Abstract
Structural dynamics of a protein molecule is often critical to its function. Single-molecule methods provide efficient ways to investigate protein dynamics, although it is very challenging to achieve a millisecond or higher temporal resolution. Here we report spontaneous structural dynamics of the histone protein core in the nucleosome based on a single-molecule method that can reveal submillisecond dynamics by combining maximum likelihood estimation and fluorescence correlation spectroscopy. The nucleosome, comprising ~147 bp DNA and an octameric histone protein core consisting of H2A, H2B, H3, and H4, is the fundamental packing unit of the eukaryotic genome. The nucleosome imposes a physical barrier that should be overcome during various DNA-templated processes. Structural fluctuation of the nucleosome in the histone core has been hypothesized to be required for nucleosome disassembly but has yet to be directly probed. Our results indicate that at 100 mM NaCl the histone H2A–H2B dimer dissociates from the histone core transiently once every 3.6 ± 0.6 ms and returns to its position within 2.0 ± 0.3 ms. We also found that the motion is facilitated upon H3K56 acetylation and inhibited upon replacing H2A with H2A.Z. These results provide the first direct examples of how a localized post-translational modification or an epigenetic variation affects the kinetic and thermodynamic stabilities of a macromolecular protein complex, which may directly contribute to its functions.
Graphical abstract
INTRODUCTION
The nucleosome, the fundamental building block of the eukaryotic genome, imposes a physical barrier that needs to be overcome during various DNA-templated processes.1–5 The nucleosome comprises an octameric histone protein core consisting of two copies of four core histones (H2A, H2B, H3, and H4) that is wrapped by ~147 bp DNA. The stable subcomplexes of the core histones under a physiological buffer condition are the H2A–H2B dimer and the (H3–H4)2 tetramer. Two H2A–H2B dimers and one (H3–H4)2 tetramer form the octameric histone core (Figure 1A). Spontaneous structural fluctuations of the nucleosome at the DNA termini and at the histone dimer–tetramer interface would trigger or facilitate nucleosome disassembly, thereby having critical implications for gene access and regulation mechanisms. The mechanism of spontaneous nucleosome disassembly and the effects of a histone H3 variant CENP-A on the histone dynamics have recently been investigated using coarse-grained and atomistic modeling.6,7 We have recently reported spontaneous opening motions of the nucleosomal DNA termini based on a hybrid single-molecule approach that allowed detection and characterization of unsynchronized submillisecond dynamics.8,9 Transient and repetitive disruption of the histone dimer–tetramer interface (Figure 1B) would trigger dimer eviction catalyzed by a histone chaperone or other enzymes with a chaperone function that will eventually lead to nucleosome disassembly.10 The existence of this spontaneous positional fluctuation of the histone dimer is essential to validating this hypothesis.
Figure 1.
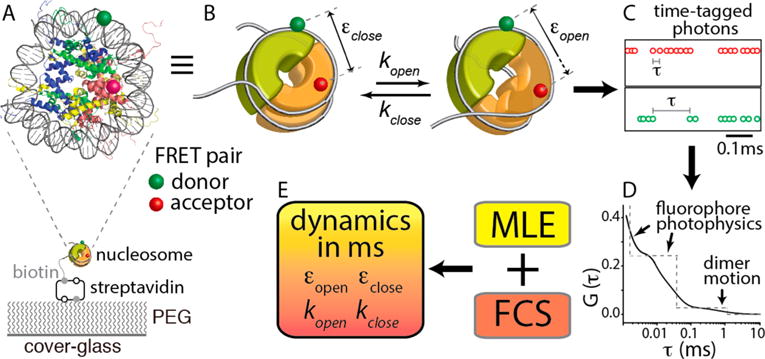
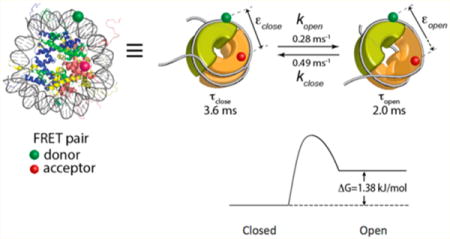
Schematic representation of the hybrid single-molecule method that revealed histone core dynamics on a millisecond time scale. (A) Nucleosome made with X. laevis core histones and a 147 bp human α-satellite sequence and labeled with a FRET donor [Cy3 (green)] at a DNA region fixed to the (H3–H4)2 tetramer (blue and green proteins) and an acceptor [ATTO647N (red)] at a H2A–H2B dimer (yellow and red proteins) at H2B T112C. The FRET pair reports the positional changes of the dimer relative to the tetramer upon opening and closing of the interface around where the H2A docking domain interacts with the tetramer (purple circle). The nucleosome was immobilized through biotin–streptavidin conjugation on a polyethylene glycol (PEG)-coated cover glass. (B) Kinetic scheme of spontaneous positional dynamics of a histone dimer (the front half-toroid colored orange) relative to the tetramer (the thick half-toroid colored green) between the closed and open states of the dimer–tetramer interface around which the H2A docking domain interacts with the tetramer (purple circle). The gray line represents the nucleosomal DNA. The green and red spheres represent the FRET donor (Cy3) and the acceptor (ATTO647N), respectively. (C) Example of streams of time-tagged fluorescence photons from a FRET pair. (D) Schematic representation of the fluorescence correlation spectrum G(τ) obtained from the intervals between two consecutive photons (τ) as shown in panel C. (E) Hybrid approach combining maximum likelihood estimation (MLE) and fluorescence correlation spectroscopy (FCS) to characterize the histone core dynamics at the dimer–tetramer interface on a millisecond time scale. Further details are described in Experimental Methods.
Post-translational modifications and variations of core histones are tied to gene regulation mechanisms. Acetylation at H3 lysine 56 (H3K56) that attenuates the structural integrity of the nucleosome near the DNA termini9,11,12 has been associated with transcription regulation, DNA damage repair, and cell proliferation.13–17 How this acetylation may affect the structure or structural dynamics of the histone core has never been directly probed. H2A.Z is a highly conserved (nearly >85%) histone H2A variant implicated in genome integrity and transcription activation.18–22 H2A.Z is enriched in the nucleosomes proximal to promoter nucleosome free regions (NFR) (i.e., +1 and −1 nucleosomes to a promoter NFR), and its acetylation is tied to gene activation.13,14,23–31 These +1 and −1 nucleosomes have very high nucleosome occupancy levels and rigid positioning, serving as physical barriers to help maintain genome integrity. There has not been any direct biophysical study to reveal any difference in the structural dynamics or flexibility between the H2A.Z nucleosome and the H2A nucleosome at a physiological salt level.
We have recently developed a single-molecule approach with which we directly probed the submillisecond spontaneous DNA terminus opening motion in the nucleosome that is dependent on the monovalent salt level, H3K56ac, and a histone H3 variant CENP-A.8,9 Here, we employed this approach to monitor the spontaneous structural fluctuations of the histone H2A–H2B dimer in the nucleosome. Our measurements revealed spontaneous positional fluctuations of the histone H2A–H2B dimer in the nucleosome on a millisecond time scale. This dynamics is significantly facilitated upon H3K56 acetylation that should further open the dimer–tetramer interface. We found that replacement of H2A with H2A.Z decelerates this dynamics, inhibiting the opening motion of the dimer–tetramer interface. These findings are the first direct evidence of a localized chemical change in a protein complex regulating its structural dynamics and potentially its functions.
EXPERIMENTAL METHODS
Protein Expression, Purification, and Modifications
Histones were expressed and purified as previously described.8 The H2B T112C mutant was prepared by substituting the T112 residue with cysteine. The T112C residue was labeled with ATTO647N via thiol-maleimide conjugation in the folded H2A–H2B dimer form. The H3 K56C/C110A double mutant was used for semisynthesis of the acetylated histone in a fully unfolded form.
H3Ks56ac (H3K56ac in this paper) was prepared by thiol–ene coupling between the cysteine thiol and N-vinyl-acetamide (NVA), which is functionally equivalent to natural histone acetylation.32 The H3 K56C/C110A double mutant at a concentration 1 mM was incubated in a buffer with a total volume of 200 μL containing 0.2 M sodium acetate (pH 4), 6 M guanidine hydrochloride, 7 mM L-glutahione, 50 mM N-vinyl-acetamide, 100 mM dimethyl sulfide, and 5 mM VA-044 {2,2′-[azobis(dimethylmethlene)]bis(2-imidazoline)-dihydrochoride} for 2 h at 70 °C. The reaction mixture was dialyzed against deionized water and then lyophilized overnight for storage at −80 °C.
Nucleosome Reconstitution
The nucleosome samples were prepared as previously described.8 For nucleosomal DNA, a 147 bp human α-satellite sequence with a single-strand 20-base linker with a biotin at its 5′ end was prepared by ligating six fragments in which the 54th nucleotide of the Watson (forward) strand was replaced with Cy3 (Integrated DNA Technology Inc., Coralville, IA). See Figure S1 for details on the DNA sequence and the labeling position.
Determination of the Förster Resonance Energy Transfer (FRET) Efficiencies and the Kinetic Rates of the Open and Closed Dimer–Tetramer Interface
Single-molecule FRET measurements on a scanning confocal microscope system were performed as previously described.8,9 The obtained data were analyzed with maximum likelihood estimation (MLE) and fluorescence correlation spectroscopy (FCS). The high and low FRET efficiencies (εclose and εopen, respectively) were obtained via MLE. The decay time constant (1/λ in eq 1) and the amplitude (A0 in eq 1) of the slowest dynamics component in the FCS spectra were used in conjunction with the FRET efficiencies to obtain the kinetic rates, kopen and kclose, with the following equation.
| (1) |
where G is the second-order correlation function of the fluorescence intensity from single-nucleosome particles, εclose and εopen are the FRET efficiencies of the closed and open states, respectively, kopen and kclose are the rates of opening and closing motions of the nucleosome, respectively, and λ is the decay rate of the correlation function. Note that the FCS spectra were fit with a triple-exponential decay function; its two fastest components are due to fluorophore photophysics.8 See Tables S1 and S2 for the fitting results of the data. The results for the fastest component may contain large errors because the decay time constants are outside of the range of the data. Consequently, the second fastest component may also contain large errors. However, the slowest component, the dynamics in which we are interested, is highly unlikely affected by these errors because the decay time constant is within the data range and is greatly separated from the other two components.
RESULTS AND DISCUSSION
Single-Molecule Observation Reveals Spontaneous Structural Fluctuations of the Histone Dimer in the Nucleosome
We employed a single-molecule FRET (smFRET) microscope in a confocal geometry with time-tagged single-photon counting capability to investigate the positional fluctuations of a histone H2A–H2B dimer relative to the histone (H3–H4)2 tetramer in the nucleosome on a time scale of milliseconds or less.8 The nucleosome assembled with Xenopus laevis core histones on a 147 bp human α-satellite sequence33 was labeled with a FRET pair. The donor, Cy3, was labeled at the 54th base of the forward strand DNA (Figure S1) replacing the nucleotide, and the acceptor, ATTO647N, was labeled at histone H2B T112C34 via thiol–maleimide conjugation (Figure 1A). A dimer labeling efficiency of 50% was achieved by mixing labeled and unlabeled H2A–H2B dimers. As the donor-labeled DNA region should be fixed to the (H3–H4)2 tetramer, changes in the FRET efficiency reflect the positional dynamics of the H2A–H2B dimer relative to the tetramer (Figure 1A,B). This dynamics represents the opening and closing motions of the dimer–tetramer interface likely around the region where the H2A docking domain interacts with the tetramer19 (Figure 1A,B). The closed state is defined as the state in which the dimer is intact in the nucleosome whose FRET efficiency is designated by εclose. The open state is defined as the state in which the dimer–tetramer interface is open as schematized in Figure 1B, whose FRET efficiency is designated by εopen. These FRET efficiencies cannot be converted to absolute distances because there are multiple unknown factors in our experimental system. However, they can be used for internal comparison between two distances (e.g., a lower FRET efficiency means a longer distance). This is possible because one of the two fluorophores is labeled via a freely rotating linker, making the FRET efficiency angle-averaged. Consequently, the longer distance between the dimer and the tetramer in the open state should result in an εopen that is lower than εclose. The kinetic rates between the two states are designated by kopen and kclose for the opening and closing motions, respectively. The fluorescence photons from a FRET pair were time-tagged (Figure 1C), and the photon arrival times were integrated to construct an smFRET time trace at a millisecond time resolution that revealed only a single FRET population per trace (Figure 2A). The single FRET population within one FRET trace indicates that the histone dimer is fixed relative to the tetramer or that the dynamics is too fast to be resolved on this time scale. We observed three nucleosome populations with three different average FRET efficiency ranges of 0.3–0.5, 0.5–0.7, and >0.8. We selected the population with the average FRET efficiency of 0.5–0.7 for further analysis because we expected a FRET efficiency of ~0.6 based on the FRET labeling positions (distance of ~48 Å) in a crystal structure (Protein Data Bank entry 1KX5). The FRET population of 0.3–0.5 likely represents the nucleosome with the off-target dimer labeled (the back half-toroid colored orange in Figure 1B), and the FRET population of >0.8 should represent the nucleosome with both H2A–H2B dimers labeled. The FRET population of 0.5–0.7 constitutes 25–30% of the nucleosome particles that we observed in the cases of the wild-type nucleosome, the H3K56-acetylated nucleosome, and the H2A.Z nucleosome. On the basis of a labeling efficiency of the dimer of 50%, 33% is the maximal density of this population because we did not collect data from nucleosomes without a FRET signal. Therefore, our analyses were performed on 76–91% of the properly labeled nucleosomes.
Figure 2.
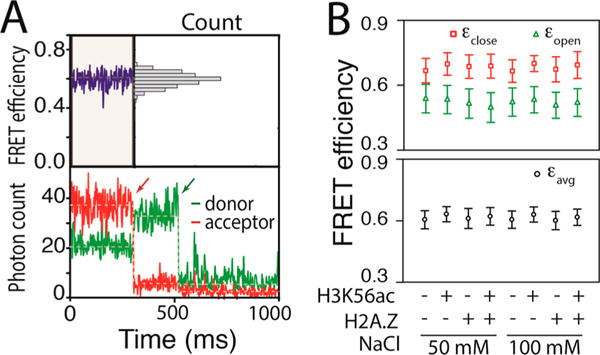
Maximum likelihood estimation reveals spontaneous structural dynamics of the histone core. (A) Representative fluorescence intensity time traces of a FRET pair (bottom) and the corresponding FRET efficiency trace (top) showing an average apparent FRET efficiency of ~0.6 with a unimodal distribution (top right). The apparent FRET efficiency was calculated with Iacceptor/(Idonor + Iacceptor), where I is the fluorescence intensity. The red and green arrows in the intensity traces indicate time points at which the acceptor and the donor, respectively, were photobleached. (B) MLE results revealing FRET efficiencies of the closed (εclose) and open (εopen) states (top) along with the average FRET efficiency (εavg) (bottom) for the nucleosomes made with the wild-type histone core (WT), the H3K56-acetylated histone core (H3K56ac), the H2A.Z-substituted histone core (H2A.Z), and the H3K56-acetylated and H2A.Z-substituted histone core (H2A.Z/H3K56ac) under two salt conditions (50 and 100 mM NaCl). The plus and minus signs represent the presence and absence, respectively, of the modification (H3K56ac) or variation (H2A.Z).
The acceptor photon streams (ATTO647N) were analyzed with a combination of MLE and FCS to extract the FRET efficiencies of the open and closed states and the kinetic rates between the two states (Figure 1D,E).8,35 FCS analysis revealed three total dynamics components in the range of 10 μs to 10 ms, two of which are faster than 0.1 ms and, therefore, assigned to the fluorophore photophysics as previously reported (Table S1).8 The millisecond dynamics component (Figure 3) must represent the positional fluctuations of the H2A–H2B dimer relative to the (H3–H4)2 tetramer because there is no photophysical dynamics with this FRET pair on this time scale.8 At this photon emission rate (25–40 kHz detected, 32.5 kHz on average), the MLE-based algorithm to extract the FRET efficiencies would work only for dynamics on a time scale slower than a few hundred microseconds.8 Therefore, the information generated by the MLE analysis should be on the dimer dynamics. The FRET efficiencies found by the MLE method for the closed and open states (εclose and εopen, respectively) are shown in Figure 2B and Table S2. The FRET efficiencies remained constant within error under the two different salt conditions examined (50 and 100 mM NaCl) (Figure 2B). On the basis of the FRET efficiencies obtained from the MLE analysis in combination with the FCS spectra (Figure 3), we computed the kinetic rates of the opening and closing motions (kopen and kclose, respectively) (Figure 4A). The results indicate that at 100 mM NaCl the histone dimer–tetramer interface opens once every 3.6 ± 0.6 ms and closes within 2.0 ± 0.3 ms (Figure 4B). These rates are converted to a 1.38 ± 0.55 kJ/mol free energy change during the opening motion (Figure 4A). At 50 mM NaCl, these values remain constant within error.
Figure 3.
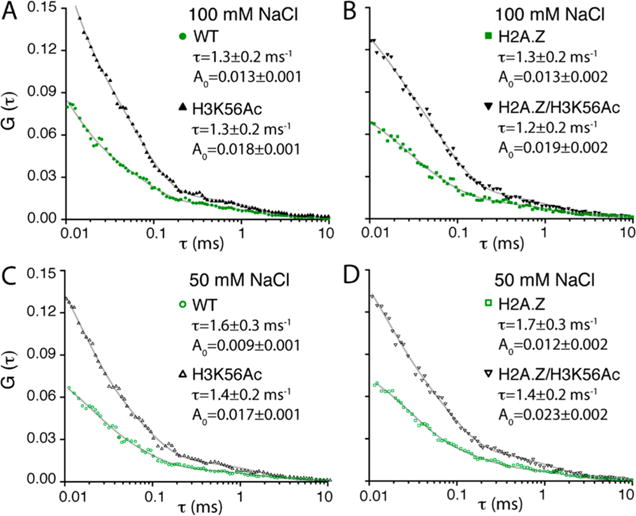
Fluorescence correlation spectroscopy (FCS) reveals the effects of H3K56ac and H2A.Z on the histone core dynamics. (A–D) FCS spectra of ATTO647N from the nucleosomes made with the wild-type histone core (WT), the H3K56-acetylated histone core (H3K56ac), the H2A.Z-substituted histone core (H2A.Z), and the H3K56-acetylated and H2A.Z-substituted histone core (H2A.Z/H3K56ac) under two salt conditions (50 and 100 mM NaCl). Each spectrum is fit to a triple-exponential decay function within the range of 10 μs to 10 ms. The fitting results of the slowest dynamics component are shown [decay times (τ) and amplitudes (□)]. The other components are due to the photophysical dynamics of the fluorophores and listed in Table S1.
Figure 4.
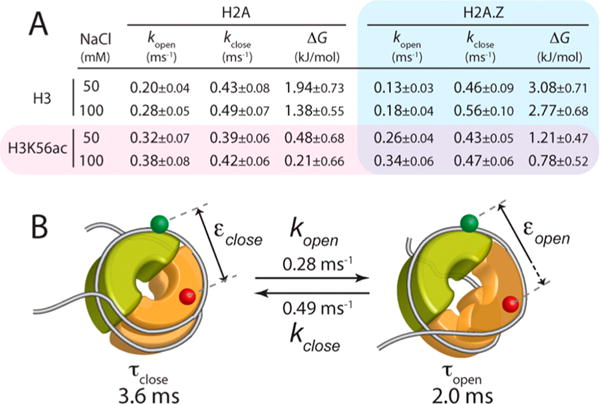
Hybrid single-molecule approach combining MLE and FCS that reveals the kinetics of the histone core dynamics at the dimer–tetramer interface. (A) Opening (kopen) and closing (kclose) rate constants, and the free energy change (ΔG) of the opening motion calculated from the rate constants for the nucleosomes made with the wild-type histone core (H2A and H3), the H3K56-acetylated histone core (H3K56ac and H3), the H2A.Z-substituted histone core (H2A.Z and H3), and the H3K56-acetylated and H2A.Z-substituted histone core (H2A.Z and H3K56ac) under two salt conditions (50 and 100 mM NaCl). (B) Quantitative summary of the histone core dynamics at the dimer–tetramer interface at 100 mM NaCl. See the legend of panels A and B of Figure 1 for the descriptions of the nucleosome structure.
This dynamic motion of the H2A–H2B dimer would result in repetitive and transient gap formation at the interface that may trigger dimer eviction and eventually nucleosome disassembly. Nucleosome disassembly in vivo is mediated by an enzyme such as a histone chaperone,36,37 which most likely requires the formation of a ternary complex of histone, DNA, and a chaperone at least transiently. This is because histones do not spontaneously dissociate from DNA under physiological salt conditions because of the strong electrostatic interaction. This mechanism implicates spontaneous fluctuations of the nucleosome structure that will expose transiently and repeatedly a site for chaperone penetration and binding. This hypothesis dictates that the nucleosome structural dynamics should impose the ultimate kinetic barrier to nucleosome disassembly. Our recent report on the existence of the spontaneous DNA opening motion at the nucleosome termini supports this hypothesis.8,9 The flexible dimer–tetramer interface presented here would further lower the barrier to chaperone binding and subsequent histone eviction.
The Dimer–Tetramer Interface Becomes More Flexible and More Open upon H3K56 Acetylation
We repeated the measurements with H3K56 acetylated. The specific acetylation was achieved with radical-initiated conjugation of N-vinylacetamide to the cysteine residue that replaced lysine 56 by site-directed mutagenesis.32 Mass spectrometric analysis confirmed acetylation (Figure S2). This acetylation method generates acetylated histones that are chemically almost identical to the naturally occurring ones.32 Acetylated histones prepared with this method have been used to reproduce the results obtained with the native chemical ligation method.38,39 The average FRET efficiencies under the two salt conditions (50 and 100 mM NaCl) remain constant within error upon H3K56 acetylation (H3K56ac) (Figure 2B and Table S2). Revealed by the MLE–FCS analysis, the dimer dynamics is facilitated upon H3K56 acetylation (increased kopen and decreased ΔG of opening from the top two rows to the bottom two rows in Figure 4A except for the H2A case at 100 mM NaCl). The larger amplitude of the FCS component upon H3K56 acetylation in all four cases (Figure 3A,B) clearly indicates that the dimer fluctuation is facilitated. In all of the cases except for H2A at 100 mM NaCl, the free energy change of opening becomes significantly lower upon inclusion of H3K56ac (ΔG values in the bottom two rows lower than those in the top two rows in Figure 4A). These findings indicate that H3K56 acetylation makes the dimer–tetramer interface more flexible and shifts the equilibrium more to the open state, resulting in a more open dimer–tetramer interface on average. This change is achieved by increasing the opening rate rather than decreasing the closing rate.
Histone acetylation at H3K56 is associated with transcription regulation, DNA damage repair, and cell proliferation.13–17 H3K56ac weakens the interactions between the nucleosomal DNA termini and the histone core9,11,12 likely by disrupting the interaction of the H3 N-terminal tail with DNA. Therefore, the terminal DNA opening energy will become lower upon H3K56 acetylation as evidenced in recent single-molecule studies.9,11 Our results reveal that this acetylation also induces a significant change in the core histone structural integrity, which may contribute to triggering nucleosome disassembly. It may not be immediately clear how the acetylation at a site not in direct contact with the dimer–tetramer interface may affect its dynamics. Resolved in a detailed structure of the nucleosome and confirmed by biochemical and biophysical studies,2,11,12 these components are all interconnected directly or indirectly, and therefore, a change in one component would affect the others. The straightforward change induced upon H3K56 acetylation is to weaken the interaction between the histone tetramer and the DNA that also interact with the dimers. As the DNA termini open, the dimers will follow the DNA and consequently disrupt the interface with the tetramer. Therefore, the biophysical roles of H3K56 acetylation may be not only in weakening the interactions between the histone core and the nucleosomal DNA but also in disintegrating the histone core at the dimer–tetramer interface, both of which will synergistically facilitate nucleosome disassembly. These results support the structural dynamics of the nucleosome on a millisecond time scale as a functional element in gene regulation mechanisms triggered by a histone modification.
The Dimer–Tetramer Interface Becomes More Rigid and Less Open upon Replacement of H2A with H2A.Z
We repeated the measurements with the wild-type nucleosome and the H3K56-acetylated nucleosome with the histone H2A replaced with H2A.Z. The FRET efficiencies of the closed and open states remain constant within error upon replacement of H2A with H2A.Z (Figure 2B and Table S2). As revealed by the MLE–FCS analysis, H2A.Z makes the dimer–tetramer interface less flexible in the unacetylated cases (decreased kopen from the left three columns to the right three columns in the top two rows of Figure 4A). The equilibrium is also possibly shifted to the closed state upon H2A.Z replacement, although the difference is only marginal. In acetylated nucleosomes, these differences become buried in errors. This is likely due to the acetylation effect of H3K56ac making the nucleosome flexible and therefore canceling the rigidification effect of H2A.Z. Notably, however, the average free energy change during the opening motion (ΔG in Figure 4A) in the unacetylated H2A.Z nucleosome case is larger than RT at 37 °C (i.e., ΔG > 2.6 kJ/mol), while it becomes far below RT in the acetylated H2A.Z nucleosome, making the acetylation effect more pronounced in the H2A.Z nucleosome than in the H2A nucleosome.
Histone variant H2A.Z is well-conserved throughout eukaryotes and implicated in maintaining genome integrity and activating transcription.19–22 The processes of H2A.Z deposition and removal are catalyzed by SWR1/SWR-C and INO80 remodelers whose activities have been linked to histone acetylations such as H3K56ac and H2A.ZK14ac.13–18,31 The conflicting roles of H2A.Z in stabilizing and destabilizing the nucleosome have been reported previously.19,30,34,40–43 There has not been any direct biophysical study to elucidate differences in the structural dynamics or flexibility between the H2A.Z nucleosome and the H2A nucleosome at a physiological salt level. Our results reveal that H2A.Z rigidifies the histone dimer–tetramer interface, supporting its role in maintaining the structural integrity of the chromatin. Another important finding is that acetylation at H3K56 brings the dimer–tetramer opening energy far below the thermal energy, making the dimer–tetramer interface significantly more open. However, the absolute free energy change is still larger than that of the H3K56-acetylated H2A nucleosome. On the basis of these results, we conclude that H2A.Z stabilizes the nucleosome by rigidifying the histone core and suggest that triggering disassembly of the H2A.Z nucleosome may require more changes than H3K56ac alone.
CONCLUSIONS
Our study provides the first examples of how histone modifications and variations affect the kinetic stability of the histone core in the nucleosome, which may directly contribute to their functions. The reported measurements were permitted by a powerful single-molecule approach that will open efficient ways to reveal how a network of multiple structural components in a protein complex may be implicated in modulating its structural integrity with the aid of localized chemical modifications.
Supplementary Material
Acknowledgments
We thank Dr. Blaine Bartholomew for providing us with yeast H2A.Z protein. This work was supported by National Institutes of Health Grant GM097286 (T.-H.L.).
Footnotes
Supporting Information
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acs.jpcb.6b06235.
Notes
The authors declare no competing financial interest.
References
- 1.Kornberg RD. Chromatin structure: a repeating unit of histones and DNA. Science. 1974;184:868–871. doi: 10.1126/science.184.4139.868. [DOI] [PubMed] [Google Scholar]
- 2.Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 3.Polach KJ, Widom J. Mechanism of protein access to specific DNA sequences in chromatin: a dynamic equilibrium model for gene regulation. J Mol Biol. 1995;254:130–149. doi: 10.1006/jmbi.1995.0606. [DOI] [PubMed] [Google Scholar]
- 4.Wolffe AP. New insights into chromatin function in transcriptional control. FASEB J. 1992;6:3354–3361. doi: 10.1096/fasebj.6.15.1464369. [DOI] [PubMed] [Google Scholar]
- 5.Simpson RT. Structure of chromatin containing extensively acetylated H3 and H4. Cell. 1978;13:691–699. doi: 10.1016/0092-8674(78)90219-2. [DOI] [PubMed] [Google Scholar]
- 6.Winogradoff D, Zhao H, Dalal Y, Papoian GA. Shearing of the CENP-A dimerization interface mediates plasticity in the octameric centromeric nucleosome. Sci Rep. 2015;5:17038. doi: 10.1038/srep17038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang B, Zheng W, Papoian GA, Wolynes PG. Exploring the free energy landscape of nucleosomes. J Am Chem Soc. 2016;138:8126–8133. doi: 10.1021/jacs.6b02893. [DOI] [PubMed] [Google Scholar]
- 8.Wei S, Falk SJ, Black BE, Lee T-H. A novel hybrid single molecule approach reveals spontaneous DNA motion in the nucleosome. Nucleic Acids Res. 2015;43:e111. doi: 10.1093/nar/gkv549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kim J, Lee J, Lee TH. Lysine acetylation facilitates spontaneous dna dynamics in the nucleosome. J Phys Chem B. 2015;119:15001–15005. doi: 10.1021/acs.jpcb.5b09734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chang H-W, Kulaeva OI, Shaytan AK, Kibanov M, Kuznedelov K, Severinov KV, Kirpichnikov MP, Clark DJ, Studitsky VM. Analysis of the mechanism of nucleosome survival during transcription. Nucleic Acids Res. 2014;42:1619–1627. doi: 10.1093/nar/gkt1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Simon M, North JA, Shimko JC, Forties RA, Ferdinand MB, Manohar, Zhang M, Fishel M, Ottesen R, Poirier JJMG. Histone fold modifications control nucleosome unwrapping and disassembly. Proc Natl Acad Sci U S A. 2011;108:12711–12716. doi: 10.1073/pnas.1106264108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Andrews AJ, Chen X, Zevin A, Stargell LA, Luger K. The histone chaperone Nap1 promotes nucleosome assembly by eliminating nonnucleosomal histone DNA interactions. Mol Cell. 2010;37:834–842. doi: 10.1016/j.molcel.2010.01.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bartholomew B. Regulating the chromatin landscape: structural and mechanistic perspectives. Annu Rev Biochem. 2014;83:671–696. doi: 10.1146/annurev-biochem-051810-093157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yu Y, Deng Y, Reed SH, Millar CB, Waters R. Histone variant Htz1 promotes histone H3 acetylation to enhance nucleotide excision repair in Htz1 nucleosomes. Nucleic Acids Res. 2013;41:9006–9019. doi: 10.1093/nar/gkt688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Papamichos-Chronakis M, Watanabe S, Rando OJ, Peterson CL. Global regulation of H2A.Z localization by the INO80 chromatin-remodeling enzyme is essential for genome integrity. Cell. 2011;144:200–213. doi: 10.1016/j.cell.2010.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Raisner RM, Hartley PD, Meneghini MD, Bao MZ, Liu CL, Schreiber SL, Rando OJ, Madhani HD. Histone variant H2A.Z marks the 5′ ends of both active and inactive genes in euchromatin. Cell. 2005;123:233–248. doi: 10.1016/j.cell.2005.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Millar CB, Xu F, Zhang K, Grunstein M. Acetylation of H2AZ Lys 14 is associated with genome-wide gene activity in yeast. Genes Dev. 2006;20:711–722. doi: 10.1101/gad.1395506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mizuguchi G, Shen X, Landry J, Wu W-H, Sen S, Wu C. ATP-Driven exchange of histone H2AZ variant catalyzed by SWR1 chromatin remodeling complex. Science. 2004;303:343–348. doi: 10.1126/science.1090701. [DOI] [PubMed] [Google Scholar]
- 19.Suto RK, Clarkson MJ, Tremethick DJ, Luger K. Crystal structure of a nucleosome core particle containing the variant histone H2A.Z. Nat Struct Biol. 2000;7:1121–1124. doi: 10.1038/81971. [DOI] [PubMed] [Google Scholar]
- 20.Hatch CL, Bonner WM, Moudrianakis EN. Minor histone 2A variants and ubiquinated forms in the native H2A:H2B dimer. Science. 1983;221:468–470. doi: 10.1126/science.6306766. [DOI] [PubMed] [Google Scholar]
- 21.Ball DJ, Slaughter CA, Hensley P, Garrard WT. Amino acid sequence of the N-terminal domain of calf thymus histone H2A.Z. FEBS Lett. 1983;154:166–170. doi: 10.1016/0014-5793(83)80896-5. [DOI] [PubMed] [Google Scholar]
- 22.West MH, Bonner WM. Histone 2A, a heteromorphous family of eight protein species. Biochemistry. 1980;19:3238–3245. doi: 10.1021/bi00555a022. [DOI] [PubMed] [Google Scholar]
- 23.Weber CM, Ramachandran S, Henikoff S. Nucleosomes are context-specific, H2A.Z-modulated barriers to RNA polymerase. Mol Cell. 2014;53:819–830. doi: 10.1016/j.molcel.2014.02.014. [DOI] [PubMed] [Google Scholar]
- 24.Monteiro FL, Baptista T, Amado F, Vitorino R, Jerónimo C, Helguero LA. Expression and functionality of histone H2A variants in cancer. Oncotarget. 2014;5:3428–3443. doi: 10.18632/oncotarget.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jacquet K, Cote J. DNA repair: Chromatin remodeling without H2A.Z? Cell Cycle. 2014;13:1059. doi: 10.4161/cc.28383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hong J, Feng H, Wang F, Ranjan A, Chen J, Jiang J, Ghirlando R, Xiao TS, Wu C, Bai Y. The catalytic subunit of the SWR1 remodeler is a histone chaperone for the H2A.Z-H2B dimer. Mol Cell. 2014;53:498–505. doi: 10.1016/j.molcel.2014.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ranjan A, Mizuguchi G, FitzGerald PC, Wei D, Wang F, Huang Y, Luk E, Woodcock CL, Wu C. Nucleosome-free region dominates histone acetylation in targeting SWR1 to promoters for H2A.Z replacement. Cell. 2013;154:1232–1245. doi: 10.1016/j.cell.2013.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Luk E, Ranjan A, Fitzgerald PC, Mizuguchi G, Huang Y, Wei D, Wu C. Stepwise histone replacement by SWR1 requires dual activation with histone H2A.Z and canonical nucleosome. Cell. 2010;143:725–736. doi: 10.1016/j.cell.2010.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Papamichos-Chronakis M, Peterson CL. The Ino80 chromatin-remodeling enzyme regulates replisome function and stability. Nat Struct Mol Biol. 2008;15:338–345. [Google Scholar]
- 30.Albert I, Mavrich TN, Tomsho LP, Qi J, Zanton SJ, Schuster SC, Pugh BF. Translational and rotational settings of H2A.Z nucleosomes across the Saccharomyces cerevisiae genome. Nature. 2007;446:572–576. doi: 10.1038/nature05632. [DOI] [PubMed] [Google Scholar]
- 31.Watanabe S, Radman-Livaja M, Rando OJ, Peterson CL. A histone acetylation switch regulates H2A.Z deposition by the SWR-C remodeling enzyme. Science. 2013;340:195–199. doi: 10.1126/science.1229758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li F, Allahverdi A, Yang R, Lua GB, Zhang X, Cao Y, Korolev N, Nordenskiold L, Liu CF. A direct method for site-specific protein acetylation. Angew Chem Int Ed. 2011;50:9611–9614. doi: 10.1002/anie.201103754. [DOI] [PubMed] [Google Scholar]
- 33.Harp JM, Uberbacher EC, Roberson AE, Palmer EL, Gewiess A, Bunick GJ. X-ray diffraction analysis of crystals containing twofold symmetric nucleosome core particles. Acta Crystallogr Sect D: Biol Crystallogr. 1996;52:283–288. doi: 10.1107/S0907444995009139. [DOI] [PubMed] [Google Scholar]
- 34.Park YJ, Dyer PN, Tremethick DJ, Luger K. A new fluorescence resonance energy transfer approach demonstrates that the histone variant H2AZ. stabilizes the histone octamer within the nucleosome. J Biol Chem. 2004;279:24274–24282. doi: 10.1074/jbc.M313152200. [DOI] [PubMed] [Google Scholar]
- 35.Gopich IV, Szabo A. Decoding the pattern of photon colors in single-molecule FRET. J Phys Chem B. 2009;113:10965–10973. doi: 10.1021/jp903671p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Luebben WR, Sharma N, Nyborg JK. Nucleosome eviction and activated transcription require p300 acetylation of histone H3 lysine 14. Proc Natl Acad Sci U S A. 2010;107:19254–19259. doi: 10.1073/pnas.1009650107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Walfridsson J, Khorosjutina O, Matikainen P, Gustafsson CM, Ekwall K. A genome-wide role for CHD remodelling factors and Nap1 in nucleosome disassembly. EMBO J. 2007;26:2868–2879. doi: 10.1038/sj.emboj.7601728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Allahverdi A, Yang R, Korolev N, Fan Y, Davey CA, Liu CF, Nordenskiold L. The effects of histone H4 tail acetylations on cation-induced chromatin folding and self-association. Nucleic Acids Res. 2011;39:1680–1691. doi: 10.1093/nar/gkq900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dhall A, Wei S, Fierz B, Woodcock CL, Lee TH, Chatterjee C. Sumoylated human histone H4 prevents chromatin compaction by inhibiting long-range internucleosomal interactions. J Biol Chem. 2014;289:33827–33837. doi: 10.1074/jbc.M114.591644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tolstorukov MY, Kharchenko PV, Goldman JA, Kingston RE, Park PJ. Comparative analysis of H2A.Z nucleosome organization in the human and yeast genomes. Genome Res. 2009;19:967–977. doi: 10.1101/gr.084830.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jin C, Felsenfeld G. Nucleosome stability mediated by histone variants H3.3 and H2A.Z. Genes Dev. 2007;21:1519–1529. doi: 10.1101/gad.1547707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Guillemette B, Bataille AR, Gevry N, Adam M, Blanchette M, Robert F, Gaudreau L. Variant histone H2A.Z is globally localized to the promoters of inactive yeast genes and regulates nucleosome positioning. PLoS Biol. 2005;3:e384. doi: 10.1371/journal.pbio.0030384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cairns BR. The logic of chromatin architecture and remodelling at promoters. Nature. 2009;461:193–198. doi: 10.1038/nature08450. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


