 Bioluminescent molecular beacons have been developed using a modular design approach that relies on BRET between the bright luciferase NanoLuc and a Cy3 acceptor.
Bioluminescent molecular beacons have been developed using a modular design approach that relies on BRET between the bright luciferase NanoLuc and a Cy3 acceptor.
Abstract
Bioluminescent molecular beacons have been developed using a modular design approach that relies on BRET between the bright luciferase NanoLuc and a Cy3 acceptor. While classical molecular beacons are hampered by background fluorescence and scattering, these BRET-beacons allow detection of low pM concentrations of nucleic acids directly in complex media.
The sequence specific detection and quantification of nucleic acids plays a vital role in life sciences ranging from molecular diagnostics, imaging and forensics to synthetic biology, genetic engineering and DNA-based molecular computing. Various strategies for nucleic acid detection have been developed. A popular approach that allows direct detection in solution is the fluorescence-based molecular beacon;1 a single-stranded oligonucleotide modified with a fluorophore–quencher pair that forms a stem-loop structure. Upon hybridization of the target oligonucleotide to the complementary loop sequence the stem opens and fluorescence increases due to separation of the fluorophore and quencher. While the output signal of the classical molecular beacon is intensiometric, FRET analogs have been developed by replacing the quencher with an acceptor fluorophore, resulting in a ratiometric output signal that is independent of the sensor concentration.2 However, the sensitivity of these fluorescence-based optical probes is inherently limited because of background fluorescence and scattering of excitation light, in particular in complex media. Given the low concentrations of target oligonucleotide that need to be detected, molecular beacon based detection of nucleic acids typically requires purification and/or pre-amplification of the analyte using PCR. Rather than amplifying the input signal, various strategies have been developed that rely on the amplification of the generated output signal, e.g. by DNA-templated assembly of (split-)protein partners such as enzyme–inhibitor pairs,3 and split-enzymes.4,5 Bioluminescent approaches using split luciferases allow for sensitive detection, as they do not require external excitation and therefore do not suffer from auto-fluorescence and background scattering. However, all of these approaches lack ratiometric detection provided by the FRET-based molecular beacons and thus require accurate calibration.
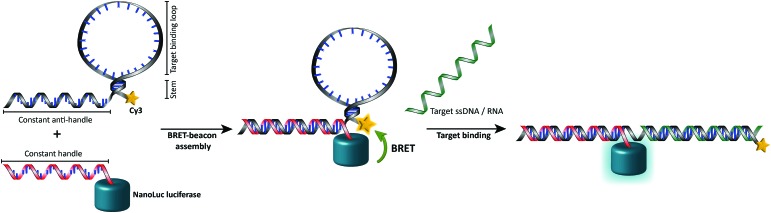
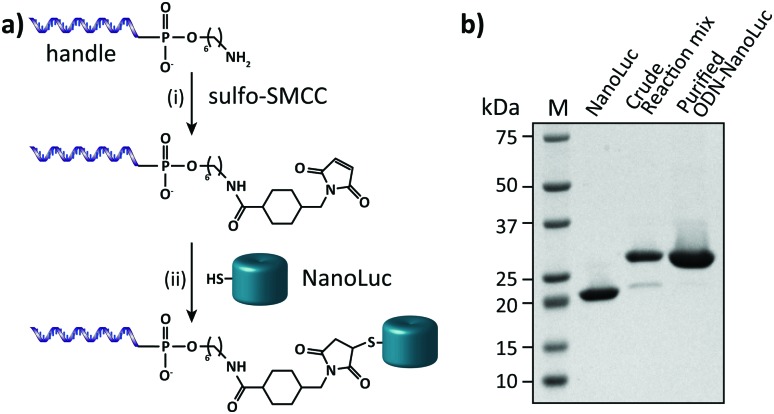
In this work we aimed to combine the robust and programmable design principles of the molecular beacon with the advantages of bioluminescence detection by developing a protein–DNA hybrid molecular sensor based on bioluminescence resonance energy transfer (BRET). In our approach the luciferase NanoLuc is conjugated to the 5′-end of a handle sequence that is complementary to an anti-handle sequence appended to a stem-loop structure, which at its 3′-end is functionalized with a Cy3 acceptor dye (Fig. 1). This separation of target recognition motif and the donor-luciferase allows the BRET-beacon to be easily adapted to different target sequences using only a single protein–DNA conjugate, thus significantly reducing synthetic efforts and costs. NanoLuc was chosen as the donor luciferase, because it exhibits superior luminescence intensity (∼150 fold brighter than firefly and Renilla luciferase) as well as high thermal and chemical stability.6 NanoLuc has already been successfully applied in BRET-based assays that allow detection of low molecular weight therapeutic compounds and antibodies directly in blood plasma and serum.7,8 The 21 nucleotide long, amine-modified handle oligonucleotide (ODN) was initially successfully conjugated to the only native cysteine (Cys164) in NanoLuc via the heterobifunctional crosslinker sulfo-succinimidyl 4-(N-maleimidomethyl)cyclohexane-1 carboxylate (Sulfo-SMCC). However, conjugation of the ODN at this site almost completely abolished the protein's activity (data not shown).9 We therefore mutated this cysteine to a serine residue and introduced a new cysteine residue at the flexible C-terminus of the protein (ESI†). Conjugation of a 3-fold excess of maleimide-functionalized ODN to the cysteine at this position did not affect the luciferase activity and resulted in a conjugation efficiency of >90% as estimated from SDS-PAGE analysis (Fig. 2b). Subsequent purification of the crude reaction mixture using consecutive Ni2+-affinity chromatography (to remove excess ODN) and anion exchange chromatography (to remove unreacted NanoLuc) yielded the ODN–NanoLuc conjugate in >99% purity.
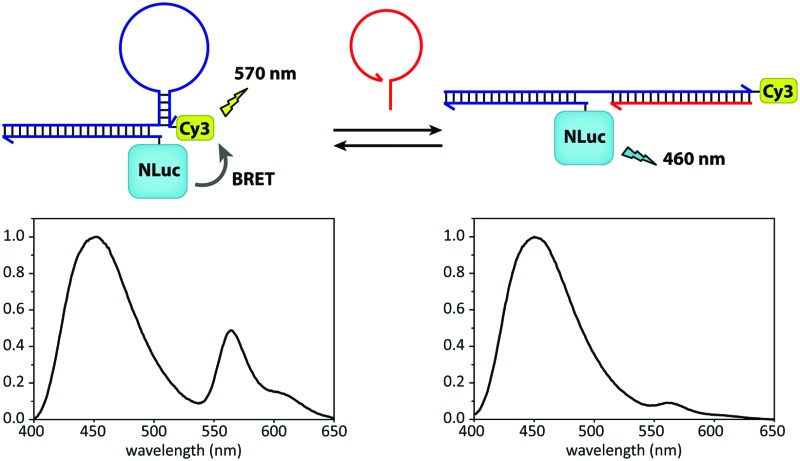
Fig. 1. General concept of the intermolecularly assembled BRET-beacon. The sensor is constructed by site-specifically conjugating a handle-oligonucleotide to NanoLuc and hybridizing it to an anti-handle sequence appended to the target-binding stem-loop structure. The stem-loop is modified with a synthetic acceptor fluorophore (Cy3) to facilitate the readout of target oligonucleotide binding by measuring BRET efficiencies. Upon the hybridization of the target oligonucleotide to the complementary stem-loop sequence a rigid double-helix is formed that disrupts the stem, hereby separating NanoLuc and the synthetic acceptor fluorophore, causing the BRET efficiency to decrease.
Fig. 2. Synthesis and characterization of the semi-synthetic protein–DNA hybrid. (a) To obtain site-specific conjugation of the oligonucleotide handle to NanoLuc a 5′-amine-modified oligonucleotide is reacted with the heterobifunctional linker Sulfo-SMCC (i). Subsequently, the maleimide-activated oligonucleotide is reacted with a cysteine that is inserted in the C-terminus of NanoLuc (ii). (b) 4–20% SDS-PAGE analysis of the conjugation reaction of the oligonucleotide handle to NanoLuc and the final ODN–NanoLuc product. Conjugation efficiency based on relative band intensities of the crude reaction mixture is estimated to be >90%. Similarly, the purity of the final product is estimated to be >99%.
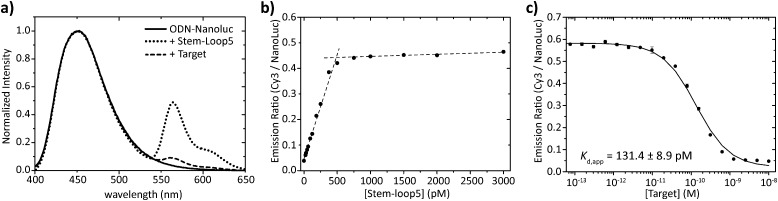
Having obtained pure ODN–NanoLuc, we next tested the assembly of the BRET-beacon using bioluminescence spectroscopy (Fig. 3a). Addition of 20 nM of a Cy3-functionalized stem-loop with a 5 bp stem to 10 nM ODN–NanoLuc resulted in efficient energy transfer from NanoLuc (λ em = 452 nm) to Cy3 (λ em = 564 nm), indicating the correct assembly of the BRET-beacon. Subsequent addition of a complementary target oligonucleotide resulted in a large, ∼13-fold decrease of Cy3/NanoLuc emission ratio, consistent with an increase in the NanoLuc–Cy3 distance due to opening of the stem-loop conformation. Similar responses were observed using 4 and 6 base pair stems, with an increase in stem length yielding a slightly higher emission ratio in the closed state (Fig. S2, ESI†). To establish the efficiency of formation of the BRET beacon, a titration experiment was performed in which increasing amounts of the stem-loop with a 5 base pair stem (stem-loop5) were added to 500 pM ODN–NanoLuc. (Fig. 3b, see Fig. S2 for similar titrations with stem-loop 4 and 6 base pairs and with anti-handle-Cy3.) The emission ratio increased linearly between 0 and 500 pM, followed by a sharp transition to a maximal emission ratio above 500 pM. These results are consistent with the formation of a tight complex between ODN–NanoLuc and the anti-handle of the stem-loop structure. In all subsequent experiments a 2-fold molar excess of stem-loop was used to avoid the presence of small and varying amounts of free ODN–NanoLuc. Having established efficient assembly of the bioluminescent molecular beacon, we next assessed its analytical performance. To allow the detection of low target concentrations, the concentration of the BRET-beacon was lowered to 4 pM (2 pM ODN–NanoLuc and 4 pM stem-loop5, see Fig. S3 for stability determination, ESI†). Titration of fully complementary target and fitting of eqn (S1) (ESI†) to the experimental data resulted in an apparent affinity (K d,app) of 131 ± 9 pM (Fig. 3c). The limit of detection (LOD) under these conditions was 5.6 pM in 50 μL sample, corresponding to 283 attomol of analyte. These concentrations are approximately 3 orders of magnitude lower than can be achieved using fluorescent molecular beacons and 2 orders of magnitude lower than previously reported BRET systems that employed less efficient luciferases and different probe designs.10,11 Please note that to reach full equilibrium of target binding at these low concentrations, incubation times of at least 4 hours are required.
Fig. 3. Characterization of the sequential assembly and target binding of the bioluminescent molecular beacon. (a) Normalized emission spectra of 10 nM ODN–NanoLuc (solid line), 10 nM ODN–NanoLuc hybridized to 20 nM stem-loop5 to form the bioluminescent molecular beacon (dotted line) and the bioluminescent molecular beacon incubated with 200 nM target oligonucleotide (dashed line). (b) Titration of various concentrations of stem-loop5 to 500 pM of ODN–NanoLuc. The stem-loop tightly binds with its anti-handle to the handle on ODN–NanoLuc as indicated by the linear increase in BRET-ratio up to stoichiometric concentrations of the stem-loop. Dotted lines represent linear fits of the experimental data from 0 pM to 500 pM and 500 pM to 3000 pM stem-loop5. (c) Titration of various concentrations of fully complementary target to 4 pM BRET-beacon5 (2 pM ODN–NanoLuc, 4 pM stem-loop5). An apparent affinity of K d,app = 131.4 ± 8.9 pM is calculated by fitting of eqn (S1) (ESI†) to the experimentally obtained data. Triplicate measurements were performed in TE/Mg/NaCl buffer supplemented with 1 mg mL–1 BSA (10 mM Tris–HCl, 1 mM EDTA, 12.5 mM MgCl2, 150 mM NaCl, 1 mg mL–1 BSA, pH 8.0, 1 : 5000 substrate).
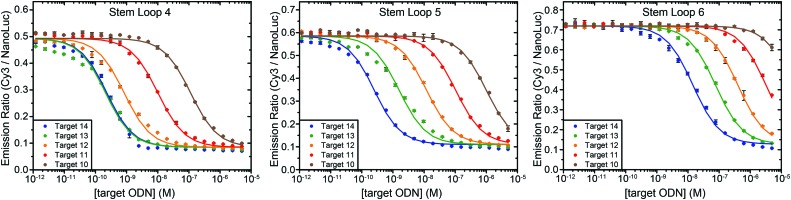
One of the attractive features of the BRET beacon design is that both the sequence of the loop and the length of the stem can be easily adapted without having to change the ODN–NanoLuc conjugate. Whereas stem lengths of 4, 5 and 6 bp yield sensors with a similar BRET response, varying the length of the stem could provide a way to tune the apparent affinity of the sensor for its intended target oligonucleotide. Fig. 4 shows experiments in which target oligonucleotides of different lengths (between 10 to 14 nucleotides) were titrated to BRET-beacons with different stem lengths (4, 5 or 6 base pairs). Using the same sensor, a 10–20 fold increase in apparent affinity is observed per nucleotide that is appended to the target oligonucleotide (Table S3, ESI†). However, when the apparent affinities of the target sequences become similar to the sensor concentration (e.g. targets 13 and 14 with stem loop 4), the titration curves of target sequences with different length start to coincide. The apparent affinity for the target oligonucleotide can be modulated by either decreasing or increasing the stability of the stem, however. Increasing the stem length by a single base pair weakens the apparent affinity for the target oligonucleotides by one order of magnitude. E.g., the affinity of the 12 nucleotide target is 1 nM, 10 nM, and 400 nM for sensors with stem loops of 4, 5 and 6 base pairs, respectively. In this way, the ability to discriminate between targets differing by only one nucleotide can be easily optimized for different target sequences, or the responsive concentration range can be tuned for the intended application.12 The 10–20 fold change in apparent affinity per additional base pair is consistent with previous studies on molecular beacons and determined by the thermodynamics of DNA hybridization, which show that each additional base pair contributes approximately 1.5 kcal mol–1 in free energy.13
Fig. 4. Tuning the response of the bioluminescent molecular beacon towards targets with varying affinities by stabilizing the stem (i.e. increasing the number of base pairs). The assays were performed using 10 pM ODN–NanoLuc, 20 pM stem-loop and 1 pM–5 μM target oligonucleotides in TE/Mg/NaCl buffer supplemented with 1 mg mL–1 BSA (10 mM Tris–HCl, 1 mM EDTA, 12.5 mM MgCl2, 150 mM NaCl, 1 mg mL–1 BSA, pH 8.0, 1 : 5000 substrate). Errorbars represent the standard error of the mean of duplicate measurements. The solid lines are global fits of eqn (S1) (ESI†) to the experimental data per stem loop length.
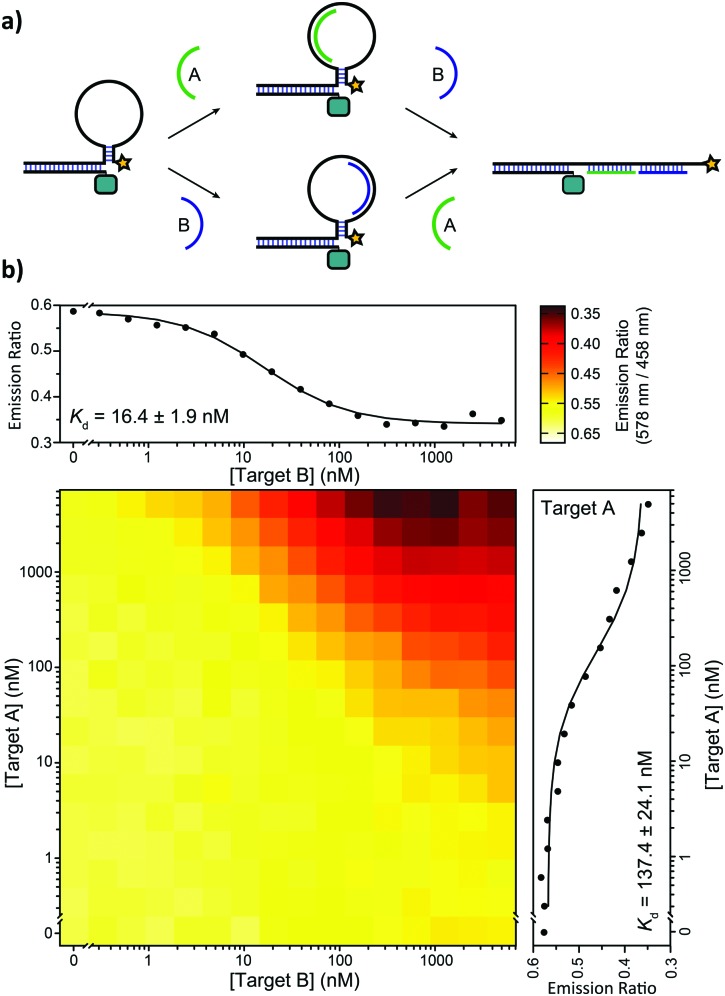
We also tested the ability of the BRET-beacon to detect the presence of two oligonucleotides in the form of an AND-gate. The development of such Boolean logic operations is important for the readout and detection of combinations of oligonucleotide triggers, which could proof useful for disease diagnosis and the characterization of DNA-based molecular circuits.14 Two small target oligonucleotides (10 nucleotides each) were designed to bind to a part of the target recognition loop (Fig. 5a). The heat map in Fig. 5b shows BRET-ratios as a function of the concentrations of both target oligonucleotides. No increase in BRET-ratio is observed when only one of the two target strands is added. This confirms that upon binding of only one of the two target strands the flexibility of the unbound region of the loop is sufficient to keep the stem in its closed conformation. The BRET-ratio decreases upon addition of both target oligonucleotides, confirming that the nicked double-helix that is formed upon addition of the second target oligonucleotide imposes sufficient mechanical strain on the stem to disrupt it. Individual binding affinities for both targets were determined in the presence of saturating concentrations of the other target (5 μM), resulting in apparent dissociation constants of K d,TargetA = 137 ± 24 nM and K d,TargetB = 16.4 ± 1.9 nM for target A and target B, respectively.
Fig. 5. Implementation of AND-gate Boolean logic. (a) Two 10 nt target oligonucleotides (A and B) bind independently in a portion of the loop region. When one of the two targets binds, the free binding region of the other target provides enough flexibility to keep the stem-loop in the closed conformation. The mechanical strain induced by binding of both targets forces the stem to open, resulting in a decrease in BRET. (b) Heatmap showing BRET-ratios in the presence of a concentration range of both target oligonucleotides. The affinity of the individual targets is determined at saturating concentrations of the other target (5 μM). The assays were performed using 10 pM ODN–NanoLuc, 20 pM stem-loop and 0 nM–5 μM of the individual target oligonucleotides in TE/Mg/NaCl buffer with 1 mg mL–1 BSA and 1 : 5000 substrate.
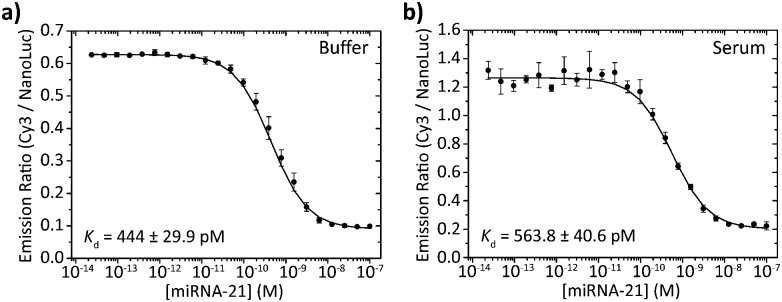
To assess the performance of the BRET beacons in a complex medium we tested their ability to detect the DNA analog of the clinically relevant micro-RNA21 sequence directly in human serum. miRNA21 is a small (22 nt), non-coding RNA sequence whose overexpression is associated with the development of various types of cancer.15 To allow the detection of this miRNA21 mimic, the loop sequence of the stem-loop structure was redesigned to be complementary to the sequence of miRNA21. Fig. 6 shows experiments in which the deoxyribose analog of miRNA21 was titrated to 50 pM of BRET-beacon in either buffer or pooled human serum. To prevent possible degradation of the BRET-beacon by nucleases present in human serum, we added 10 μM of an inert T20 oligonucleotide carrier strand. The absolute emission ratios are 2-fold higher in serum compared to buffer possibly due to reabsorption of light by serum components, but the relative change in emission ratio is very similar. Both titration experiments also reported similar apparent affinities for the miRNA21 analog of K d = 444 ± 30 pM and K d = 564 ± 41 pM in buffer and serum, respectively. These results convincingly show the excellent performance of BRET-based detection in human serum.
Fig. 6. miRNA21 detection in buffer and serum. 50 pM BRET-beacon (25 pM ODN–NanoLuc prehybridized to 50 pM stem-loop) was incubated with various concentrations of a synthetic deoxyribose analog of miRNA-21 in (a) TE/Mg/NaCl buffer supplemented with 1 mg mL–1 BSA and (b) 1 times diluted pooled human serum. To reduce degradation of the BRET-beacon by nucleases present in serum, 10 μM of a T20 oligonucleotide was added as carrier strand in both buffer and serum measurements. Errorbars represent the standard deviation of triplicate measurements.
In conclusion, BRET-beacons have been developed as a new bioluminescent sensor platform for the detection of ssDNA and RNA. By using the bright luciferase NanoLuc these BRET beacons are at least 2 orders of magnitude more sensitive than previously reported nucleic acid BRET sensors. Because of their high stability, well-defined conjugation chemistry and modular assembly, BRET-beacons represent an attractive sensor format for numerous applications in molecular diagnostics, molecular biology, and DNA-based computing.
We thank Dr Tom de Greef and Ir. Remco Arts for fruitful discussions during this research. This work was supported by an ERC starting grant (ERC-2011-StG 280255) and was partially conducted in the framework of the project NextDx, funded within the European Commission FP7 program.
Footnotes
References
- Tyagi S., Kramer F. R. Nat. Biotechnol. 1996;14:303–308. doi: 10.1038/nbt0396-303. [DOI] [PubMed] [Google Scholar]
- Zhang P., Beck T., Tan W. Angew. Chem. 2001;113:416–419. [Google Scholar]
- Janssen B. M. G., Engelen W., Merkx M. ACS Synth. Biol. 2015;4:547–553. doi: 10.1021/sb500278z. [DOI] [PubMed] [Google Scholar]
- Sancho Oltra N., Bos J., Roelfes G. ChemBioChem. 2010;11:2255–2258. doi: 10.1002/cbic.201000517. [DOI] [PubMed] [Google Scholar]
- Furman J. L., Badran A. H., Ajulo O., Porter J. R., Stains C. I., Segal D. J., Ghosh I. J. Am. Chem. Soc. 2010;132:11692–11701. doi: 10.1021/ja104395b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall M. P., Unch J., Binkowski B. F., Valley M. P., Butler B. L., Wood M. G., Otto P., Zimmerman K., Vidugiris G., Machleidt T., Robers M. B., Benink H. A., Eggers C. T., Slater M. R., Meisenheimer P. L., Klaubert D. H., Fan F., Encell L. P., Wood K. V. ACS Chem. Biol. 2012;7:1848–1857. doi: 10.1021/cb3002478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griss R., Schena A., Reymond L., Patiny L., Werner D., Tinberg C. E., Baker D., Johnsson K. Nat. Chem. Biol. 2014;10:598–603. doi: 10.1038/nchembio.1554. [DOI] [PubMed] [Google Scholar]
- Arts R., den Hartog I., Zijlema S. E., Thijssen V., van der Beelen S. H. E., Merkx M. Anal. Chem. 2016;88:4525–4532. doi: 10.1021/acs.analchem.6b00534. [DOI] [PubMed] [Google Scholar]
- Inouye S., Sasaki S. Protein Expression Purif. 2007;56:261–268. doi: 10.1016/j.pep.2007.08.002. [DOI] [PubMed] [Google Scholar]
- Kumar M., Zhang D., Broyles D., Deo S. K. Biosens. Bioelectron. 2011;30:133–139. doi: 10.1016/j.bios.2011.08.043. [DOI] [PubMed] [Google Scholar]
- Hunt E. A., Deo S. K. Chem. Commun. 2011;47:9393–9395. doi: 10.1039/c1cc13495g. [DOI] [PubMed] [Google Scholar]
- Ricci F., Vallée-Bélisle A., Simon A. J., Porchetta A., Plaxco K. W. Acc. Chem. Res. 2016;49:1884–1892. doi: 10.1021/acs.accounts.6b00276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SantaLucia J., Hicks D. Annu. Rev. Biophys. Biomol. Struct. 2004;33:415–440. doi: 10.1146/annurev.biophys.32.110601.141800. [DOI] [PubMed] [Google Scholar]
- Großhans H., Filipowicz W. Nature. 2008;451:414–416. doi: 10.1038/451414a. [DOI] [PubMed] [Google Scholar]
- Volinia S., Calin G. A., Liu C.-G., Ambs S., Cimmino A., Petrocca F., Visone R., Iorio M., Roldo C., Ferracin M., Prueitt R. L., Yanaihara N., Lanza G., Scarpa A., Vecchione A., Negrini M., Harris C. C., Croce C. M. Proc. Natl. Acad. Sci. U. S. A. 2006;103:2257–2261. doi: 10.1073/pnas.0510565103. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.