Abstract
Nucleotide excision repair (NER) comprises two damage recognition pathways: global genome NER (GG-NER) and transcription-coupled NER (TC-NER), which remove a wide variety of helix-distorting lesions including UV-induced damage. During NER, a short stretch of single-stranded DNA containing damage is excised and the resulting gap is filled by DNA synthesis in a process called unscheduled DNA synthesis (UDS). UDS is measured by quantifying the incorporation of nucleotide analogues into repair patches to provide a measure of NER activity. However, this assay is unable to quantitatively determine TC-NER activity due to the low contribution of TC-NER to the overall NER activity. Therefore, we developed a user-friendly, fluorescence-based single-cell assay to measure TC-NER activity. We combined the UDS assay with tyramide-based signal amplification to greatly increase the UDS signal, thereby allowing UDS to be quantified at low UV doses, as well as DNA-repair synthesis of other excision-based repair mechanisms such as base excision repair and mismatch repair. Importantly, we demonstrated that the amplified UDS is sufficiently sensitive to quantify TC-NER-derived repair synthesis in GG-NER-deficient cells. This assay is important as a diagnostic tool for NER-related disorders and as a research tool for obtaining new insights into the mechanism and regulation of excision repair.
INTRODUCTION
The integrity of DNA is threatened constantly by endogenous and exogenous DNA damaging agents, such as reactive oxygen species and ultraviolet light (UV), which severely affect DNA replication, transcription, and cell cycle progression. If they are not repaired correctly these DNA lesions may lead to cell death or mutagenesis, which can eventually result in accelerated aging or malignant transformation. Various DNA repair mechanisms have evolved to maintain genomic integrity, which each repair a subset of DNA lesions (1). Several key repair systems remove single-stranded DNA damage via the excision of nucleotides, including nucleotide excision repair (NER) (2), base excision repair (BER) (3) and mismatch repair (MMR) (4), and use the non-damaged DNA strand as a template for gap-filling DNA synthesis (1).
NER recognizes and repairs a wide spectrum of helix-distorting DNA lesions such as those induced by UV light from the sun (5). NER is characterized by two distinct mechanisms for damage recognition. Global genome NER (GG-NER) recognizes DNA lesions throughout the genome via the joint action of the damage sensors XPC and UV-DDB (6). Transcription-coupled NER (TC-NER) specifically recognizes DNA lesions in actively transcribed strands. Lesion-stalled RNA polymerase 2 is recognized by the proteins CSB, CSA, and UVSSA to initiate the TC-NER pathway (7,8). The damage recognition events in both NER pathways are followed by a shared pathway where excision of the damaged DNA is followed by gap-filling DNA synthesis, thereby completing the NER reaction (2,5).
NER deficits are linked to genetic disorders, which range from mild UV-sensitivity, to severe premature aging, or an extreme predisposition to cancer (5,9,10). Xeroderma Pigmentosum (XP), manifested by photosensitivity and a highly increased incidence of cancer, results mainly from GG-NER deficiency, e.g. caused by mutations in XPC (9,11,12). However, although the TC-NER linked syndromes Cockayne syndrome (CS) and UV-sensitivity syndrome (UVSS) are characterized by a failure to restart transcription following transcription blocking damage, the phenotypes of patients vary dramatically. CS is caused by inactivating mutations in the CSB or CSA genes, which lead to severe developmental, neurological, and premature aging features (11,12). By contrast, UVSS caused by UVSSA mutations or specific mutations in CSA or CSB only has mild features, thus far solely restricted to UV-sensitivity by the skin (13). A precise molecular mechanism that explains the divergent phenotypes of these TC-NER syndromes is still required (5,14).
Analyzing cells derived from NER-deficient patients has been crucial for the identification of most NER-related genes and they have provided important mechanistic insights into the NER reaction. NER-deficient syndromes are characterized by deficient DNA repair, which is used as an important diagnostic marker. The excision of an ∼30 nucleotide-long patch surrounding the DNA damage is a unique property of NER, and thus the subsequent gap-filling DNA synthesis, referred to as unscheduled DNA synthesis (UDS), provides a direct measure of the damage excision and repair efficacy (15,16). The ability to measure UDS is of great importance for diagnostic identification and classification, but it can also provide a crucial quantitative research tool for assessing the NER capacity, thereby obtaining new insights into the NER pathway and its regulation (17). NER-derived UDS is usually monitored by measuring the incorporation of traceable nucleotide analogs after UV irradiation. Originally, pulse labeling with radioactive thymidine (3H-thymidine) after UV and subsequent autoradiography was used to measure UDS (15,16). However, pulse labeling with the thymidine analogue 5-ethynyl-2-deoxyuridine (EdU) and subsequent visualization via conjugation to a fluorescent azide has been used more often recently due to its simplicity, increased sensitivity and higher dynamic range (18,19). GG-NER is responsible for approximately 90% of the total repair executed by NER, so the UDS signal mainly represents the GG-NER activity (18) (Figure 1A). The remaining ∼10% of TC-NER-derived UDS, which is close to the background signal levels, appears to be difficult to determine quantitatively, particularly at physiologically relevant UV doses (Figure 1A) (18,20). In agreement, TC-NER-deficient cells derived from CS patients are highly UV-sensitive but they have almost normal UDS levels, in contrast to cells derived from GG-NER-deficient patients, which exhibit greatly reduced and difficult to quantify UV-induced gap-filling synthesis. Hence, other techniques are currently used to measure TC-NER activity, such as strand-specific repair or recovery of RNA synthesis (RRS) for UV-induced transcription inhibition. Strand-specific repair measures the removal of cyclobutane pyrimidine dimers (CPDs), one of the most common UV-induced DNA lesions, in the transcribed strand of a specific gene by using T4 endonuclease V to incise DNA at a CPD (21). The more sensitive comet-FISH assay uses a similar approach based on fluorescent strand-specific probes combined with single-cell gel electrophoresis to quantify TC-NER repair rates (22). Both assays measure repair itself, but they are highly laborious and only yield information about the repair rate of the gene tested and not the average of all the transcribed genes. Therefore, TC-NER deficiency is routinely assessed using RRS, also for diagnostic purposes. The RRS assay quantifies the restart of transcription after the repair of DNA damage is completed thereby resolving lesion-induced transcription inhibition, which indicates the completion of TC-NER (19). However, the exact mechanism and timing of the restart of transcription following TC-NER remains unclear (23,24). Using the RRS assay, newly synthesized RNA is visualized by pulse labeling with 5-bromouridine (BrU) or 5-ethynyluridine (EU), which is incorporated into nascent RNA (19,25). However, this assay does not provide a direct measure of repair and it cannot discriminate between factors that are specifically involved in repair or in the restart of transcription. The ability to measure TC-NER mediated repair directly is very important because several factors have been described recently, such as HIRA (26), DOT1L (27), Spt16 (28) and ELL (20), that can uncouple TC-NER mediated repair from transcriptional restart (23,24).
Figure 1.
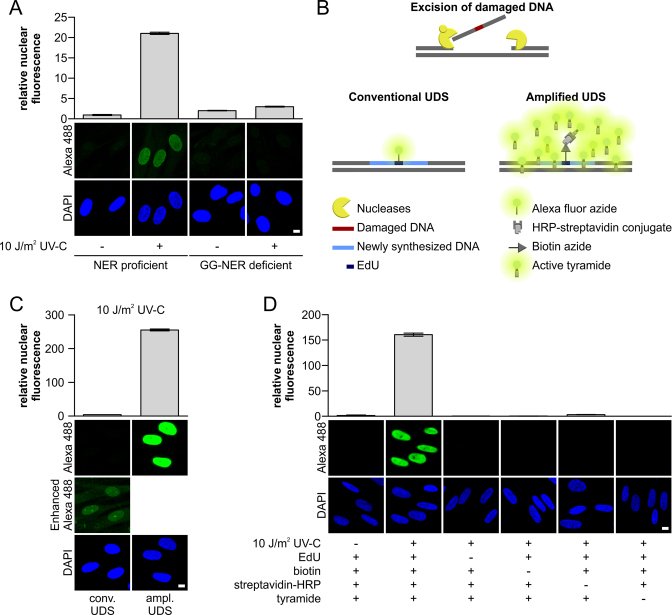
TSA-mediated amplification of UDS signals. (A) VH10 (NER-proficient) or XP186LV (XPC-deficient) cells grown on cover slips were irradiated with UV-C (10 J/m2) or mock-treated as indicated, and subsequently labeled for 3 h with EdU. The UDS signal (n > 300 cells per condition, three independent experiments) was quantified (upper panel) by confocal microscopy measurement of the total nuclear fluorescence (Alexa-Fluor 488 nm) and expressed as relative nuclear fluorescence. Representative images are shown (lower panel). Gap-filling synthesis measured by EdU-based UDS without signal amplification was only observed in GG-NER-proficient cells (VH10), whereas no TC-NER specific signal could be measured in XP-C cells (XP186LV). (B) Schematic overview of the different labeling approaches used by conventional and amplified UDS. The key difference is the number of fluorophores per incorporated nucleotide, where one fluorophore binds to one incorporated EdU in conventional UDS, multiple fluorophores can be bound in the proximity of 1 EdU in the amplified UDS, which is mediated by HRP activation of tyramide-labeled Alexa 488. (C) VH10 cells grown on a coverslip were UV-irradiated (10 J/m2) or left untreated (Supplementary Figure S1E), and subsequently labeled for 3 h with EdU. UV-induced gap-filling synthesis was measured by conventional or amplified UDS (tyramide-based signal amplification). UDS signals were quantified based on the total signal intensity of Alexa-Fluor 488 nm per nucleus. Comparing the amplified UDS assay with the conventional UDS indicated amplification of the UDS signal by >60 times (n > 380 cells per condition, two independent experiments). Representative images are shown (lower panel). To visualize the UDS signal in the conventional UDS assay, the fluorescent signal was digitally amplified for the image labeled as ‘Enhanced Alexa 488’. (D) Quantification of the amplified UDS signal in VH10 cells (n > 250 cells per condition, two independent experiments) where essential components of the EdU-based Click-it chemistry reaction and TSA amplification were omitted as indicated by (–). Representative images are shown (lower panel). Nuclei were visualized using the DNA marker DAPI. SEM is shown. Scale bar: 10 μm.
To measure the TC-NER repair activity, we developed a user-friendly, highly sensitive single-cell assay. For this purpose, we amplified the EdU-mediated UDS assay in GG-NER-deficient cells by using a tyramide-based signal amplification (TSA) procedure. The TSA procedure is a widely used technique for enhancing immunofluorescence signals (29,30) with an HRP-based detection method. Briefly, using Click-chemistry, biotin-azides are coupled to EdU that are incorporated as a consequence of NER-derived DNA synthesis. Subsequently, HRP–streptadivin is bound to the biotin-azide and the oxidation of fluorescent-labeled tyramide by HRP in the presence of hydrogen peroxide generates short-lived fluorescent-labeled tyramide radicals, which are covalently coupled to local nucleophilic residues such as protein tyrosine residues (31) (Figure 1B).
The proposed amplified UDS method for obtaining sensitive measurements of excision-based DNA repair obtained a 60-fold increase in the signal compared with conventional UDS protocols. This signal amplification method allowed us to detect UDS induced by low physiologically relevant UV-C doses, but more importantly, it also enabled us to quantify DNA repair derived only from TC-NER in XPC-deficient cells. Interestingly, the DNA repair synthesis induced by oxidative and alkylating agents can also be measured using this amplified UDS method.
MATERIALS AND METHODS
Cell culture
Human hTert immortalized NER proficient VH10 fibroblasts, GG-NER deficient XP-C-fibroblasts XP186LV, XP3MA and XP20MA, NER deficient XP-A (XP25RO) and TC-NER deficient CS-B fibroblasts (CS1AN) were cultured in Ham's F10 medium (Lonza) supplemented with 15% fetal calf serum (Biowest) and 1% penicillin–streptomycin (Sigma-Aldrich) at 37°C with 5% CO2 in a humidified incubator. All NER deficient cells were characterized either by complementation studies or by mutation analysis (32,33). For transcription inhibition, cells were pretreated either with 25 μg/ml α-amanitin (Sigma-Aldrich) for 16 h or 100 μM 5,6-dichloro-1-β-d-ribofuranosylbenzimidazole (DRB, Calbiochem) for 1 h before DNA damage infliction. For inhibiting DNA polymerase ß, pamoic acid (PA, Sigma-Aldrich) dissolved in 200 mM NaCl and 20 mM Tris–HCl (pH 7) was added to the cells in a final concentration of 500 μM 16 h before the experiment was started. Fresh PA was added during EdU labeling.
DNA damage induction
Prior to DNA damage induction, cells were washed with PBS. For global or local UV-C exposure a 254 nm germicidal lamp (Philips) was used to irradiate cells with indicated UV-C doses. Local UV irradiation was induced using isopore membranes (Millipore) with 5 μm pores (34). Hydrogen peroxide solution (500 μM) (Sigma-Aldrich) was added freshly each hour during the 3h EdU labeling, 100 μM 1-methly-3-nitro-1-nitroguanidine (MNNG, Tokyo Chemical Industry Co., Ltd) was added to culture medium at the same moment it was started with EdU labeling for 3 h.
EdU incorporation
Cells were cultured on 24-mm cover slips and were serum starved (0.5% fetal calf serum) for 2 days to accumulate cells in G0 phase (∼98%). After damage induction, cells were directly labeled with EdU for 3 h (wt cells) or 7 h (GG-NER deficient cells) using Ham's F10 supplemented with 0.5% dialyzed fetal calf serum containing 20 μM 5-ethynyl-2΄-deoxyuridine (EdU, ThermoFisher Scientific). 2΄-Deoxy-5-fluorouridine (1 μM) (Floxuridine, Sigma-Aldrich) was added to inhibit the thymidylate synthase to prevent the generation of endogenous thymidine. After Edu labeling, medium was changed to Ham's F10 supplemented with 0.5% dialyzed fetal calf serum containing 10 μM thymidine (Sigma-Aldrich) for 15 min to deplete unincorporated EdU in the cell. Cells were fixed by incubation with 3.6% formaldehyde (Sigma-Aldrich) in PBS with 0.5% Triton X-100 (Sigma-Aldrich) for 15 min at room temperature. After permeabilization in 0.5% Triton for 20 min, cells were blocked with 3% BSA (Sigma-Aldrich) in PBS.
Fluorescent-based UDS assay with incorporation of EdU
The procedure of the UDS assay was performed as described previously (19,35). Incorporated EdU was visualized using Click-chemistry based coupling of Alexa-Fluor 488 nm azide (ThermoFisher Scientific) according to the manufactures protocol. In short, after fixation, cells were permeabilized for 20 min with 0.5% triton X-100 in PBS at room temperature and subsequently washed twice with 3% BSA in PBS. The Click-it reaction was performed by incubating cells for 30 min at room temperature with 100 μl Click-it reaction Cocktail, containing 1× Click-it reaction buffer (ThermoFisher Scientific), copper(III) sulfate (0.1 M), Alexa Fluor 488 Azide (Click-iT EdU Imaging Kit by ThermoFisher Scientific) and 10× reaction buffer additive (ThermoFisher Scientific). After washing the cells twice in 3% BSA in PBS and twice in PBS, cells were mounted using Vectashield (Vector Laboratories) with DAPI to stain DNA and cover slips were sealed using nail polish.
Amplified UDS assay
After blocking, endogenous peroxidases were quenched to reduce non-specific activation of tyramide using 3% hydrogen peroxide solution (Sigma-Aldrich) in PBS for 1 h. Subsequently, DNA was denaturated using freshly prepared 0.07 M NaOH in PBS for 5 min at room temperature, after which samples were incubated with PBS containing 0.5% BSA and 0.15% glycine for 1 h. The Click-it reaction was performed using 100 μl of the Click it reaction cocktail containing Azide-PEG3-Biotin Conjugate (20 μM in DMSO, Jena Bioscience), 1× Click-it reaction buffer (ThermoFisher Scientific), copper(III) sulfate (0.1 M) and 10× reaction buffer additive (ThermoFisher Scientific). Subsequently, for tyramide-based amplification, samples were incubated with 100 μl HRP–streptavidin conjugate (500 μg/ml) in 1% BSA for 1 h. Subsequently, samples were incubated for 10 min at room temperature with 100 μl amplification buffer containing 0.0015% H2O2 and 1:100 dilution of Alexa-Fluor 488 nm labeled tyramide, which was dissolved in 150 μl DMSO (ThermoFisher Scientific). The addition of 0.0015% H2O2 is required for the HRP-mediated oxidation and consecutive activation of tyramide. After each incubation step, samples were washed three times with PBS. Finally, samples were rinsed twice in PBS containing 0.5% BSA and 0.15% glycine, then three times shortly and two times for 10 min with 0.1%Triton X-100 in PBS. If an additional CPD staining was needed, cover slips were incubated with anti-CPD antibody (1:1000; TDM-2 clone; COSMO BIO Co., Ltd) for 1 h at room temperature. After washing five times with 0.1% triton X-100, of which the last two were washed for 10 min, and once with PBS containing 0.15% glycine and 0.5% BSA, cover slips were incubated for 1 h with the antibody donkey anti mouse Alexa-Fluor 594 nm (ThermoFisher Scientific). After washing the cover slips with 0.1% Triton X-100 in PBS, cover slips were embedded in DAPI containing Vectashield mounting medium (VectorLaboratories) and sealed using nail polish.
Western blotting
For Immunoblotting, cells lysed in 100 μl 2× Laemmli sample buffer were boiled for 5 min prior to protein size-fractionation by SDS-PAGE and subsequent electro-transfer to a PVDF membrane (0.45 μm) as described (35). Membranes were blocked in 3% BSA in PBS for 1 h at room temperature and after washing three times with PBS containing 0.05% Tween the membranes were subsequently incubated with primary antibodies against CSB (E-18, sc-10459, Santa Cruz Biotechnology, Inc.) and Tubulin (B512, Sigma-Aldrich) that were used in combination with Alexa-Fluor 795 donkey anti-goat (SAB4600375, Sigma-Aldrich) or Alexa-Fluor 795 goat anti-mouse (SAB4600214, Sigma-Aldrich) respectively. Western blots were analyzed and quantified using the Odyssey CLx Infrared Imaging System (LI-COR Biosciences).
RNA interference
siRNA was transfected using RNAiMax (ThermoFisher Scientific) 2 days before the experiment according to manufacturer's protocol. siRNAs used in this study were purchased from Thermo Scientific Dharmacon. CTRL: siGENOME Non_Targeting siRNA#5 5΄-UGGUUUACAUGUCGACUAA-3΄ (D-001210-05), CSB: ON-Target plus Human ERCC6 (CSB) A) 5΄- GCAUGUGUCUUACGAGAUA-3΄ (J-004888-10), B) 5΄-CAAACAGAGUUGUCAUCUA-3΄ (J-004888-09)).
Image acquisition
Images were obtained using a LSM700 microscope equipped with a 40× oil Plan-Apochromat 40 0.6–1.3 numerical aperture (NA) oil immersion lens (Carl Zeiss Micro imaging Inc.). The ImageJ software (Version 1.48) (36) was used for quantification purposes and to identify cell nuclei using the DAPI staining in combination with the particle analysis tool. An average signal outside nuclei was determined for each field and was used for background correction. At least 10 fields for each condition were measured. S-phase cells were identified by the very strong and distinct S-phase specific patterns of the 488 nm fluorescent signal and were excluded from the analysis. All experiments were conducted twice and data points of the two independent experiments were pooled, unless stated otherwise. The background corrected total Alexa 488 nm fluorescence signal (nuclear fluorescence) in the nucleus was determined of each cell. The mean nuclear fluorescence is the average of the relative nuclear fluorescence of all cells measured across the independent experiments. The standard error of the mean was calculated using n = the total number of cells across the independent experiments as indicated in the figure legends. All figures show the mean nuclear fluorescence signal ± standard error of the mean. Statistical analysis was performed by a two-way analysis of variance (ANOVA) and the multiple post hoc Bonferroni-corrected t tests to compare differences between the various treatments. In all cases, a level of 5% was considered statistically significant (P < 0.05).
RESULTS
Tyramide-based signal amplification combined with conventional UDS
Previously, UDS was not sufficiently sensitive to quantify TC-NER-derived gap filling synthesis (Figure 1A). To overcome this technical limitation, we combined TSA with the Click-chemistry-based UDS protocol (Figure 1B) (19). Tyramide can be activated nonspecifically by endogenous peroxidases, thereby yielding nonspecific fluorescent signals throughout the cell. This nonspecific tyramide activation was inhibited successfully by quenching the activity of endogenous peroxidases with 3% hydrogen peroxide (Supplementary Figure S1A). The addition of 3% hydrogen peroxide after cell fixation did not induce DNA damage-induced DNA synthesis (Supplementary Figure S1B). Furthermore, when the amplified-UDS signal was quantified after treatment with NaOH (0.07 M for 5 min), which is required for staining DNA damage with α-CPD antibodies, a pronounced increase in the amplified UDS signal was observed (Supplementary Figure S1C). Therefore, all of the amplified UDS experiments were executed with successive hydrogen peroxide and NaOH incubation steps. A flowchart illustrating the different steps in the amplified UDS assay is shown in Supplementary Figure S1D.
To quantify the amplification levels of the tyramide-based signal amplification, we compared the standard UDS signal with the amplified UDS signal in NER-proficient cells after irradiation with 10 J/m2 UV-C. We measured enhanced amplification of the UDS signal by at least 60-fold (Figure 1C), which agreed with the previously described 30-fold amplification of the signal obtained by the TSA procedure (31). However, the actual signal amplification levels might have been underestimated due to limitations on the dynamic range of the photomultiplier employed. The signals measured in non-irradiated samples probably represented background signals because similar signal levels were obtained when essential components of the TSA or Click-chemistry-based UDS reaction were excluded (Figure 1D). In contrast to the UV-induced samples, the background signal detected in non-irradiated samples was amplified by less than threefold using the TSA procedure (Supplementary Figure S1E). Overall, these results indicate that the UDS signal can be amplified specifically by the tyramide-based amplification.
Quantitative measurements of NER at low UV-C doses
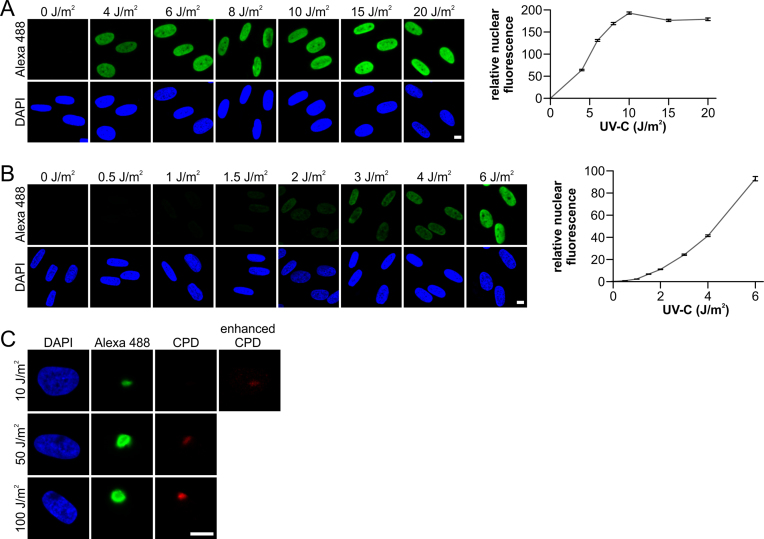
Next, we tested the UV doses at which the amplified UDS assay procedure could be used in a quantitative manner to measure gap-filling DNA synthesis. A dose-dependent amplified UDS signal was observed for UV-C doses up to 10 J/m2, after which the signal plateaued (Figure 2A). This leveling off might have been caused by a limitation imposed by the tyramide concentration at greater UV doses because higher UDS signals were observed as the tyramide concentration increased (Supplementary Figure S1F). More importantly, these results suggest that this amplification procedure can detect UDS at low physiologically relevant UV doses. Indeed, the amplified UDS signal obtained with UV-C doses ranging from 0.5 to 6 J/m2 exhibited a quantitative dose-dependent relationship (Figure 2B), thereby indicating that this amplified UDS procedure is capable of detecting gap-filling synthesis at UV-C doses as low as 1 J/m2 in a reproducible manner. This amplified UDS assay is also suitable for detecting UDS signals at sites of UV-induced DNA damage in a sub-nuclear region (local UV damage) in a dose-dependent manner (Figure 2C). The detection of local UV damage using α-CPD staining indicates that the amplified UDS procedure is compatible with immunofluorescence procedures. It should be noted that at a relatively low dose of 10 J/m2, a distinct UDS signal was observed with a concomitant barely visible CPD-derived signal, which clearly illustrates the sensitivity of this amplification procedure.
Figure 2.
Amplified UDS assay facilitates quantitative measurement of NER. (A and B) Representative images (left) and quantification (right) of amplified UDS signals, as measured by fluorescence (Alexa-Fluor 488 nm, relative total signal intensity per nucleus) in VH10 cells (n > 340 cells per data point, two independent experiments) irradiated with the indicated UV-C dose and labeled for 3 h with EdU. (C) Representative images of the amplified UDS assay in cells irradiated locally with the indicated UV-C doses and subsequently labeled for 3 h with EdU. Sub-nuclear UV-damaged regions were identified by antibody staining for cyclobutane pyrimidine dimers (CPDs). To visualize the CPD signal in samples irradiated with 10 J/m2 UV-C, the fluorescent signal was digitally amplified using the image labeled as ‘enhanced CPD’. Nuclei were visualized using the DNA marker DAPI. SEM is shown. Scale bar: 10 μm.
TC-NER can be measured quantitatively by the amplified UDS assay
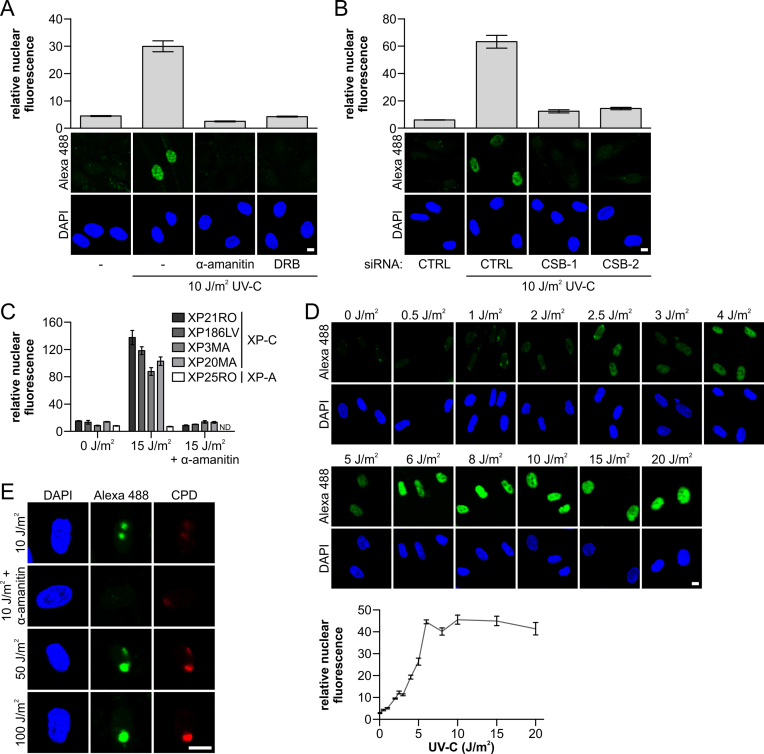
The greatly increased sensitivity of the amplified UDS procedure might facilitate the detection of TC-NER-mediated DNA repair synthesis using this assay. To test this, we employed noncycling XP-C cells, which are defective for GG-NER. In these XP-C cells, the complete NER activity, and thus gap-filling DNA synthesis, can be assigned specifically to TC-NER (2,5). Using the amplified UDS procedure, we detected a clear UV-induced and TC-NER-derived UDS signal in XPC-deficient cells (Figure 3A). The specificity of the TC-NER-dependent amplified UDS signal was demonstrated by the omission of essential components of the amplified UDS assay, which resulted in the complete absence of the signal (Supplementary Figure S2A). Importantly, we also performed this assay in the presence of transcription inhibitors to confirm that the detected UDS signal was indeed attributable to TC-NER-mediated repair of the transcribed DNA strand. Following treatment with α-amanitin (an RNA polymerase II inhibitor, 16 h) or DRB (a CDK9 inhibitor, 1 h) (37) prior to UV-C-induced DNA damage, the UDS signals were comparable to those from non-irradiated samples (Figure 3A), indicating that the amplified UDS signal was completely dependent on active transcription. In addition, siRNA-mediated knockdown of the essential TC-NER protein CSB using two different siRNAs (Supplementary Figure S2B), showed that the UV-induced gap filling synthesis measured in these XPC-deficient cells was dependent on active TC-NER (Figure 3B).
Figure 3.
Gap-filling activity of TC-NER can be measured quantitatively with amplified UDS. (A) GG-NER-deficient XP186LV (XP-C) cells were treated with transcription inhibitors (α-amanitin: 25 μg/ml; 16 h before UV-C treatment, DRB: 100 μM; 1 h before UV-C treatment) or mock treated; or (B) transfected with non-targeting control siRNA (CTRL) and two independent siRNAs that targeting CSB (CSB-1 and -2), as indicated. Cells were labeled directly with EdU for 7 h after irradiation with UV-C (10 J/m2), as indicated. (A and B) Amplified UDS signals were quantified (upper panel) by measuring the total nuclear fluorescence (Alexa-Fluor 488 nm, n > 80 cells for each condition, two independent experiments) and representative images (lower panel) are shown. (C) Quantification of the amplified UDS signal measured by the total nuclear Alexa-Fluor 488 nm fluorescence in the indicated XP-C cells (n > 50 cells per condition). The TC-NER specificity of the signal was shown by the loss of signal after transcription inhibition with α-amanitin (25 μg/ml, added 16 h prior to EdU labeling for 7 h) or when amplified UDS signals were measured in an NER-deficient XP-A cell line (XP25RO). ND indicates not determined. Representative images are shown in Supplementary Figure S2C. (D) Representative images (upper panel) of XP-C (XP186LV) cells (n > 170 cells per condition, two independent experiments) irradiated with indicated UV-C doses. Amplified UDS signals were quantified (lower panel) based on total nuclear fluorescence (Alexa-Fluor 488 nm) after EdU labeling for 7 h and they exhibit dose-dependent UDS signals. (E) Representative images of XPC-deficient XP186LV cells where UV-C damage was induced locally with the indicated UV-C doses. Transcription was inhibited by 25 μg/ml α-amanitin at 16 h prior to UV-C damage and during labeling with EdU for 7 h. Sub-nuclear UV-exposed regions were identified by α-CPD staining. Nuclei were identified by DAPI staining. SEM is shown. Scale bar: 10 μM.
In order to exclude cell-type specific effects, we irradiated different XP-C patient cell lines with UV-C and obtained comparable amplified UDS signals in the cell lines using the amplified UDS procedure (Figure 3C). As expected, those signals were reduced to background signal levels when cells were irradiated in the presence of the transcription inhibitor α-amanitin, and when we used XP-A cells deficient in both GG-NER and TC-NER (Figure 3C and Supplementary Figure S2C).
Moreover, TC-NER-mediated repair could be measured by the amplified UDS procedure in a dose-dependent manner below 6 J/m2 UV-C (Figure 3D). Furthermore, we detected dose-dependent TC-NER-induced gap-filling synthesis at ≥10 J/m2 after local UV-C damage induction (Figure 3E). As expected, this local amplified UDS signal was lost completely when prior to DNA damage infliction transcription was inhibited by α-amanitin. These results clearly indicate that tyramide-based UDS amplification in GG-NER-deficient cells is a highly sensitive quantitative assay for measuring TC-NER at the single-cell level.
DNA repair induced by various types of damage can be detected using the amplified UDS method
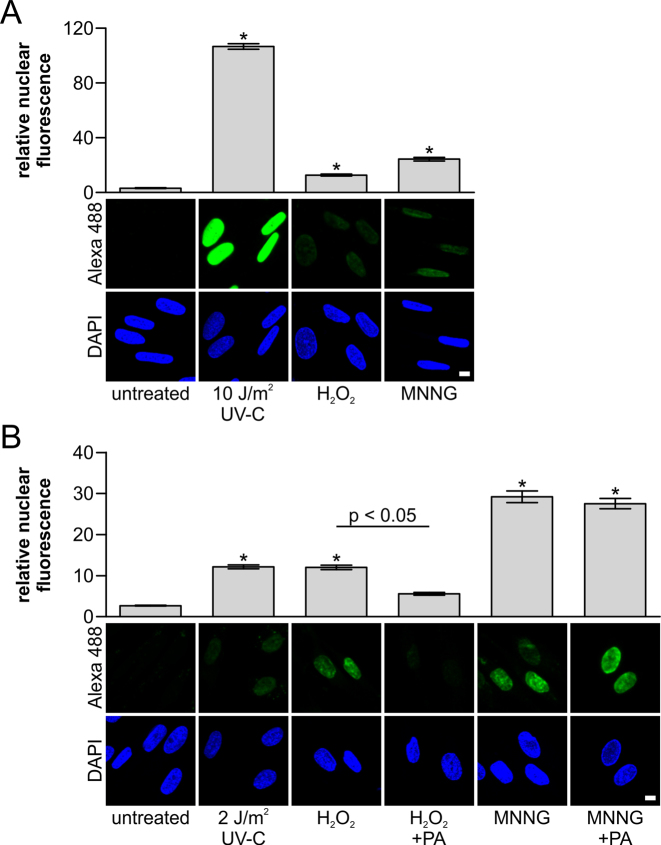
In addition to NER, base-excision repair (BER) and mismatch repair (MMR) are DNA repair pathways based on the excision of damaged DNA fragments. The great increase in the sensitivity of the amplified UDS assay prompted us to test whether excision repair initiated by BER and MMR could also be detected. After inducing damage with oxidative (hydrogen peroxide, 500 μM) or alkylating (MNNG, 100 μM) agents, we could detect amplified UDS signals (Figure 4A), thereby suggesting that the induced damage was removed by excision repair. BER is known to be involved in the repair of oxidized DNA bases (38). During BER, a damaged base is removed by specific glycosylases to create apurinic/apyrimidinic (AP) sites, which are incised by AP-endonuclease. The resulting 5-deoxyribose phosphate (dRP) is removed by DNA polymerase β, which also catalyzes DNA synthesis to fill the single nucleotide gap (39). To verify that we could actually measure BER-mediated DNA synthesis using the amplified UDS assay, we added a DNA polymerase β inhibitor (pamoic acid, PA) (40,41). The amplified UDS signals in samples treated with H2O2 were decreased greatly after PA treatment (Figure 4B), thereby indicating that the majority of the UDS signal after H2O2 treatment was indeed dependent on BER. Interestingly, it is assumed that the DNA damage induced by MNNG is a target for BER (42), but the amplified UDS signal measured after DNA damage induction by MNNG could not be decreased using PA (Figure 4B). This indicates that the UDS signal due to alkylated DNA damage induced by MNNG was probably caused by the activity of MMR in these noncycling cells (43,44). Previously, MNNG-induced UDS was also observed using the classical 3H-TdR procedure (45), but quantifying the UDS signal is much more laborious and less reproducible with this approach. Overall, these results show that combining the tyramide signal amplification procedure with the UDS assay greatly increases its sensitivity and allows quantification of repair by TC-NER and other excision repair pathways in a user-friendly, single-cell assay.
Figure 4.
Amplified UDS enables the measurement of excision repair. (A and B) Representative images of the amplified UDS assay (lower panel) and quantification (upper panel) of VH10 cells (n > 420 cell per condition, two independent experiments) after DNA damage. (A) DNA damage was inflicted with UV-C (10 J/m2), H2O2 (500 μM) or MNNG (100 μM), or cells were mock-treated, before EdU labeling for 3 h (n > 420 cells per condition, two independent experiments). (B) DNA damage was inflicted with UV-C (2 J/m2), H2O2 (500 μM) or MNNG (100 μM), or cells were mock-treated, before EdU labeling for 3 h (n > 180 cells per condition, two independent experiments, except for 2 J/m2: one experiment, n = 81 cells). At 16 h prior to and during EdU labeling, 500 μM pamoic Acid (PA, a DNA polymerase β inhibitor) was added to samples treated with H2O2 or MNNG, as indicated. Nuclei were identified by DAPI staining. SEM is shown, * indicates P < 0.05 compared with untreated samples. Scale bar: 10 μM.
DISCUSSION
For about four decades, UV-induced UDS has been employed successfully for monitoring the cellular NER activity using radio-labeled thymidine followed by autoradiography (15,17). This procedure provides sensitive and reproducible information regarding NER performance, but the dependence on non-user-friendly tritiated thymidine and the fact that the assay is laborious and requires long autoradiographic exposure times, means that its application was limited to only a few laboratories worldwide. The recent replacement of radioactive labeled thymidine with EdU, which can be labeled using simple in situ Click-chemistry with azide-coupled fluorescent dyes of choice, makes the assay much more accessible, faster, and easier to quantify using fluorescence microscopy (19,46). In the present study, we developed an amplified UDS assay by combining the EdU-based UDS assay with a tyramide signal amplification. This combination resulted in a signal amplification by at least 60-fold-specific signal amplification. This facilitates a more easy visualization and quantification of NER-mediated repair at more relevant, low physiological UV doses (1 J/m2). Previously, conventional fluorescent UDS was generally performed with UV-C doses ranging from 16 J/m2 (28) to 20 J/m2 (18,46). The use of much lower UV doses might be crucial for identifying the more subtle effects of novel regulators of the NER reaction based on its repair activity, which can only be identified at more physiologically relevant UV doses. Furthermore, the amplified UDS assay allows us to measure the gap-filling synthesis induced by DNA-damaging agents such as hydrogen peroxide and MNNG. The UDS signals measured after exposure to these genotoxic agents, which are not recognized by NER, are probably attributable to the excision activity of BER (47) or MMR (44), respectively. This indicates that the amplified UDS assay can be employed to measure the repair activity of excision-based DNA repair pathways, for which no user-friendly cellular gap-filling synthesis assays are available at this moment.
Importantly, the amplified UDS method can be applied as a quantitative, single-cell assay to measure the TC-NER-induced repair activity at relatively low doses using GG-NER-deficient cells. The induced signal is TC-NER-specific because the detected UV-induced repair signal is dependent completely on the crucial TC-NER protein CSB and active transcription. Interestingly, we observed a leveling off of the TC-NER-induced amplified UDS signal for UV-C doses above 6 J/m2 (Figure 3D). It is unlikely that this was caused by limitations of compounds used in either the tyramide-based signal amplification or Click-it chemistry because in GG-NER proficient cells, which have much higher UDS levels, the signal could be measured quantitatively for UV-doses up to 10 J/m2 before the signal plateaued (Figure 2A). The plateauing of the amplified UDS signal for TC-NER suggests that a maximum number of lesions can be processed by TC-NER within the given time of a UDS experiment. Further research should be conducted to understand the underlying mechanism involved. Together this shows that the amplified UDS assay can obtain important new molecular, mechanistic, and/or kinetic information about TC-NER.
Our TC-NER assay based on the amplified UDS method has several key advantages compared with other TC-NER assays. Strand-specific repair assays (21), including the novel comet-FISH method (22), measure TC-NER-mediated repair by quantifying the specific removal of CPDs from the transcribed strand and by comparing this repair rate with the non-transcribed strand. The disadvantage of these elegantly designed strand-specific repair assays is that they are highly laborious and they can only study repair in one or a few specific gene(s) at a time. However, due to specific transcriptional programs or differences in the chromatin environment, the repair rate of specific genes might not be representative for the overall TC-NER activity (5,48). By contrast, our amplified UDS-based TC-NER assay determines the average repair activity in all actively transcribed genes in a quantitative manner using single cells. Furthermore, unlike our amplified UDS method, the routinely employed RRS assay provides an indirect measure of the TC-NER dependent repair activity because it only determines the restart of transcription (25). The exact mechanism of transcription restart after the removal of transcription blocking lesions by TC-NER is currently unknown. However, recent research has shown that several factors are specifically required to restart transcription, although they are not considered to be involved in TC-NER-mediated repair itself, including ELL (20), HIRA (26), Spt16 (28), and DOT1L1 (27). This indicates that a specific group of proteins might be involved in facilitating transcription recovery after genotoxic stress and that transcription restart is a specific form of transcription that has not been extensively studied (23,24). Our single cell, fluorescence-based, amplified UDS TC-NER assay can be applied easily to high content screening approaches using genomic libraries (shRNA and sgRNA) to identify factors that are involved directly in TC-NER activity. By comparing amplified UDS data with previously obtained RRS data from TC-NER screening (49), or even simultaneously conducting amplified UDS and RRS assays, it may be possible to discriminate between proteins involved in repair and those involved specifically in the transcription restart process. This approach will be important for understanding the transcription restart process after genotoxic stress.
Furthermore, this user-friendly single-cell assay facilitates quantitative analyses of TC-NER repair capacity, which is crucial for identifying TC-NER mutants, chromatin remodelers, or chemotherapeutics that might affect TC-NER. In conclusion, we describe an important improvement to the conventional UDS assay by using tyramide-based signal amplification, thereby allowing the measurement of GG-NER-mediated gap-filling synthesis at low physiologically relevant UV doses, as well as the detection of TC-NER-mediated repair in a quantitative manner. In addition, we showed that this amplified form of UDS can be used to measure other excision-based DNA repair pathways induced by other types of damage than UV-C (50).
Supplementary Material
ACKNOWLEDGEMENTS
We thank the optical imaging centre (OIC) at Erasmus MC for support with microscopes.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Dutch Organization for Scientific Research ZonMW TOP Grant [912.12.132]; Horizon Zenith [935.11.042]; VIDI ALW [864.13.004]; European Research Council Advanced Grant [340988‐ERC‐ID]; Erasmus MC fellowship. Funding for open access charge: Dutch Science Organization (NWO).
Conflict of interest statement. None declared.
REFERENCES
- 1. Hoeijmakers J.H. Genome maintenance mechanisms for preventing cancer. Nature. 2001; 411:366–374. [DOI] [PubMed] [Google Scholar]
- 2. Scharer O.D. Nucleotide excision repair in eukaryotes. Cold Spring Harb. Perspect. Biol. 2013; 5:a012609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Kim Y.J., Wilson D.M. 3rd. Overview of base excision repair biochemistry. Curr. Mol. Pharmacol. 2012; 5:3–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Groothuizen F.S., Sixma T.K.. The conserved molecular machinery in DNA mismatch repair enzyme structures. DNA Repair (Amst.). 2016; 38:14–23. [DOI] [PubMed] [Google Scholar]
- 5. Marteijn J.A., Lans H., Vermeulen W., Hoeijmakers J.H.. Understanding nucleotide excision repair and its roles in cancer and ageing. Nat. Rev. Mol. Cell. Biol. 2014; 15:465–481. [DOI] [PubMed] [Google Scholar]
- 6. Gillet L.C., Scharer O.D.. Molecular mechanisms of mammalian global genome nucleotide excision repair. Chem Rev. 2006; 106:253–276. [DOI] [PubMed] [Google Scholar]
- 7. Vermeulen W., Fousteri M.. Mammalian transcription-coupled excision repair. Cold Spring Harb. Perspect. Biol. 2013; 5:a012625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Hanawalt P.C., Spivak G.. Transcription-coupled DNA repair: two decades of progress and surprises. Nat. Rev. Mol. Cell. Biol. 2008; 9:958–970. [DOI] [PubMed] [Google Scholar]
- 9. Kraemer K.H., Patronas N.J., Schiffmann R., Brooks B.P., Tamura D., DiGiovanna J.J.. Xeroderma pigmentosum, trichothiodystrophy and Cockayne syndrome: a complex genotype-phenotype relationship. Neuroscience. 2007; 145:1388–1396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Nouspikel T. DNA repair in mammalian cells: nucleotide excision repair: variations on versatility. Cell. Mol. Life Sci. 2009; 66:994–1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. de Boer J., Hoeijmakers J.H.. Nucleotide excision repair and human syndromes. Carcinogenesis. 2000; 21:453–460. [DOI] [PubMed] [Google Scholar]
- 12. Lehmann A.R. DNA repair-deficient diseases, xeroderma pigmentosum, Cockayne syndrome and trichothiodystrophy. Biochimie. 2003; 85:1101–1111. [DOI] [PubMed] [Google Scholar]
- 13. Spivak G. UV-sensitive syndrome. Mut. Res./Fundam. Mol. Mech. Mutagen. 2005; 577:162–169. [DOI] [PubMed] [Google Scholar]
- 14. Schwertman P., Vermeulen W., Marteijn J.A.. UVSSA and USP7, a new couple in transcription-coupled DNA repair. Chromosoma. 2013; 122:275–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Cleaver J.E. Defective repair replication of DNA in xeroderma pigmentosum. Nature. 1968; 218:652–656. [DOI] [PubMed] [Google Scholar]
- 16. Friedberg E.C. The discovery that xeroderma pigmentosum (XP) results from defective nucleotide excision repair. DNA Repair (Amst.). 2004; 3:183–195. [PubMed] [Google Scholar]
- 17. Latimer J.J., Kelly C.M.. Unscheduled DNA synthesis: the clinical and functional assay for global genomic DNA nucleotide excision repair. Methods Mol. Biol. 2014; 1105:511–532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Limsirichaikul S., Niimi A., Fawcett H., Lehmann A., Yamashita S., Ogi T.. A rapid non-radioactive technique for measurement of repair synthesis in primary human fibroblasts by incorporation of ethynyl deoxyuridine (EdU). Nucleic Acids Res. 2009; 37:e31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Nakazawa Y., Yamashita S., Lehmann A.R., Ogi T.. A semi-automated non-radioactive system for measuring recovery of RNA synthesis and unscheduled DNA synthesis using ethynyluracil derivatives. DNA Repair. 2010; 9:506–516. [DOI] [PubMed] [Google Scholar]
- 20. Mourgues S., Gautier V., Lagarou A., Bordier C., Mourcet A., Slingerland J., Kaddoum L., Coin F., Vermeulen W., Gonzales de Peredo A. et al. ELL, a novel TFIIH partner, is involved in transcription restart after DNA repair. Proc. Natl. Acad. Sci. U.S.A. 2013; 110:17927–17932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Bohr V.A., Smith C.A., Okumoto D.S., Hanawalt P.C.. DNA repair in an active gene: removal of pyrimidine dimers from the DHFR gene of CHO cells is much more efficient than in the genome overall. Cell. 1985; 40:359–369. [DOI] [PubMed] [Google Scholar]
- 22. Guo J., Hanawalt P.C., Spivak G.. Comet-FISH with strand-specific probes reveals transcription-coupled repair of 8-oxoGuanine in human cells. Nucleic Acids Res. 2013; 41:7700–7712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Mandemaker I.K., Vermeulen W., Marteijn J.A.. Gearing up chromatin: A role for chromatin remodeling during the transcriptional restart upon DNA damage. Nucleus (Austin, Tex.). 2014; 5:203–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Adam S., Polo S.E.. Blurring the line between the DNA damage response and transcription: the importance of chromatin dynamics. Exp. Cell Res. 2014; 329:148–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Mayne L.V., Lehmann A.R.. Failure of RNA synthesis to recover after UV irradiation: an early defect in cells from individuals with Cockayne's syndrome and xeroderma pigmentosum. Cancer Res. 1982; 42:1473–1478. [PubMed] [Google Scholar]
- 26. Adam S., Polo S.E., Almouzni G.. Transcription recovery after DNA damage requires chromatin priming by the H3.3 histone chaperone HIRA. Cell. 2013; 155:94–106. [DOI] [PubMed] [Google Scholar]
- 27. Oksenych V., Zhovmer A., Ziani S., Mari P.-O., Eberova J., Nardo T., Stefanini M., Giglia-Mari G., Egly J.-M., Coin F.. Histone methyltransferase DOT1L drives recovery of gene expression after a genotoxic attack. PLoS Genet. 2013; 9:e1003611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Dinant C., Ampatziadis-Michailidis G., Lans H., Tresini M., Lagarou A., Grosbart M., Theil A.F., van Cappellen W.A., Kimura H., Bartek J. et al. Enhanced chromatin dynamics by FACT promotes transcriptional restart after UV-induced DNA damage. Mol. Cell. 2013; 51:469–479. [DOI] [PubMed] [Google Scholar]
- 29. Stack E.C., Wang C., Roman K.A., Hoyt C.C.. Multiplexed immunohistochemistry, imaging, and quantitation: a review, with an assessment of Tyramide signal amplification, multispectral imaging and multiplex analysis. Methods. 2014; 70:46–58. [DOI] [PubMed] [Google Scholar]
- 30. Faget L., Hnasko T.S.. Tyramide signal amplification for immunofluorescent enhancement. Methods Mol. Biol. 2015; 1318:161–172. [DOI] [PubMed] [Google Scholar]
- 31. Clutter M.R., Heffner G.C., Krutzik P.O., Sachen K.L., Nolan G.P.. Tyramide signal amplification for analysis of kinase activity by intracellular flow cytometry. Cytometry A. 2010; 77:1020–1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Klein B., Pastink A., Odijk H., Westerveld A., van der Eb A.J.. Transformation and immortalization of diploid xeroderma pigmentosum fibroblasts. Exp. Cell. Res. 1990; 191:256–262. [DOI] [PubMed] [Google Scholar]
- 33. Troelstra C., van Gool A., de Wit J., Vermeulen W., Bootsma D., Hoeijmakers J.H.. ERCC6, a member of a subfamily of putative helicases, is involved in Cockayne's syndrome and preferential repair of active genes. Cell. 1992; 71:939–953. [DOI] [PubMed] [Google Scholar]
- 34. Marteijn J.A., Bekker-Jensen S., Mailand N., Lans H., Schwertman P., Gourdin A.M., Dantuma N.P., Lukas J., Vermeulen W.. Nucleotide excision repair-induced H2A ubiquitination is dependent on MDC1 and RNF8 and reveals a universal DNA damage response. J. Cell Biol. 2009; 186:835–847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. van Cuijk L., van Belle G.J., Turkyilmaz Y., Poulsen S.L., Janssens R.C., Theil A.F., Sabatella M., Lans H., Mailand N., Houtsmuller A.B. et al. SUMO and ubiquitin-dependent XPC exchange drives nucleotide excision repair. Nat. Commun. 2015; 6:7499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B. et al. Fiji: an open-source platform for biological-image analysis. Nat. Methods. 2012; 9:676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Bensaude O. Inhibiting eukaryotic transcription: Which compound to choose? How to evaluate its activity?. Transcription. 2011; 2:103–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. David S.S., O'Shea V.L., Kundu S.. Base-excision repair of oxidative DNA damage. Nature. 2007; 447:941–950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Beard W.A., Wilson S.H.. Structure and mechanism of DNA polymerase Beta. Chem. Rev. 2006; 106:361–382. [DOI] [PubMed] [Google Scholar]
- 40. Hazan C., Boudsocq F., Gervais V., Saurel O., Ciais M., Cazaux C., Czaplicki J., Milon A.. Structural insights on the pamoic acid and the 8 kDa domain of DNA polymerase beta complex: towards the design of higher-affinity inhibitors. BMC Struct. Biol. 2008; 8:22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Hu H.Y., Horton J.K., Gryk M.R., Prasad R., Naron J.M., Sun D.A., Hecht S.M., Wilson S.H., Mullen G.P.. Identification of small molecule synthetic inhibitors of DNA polymerase beta by NMR chemical shift mapping. J. Biol. Chem. 2004; 279:39736–39744. [DOI] [PubMed] [Google Scholar]
- 42. Wyatt M.D., Pittman D.L.. Methylating agents and DNA repair responses: methylated bases and sources of strand breaks. Chem. Res. Toxicol. 2006; 19:1580–1594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Chen X., Zhao Y., Li G.M., Guo L.. Proteomic analysis of mismatch repair-mediated alkylating agent-induced DNA damage response. Cell Biosci. 2013; 3:37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Pena-Diaz J., Bregenhorn S., Ghodgaonkar M., Follonier C., Artola-Boran M., Castor D., Lopes M., Sartori A.A., Jiricny J.. Noncanonical mismatch repair as a source of genomic instability in human cells. Mol. Cell. 2012; 47:669–680. [DOI] [PubMed] [Google Scholar]
- 45. Molinete M., Vermeulen W., Burkle A., Menissier-de Murcia J., Kupper J.H., Hoeijmakers J.H., de Murcia G.. Overproduction of the poly(ADP-ribose) polymerase DNA-binding domain blocks alkylation-induced DNA repair synthesis in mammalian cells. EMBO J. 1993; 12:2109–2117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Jia N., Nakazawa Y., Guo C., Shimada M., Sethi M., Takahashi Y., Ueda H., Nagayama Y., Ogi T.. A rapid, comprehensive system for assaying DNA repair activity and cytotoxic effects of DNA-damaging reagents. Nat. Protoc. 2015; 10:12–24. [DOI] [PubMed] [Google Scholar]
- 47. Bauer N.C., Corbett A.H., Doetsch P.W.. The current state of eukaryotic DNA base damage and repair. Nucleic Acids Res. 2015; 43:10083–10101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Lans H., Marteijn J.A., Vermeulen W.. ATP-dependent chromatin remodeling in the DNA-damage response. Epigenet. Chromatin. 2012; 5:4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Boeing S., Williamson L., Encheva V., Gori I., Saunders R.E., Instrell R., Aygun O., Rodriguez-Martinez M., Weems J.C., Kelly G.P. et al. Multiomic Analysis of the UV-Induced DNA Damage Response. Cell Rep. 2016; 15:1597–1610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Newman J.C., Bailey A.D., Fan H.-Y., Pavelitz T., Weiner A.M.. An abundant evolutionarily conserved CSB-PiggyBac fusion protein expressed in Cockayne syndrome. PLoS Genet. 2008; 4:e1000031. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.