Abstract
Until recently, our knowledge of the host range and diversity of members of the Chlamydiaceae, obligate intracellular bacterial pathogens of humans and animals, was thought to be nearly complete. Aided by advances in molecular diagnostics, a new picture is emerging, however, that the host barriers may be looser than previously thought for many chlamydial species. While cross-host transmission of chlamydial species is a concern for animal health, new reports highlight an emerging zoonotic risk for several species associated with intensification of farming and the widespread popularity of companion animals. The description of an expanded cohort of new species within this family from avian and reptilian hosts has also highlighted how much we still have to learn about the biology and pathogenicity of the Chlamydiaceae as a whole. Reports emerging about these relatives of the traditional chlamydial pathogens are matched by the continued identification of novel Chlamydia-related bacteria in the phylum Chlamydiae, providing evidence that many may be pathogenic to humans or animals and pose a zoonotic or vector-borne risk. The review examines the new hosts described for well-characterized chlamydial veterinary pathogens, emerging novel chlamydial species and the potential for these to cause disease in their respective hosts.
Keywords: Chlamydia, Chlamydia-related bacteria, chlamydiosis, disease, epidemiology, infection, pathogen, zoonosis
Introduction
Bacteria in the family Chlamydiaceae are globally significant human and animal pathogens. Until recently, the Chlamydiaceae, the best-characterized family in the phylum Chlamydiae, comprised nine taxonomically recognized, well-defined species belonging to the genus Chlamydia: C. trachomatis, C. muridarum, C. suis, C. psittaci, C. abortus, C. caviae, C. felis, C. pneumoniae and C. pecorum [1]. This view has begun to rapidly change with the discovery and description of two novel chlamydial species and two Candidatus species in avian and reptile hosts: C. avium, C. gallinacea [2] and Candidatus C. ibidis [3] from domestic and wild birds and Ca. C. sanzinia from a captive snake [4]. Outside of this well-described family, wider sampling and advances in molecular methods have revealed a breadth of novel families within the phylum [5], collectively referred to as Chlamydia-related bacteria (CRBs) because of their phenotypic and genetic similarities but phylogenetic separation from the Chlamydiaceae.
The detection of emerging infectious diseases has been steadily increasing over the last 70 years and predominantly comprises zoonoses from wildlife [6], with wildlife species richness being a predictor for the emergence of zoonotic diseases with a wildlife origin [6]. The intensification of farming and widespread popularity of companion animals also present ongoing opportunities for emerging zoonoses [7]. Whilst improvements in surveillance and diagnostics have contributed to a wider recognition of the emergence of these pathogens [7], anthropogenic factors such as antimicrobial use, agricultural practices and human population density are drivers of emerging infectious diseases in general [6].
This review examines the new animal hosts described for well-characterized chlamydial veterinary pathogens, emerging novel chlamydial species and their potential to cause disease in humans, animals or both. We have limited our review to the last 4 years, since the isolation and description of Ca. C. ibidis, to highlight the recent and rapid changes in our understanding of the diversity of relationships between chlamydial species and their hosts.
Old Chlamydia Species, New Infections
A series of recent discoveries have cast doubts over our complete understanding of the natural host range and transmission networks of the established species described in the family Chlamydiaceae.
From a public health perspective, the most important of these new discoveries relate to C. psittaci, traditionally recognized as an avian pathogen and potential cause of zoonotic disease. Most human psittacosis cases, which manifest as atypical pneumonia, are confined to bird keepers, poultry workers and healthcare workers [8], [9]. Human-to-human transmission appears possible but is exceptionally rare [10], [11], while there is growing evidence for transmission from birds to other mammalian species such as livestock [12], [13]. In some of these cases, farm environments possibly resulted in direct contact between birds and humans [13].
An interesting recent case of equine reproductive loss has placed a renewed spotlight on this pathogen [14]. In this case, the strain isolated from an abnormal horse placenta was most closely related to the C. psittaci sequence type 24/6BC clade, a clonally related and highly virulent cluster of strains thought to be shed primarily by infected parrots but also linked to indirect transmission to humans via environmental contamination [15]. Interestingly, while the human cases were not confirmed, contact with the infected reproductive material from the affected horse in this case was subsequently linked to a cluster of human psittacosis [14], [15], highlighting a new potential route of transmission for this zoonotic agent. Historically, reports of C. psittaci infections in horses are incredibly rare, although there is some evidence that these infections may be an important underdiagnosed cause of equine reproductive disease [14]. These reports and others highlight the need for environmental monitoring to limit the exposure of people involved in agricultural practises and animal care and husbandry.
In terms of new zoonotic threats, there is also growing molecular evidence that C. suis, a chlamydial species endemic to pigs, can infect humans [12]. Recently, C. suis was detected in the air and on surfaces at a slaughterhouse in Belgium, where swabs from two human conjunctivae were also positive for this pathogen [16]. At the same time, a broad study of chlamydial agents in Nepal detected C. suis DNA in the eyes of patients with trachoma, a debilitating ocular disease traditionally linked to the human chlamydial pathogen C. trachomatis [17]. While the zoonotic potential of C. suis exposure is unclear, what is troubling is that C. suis is the only known chlamydial species to naturally harbour a tetracycline resistance gene cassette [18], which, in laboratory conditions, can be transferred from C. suis to C. trachomatis and C. muridarum by recombination [19], highlighting the potential threat that this veterinary pathogen may pose to human health.
The expansion of the host range of members of the Chlamydiaceae is not just limited to humans. The veterinary pathogen C. pecorum is linked to a range of diseases in cattle, sheep, pigs and goats, including polyarthritis, encephalomyelitis and conjunctivitis [20]. Rare cases of C. pecorum–associated abortion in ruminants have also been reported [21]. While this pathogen has been long recognized as a major threat to the survival of Australia's iconic native marsupial, the koala [22], recent molecular and serologic studies have shown other wildlife globally may harbour strains of this pathogen. These hosts include ibex, red deer, buffalo and wild boar [22]. The impact that these infections have is currently unclear, however [22]. Two studies from Argentina and Japan also suggest that C. pecorum may be present in low levels in passeriform, psittaform and columbrid birds [23], [24], thus potentially representing one method by which C. pecorum is spread between such diverse hosts.
New Chlamydia Species, New Threats?
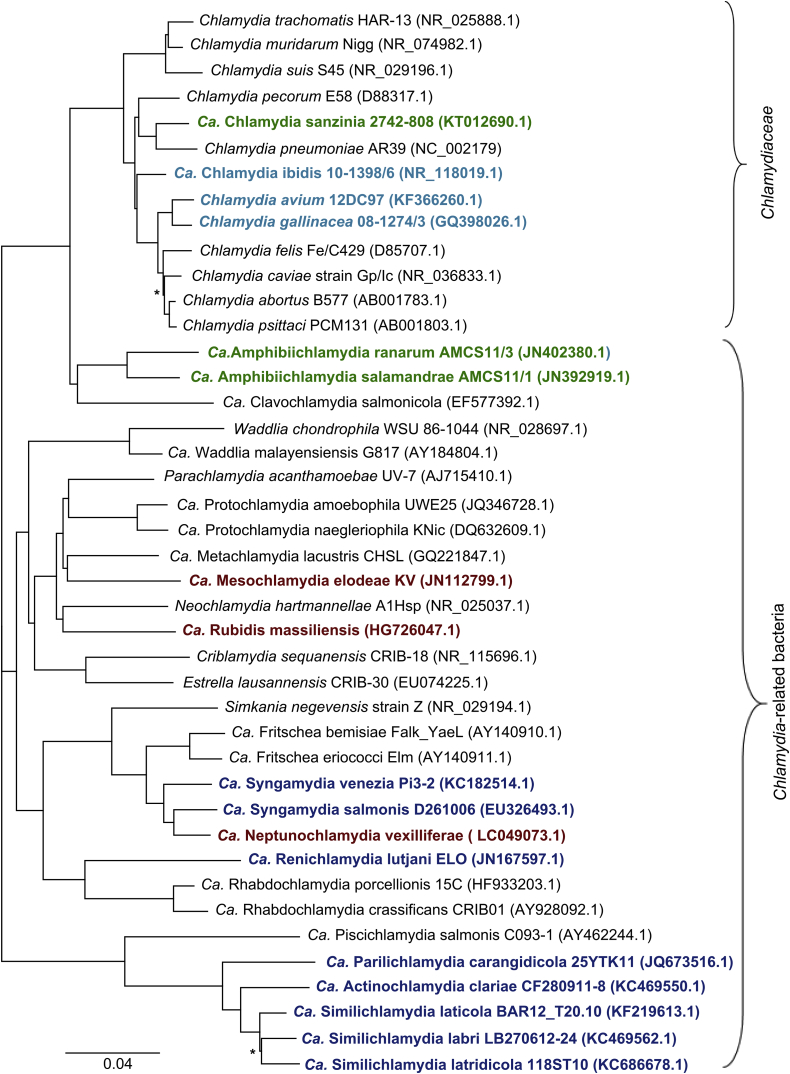
Advances in molecular methods, combined with improved sampling techniques and an expansion of animal studies, has continued to uncover novel diversity in the phylum Chlamydiae, including the identification of potential new chlamydial pathogens of humans and animals. Fig. 1 illustrates the novel species that have been identified in the last 4 years within the Chlamydiaceae family and more broadly across the phylum Chlamydiae.
Fig. 1.
16S rRNA gene-based phylogenetic tree depicting evolutionary relationships of Chlamydiales. Species described in last 4 years are in bold type. Species descriptions during this time have been limited to reptilian and amphibian (green), avian (pale blue), piscine (dark blue) and protozoan (dark red) hosts. Sequences were downloaded from GenBank and aligned using MAFFT before tree construction using FastTree in Geneious v7.1.9 [36]. Asterisks denote branches with support values <50%.
Perhaps the biggest shakeup to the chlamydial field in the last few years was the discovery and description of three new taxa in avian hosts: C. avium, comprising strains from pigeons and psittacine birds [2]; C. gallinacea, comprising strains from poultry [2]; and Ca. C. ibidis, from the digestive tracts of feral ibises in France [3]. Although these novel species were only initially described recently in Europe, C. avium and C. gallinacea are now considered to be widespread in both European and Asian countries [25], [26].
Interestingly, whilst endemic to poultry, C. gallinacea does not appear to be restricted to these hosts, with a recent study reporting its presence in 88.9% of Chlamydia-positive vaginal swabs from cattle throughout China [27]. The genotypes described were identical to poultry strains from a sympatric area, suggesting a potential cross-host transmission event and/or a genotype specific to this region. Adding to this list of novel avian chlamydial species, several novel unclassified chlamydial species were also reported in faecal specimens in wild seabirds such as penguins and gulls [28], [29]. These taxa are closely related despite their geographically distinct locations.
Reptiles also appear to harbour an untapped diversity of chlamydia with the identification of a novel Candidatus species in a pet Madagascan tree boa in Switzerland (Ca. C. sanzinia) [4] closely related to C. pneumoniae. This sample originated from a broader study of chlamydial agents in captive snakes, with molecular evidence suggesting that additional novel C. pneumoniae–like strains may also be present [30]. In other studies, phylogenetic analysis of 16S rRNA and ompA sequences from lung, liver, spleen and brain tissues from crocodiles showed three genotypes within a novel clade related to C. caviae and C. felis [31]. A separate study identified a 55% prevalence of Chlamydiaceae in Australian crocodiles with conjunctivitis and/or pharyngitis syndrome, but no sequencing was conducted to further identify the agent or agents [32].
Lastly, a novel lineage has been described in female roe deer from France. The lineage comprises vaginal and faecal samples and branches close to C. trachomatis and C. suis [33]. The pathogenic potential is unknown, but these novel taxa highlight how little we know about emerging chlamydial infections in wild animals.
Beyond the family Chlamydiaceae, the identification of potentially pathogenic CRBs has been an ongoing trend for the last 20 years, since their first reports [5]. Probably the best studied of these is Waddlia chondrophila, which is considered to be an emerging threat to the reproductive health of humans and cattle [34]. W. chondrophila was first associated with pregnancy failure in a dairy herd by serology and was subsequently been detected in the vaginal swabs from cows that had aborted [34]. In the first step to conclusively evaluating the pathogenic potential of this agent in cattle, a recent study revealed that experimental infection with W. chondrophila failed to cause abortion in a similar way to the other chlamydial abortigenic agent, C. abortus [35]. While placental infection was observed, this only occurred in one of the animals challenged, with the conclusions that first, W. chondrophila infection is opportunistic, and second that a role in abortion remains to be confirmed. With questions over the role of this pathogen in animals, it is interesting to note that several studies have recently postulated that this agent may be zoonotic, with evidence of its presence in the placentas of women with adverse pregnancy outcomes [34].
The most convincing evidence for a pathogenic role in the CRBs comes from species in the recently described Ca. Parilichlamydiaceae family [37] (Fig. 1). These bacteria, as well as several other species in the families Simkaniaceae, Ca. Piscichlamydiaceae, and Ca. Clavochlamydiaceae, have been described in association with epitheliocystis, the common gill disease of wild and cultured fish [37]. Whilst this disease is by no means new, our understanding of the aetiologic agents has only been possible through advances in molecular methods. It may also be that the incidence of epitheliocystis may be on the rise as a result of increased aquaculture activity globally, including the introduction of new wild fish species into culture, to address the growing demand for sustainable food sources.
Recent studies in arthropods and other well-recognized vectors have offered a richness of emergent groups of chlamydial species. Ticks are prevalent vectors for other pathogens. Molecular studies in Europe, Africa and Australia have recently revealed that these arthropods also harbour CRBs [38], [39], [40], [41]. Notably, the chlamydial species identified in these ticks primarily belonged to the Ca. Rhabdochlamydiaceae, Parachlamydiaceae, and Simkaniaceae, with Ca. Rhabdochlamydiaceae DNA being most abundant, suggesting that ticks are the natural hosts of the latter agents [41]. Although the pathogenic potential of these CRBs is unclear, a recent study examining skin biopsy samples from tick bites identified tick-related chlamydial sequences in the samples but not from samples taken from controls [39], highlighting the importance of studies to evaluate the role that these CRBs might play in human and animal health.
Along with arthropods, bats are well-characterized vectors for a plethora of human and animal pathogens. So far, two novel Waddlia species have been isolated from or detected in the urine [42] or tissue [43] of bats in Malaysia and Mexico, respectively. Building on a metagenomic study that identified Chlamydiae in bat faeces, a recent screening study reported both novel Chlamydia lineages as well as novel Ca. Rhabdochlamydiaceae members in bat droppings in Finland [44]. Interestingly, insects collected at feeding sites also harboured a range of Chlamydiales, but none were identical to the sequences from bats. The significance of these findings to human and animal health is still unclear.
Challenges and Future Directions
Reports describing the emergence of previously described chlamydial species in new hosts and novel chlamydial species have only been possible with advances in culture-independent molecular methods and diagnostics [45]. While these methods have played a significant role in expanding our understanding of chlamydial diversity and in detecting new and emerging chlamydial disease threats, the significance of detecting chlamydial DNA in specimens in the absence of other methods raises questions over whether these reports indicate exposure to these organisms rather than actual evidence of viable bacterial replication. Thus, discrepancies between detection methods must be addressed. In the absence of challenge studies, which are few and far between in recent years, serologic recognition of a specific chlamydial antigen or isolation of a viable organism would ideally enable differentiation between whether these detection events are representative of sporadic, endemic or epidemic infections and assist stakeholders in understanding the significance of these reports. Such studies are sorely needed but are also obviously challenging, given the range of confounding factors including the lack of commercially available reagents for nonmodel organisms and the logistical and economic challenges associated with the collection of the appropriate specimens for such studies.
On a positive note, while updating chlamydial taxonomy has not been without its challenges, the recent acceptance of a new taxonomic system for this group of intracellular pathogens [1], as well as the identification and use of an expanded gene set encompassing phylogenetic markers capable of differentiating taxa at the species, genus and family level [46], will support the efforts of researchers to describe new and emerging chlamydial pathogens. With the increasing ease and decreasing cost of culture-independent genome sequencing, it is anticipated that these methods will come into use more frequently for emerging chlamydial pathogens. However, culture of viable organisms and challenge experiments as that recently performed [2], [3], [36] will nevertheless remain the reference standard for assessing the true pathogenic potential of novel chlamydial infections.
Conflict of Interest
None declared.
References
- 1.Sachse K., Bavoil P.M., Kaltenboeck B., Stephens R.S., Kuo C.C., Rossello-Mora R. Emendation of the family Chlamydiaceae: proposal of a single genus, Chlamydia, to include all currently recognized species. Syst Appl Microbiol. 2015;38:99–103. doi: 10.1016/j.syapm.2014.12.004. [DOI] [PubMed] [Google Scholar]
- 2.Sachse K., Laroucau K., Riege K., Wehner S., Dilcher M., Creasy H.H. Evidence for the existence of two new members of the family Chlamydiaceae and proposal of Chlamydia avium sp. nov. and Chlamydia gallinacea sp. nov. Syst Appl Microbiol. 2014;37:79–88. doi: 10.1016/j.syapm.2013.12.004. [DOI] [PubMed] [Google Scholar]
- 3.Vorimore F., Hsia R.C., Huot-Creasy H., Bastian S., Deruyter L., Passet A. Isolation of a new Chlamydia species from the feral sacred ibis (Threskiornis aethiopicus): Chlamydia ibidis. PLoS One. 2013;8:e74823. doi: 10.1371/journal.pone.0074823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Taylor-Brown A., Bachmann N.L., Borel N., Polkinghorne A. Culture-independent genomic characterisation of Candidatus Chlamydia sanzinia, a novel uncultivated bacterium infecting snakes. BMC Genomics. 2016;17:710. doi: 10.1186/s12864-016-3055-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Taylor-Brown A., Vaughan L., Greub G., Timms P., Polkinghorne A. Twenty years of research into Chlamydia-like organisms: a revolution in our understanding of the biology and pathogenicity of members of the phylum Chlamydiae. Pathog Dis. 2015;73:1–15. doi: 10.1093/femspd/ftu009. [DOI] [PubMed] [Google Scholar]
- 6.Jones K.E., Patel N.G., Levy M.A., Storeygard A., Balk D., Gittleman J.L. Global trends in emerging infectious diseases. Nature. 2008;451(7181):990–993. doi: 10.1038/nature06536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cantas L., Suer K. Review: the important bacterial zoonoses in ‘one health’ concept. Front Public Health. 2014;2:144. doi: 10.3389/fpubh.2014.00144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Laroucau K., Aaziz R., Meurice L., Servas V., Chossat I., Royer H. Outbreak of psittacosis in a group of women exposed to Chlamydia psittaci–infected chickens. Euro Surveill. 2015;(24):20. doi: 10.2807/1560-7917.es2015.20.24.21155. [DOI] [PubMed] [Google Scholar]
- 9.Ling Y., Chen H., Chen X., Yang X., Yang J., Bavoil P.M. Epidemiology of Chlamydia psittaci infection in racing pigeons and pigeon fanciers in Beijing, China. Zoonoses Public Health. 2015;62:401–406. doi: 10.1111/zph.12161. [DOI] [PubMed] [Google Scholar]
- 10.McGuigan C.C., McIntyre P.G., Templeton K. Psittacosis outbreak in Tayside, Scotland, December 2011 to February 2012. Euro Surveill. 2012;(22):17. doi: 10.2807/ese.17.22.20186-en. [DOI] [PubMed] [Google Scholar]
- 11.Wallensten A., Fredlund H., Runehagen A. Multiple human-to-human transmission from a severe case of psittacosis, Sweden, January–February 2013. Euro Surveill. 2014;(42):19. doi: 10.2807/1560-7917.es2014.19.42.20937. [DOI] [PubMed] [Google Scholar]
- 12.Schautteet K., Vanrompay D. Chlamydiaceae infections in pig. Vet Res. 2011;42:29. doi: 10.1186/1297-9716-42-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Osman K.M., Ali H.A., ElJakee J.A., Galal H.M. Prevalence of Chlamydophila psittaci infections in the eyes of cattle, buffaloes, sheep and goats in contact with a human population. Transbound Emerg Dis. 2013;60:245–251. doi: 10.1111/j.1865-1682.2012.01337.x. [DOI] [PubMed] [Google Scholar]
- 14.Jelocnik M., Branley J., Heller J., Raidal S., Alderson S., Galea F. Multilocus sequence typing identifies an avian-like Chlamydia psittaci strain involved in equine placentitis and associated with subsequent human psittacosis. Emerg Microbes Infect. 2017;6:e7. doi: 10.1038/emi.2016.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Branley J., Bachmann N.L., Jelocnik M., Myers G.S., Polkinghorne A. Australian human and parrot Chlamydia psittaci strains cluster within the highly virulent 6BC clade of this important zoonotic pathogen. Sci Rep. 2016;6:30019. doi: 10.1038/srep30019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.De Puysseleyr K., De Puysseleyr L., Dhondt H., Geens T., Braeckman L., Morre S.A. Evaluation of the presence and zoonotic transmission of Chlamydia suis in a pig slaughterhouse. BMC Infect Dis. 2014;14:560. doi: 10.1186/s12879-014-0560-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dean D., Rothschild J., Ruettger A., Kandel R.P., Sachse K. Zoonotic Chlamydiaceae species associated with trachoma, Nepal. Emerg Infect Dis. 2013;19:1948–1955. doi: 10.3201/eid1912.130656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Joseph S.J., Marti H., Didelot X., Read T.D., Dean D. Tetracycline selective pressure and homologous recombination shape the evolution of Chlamydia suis: a recently identified zoonotic pathogen. Genome Biol Evol. 2016;8:2613–2623. doi: 10.1093/gbe/evw182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Suchland R.J., Sandoz K.M., Jeffrey B.M., Stamm W.E., Rockey D.D. Horizontal transfer of tetracycline resistance among Chlamydia spp. in vitro. Antimicrob Agents Chemother. 2009;53:4604–4611. doi: 10.1128/AAC.00477-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Walker E., Lee E.J., Timms P., Polkinghorne A. Chlamydia pecorum infections in sheep and cattle: a common and under-recognised infectious disease with significant impact on animal health. Vet J. 2015;206:252–260. doi: 10.1016/j.tvjl.2015.09.022. [DOI] [PubMed] [Google Scholar]
- 21.Giannitti F., Anderson M., Miller M., Rowe J., Sverlow K., Vasquez M. Chlamydia pecorum: fetal and placental lesions in sporadic caprine abortion. J Vet Diagn Invest. 2016;28:184–189. doi: 10.1177/1040638715625729. [DOI] [PubMed] [Google Scholar]
- 22.Burnard D., Polkinghorne A. Chlamydial infections in wildlife-conservation threats and/or reservoirs of ‘spill-over’ infections? Vet Microbiol. 2016;196:78–84. doi: 10.1016/j.vetmic.2016.10.018. [DOI] [PubMed] [Google Scholar]
- 23.Frutos M.C., Venezuela F., Kiguen X., Re V., Cuffini C. Detection of the ompA gene of Chlamydophila pecorum in captive birds in Argentina. Rev Argent Microbiol. 2012;44:65–68. [PubMed] [Google Scholar]
- 24.Tanaka C., Miyazawa T., Watarai M., Ishiguro N. Bacteriological survey of feces from feral pigeons in Japan. J Vet Med Sci. 2005;67:951–953. doi: 10.1292/jvms.67.951. [DOI] [PubMed] [Google Scholar]
- 25.Zocevic A., Vorimore F., Marhold C., Horvatek D., Wang D., Slavec B. Molecular characterization of atypical Chlamydia and evidence of their dissemination in different European and Asian chicken flocks by specific real-time PCR. Environ Microbiol. 2012;14:2212–2222. doi: 10.1111/j.1462-2920.2012.02800.x. [DOI] [PubMed] [Google Scholar]
- 26.Guo W., Li J., Kaltenboeck B., Gong J., Fan W., Wang C. Chlamydia gallinacea, not C. psittaci, is the endemic chlamydial species in chicken (Gallus gallus) Sci Rep. 2016;6:19638. doi: 10.1038/srep19638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li J., Guo W., Kaltenboeck B., Sachse K., Yang Y., Lu G. Chlamydia pecorum is the endemic intestinal species in cattle while C. gallinacea, C. psittaci and C. pneumoniae associate with sporadic systemic infection. Vet Microbiol. 2016;193:93–99. doi: 10.1016/j.vetmic.2016.08.008. [DOI] [PubMed] [Google Scholar]
- 28.Aaziz R., Gourlay P., Vorimore F., Sachse K., Siarkou V.I., Laroucau K. Chlamydiaceae in North Atlantic seabirds admitted to a wildlife rescue center in Western France. Appl Environ Microbiol. 2015;81:4581–4590. doi: 10.1128/AEM.00778-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Isaksson J., Christerson L., Blomqvist M., Wille M., Alladio L.A., Sachse K. Chlamydiaceae-like bacterium, but no Chlamydia psittaci, in sea birds from Antarctica. Polar Biol. 2015;38:1931–1936. [Google Scholar]
- 30.Taylor-Brown A., Ruegg S., Polkinghorne A., Borel N. Characterisation of Chlamydia pneumoniae and other novel chlamydial infections in captive snakes. Vet Microbiol. 2015;178:88–93. doi: 10.1016/j.vetmic.2015.04.021. [DOI] [PubMed] [Google Scholar]
- 31.Sariya L., Kladmanee K., Bhusri B., Thaijongrak P., Tonchiangsai K., Chaichoun K. Molecular evidence for genetic distinctions between Chlamydiaceae detected in Siamese crocodiles (Crocodylus siamensis) and known Chlamydiaceae species. Jpn J Vet Res. 2015;63:5–14. [PubMed] [Google Scholar]
- 32.Shilton C.M., Jerrett I.V., Davis S., Walsh S., Benedict S., Isberg S.R. Diagnostic investigation of new disease syndromes in farmed Australian saltwater crocodiles (Crocodylus porosus) reveals associations with herpesviral infection. J Vet Diagn Invest. 2016;28:279–290. doi: 10.1177/1040638716642268. [DOI] [PubMed] [Google Scholar]
- 33.Aaziz R., Vorimore F., Verheyden H., Picot D., Bertin C., Ruettger A. Detection of atypical Chlamydiaceae in roe deer (Capreolus capreolus) Vet Microbiol. 2015;181:318–322. doi: 10.1016/j.vetmic.2015.10.018. [DOI] [PubMed] [Google Scholar]
- 34.de Barsy M., Greub G. Waddlia chondrophila: from biology to pathogenicity. Microbes Infect. 2013;15:1033–1041. doi: 10.1016/j.micinf.2013.09.010. [DOI] [PubMed] [Google Scholar]
- 35.Wheelhouse N., Flockhart A., Aitchison K., Livingstone M., Finlayson J., Flachon V. Experimental challenge of pregnant cattle with the putative abortifacient Waddlia chondrophila. Sci Rep. 2016;6:37150. doi: 10.1038/srep37150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kearse M., Moir R., Wilson A., Stones-Havas S., Cheung M., Sturrock S. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 2012;28:1647–1649. doi: 10.1093/bioinformatics/bts199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stride M.C., Polkinghome A., Nowak B.F. Chlamydial infections of fish: diverse pathogens and emerging causes of disease in aquaculture species. Vet Microbiol. 2014;171:258–266. doi: 10.1016/j.vetmic.2014.03.022. [DOI] [PubMed] [Google Scholar]
- 38.Burnard D., Weaver H., Gillett A., Loader J., Flanagan C., Polkinghorne A. Novel Chlamydiales genotypes identified in ticks from Australian wildlife. Parasit Vectors. 2017;10:46. doi: 10.1186/s13071-017-1994-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hokynar K., Sormunen J.J., Vesterinen E.J., Partio E.K., Lilley T., Timonen V. Chlamydia-like organisms (CLOs) in Finnish Ixodes ricinus ticks and human skin. Microorganisms. 2016;4(3):E28. doi: 10.3390/microorganisms4030028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pilloux L., Aeby S., Gaumann R., Burri C., Beuret C., Greub G. The high prevalence and diversity of Chlamydiales DNA within Ixodes ricinus ticks suggest a role for ticks as reservoirs and vectors of Chlamydia-related bacteria. Appl Environ Microbiol. 2015;81:8177–8182. doi: 10.1128/AEM.02183-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Croxatto A., Rieille N., Kernif T., Bitam I., Aeby S., Peter O. Presence of Chlamydiales DNA in ticks and fleas suggests that ticks are carriers of Chlamydiae. Ticks Tick Borne Dis. 2014;5:359–365. doi: 10.1016/j.ttbdis.2013.11.009. [DOI] [PubMed] [Google Scholar]
- 42.Chua P.K., Corkill J.E., Hooi P.S., Cheng S.C., Winstanley C., Hart C.A. Isolation of Waddlia malaysiensis, a novel intracellular bacterium, from fruit bat (Eonycteris spelaea) Emerg Infect Dis. 2005;11:271–277. doi: 10.3201/eid1102.040746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pierle S.A., Morales C.O., Martinez L.P., Ceballos N.A., Rivero J.J., Diaz O.L. Novel Waddlia intracellular bacterium in Artibeus intermedius fruit bats, Mexico. Emerg Infect Dis. 2015;21:2161–2163. doi: 10.3201/eid2112.150002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hokynar K., Vesterinen E.J., Lilley T.M., Pulliainen A.T., Korhonen S.J., Paavonen J. Molecular evidence of Chlamydia-like organisms in the feces of Myotis daubentonii bats. Appl Environ Microbiol. 2017;83 doi: 10.1128/AEM.02951-16. e02951–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sachse K., Vretou E., Livingstone M., Borel N., Pospischil A., Longbottom D. Recent developments in the laboratory diagnosis of chlamydial infections. Vet Microbiol. 2009;135:2–21. doi: 10.1016/j.vetmic.2008.09.040. [DOI] [PubMed] [Google Scholar]
- 46.Pillonel T., Bertelli C., Salamin N., Greub G. Taxogenomics of the order Chlamydiales. Int J Syst Evol Microbiol. 2015;65(Pt 4):1381–1393. doi: 10.1099/ijs.0.000090. [DOI] [PubMed] [Google Scholar]