Abstract
The nucleosome, comprising a histone protein core wrapped around by DNA, is the fundamental packing unit of DNA in cells. Lysine acetylation at the histone core elevates DNA accessibility in the nucleosome, the mechanism of which remains largely unknown. By employing our recently developed hybrid single molecule approach, here we report how the structural dynamics of DNA in the nucleosome is altered upon acetylation at histone H3 lysine 56 (H3K56) that is critical for elevated DNA accessibility. Our results indicate that H3K56 acetylation facilitates the structural dynamics of the DNA at the nucleosome termini that spontaneously and repeatedly open and close on a ms time scale. The results support a molecular mechanism of histone acetylation in catalyzing DNA unpacking whose efficiency is ultimately limited by the spontaneous DNA dynamics at the nucleosome temini. This study provides the first and unique experimental evidence revealing a role of protein chemical modification in directly regulating the kinetic stability of the DNA packing unit.
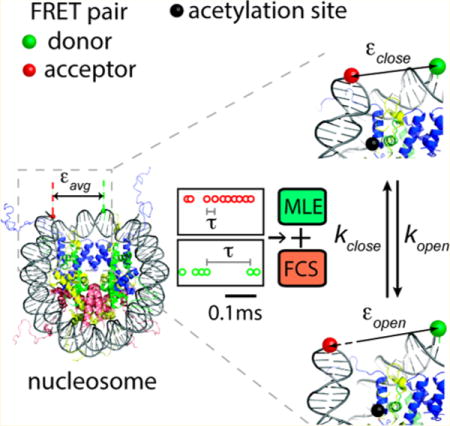
Graphical abstract
INTRODUCTION
Chemical modifications of protein such as acetylation, methylation, and phosphorylation play important roles in many vital cellular processes.1–4 These modifications may act as biochemical markers to recruit binding partners or directly impact on the physical properties of protein.5–7 In particular, modifications with small chemical groups such as lysine ε-amine acetylation often have only weak binding partners,8–10 suggesting that changes in the physical properties of protein may be implicated in their mechanisms of action.
The nucleosome is the fundamental packing unit of DNA in eukaryotic cells.11 Dynamic changes in the structure and structural flexibility of the nucleosome are associated with various mechanisms of DNA accessibility regulation.12–14 The nucleosome is composed of an octameric histone protein core wrapped around by ∼147 bp DNA.15 It is now widely accepted that chemical modifications of DNA and histones are directly involved in regulating various vital cellular processes.16,17
Lysine ε-amine acetylation in the nucleosome is typically associated with elevated DNA accessibility, the mechanism of which remains largely unknown. Binding of enzymes that can specifically recognize acetylated lysine is too weak to form a stable complex for cascading events.9,10 Therefore, physical changes to the nucleosome might be implicated in elevating DNA accessibility upon lysine acetylation. It has been hypothesized that the structural integrity of the nucleosome would be compromised upon lysine acetylation so that DNA unwrapping and nucleosome disassembly become facilitated.14 In particular, histone H3 lysine 56 (H3K56) is in proximal contact with the nucleosomal DNA termini (Figure 1A) via electrostatic interaction which would be considerably weakened by removal of the positive charge of the ε-amine upon its acetylation. The weakened DNA-histone interaction would result in flexible nucleosome termini that would facilitate DNA unwrapping and subsequent nucleosome disassembly.18–20 Previous ensemble measurements have demonstrated facilitated nucleosomal DNA unwrapping or protein binding upon H3K56 acetylation.18–20 However, these measurements depended on large-scale unwrapping induced by factor binding or thermodynamic equilibrium between wrapped and unwrapped states. Information on the kinetics of spontaneous transient breathing motion of the nucleosomal DNA termini is critical to understanding the molecular mechanism and dynamics of the large-scale unwrapping and nucleosome disassembly. This is because the spontaneous breathing dynamics should be what eventually triggers the large-scale unwrapping and, therefore, imposes the upper limit to the protein binding rate to DNA wrapped in the nucleosome.13
Figure 1.
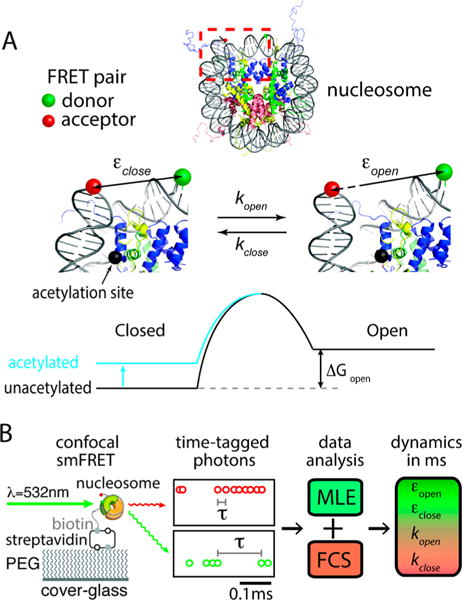
Schematic description of the hybrid single molecule method that revealed lysine acetylation effect on DNA dynamics in the nucleosome. (A) The nucleosome is labeled with a FRET donor (Cy3, green sphere) and an acceptor (ATTO647N, red sphere) at the DNA dyad and at a terminus, respectively. Note that these two states (closed and open) still have the DNA in a traditionally defined wrapped state. Lysine acetylation may shift the free energy of the DNA opening motion at the terminus (ΔGopen). (B) The nucleosome was immobilized on a poly(ethylene glycol) (PEG) coated cover glass via biotin–streptavidin conjugation at the terminus opposite to where ATTO647N is labeled (see Figure S2 for labeling positions). An example of streams of time-tagged photons from a FRET pair (Cy3, green; ATTO647N, red) is shown. A hybrid approach combining maximum likelihood estimation (MLE) and fluorescence correlation spectroscopy (FCS) was employed to investigate lysine acetylation effects on DNA dynamics in the nucleosome on a ms time scale.
We previously reported spontaneous and repetitive DNA opening motion at the nucleosome termini on a ms time scale.21 By employing our recently developed single molecule approach, here we report the first direct experimental evidence of the effect of H3K56 acetylation (H3K56ac) on the kinetics and thermodynamics of this fast transitional fluctuation. According to our results, H3K56ac makes the nucleosome termini more flexible and drives the equilibrium more to the open metastable state where the DNA is still in a traditionally defined wrapped state, which is in line with its role in facilitating nucleosome disassembly. This study provides unprecedented experimental evidence pointing to a molecular mechanism of protein chemical modification in directly regulating the kinetic stability of the fundamental DNA packing unit.
EXPERIMENTAL METHODS
Protein Expression, Purification, and Acetylation
The histone proteins were expressed and purified as previously described.21 H3K56C/C110A double mutant was used for acetylating histone H3 in a fully unfolded state. Acetylation at H3K56 (H3K56ac) was through thiol–ene coupling between cysteine thiol and N-vinylacetamide (NVA), which is functionally equivalent to the natural acetylation at histone.22 See Figure S1 for the mass-spectrometric analysis to confirm acetylation. The H3 K56C/C110A double mutant was dissolved in a buffer of 200 μL total volume containing 0.2 M sodium acetate (pH 4), 6 M guanidine–HCl, 7 mM L-glutahione, 50 mM N-vinylacetamide, 100 mM dimethyl sulfide, and 5 mM VA-044 (2,2′-[azobis(dimethylmethlene)]bis(2-imidazoline)-dihydrochoride), resulting in the final protein concentration of 1 mM. The reaction mixture was incubated for 2 h at 70 °C and dialyzed against deionized water and then lyophilized overnight for storage at −80 °C.
Nucleosome Reconstitution
The nucleosome samples were prepared by salt step dialysis as previously described.21 For nucleosomal DNA, a single strand 20 base linker with a biotin at the 5′ end was added to the Watson (forward) strand of a 147 bp human α-satellite sequence. Ligation of the six fragments yields the DNA substrates where Cy3 was labeled by replacing the 76th nucleotide of the Watson (forward) strand and ATTO647N was conjugated to the 5′ terminus of the Crick (reverse) strand. See Figure S2 for the details on the DNA sequence and the labeling positions.
FRET Data Analysis
Single molecule FRET measurements were performed on a scanning confocal microscope as previously described.21 Two streams of photon arrival times for Cy3 and ATTO647N were recorded at a resolution of 50 ns. The photon arrival times were processed with maximum likelihood estimation (MLE) to estimate the FRET efficiencies of the closed and open states which were further combined with fluorescence correlation spectroscopy (FCS) to reveal the kinetic rates between the two states. The detailed analysis procedure has been published previously.21 The MLE analysis resulted in the high, low, and average FRET efficiencies (εclose, εopen, and εavg respectively) that are not corrected with possible variations of the quantum efficiencies of the fluorophores, and therefore, are only approximate. Interpreting the approximate high and low FRET efficiencies as closed and open nucleosomal states has already been validated previously with a FRET-labeled DNA control that did not show this dynamics.21 In combination with the high and low FRET efficiencies, the decay time constant and the amplitude of the ms component in the FCS spectra were utilized to compute the kinetic rates, kopen and kclose, with the following equation. Note that the FCS spectra were fit to a double exponential decay function, the faster component of which is due to fluorophore photophysics.21 The details of the fitting procedure have been previously published.21 Briefly, we made sure that the dynamics component of our interest is not affected by arbitrarily set fitting boundaries. See Tables S1 and S2 for further details on the fitting results.
RESULTS AND DISCUSSION
Our experimental system is based on single molecule fluorescence resonance energy transfer (smFRET). The FRET efficiency time traces of the nucleosome particles report the spontaneous opening motion of the nucleosomal DNA termini (Figure 1). Note that both the closed and open states still have the DNA in a traditionally defined wrapped state. The nucleosome is made of Xenopus laevis core histones and 147 bp human α satellite DNA sequence with tandem 20 base-long single stranded DNA linker for surface immobilization (Figure 1B). Site-specific acetylation at histone H3K56 was achieved with thiol–ene coupling between cysteine thiol and N-vinylacetamide (NVA) following a published protocol.22 Histones acetylated with this strategy have been successfully used to reproduce the results obtained with histones acetylated23,24 with native chemical ligation.24,25 The end of the linker DNA is labeled with biotin. The nucleosome particles were immobilized on a poly(ethylene glycol) coated microscope cover glass via streptavidin–biotin conjugation.21 The FRET donor (Cy3) was excited with a 532 nm laser in a confocal geometry at a power level inducing a 25–40 kHz average photon detection rate from a FRET pair. All the fluorescence photons from a FRET pair were time-tagged at a 100 ns resolution and analyzed to obtain the parameters of the smFRET dynamics (Figure 1B). Data was collected only when the average FRET efficiency is within a range of 0.6–0.8 in order to avoid partially disassembled or noncanonical nucleosomes because we estimated the FRET efficiency from the canonical nucleosome to be 0.7 (see Supporting Information for DNA sequence and fluorophore positions). We also excluded a small (<5%) population showing fluorophore blinking on a few hundred ms time scale. These are likely due to abnormal photophysical behavior of fluorophore impurities as previously reported.26
As shown in Figure 2A, 1 ms integration of the fluorescence photons did not show any sign of closed and open states that are apparently separable from each other. To this end, we used all the collected fluorescence photons to identify the FRET efficiencies of the open and the closed states based on maximum likelihood estimation (MLE) (Figure 2B).21,27,28 The two FRET efficiencies are 0.76–0.79 and 0.60–0.62 respectively for the closed and the open states in the acetylated and the unacetylated nucleosomes. The average FRET efficiency is 0.70–0.73 that is in a good agreement with the estimated value. The average FRET efficiency stays constant within error at 50 and 100 mM NaCl regardless of the acetylation status, indicating that H3K56ac does not induce any change in the time-averaged nucleosome structure that is detectable with our setup. The FRET efficiency values of the open and the closed states are significantly different from each other (Figure 2B), validating the termini opening motion. These values stay constant across the samples at 50 and 100 mM NaCl, indicating that the structures of the open and the closed states of the acetylated nucleosome are identical within error to those of the unacetylated nucleosome at these salt levels.
Figure 2.
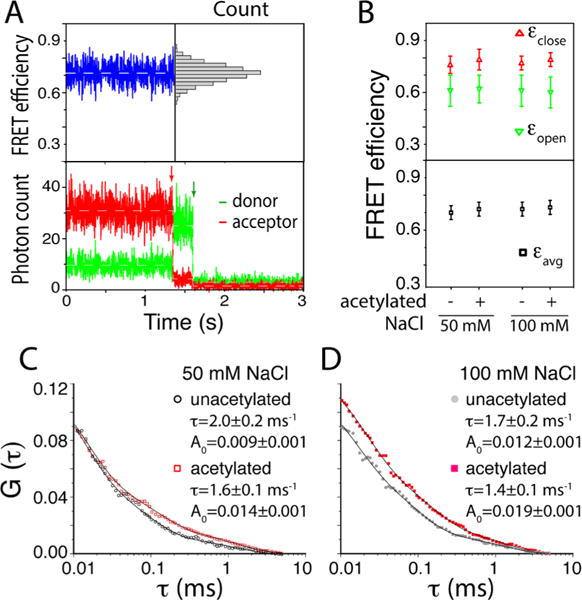
Hybrid single molecule approach combining maximum likelihood estimation (MLE) and fluorescence correlation spectroscopy (FCS) revealing the effects of lysine acetylation on spontaneous DNA dynamics in the nucleosome. (A) Representative fluorescence intensity time traces of a FRET pair (bottom) and the corresponding FRET efficiency trace (top) showing an approximate average FRET efficiency of 0.7 with a unimodal distribution (top right). The approximate FRET efficiency was calculated with Iacceptor/(Idonor + Iacceptor) where I is the fluorescence intensity. The red and green arrows in the intensity traces indicate time points when the acceptor and the donor, respectively, were photobleached. The total numbers of nucleosome particles analyzed are 442 and 194 respectively for acetylated and unacetylated cases at 50 mM NaCl and 250 and 229 respectively for acetylated and unacetylated cases at 100 mM NaCl. (B) MLE results revealing FRET efficiencies of the closed (εclose) and the open (εopen) states (top) along with the average FRET efficiency (εavg) (bottom) for the nucleosomes made with the unacetylated and acetylated histone cores. (C) FCS spectra of ATTO647N from unacetylated and acetylated nucleosomes at two salt concentrations. The fitting results for the DNA dynamics component are shown (the decay time (τ) and the amplitude (A0)).
The kinetic rates for the opening and the closing motions obtained from MLE contain large errors far exceeding the differences between the acetylated and unacetylated cases, making direct interpretation of the results impossible. To resolve this issue, we employed a hybrid MLE-FCS analysis as we published recently.21 Fluorescence emission from the FRET acceptor (ATTO 647N) was analyzed to construct FCS spectra as shown in Figure 2, parts C and D. The spectra were fit with exponential decay functions to obtain the time constants of the dynamics. The hybrid analysis with the FRET efficiency values from MLE and the time constants and amplitudes from FCS (Supporting Information) revealed the kinetic rates of the opening and the closing motions with reasonable uncertainties (Figure 3A). The FCS spectra contain more than a single decay constant and the curves were fit with a double exponential decay function in the range of 0.1–7 ms. The faster decay component should be due to the dynamics of the fluorophores as we previously reported.21 The component in the 1–2 ms range should be due to the DNA opening motion because no dynamics in this range was identified with the DNA-only control as previously reported.21
Figure 3.
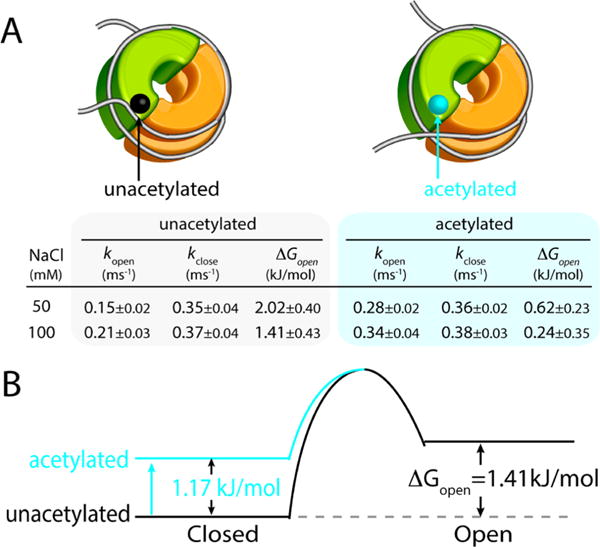
Hybrid single molecule approach combining MLE and FCS, which reveals the kinetics and the thermodynamics of the DNA termini opening motion in the unacetylated and acetylated nucleosomes. (A) Opening (kopen) and closing (kclose) rate constants, and the free energy change (ΔGopen) of the spontaneous DNA opening calculated from the rate constants for the acetylated and unacetylated nucleosomes at two salt concentrations (50 and 100 mM NaCl). (B) The free energy of the closed state increases upon acetylation and ΔGopen consequently decreases. The values shown are for the 100 mM NaCl cases. Note that K = kopen/kclose = [open]/[closed].
As shown in Figure 3A, the unacetylated nucleosomal DNA termini open once every 7 ms and close within 3 ms at 50 mM NaCl. At 100 mM, they open more frequently (once every 5 ms) and close within 3 ms. These results confirm that the interaction between the histone core and the DNA is electrostatic and consequently the increased ionic strength of the environment stabilizes the open state. The stabilization effect is mainly by increasing the opening rate while maintaining a constant closing rate. According to the results (Figure 3A), H3K56ac facilitates the opening motion at both 50 and 100 mM NaCl while it does not affect the closing rate significantly. Between the acetylated cases, the DNA termini are also more dynamics at 100 mM NaCl than at 50 mM NaCl, further confirming the electrostatic nature of the histone–DNA termini interaction. The acetylation effect in the opening rate is more pronounced at 50 mM NaCl likely because the motion is already somewhat facile at 100 mM NaCl. At both salt levels, the equilibrium (ΔGopen in Figure 3A) is significantly shifted to the open conformation upon H3K56 acetylation. Note that the open state is a metastable transitional state that has the DNA termini still in a traditionally defined wrapped state.
On the basis of these results, we constructed a kinetic model for the effect of H3K56 acetylation on the spontaneous DNA termini opening motion (Figure 3B). The reason why we modeled the transition state energy to be constant regardless of H3K56ac is as following. We assumed that the open state stability is likely constant regardless of H3K56ac because the interaction between DNA termini and H3K56 should be very weak or nonexisting in the open state. Consequently, the transition state energy should stay constant and the closed state energy should be elevated in order to account for our experimental results of constant closing rate and increased opening rate upon acetylation. Figure 3B shows that the effect of facilitated opening motion upon H3K56 acetylation is mainly achieved by destabilizing the closed state and the equilibrium is consequently shifted to the open state (decrease in ΔGopen, Figure 3A). This change is likely due to weakened histone core interaction with DNA termini upon removal of the positive charge at the lysine residue (Figure 1A) as previously suggested.18–20
Previous reports based on ensemble measurements demonstrated that the dynamics of large-scale DNA unwrapping induced by factor binding is facilitated upon H3K56ac.19,20 Our results on the salt dependence and the H3K56ac effect of the DNA termini dynamics suggest that the transient spontaneous DNA motion we report here is what eventually triggers the large-scale unwrapping. The large-scale unwrapping if it were spontaneous must happen on a time scale longer than at least a few seconds because we did not observe this motion from the majority of the particles within a few seconds. Therefore, spontaneous DNA unwrapping on this length scale is an unlikely mechanism of DNA access in cells because the time scale is too long. Instead, faster smaller-scale DNA dynamics that may open DNA transiently out of the nucleosome is more likely important for DNA access. We suggest that transient ternary complex formation among DNA, histone core, and a DNA/histone binding protein may be triggered by fast DNA opening motion and should precede large-scale DNA unwrapping. Consequently, the rate of protein binding and DNA unwrapping would be fundamentally limited by the opening rate we report here. H3K56ac would push this limit, eventually facilitating DNA access.
CONCLUSIONS
We report small, fast, and spontaneous DNA opening motion at the nucleosome termini on a ms time scale that is shortened upon H3K56 acetylation, supporting a mechanism of DNA access wrapped in the nucleosome. This study provides unprecedented experimental evidence revealing a role of protein chemical modification in directly regulating the kinetic stability of the fundamental DNA packing structure.
Supplementary Material
Acknowledgments
This work was supported by NIH Grant GM097286 (T.-H.L.).
Footnotes
Supporting Information
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acs.jpcb.5b09734.
Notes
The authors declare no competing financial interest.
References
- 1.Fierz B, Muir TW. Chromatin as an Expansive Canvas for Chemical Biology. Nat Chem Biol. 2012;8(5):417–427. doi: 10.1038/nchembio.938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Valkevich EM, Guenette RG, Sanchez NA, Chen YC, Ge Y, Strieter ER. Forging Isopeptide Bonds Using Thiol-ene Chemistry: Site-Specific Coupling of Ubiquitin Molecules for Studying the Activity of Isopeptidases. J Am Chem Soc. 2012;134(16):6916–6919. doi: 10.1021/ja300500a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Li X, Foley EA, Molloy KR, Li Y, Chait BT, Kapoor TM. Quantitative Chemical Proteomics Approach to Identify Post-Translational Modification-Mediated Protein-Protein Interactions. J Am Chem Soc. 2012;134(4):1982–1985. doi: 10.1021/ja210528v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Muller MM, Muir TW. Histones: At the Crossroads of Peptide and Protein Chemistry. Chem Rev. 2015;115(6):2296–2349. doi: 10.1021/cr5003529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tan Y, Xue Y, Song C, Grunstein M. Acetylated Histone H3K56 Interacts with Oct4 to Promote Mouse Embryonic Stem Cell Pluripotency. Proc Natl Acad Sci USA. 2013;110(28):11493–11498. doi: 10.1073/pnas.1309914110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lee JY, Lee TH. Effects of DNA Methylation on the Structure of Nucleosomes. J Am Chem Soc. 2012;134(1):173–175. doi: 10.1021/ja210273w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kornacki JR, Stuparu AD, Mrksich M. Acetyltransferase p300/CBP Associated Factor (PCAF) Regulates Crosstalk-Dependent Acetylation of Histone H3 by Distal Site Recognition. ACS Chem Biol. 2015;10(1):157–164. doi: 10.1021/cb5004527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Winogradoff D, Echeverria I, Potoyan DA, Papoian GA. The Acetylation Landscape of the H4 Histone Tail: Disentangling the Interplay between the Specific and Cumulative Effects. J Am Chem Soc. 2015;137(19):6245–6253. doi: 10.1021/jacs.5b00235. [DOI] [PubMed] [Google Scholar]
- 9.Jacobson RH, Ladurner AG, King DS, Tjian R. Structure and Function of a Human TAFII250 Double Bromodomain Module. Science. 2000;288(5470):1422–1425. doi: 10.1126/science.288.5470.1422. [DOI] [PubMed] [Google Scholar]
- 10.Singh M, Popowicz GM, Krajewski M, Holak TA. Structural Ramification for Acetyl-Lysine Recognition by the Bromodomain of Human BRG1 Protein, a Central ATPase of the SWI/SNF Remodeling Complex. ChemBioChem. 2007;8(11):1308–1316. doi: 10.1002/cbic.200600562. [DOI] [PubMed] [Google Scholar]
- 11.Kornberg RD. Chromatin Structure: a Repeating Unit of Histones and DNA. Science. 1974;184(139):868–871. doi: 10.1126/science.184.4139.868. [DOI] [PubMed] [Google Scholar]
- 12.Gao M, Nadaud PS, Bernier MW, North JA, Hammel PC, Poirier MG, Jaroniec CP. Histone H3 and H4 N-Terminal Tails in Nucleosome Arrays at Cellular Concentrations Probed by Magic Angle Spinning NMR Spectroscopy. J Am Chem Soc. 2013;135(41):15278–15281. doi: 10.1021/ja407526s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Polach KJ, Widom J. Mechanism of Protein Access to Specific DNA Sequences in Chromatin: a Dynamic Equilibrium Model for Gene Regulation. J Mol Biol. 1995;254(2):130–149. doi: 10.1006/jmbi.1995.0606. [DOI] [PubMed] [Google Scholar]
- 14.Bowman GD, Poirier MG. Post-Translational Modifications of Histones that Influence Nucleosome Dynamics. Chem Rev. 2015;115(6):2274–2295. doi: 10.1021/cr500350x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ. Crystal Structure of the Nucleosome Core Particle at 2.8 A Resolution. Nature. 1997;389(6648):251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 16.Grunstein M. Histone Acetylation in Chromatin Structure and Transcription. Nature. 1997;389(6649):349–352. doi: 10.1038/38664. [DOI] [PubMed] [Google Scholar]
- 17.Venkatesh S, Workman JL. Histone Exchange, Chromatin Structure and the Regulation of Transcription. Nat Rev Mol Cell Biol. 2015;16(3):178–189. doi: 10.1038/nrm3941. [DOI] [PubMed] [Google Scholar]
- 18.Neumann H, Hancock SM, Buning R, Routh A, Chapman L, Somers J, Owen-Hughes T, van Noort J, Rhodes D, Chin JW. A Method for Genetically Installing Site-Specific Acetylation in Recombinant Histones Defines the Effects of H3 K56 Acetylation. Mol Cell. 2009;36(1):153–163. doi: 10.1016/j.molcel.2009.07.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.North JA, Shimko JC, Javaid S, Mooney AM, Shoffner MA, Rose SD, Bundschuh R, Fishel R, Ottesen JJ, Poirier MG. Regulation of the Nucleosome Unwrapping Rate Controls DNA Accessibility. Nucleic Acids Res. 2012;40(20):10215–10227. doi: 10.1093/nar/gks747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shimko JC, North JA, Bruns AN, Poirier MG, Ottesen JJ. Preparation of Fully Synthetic Histone H3 Reveals that Acetyl-Lysine 56 Facilitates Protein Binding Within Nucleosomes. J Mol Biol. 2011;408(2):187–204. doi: 10.1016/j.jmb.2011.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wei S, Falk SJ, Black BE, Lee T-H. A Novel Hybrid Single Molecule Approach Reveals Spontaneous DNA Motion in the Nucleosome. Nucleic Acids Res. 2015;43(17):e111. doi: 10.1093/nar/gkv549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li F, Allahverdi A, Yang R, Lua GB, Zhang X, Cao Y, Korolev N, Nordenskiold L, Liu CF. A Direct Method for Site-Specific Protein Acetylation. Angew Chem Int Ed. 2011;50(41):9611–9614. doi: 10.1002/anie.201103754. [DOI] [PubMed] [Google Scholar]
- 23.Allahverdi A, Yang RL, Korolev N, Fan YP, Davey CA, Liu CF, Nordenskiold L. The Effects of Histone H4 Tail Acetylations on Cation-Induced Chromatin Folding and Self-Association. Nucleic Acids Res. 2011;39(5):1680–1691. doi: 10.1093/nar/gkq900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dhall A, Wei S, Fierz B, Woodcock CL, Lee TH, Chatterjee C. Sumoylated Human Histone H4 Prevents Chromatin Compaction by Inhibiting Long-Range Internucleosomal Interactions. J Biol Chem. 2014;289(49):33827–33837. doi: 10.1074/jbc.M114.591644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dawson PE, Muir TW, Clarklewis I, Kent SBH. Synthesis of Proteins by Native Chemical Ligation. Science. 1994;266(5186):776–779. doi: 10.1126/science.7973629. [DOI] [PubMed] [Google Scholar]
- 26.Ha T, Tinnefeld P. Photophysics of Fluorescent Probes for Single-Molecule Biophysics and Super-Resolution Imaging. Annu Rev Phys Chem. 2012;63:595–617. doi: 10.1146/annurev-physchem-032210-103340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gopich IV, Szabo A. Decoding the Pattern of Photon Colors in Single-Molecule FRET. J Phys Chem B. 2009;113(31):10965–10973. doi: 10.1021/jp903671p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Watkins LP, Yang H. Information Bounds and Optimal Analysis of Dynamic Single Molecule Measurements. Biophys J. 2004;86(6):4015–4029. doi: 10.1529/biophysj.103.037739. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


