA synthesis, consistent with the documented association of SGS3 with slicing-defective RISC in the presence of uncut TAS2 transcript (32). Perhaps SGS3, besides its proposed RNA-protecting role, is also important for the proper localization of TAS transcripts in subcellular locales where RDR6 is active and/or for attracting RDR6 to TAS ncRNAs. Both hypotheses are supported by the finding that SGS3 and RDR6 interact with each other and co-localize, together with AGO7, in specialized cytoplasmic bodies (33,34). Clearly, more work will be required to address this specific issue.
Constraints and advantages of various modes of tasiRNA biogenesis
Genomic studies suggest that ‘cleavable single-hit’ situations, in which miRNA-mediated slicing sets the termination point of complementary-strand neo-synthesis and the initiation of siRNA processing/phasing, dominates the mode of tasiRNA and phasiRNA biogenesis across many plant species (26,27). Our finding that ‘non-cleaved one-hit’ events are in fact probably sufficient for functional secondary siRNA synthesis has implications in terms of the possible origins of TAS ncRNAs. It may also help rationalizing the ‘two-hit’ and ‘dual-cleavage’ processes in A. thaliana TAS3, and in P. patens and P. taeda, respectively, as providing examples of further refined tasiRNA biogenesis mechanisms presenting both advantages and constraints, as discussed below.
‘non-cleaved one-hit’ and ‘cleavable one-hit’ modes of tasiRNA production
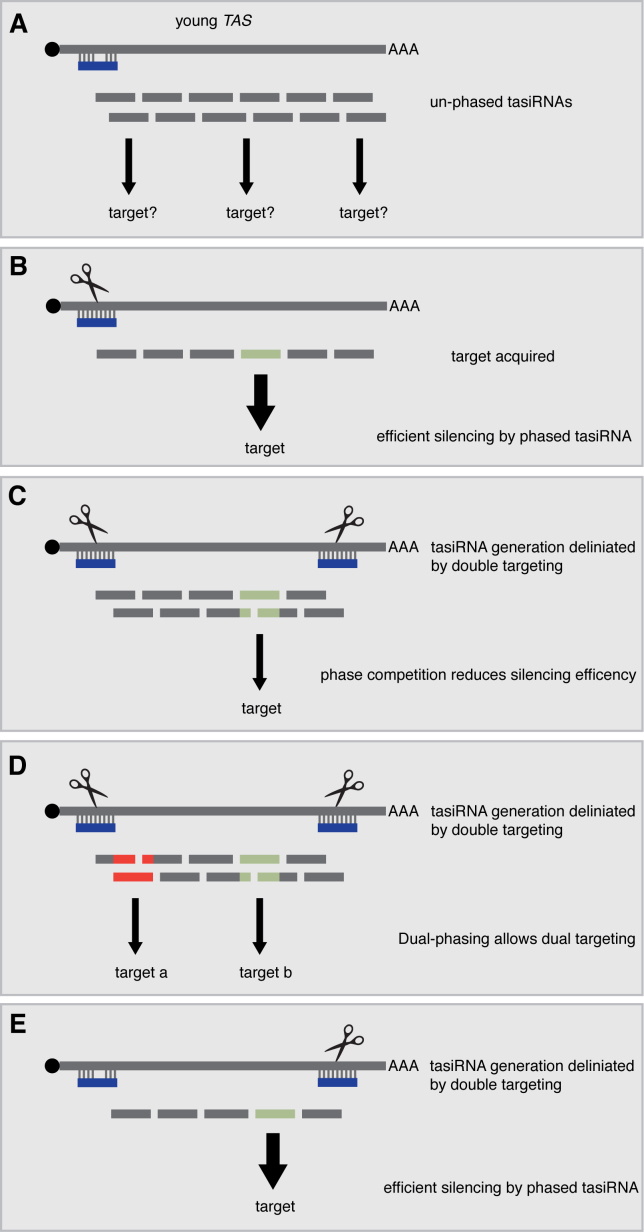
We contend that any mRNA or non-coding RNA might in principle become a tasiRNA-generating transcript following neo-targeting by an appropriate miRNA/AGO combination (Figure 6A). The initial miRNA-TAS pair would possibly display sequence mismatches but, as uncovered in this study, miRNA-mediated cleavage would not be an expected requirement for RDR6 recruitment: presumably, lack of slicing would merely result in predominantly un-phased tasiRNA production from such transcripts. In the event that one of the ensuing secondary siRNAs would acquire a target mRNA, and assuming positive selection, phasing could become important for optimal trans-silencing of the target. Consequently, random mutations enabling miRNA-mediated cleavage could be selected and fixed, causing a transition from a ‘non-cleaved one-hit’ to a ‘cleavable one-hit’ configuration (Figure 6B). In other instances, this transition would not be necessary, including in cases of trans-regulation of related multigene family members displaying extensive nucleotide-sequence homology, which would make phasing superfluous. Incidentally, only low levels of phasing are detected in the many examples of phasiRNAs that have now been uncovered in various plant species (26,27,35).
Figure 6.
Different strategies for tasiRNA production and their possible consequences. (A) The ‘non-cleavable one-hit’ configuration would result in production of un-phased siRNAs, and initially would possibly involve mismatched miRNA-target base-pairing. (B) miRNA cleavage enabling phasing could be positively selected to provide efficient silencing by tasiRNAs via the ‘cleavable one-hit’ configuration. (C) The ‘dual-cleavage’ configuration allows delineating a small region for production of tasiRNAs, avoiding possible off-targeting events; however, silencing efficiency may be compromised by the ensuing phase conflict. (D) Alternatively, dual phase registers enabled by ‘dual-cleavage’ could allow targeting of distinct mRNAs from the same TAS locus. E) The ‘Two-hit, one-cleavage’ configuration enables tasiRNA phasing from the 3΄ cleavage site, while the non-cleavable 5΄ site delineate a small region of tasiRNA production, leading to putative optimal silencing.
‘two-hit’ and ‘dual-cleavage’ modes of tasiRNA production
tasiRNA production naturally occurring via the ‘two-hit’ process could constitute a convergent evolution presenting a key added value compared to the single-hit process, in that tasiRNA biogenesis would be limited to a small, defined region, thereby reducing undesirable off-targeting events. A potential drawback of having two cleavable miRNA target sites in the ‘two-hit’ configuration, however, would be the generation of two well-defined dsRNA ends both available for processing by DCL4. If the target sites are not in the same phase, tasiRNAs would be produced in at least two different registers, a conflict leading to less secondary siRNAs carrying the active nucleotide sequence; accordingly, their trans-silencing potential would be reduced (Figure 6C).
A phase conflict in tasiRNA biogenesis caused by efficient dual-cleavage events is probably naturally encountered in P. patens in which the TAS3a-d ncRNA atypically generates two groups of sequence-unrelated regulatory siRNAs from the 5΄ and 3΄ end, respectively, following their cleavage by miR390. The 5΄ proximal cleavage event sets a preferred register for block 1(+) tasiRNAs, which target three AP2 domain-containing mRNAs, while the 3΄ proximal cleavage event promotes production of tasi-ARFs targeting Arabidopsis ARF-like transcripts. Interestingly, the phase register for each tasiRNA is poorly defined, as expected from a ncRNA having evolved under the constraint of opposing and functional phases. Moreover, in both cases, a second, moss-specific miRNA has evolved to presumably strengthen the action of each tasiRNA in AP2 and ARF regulation, respectively, suggesting that, due to its configuration, P. patens TAS3a-d might mediate suboptimal silencing on its own (36). Dual cleavage leading to distinct species of functional tasiRNAs might be beneficial, however, if each slicing event involves a different miRNA species produced in a temporally and/or spatially distinct manner: this would increase the regulatory potential from a single TAS locus and simultaneously reduce or alleviate the phase conflict issue (Figure 6D).
‘two-hit; one-cleavage’ as an ultimate tasiRNA production process?
We have shown here that, unlike a prevalent yet previously untested assumption, artificially engineering a cleavable 5΄ miR390 target site does allow authentic tasiRNA production from the original A. thaliana TAS3 (Figure 1, construct 390c_5D_390c). This result suggests that this ncRNA and its natural dual-cleavage counterparts in other species do not differ fundamentally in their ability to trigger transitive silencing. Instead, we propose that A. thaliana TAS3 and perhaps similar loci in other plants might represent an embodiment of optimal tasiRNA biogenesis in which tasiRNA processing/phasing is prominently set from a unique 3΄ cleavage site eliminating the dual-phase conflict, and is simultaneously bracketed via an upstream non-cleavable site occupied by the miR390-AGO7 RISC that limits or precludes off-targeting events (Figure 6E).
Strong support for the different scenarios summarized in Figure 6 showing that tasiRNA loci are constantly evolving and under selection, comes from the discovery of a second TAS3-related gene (37,38). This new locus, referred to as TAS3-2 and found in many dicots (except Arabidopsis), conifers and cycads resembles the original TAS3 gene because it is targeted twice by the miR390 in most species but, unlike for Arabidopsis TAS3, miRNA-mediated cleavage of both target sites is usually observed. Nonetheless, in some species of citrus, chicories and populous, TAS3-2 possesses only one miR390 target site, supporting our findings that ‘one-hit’ is sufficient for tasiRNA production from TAS3. Another interesting example of the plasticity of tasiRNA loci comes from the study of sRNAs in Spruce (19) in which TAS3 encompasses a large family of 18 members. In addition to the ‘classical’ configuration with two miR390 target sites required for production of two tasiARFs sRNAs, other spruce TAS3 members are much more diverse, including examples of cleavable and non-cleavable, single, double and even triple miR390 sites.
Outlook
The progresses in large-scale sequencing have resulted in the characterization of sRNA populations from many new species, including new secondary siRNA-producing loci (2,3) mostly identified based on phased patterns of tasi- and phasiRNA production. Our observation that secondary siRNA production can be triggered without cleavage and, thus, without clearly discernible phasing, has therefore the potential to increase the number of such loci, which might have been overlooked in prior analyses. A second prevalent criterion for in silico identification of new secondary siRNA-producing loci is inspired by the ‘two-hit’ model, and relies on identification of separate cleavable sites for two discrete 21-nt-long sRNA species (referred as to the 221 configuration), or, as proposed more recently, by the detection of double (non-necessarily cleavable) hits for 22-nt long sRNAs (222) (26). We suspect based on our findings, that in many of these cases, generation of secondary siRNAs could be triggered following a ‘one-hit’ model. Altogether, the data presented here could prompt further refinements in the predictive tools currently available for secondary siRNA identification.
Supplementary Material
ACKNOWLEDGEMENTS
We thank the Voinnet laboratory for useful feedback and Chris Brosnan and Arturo Marí-Ordóñez for critical reading of the manuscript.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
European Commission (Marie Curie Intra-European Career Development) [300974 to F.F.F.]; European Molecular Biology Organization [EMBO long-term fellowships ALTF-1301-2011 to F.F.F.]; ETH-Z (to O.V.); European Research Council (Frontiers of RNAi-II) [323071 to O.V.].
Conflict of interest statement. None declared.
REFERENCES
- 1. Bologna N.G., Voinnet O.. The diversity, biogenesis, and activities of endogenous silencing small RNAs in Arabidopsis. Annu. Rev. Plant Biol. 2014; 65:473–503. [DOI] [PubMed] [Google Scholar]
- 2. Fei Q., Xia R., Meyers B.C.. Phased, secondary, small interfering RNAs in posttranscriptional regulatory networks. Plant Cell. 2013; 25:2400–2415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Vazquez F., Hohn T.. Biogenesis and biological activity of secondary siRNAs in plants. Scientifica (Cairo). 2013; 2013:783253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Allen E., Xie Z., Gustafson A.M., Carrington J.C.. microRNA-directed phasing during trans-acting siRNA biogenesis in plants. Cell. 2005; 121:207–221. [DOI] [PubMed] [Google Scholar]
- 5. Yoshikawa M., Peragine A., Park M.Y., Poethig R.S.. A pathway for the biogenesis of trans-acting siRNAs in Arabidopsis. Genes Dev. 2005; 19:2164–2175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Rajagopalan R., Vaucheret H., Trejo J., Bartel D.P.. A diverse and evolutionarily fluid set of microRNAs in Arabidopsis thaliana. Genes Dev. 2006; 20:3407–3425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Axtell M.J., Jan C., Rajagopalan R., Bartel D.P.. A two-hit trigger for siRNA biogenesis in plants. Cell. 2006; 127:565–577. [DOI] [PubMed] [Google Scholar]
- 8. Montgomery T.A., Howell M.D., Cuperus J.T., Li D., Hansen J.E., Alexander A.L., Chapman E.J., Fahlgren N., Allen E., Carrington J.C.. Specificity of ARGONAUTE7-miR390 interaction and dual functionality in TAS3 trans-acting siRNA formation. Cell. 2008; 133:128–141. [DOI] [PubMed] [Google Scholar]
- 9. Mi S., Cai T., Hu Y., Chen Y., Hodges E., Ni F., Wu L., Li S., Zhou H., Long C. et al. Sorting of small RNAs into Arabidopsis argonaute complexes is directed by the 5΄ terminal nucleotide. Cell. 2008; 133:116–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Montgomery T.A., Yoo S.J., Fahlgren N., Gilbert S.D., Howell M.D., Sullivan C.M., Alexander A., Nguyen G., Allen E., Ahn J.H. et al. AGO1-miR173 complex initiates phased siRNA formation in plants. Proc. Natl. Acad. Sci. U.S.A. 2008; 105:20055–20062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Felippes F.F., Weigel D.. Triggering the formation of tasiRNAs in Arabidopsis thaliana: the role of microRNA miR173. EMBO Rep. 2009; 10:264–270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Chen H.M., Chen L.T., Patel K., Li Y.H., Baulcombe D.C., Wu S.H.. 22-Nucleotide RNAs trigger secondary siRNA biogenesis in plants. Proc. Natl. Acad. Sci. U.S.A. 2010; 107:15269–15274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Cuperus J.T., Carbonell A., Fahlgren N., Garcia-Ruiz H., Burke R.T., Takeda A., Sullivan C.M., Gilbert S.D., Montgomery T.A., Carrington J.C.. Unique functionality of 22-nt miRNAs in triggering RDR6-dependent siRNA biogenesis from target transcripts in Arabidopsis. Nat. Struct. Mol. Biol. 2010; 17:997–1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Manavella P.A., Koenig D., Weigel D.. Plant secondary siRNA production determined by microRNA-duplex structure. Proc. Natl. Acad. Sci. U.S.A. 2012; 109:2461–2466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Fahlgren N., Montgomery T.A., Howell M.D., Allen E., Dvorak S.K., Alexander A.L., Carrington J.C.. Regulation of AUXIN RESPONSE FACTOR3 by TAS3 ta-siRNA affects developmental timing and patterning in Arabidopsis. Curr. Biol. 2006; 16:939–944. [DOI] [PubMed] [Google Scholar]
- 16. Williams L., Carles C.C., Osmont K.S., Fletcher J.C.. A database analysis method identifies an endogenous trans-acting short-interfering RNA that targets the Arabidopsis ARF2, ARF3, and ARF4 genes. Proc. Natl. Acad. Sci. U.S.A. 2005; 102:9703–9708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hunter C., Willmann M.R., Wu G., Yoshikawa M., de la Luz Gutierrez-Nava M., Poethig S.R.. Trans-acting siRNA-mediated repression of ETTIN and ARF4 regulates heteroblasty in Arabidopsis. Development. 2006; 133:2973–2981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hou C.Y., Lee W.C., Chou H.C., Chen A.P., Chou S.J., Chen H.M.. Global analysis of truncated RNA ends reveals new insights into ribosome stalling in plants. Plant Cell. 2016; 28:2398–2416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Xia R., Xu J., Arikit S., Meyers B.C.. Extensive families of miRNAs and PHAS loci in Norway spruce demonstrate the origins of complex phasiRNA networks in seed plants. Mol. Biol. Evol. 2015; 32:2905–2918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Peragine A., Yoshikawa M., Wu G., Albrecht H.L., Poethig R.S.. SGS3 and SGS2/SDE1/RDR6 are required for juvenile development and the production of trans-acting siRNAs in Arabidopsis. Genes Dev. 2004; 18:2368–2379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Clough S.J., Bent A.F.. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998; 16:735–743. [DOI] [PubMed] [Google Scholar]
- 22. Deleris A., Gallego-Bartolome J., Bao J., Kasschau K.D., Carrington J.C., Voinnet O.. Hierarchical action and inhibition of plant Dicer-like proteins in antiviral defense. Science. 2006; 313:68–71. [DOI] [PubMed] [Google Scholar]
- 23. Chen C.J., Servant N., Toedling J., Sarazin A., Marchais A., Duvernois-Berthet E., Cognat V., Colot V., Voinnet O., Heard E. et al. ncPRO-seq: a tool for annotation and profiling of ncRNAs in sRNA-seq data. Bioinformatics. 2012; 28:3147–3149. [DOI] [PubMed] [Google Scholar]
- 24. Langmead B., Salzberg S.L.. Fast gapped-read alignment with Bowtie 2. Nat. Methods. 2012; 9:357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Koncz C., Mayerhofer R., Koncz-Kalman Z., Nawrath C., Reiss B., Redei G.P., Schell J.. Isolation of a gene encoding a novel chloroplast protein by T-DNA tagging in Arabidopsis thaliana. EMBO J. 1990; 9:1337–1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Zhai J., Jeong D.H., De Paoli E., Park S., Rosen B.D., Li Y., Gonzalez A.J., Yan Z., Kitto S.L., Grusak M.A. et al. MicroRNAs as master regulators of the plant NB-LRR defense gene family via the production of phased, trans-acting siRNAs. Genes Dev. 2011; 25:2540–2553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Johnson C., Kasprzewska A., Tennessen K., Fernandes J., Nan G.L., Walbot V., Sundaresan V., Vance V., Bowman L.H.. Clusters and superclusters of phased small RNAs in the developing inflorescence of rice. Genome Res. 2009; 19:1429–1440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Wan L.C., Wang F., Guo X., Lu S., Qiu Z., Zhao Y., Zhang H., Lin J.. Identification and characterization of small non-coding RNAs from Chinese fir by high throughput sequencing. BMC Plant Biol. 2012; 12:146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. de Felippes F.F., Ott F., Weigel D.. Comparative analysis of non-autonomous effects of tasiRNAs and miRNAs in Arabidopsis thaliana. Nucleic Acids Res. 2011; 39:2880–2889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Arribas-Hernandez L., Marchais A., Poulsen C., Haase B., Hauptmann J., Benes V., Meister G., Brodersen P.. The slicer activity of ARGONAUTE1 is required specifically for the phasing, not production, of trans-acting short interfering RNAs in arabidopsis. Plant Cell. 2016; 28:1563–1580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Vazquez F., Vaucheret H., Rajagopalan R., Lepers C., Gasciolli V., Mallory A.C., Hilbert J.L., Bartel D.P., Crete P.. Endogenous trans-acting siRNAs regulate the accumulation of Arabidopsis mRNAs. Mol. Cell. 2004; 16:69–79. [DOI] [PubMed] [Google Scholar]
- 32. Yoshikawa M., Iki T., Tsutsui Y., Miyashita K., Poethig R.S., Habu Y., Ishikawa M.. 3΄ fragment of miR173-programmed RISC-cleaved RNA is protected from degradation in a complex with RISC and SGS3. Proc. Natl. Acad. Sci. U.S.A. 2013; 110:4117–4122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kumakura N., Takeda A., Fujioka Y., Motose H., Takano R., Watanabe Y.. SGS3 and RDR6 interact and colocalize in cytoplasmic SGS3/RDR6-bodies. FEBS Lett. 2009; 583:1261–1266. [DOI] [PubMed] [Google Scholar]
- 34. Jouannet V., Moreno A.B., Elmayan T., Vaucheret H., Crespi M.D., Maizel A.. Cytoplasmic Arabidopsis AGO7 accumulates in membrane-associated siRNA bodies and is required for ta-siRNA biogenesis. EMBO J. 2012; 31:1704–1713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Song X., Li P., Zhai J., Zhou M., Ma L., Liu B., Jeong D.H., Nakano M., Cao S., Liu C. et al. Roles of DCL4 and DCL3b in rice phased small RNA biogenesis. Plant J. 2012; 69:462–474. [DOI] [PubMed] [Google Scholar]
- 36. Axtell M.J., Snyder J.A., Bartel D.P.. Common functions for diverse small RNAs of land plants. Plant Cell. 2007; 19:1750–1769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Krasnikova M.S., Milyutina I.A., Bobrova V.K., Ozerova L.V., Troitsky A.V., Solovyev A.G., Morozov S.Y.. Novel miR390-dependent transacting siRNA precursors in plants revealed by a PCR-based experimental approach and database analysis. J. Biomed. Biotechnol. 2009; 952304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Xia R., Zhu H., An Y.Q., Beers E.P., Liu Z.. Apple miRNAs and tasiRNAs with novel regulatory networks. Genome Biol. 2012; 13:R47. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.