Abstract
Hydrogen peroxide (H2O2) controls signaling pathways in cells by oxidative modulation of the activity of redox sensitive proteins denominated redox switches. Here, quantitative biology concepts are applied to review how H2O2 fulfills a key role in information transmission. Equations described lay the foundation of H2O2 signaling, give new insights on H2O2 signaling mechanisms, and help to learn new information from common redox signaling experiments. A key characteristic of H2O2 signaling is that the ratio between reduction and oxidation of redox switches determines the range of H2O2 concentrations to which they respond. Thus, a redox switch with low H2O2-dependent oxidability and slow reduction rate responds to the same range of H2O2 concentrations as a redox switch with high H2O2-dependent oxidability, but that is rapidly reduced. Yet, in the first case the response time is slow while in the second case is rapid. H2O2 sensing and transmission of information can be done directly or by complex mechanisms in which oxidation is relayed between proteins before oxidizing the final regulatory redox target. In spite of being a very simple molecule, H2O2 has a key role in cellular signaling, with the reliability of the information transmitted depending on the inherent chemical reactivity of redox switches, on the presence of localized H2O2 pools, and on the molecular recognition between redox switches and their partners.
Keywords: Redox switches, Kinetics, Steady-state, Dynamic range, Response time, Information transmission
Graphical abstract
Highlights
-
•
Hydrogen peroxide signaling proceeds through oxidation of redox switches.
-
•
Oxidation of redox switches can be direct or mediated by highly reactive sensors.
-
•
Response of redox switches is controlled by their oxidability and reduction rate.
-
•
Localized protein interactions ensure the accuracy of information transmission.
1. Introduction
Hydrogen peroxide (H2O2) is a non-radical oxidant present in virtually all aerobic organisms. Viewed initially as a detrimental byproduct of oxidative metabolism, today H2O2 is recognized to play important roles in cellular physiology [1]. The cellular function of H2O2 is supported by coupling of cellular signals with its production. Many enzymatic sources have been identified that produce H2O2 directly or produce superoxide radical, which is subsequently dismutated into water and H2O2, a process that is accelerated many orders of magnitude by the action of superoxide dismutases. A particularly relevant source of H2O2 is NADPH oxidases because their sole function seems to be the tightly-regulated production of superoxide/H2O2 [2].
Production of H2O2 is balanced by the action of antioxidant enzymatic systems, such as catalase, glutathione peroxidases, and peroxiredoxins, that remove H2O2 very rapidly [3], [4]. An homeostatic steady-state level of 10−7−10−8 M results under physiological conditions [5], and changes around this background steady-state level will trigger cellular responses. If these concentration shifts are moderated, transient or localized in space, being a result of for example signaling processes, a physiological stress response – or eustress – is observed [3]. If variations in the H2O2 concentration are large, sustained or affect H2O2 bulk levels, a pathological stress with deleterious effects for the organism materializes [3]. Thus, oxidative effects are inherently non-linear and biphasic with threshold levels separating the physiological and the pathological domains [6], [7]. In addition, eustress and pathological stress can either be oxidative or reductive, depending on whether they are caused by an increase or decrease of H2O2 around its background steady-state level.
In this review, quantitative biology concepts are introduced to analyze the transmission of information mediated by H2O2 in the oxidative eustress setting.
1.1. H2O2 signaling
Signaling pathways are regulated by the reaction of H2O2 with proteins harboring redox sensitive moieties, like metal centers or cysteine residues, whose oxidation controls their activity. These proteins denominated redox switches are key players in the regulation of biochemical pathways, including protein phosphatases, kinases or transcription factors [8]. Thus, a change in the concentration of H2O2 is matched by a change in the oxidation state of a redox switch, regulating a downstream pathway and transducing the information encoded in the H2O2 concentration profile along a signaling cascade.
Chemically, most previously identified redox-controlled switches are thiol proteins [9], [10], [11], but metal switches have also been described [12], [13]. Thiol switches are proteins with cysteine residues with low pKa that favors their proton dissociation to form a thiolate at physiological pH. Thiolates have a higher reactivity towards H2O2, but the pKa of the cysteine residue is not the only determinant of the reactivity of the thiol protein with H2O2. Rather, stabilization of the transition state between H2O2 and the cysteine residue is critical to achieve high catalytic rates with the protein environment affecting the reactivity of the cysteine group [14]. Thus, reactivity of thiol proteins towards H2O2 spans several orders of magnitude, from the low 20 M−1s−1 for some protein tyrosine phosphatases, like PTP1B and SHP-2, to the high 107 M−1s−1 for peroxiredoxin 2 [15].
The chemical reactivity of redox switches is a potential mechanism underlying specific biological effects caused by different concentrations of H2O2. At low H2O2 concentrations only the most reactive switches will sense H2O2, while less reactive switches will sense H2O2 at high concentrations. As will be described below, such chemical specificity based on the oxidability of the redox switch is just one of several regulatory mechanisms in H2O2 signaling.
2. Quantitative analysis of H2O2 signal processing
2.1. The steady-state approximation
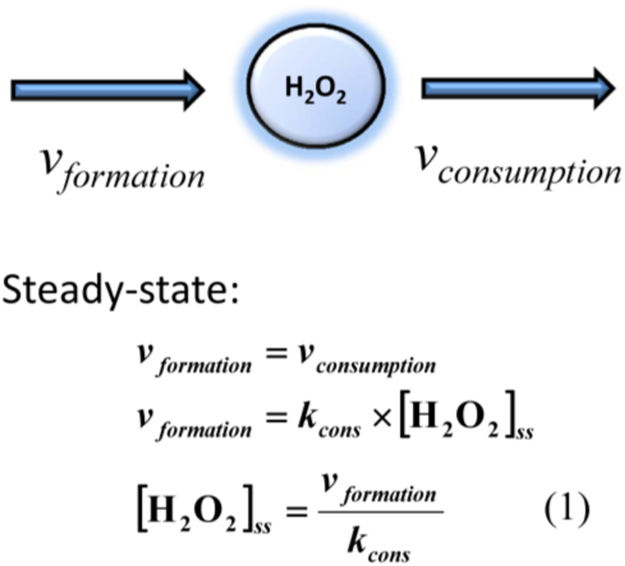
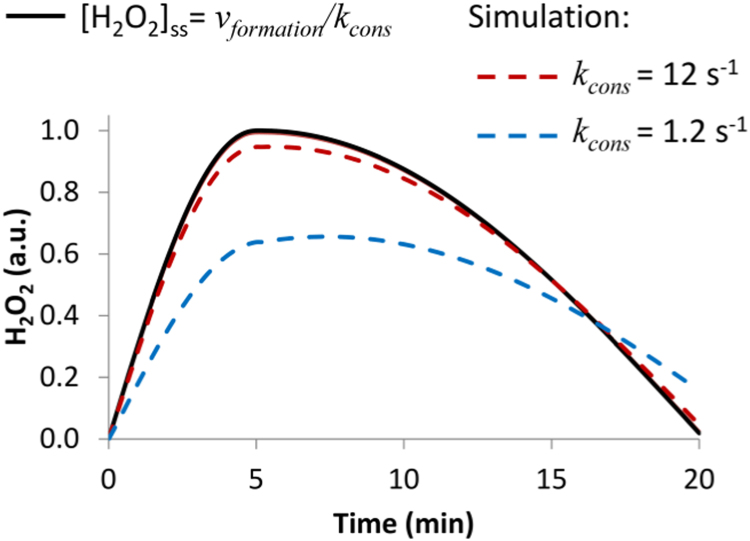
The steady-state concept is central to quantitative analyses in redox biology [16], [17], [18]. As a result of continuous formation and elimination, H2O2 settles to a near steady-state given by Eq. 1 in Fig. 1. It is important to test the validity of the steady-state approximation during signaling events when variations of the H2O2 concentration are observed. In other words, does the steady-state approximation hold when H2O2 is not steady? When H2O2 production is increased, for example due to the activation of an NADPH oxidase, the steady-state approximation can be used to calculate the transient dynamics of H2O2 because the very fast elimination of H2O2 by antioxidants systems has a reaction time much quicker than the transient responses formed during signaling events (Fig. 2). Thus, the steady-state approximation is valid even when H2O2 levels change during signaling events.
Fig. 1.
The steady-state of H2O2. A steady-state is reached when the rates of formation (vformation) are balanced by the rates of elimination (vconsumption). The rate of H2O2 elimination is assumed to follow first-order kinetics because in the eustress domain H2O2 does not overload the antioxidant systems. Thus, vconsumption= kcons×[H2O2] with kcons being the pseudo first-order rate constant for the overall consumption of H2O2. The steady-state Eq. 1 is deduced from the equality between the rates of formation and elimination of H2O2.
Fig. 2.
Application of the steady-state approximation to H2O2 dynamics during signaling events. To reproduce a transient H2O2 increase, the rate of H2O2 formation was assumed to peak at 5 min and to decay to zero at 20 min as observed in Ref. [19]. Three H2O2 profiles are shown: one was calculated according to steady-state Eq. 1 and two according to simulations reproducing the cell behavior for two values of consumption rate constants – 1.2 s−1 and 12 s−1. Simulated H2O2 profiles approach that calculated from Eq. 1 when the value of kcons increases, and for kcons=120 s−1 or higher, simulation curves coincide with the steady-state curve (not shown). This trend is justified by the very fast time scale of the kcons rate constant. A time scale of 0.06 s is calculated according to the formula t1/2= ln(2)/kcons, with ln(2) being the natural logarithm of 2, for a kcons =12 s−1, a lower limit for the value of the kcons rate constant (see Table 1 below). A t1/2 value of 0.06 s is much faster than the time scale associated with the variation of H2O2 formation during signaling events, which is in the minute range, and thus the steady-state approximation is valid. In general, the steady-state approximation is a reasonable assumption when analyzing processes in the minute range or slower because antioxidant systems are usually fast enough.
To make a quantitative analysis of H2O2 signal processing, the simple steady-state scheme of Fig. 1 was extended to include a signaling reaction. The formation of H2O2 is now balanced by two elimination reactions, one being the consumption of H2O2 by antioxidant systems and the other the oxidation of redox switches (Fig. 3). When first-order kinetics are assumed for these elimination processes, H2O2 steady-state is given by Eq. 2 in Fig. 3.
Fig. 3.
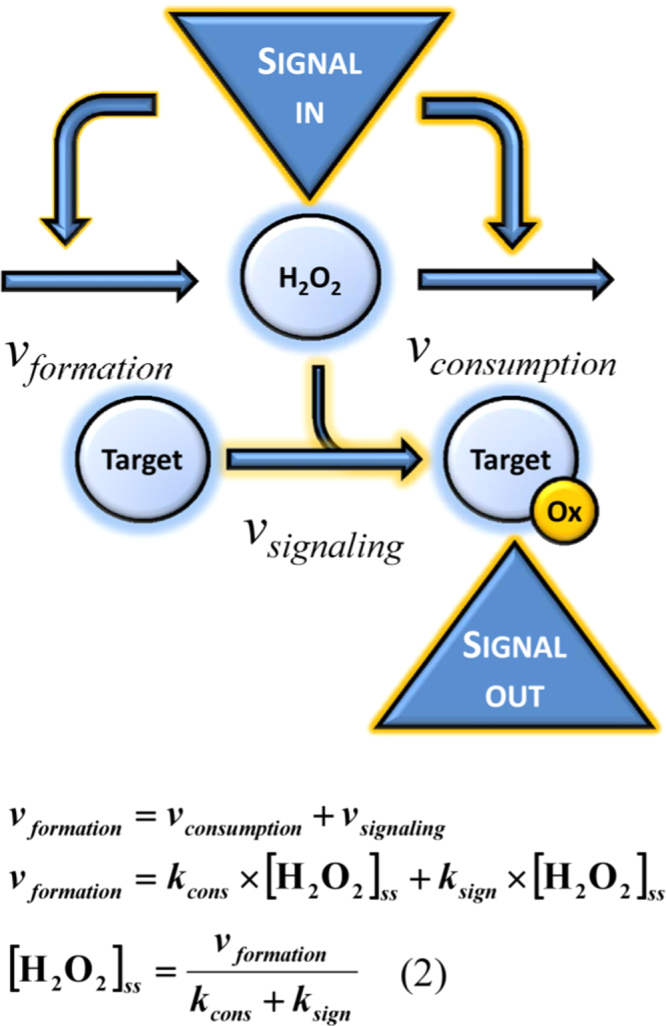
The H2O2 steady-state in the presence of signaling. In principle, a signal (Signal in) can modulate either the production or the removal of H2O2, the activation of a NADPH oxidase being a common mechanism. The subsequent change in H2O2 concentration is sensed by a redox switch (Target) that upon oxidation (vsignaling) transmits information downstream the signaling cascade (Signal out). Similarly to the rate of H2O2 consumption by antioxidant systems, the signaling reaction also follows first-order kinetics, being vsignaling=ksign×[H2O2] with ksign referring to the rate constant for the reaction of H2O2 with the redox target. The resulting steady-state H2O2 concentration is given by Eq. 2.
Eq. 2 shows the relative magnitude of kcons and ksign only is needed to predict whether signaling processes affect directly the H2O2 steady-state. According to published data, kcons is five to six orders of magnitude higher than ksign (Table 1), implying that antioxidant reactions vastly outcompete signaling reactions for H2O2. Thus, a kinetic bottleneck for H2O2 signaling is established [10], [14], [15], [20]. If highly efficient antioxidant systems divert more than 99.999% of H2O2 from signaling reactions, how are H2O2 variations sensed? The rate of signaling is calculated as the product of the rate constant ksign by the concentration of H2O2 (Fig. 3). So, the rate of the signaling reaction will match the variations of H2O2, and the information encoded in the H2O2 concentration profile can, in principle, be transmitted downstream the signaling cascade. The key question is whether the information is transferred fast enough when vsignaling is very slow.
Table 1.
Competition between enzymatic antioxidants and redox switches for H2O2. kcons and ksign are pseudo first-order rate constants calculated as the product between ktarget+H2O2, the chemical rate constant between H2O2 and its target protein, and the concentration of the target protein, either an enzymatic antioxidant or a redox switch. For protein concentrations and rate constant values see [15], [8]. The rather high value for catalase concentration refers to the peroxisome [21].
|
ktarget+H2O2 (M−1s−1) |
[protein] (µM) |
ktarget+H2O2× protein (s−1) | |
|---|---|---|---|
| Antioxidant | kcons | ||
| Prx | 105–107 | 10 | 1–100 |
| GPx | 6×107 | 0.2–10 | 12–600 |
| Catalase | 107 | 103 | 104 |
| Redox switch | ksign | ||
| PTP1B | 24 | 0.01 | 2.4×10−7 |
| SHP‐2 | 20 | 0.01 | 2.0×10−7 |
| Keap1 | 140 | 1 | 1.4×10−4 |
Prx – Peroxiredoxin; GPx – Glutathione peroxidase 1.
2.2. Equations governing H2O2 signaling
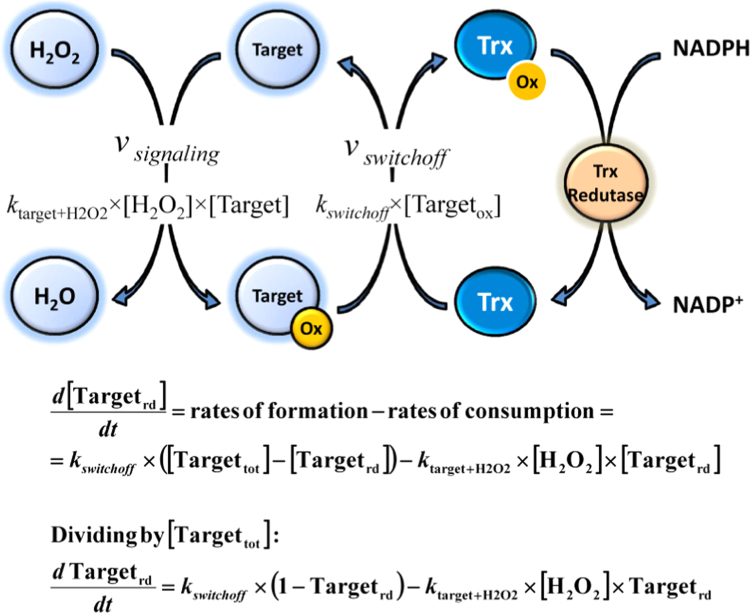
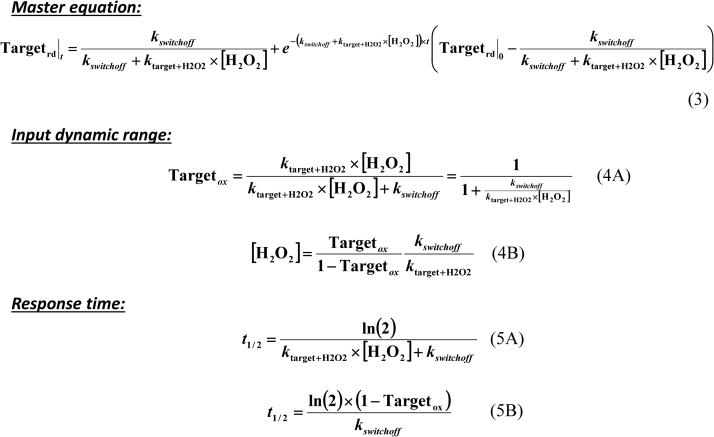
The issue whether a slow chemical reaction between H2O2 and a redox switch ensures timely information transmission during signal processing may be addressed with the help of the minimal mathematical model shown in Fig. 4. This model is formed by the oxidation-reduction cycle of a redox switch, which may be viewed as a switch-on switch-off sequence [22] with the on state – oxidized form of the redox switch – relaying the information encoded in H2O2 down the signaling cascade. The solution of this model yields a master equation (Eq. 3 in Fig. 5) describing the time course of the fraction of the redox switch in the reduced form [23]. From Eq. 3 two sets of simpler equations are deduced (Fig. 5), namely (i) Eqs. 4 A and 4B describing the effect of input H2O2 concentrations on the signaling response, and (ii) Eqs. 5 A and 5B characterizing the time-dependent H2O2 signaling properties. The input H2O2 concentrations and the response time are two important quantitative measures of redox signaling proposed before [18].
Fig. 4.
A minimal model of H2O2 signaling mediated by redox switches. A redox switch (Target) is oxidized by H2O2 and then reduced by, for example, a member of the thioredoxin family (Trx). The rate of target oxidation is equaled to the rate of the chemical reaction between H2O2 and the reduced form of the target (ktarget+H2O2×[H2O2]×[Targetrd]), and the rate of reduction is set to kswitchoff×[Targetox], with kswitchoff being a pseudo first-order rate constant. The differential equation is built, simplified by dividing by the total concentration of target [Targettot], and solved with a software like Maxima [24]. The resulting solution describes the time course of the target in terms of its molar fraction in the reduced state (Targetrd), thus avoiding the utilization of absolute concentrations, a measure that is difficult to measure experimentally.
Fig. 5.
Governing equations of H2O2 signaling. The master equation (Eq. 3) includes the dependence on the sustained H2O2 signaling concentration [H2O2] attained in the vicinity of the redox switch during the signaling process, as well as the rate constants for oxidation (ktarget+H2O2) and reduction (kswitchoff) of the redox switch, and the fraction of the redox switch in the reduced form at time 0 (). The key features of H2O2 signaling are described by two sets of simpler equations deduced from the master Eq. 3 [8], [23]. Eq. 4A is deduced by letting t tend to infinite and represents the steady-state fraction of the redox switch in the oxidized form (Targetox), which is a measure of the amount of information transmitted from H2O2 to the redox target. Calculation of the H2O2 signaling concentration causing a certain steady-state value of target oxidation is done with Eq. 4B, which results from an arrangement of Eq. 4A. The second set of equations (Eqs. 5A and 5B) calculates the response time of the redox switch to H2O2, giving the time (t1/2) needed for oxidizing half of the target present initially, i.e., indicating whether transmission of information proceeds rapid enough. Eq. 5A is deduced by replacing by and t by t1/2 in Eq. 3 and calculates t1/2 as a function of H2O2 concentration. Eq. 5B is deduced by replacing Eq. 4B in Eq. 5A and calculates t1/2 as a function of the steady-state fraction of the redox switch in the oxidized form and on the value of kswitchoff.
2.3. H2O2 dynamic range
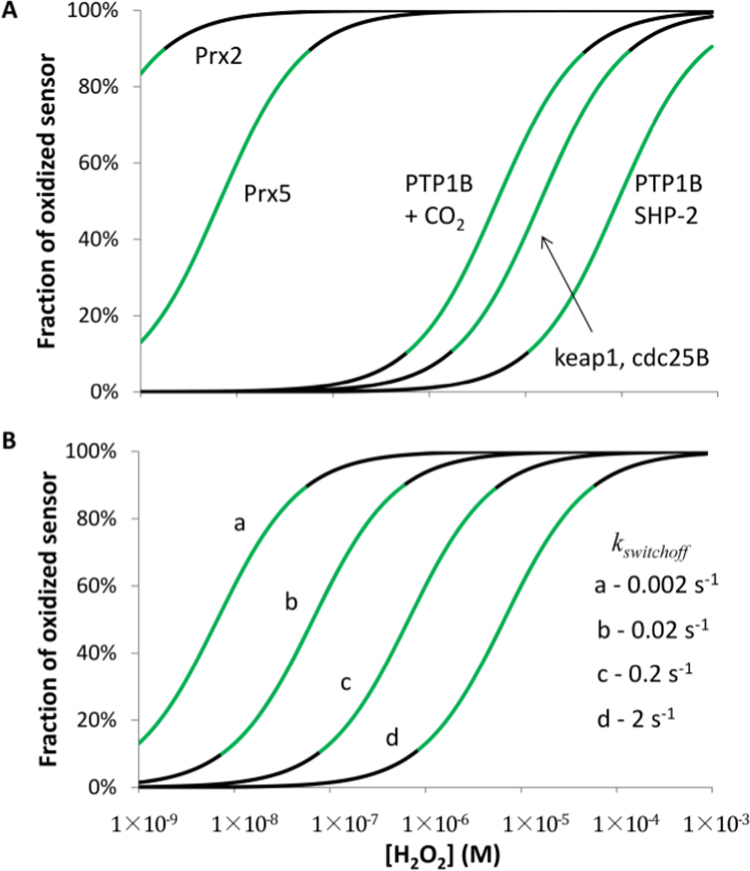
The input dynamic range, i.e. the range of H2O2 concentrations for which redox switches act as sensors of H2O2, depends on the reactivity of the redox switch towards H2O2, being inversely proportional to ktarget+H2O2 (Fig. 6A). One source of uncertainty in the plot of Fig. 6A is the ktarget+H2O2 value because, among other factors, H2O2 reactivity increases several orders of magnitude upon reaction with phosphate and carbon dioxide yielding peroxymonophosphate [25] and peroxymonocarbonate [26], [27], respectively. The reactivity of PTP1B with peroxymonophosphate is 7000-fold higher than with H2O2 itself [25], and at pH 7, the presence of carbonate at 25 mM, a physiological level, accelerates the reaction between PTP1B and H2O2 from 24 M−1s−1 to 202 M−1s−1 at 25 °C and to 396 M−1s−1 at 37 °C. The formation of these derivatives is not immediate, taking 5–8 min to reach an equilibrium with H2O2 [25], [27]. Nevertheless, for peroxymonocarbonate, PTP1B accelerates this equilibration to a few seconds or faster [26]. This effect was attributed to oxidation of the active-site cysteine by peroxymonocarbonate possibly formed in the active center of the enzyme [26]. Zn(II) complexes and other Lewis acids increase the rate of peroxymonocarbonate formation [27], and one may speculate that Arg221, being present in the active site of PTP1B and being essential for catalysis [28], can act as a Lewis acid catalyzing the formation of peroxymonocarbonate. Therefore, in Fig. 6A the input dynamic range was also calculated for PTP1B in the presence of CO2.
Fig. 6.
H2O2 input dynamic range for redox switches. Plots of Eq. 4A show the range of H2O2 concentrations to which thiol proteins with different reactivity respond. The following ktarget+H2O2 values were used: peroxiredoxin 2 (Prx2) – 1×107 M−1s−1; peroxiredoxin 5 (Prx5) – 3×105 M−1s−1; Kelch-like ECH-associated protein 1 (Keap1) – 140 M−1s−1; cell division cycle 25B (cdc25B) – 140 M−1s−1; protein tyrosine phosphatase 1B (PTP1B) – 24 M−1s−1; PTP1B in presence of bicarbonate (PTP1B + CO2) – 396 M−1s−1; and, src-homology 2 containing tyrosine phosphatase (SHP-2) – 20 M−1s−1. In (A) kswitchoff value was 2×10−3 s−1, while in (B) the influence of a range of kswitchoff values is shown for peroxiredoxin 5. Input H2O2 concentrations sustaining information transmission by redox switches are shown in green and were defined as the input eliciting a 10–90% oxidation of the protein. Below 10% oxidation, redox switch response is considered too weak to transmit efficiently the H2O2 signal, while above 90% oxidation, the response is near saturated to further increase in the H2O2 concentration.
In addition to the reactivity of the redox protein towards H2O2, the input dynamic range also depends on the rate of reduction of the redox switch, increasing for high kswitchoff rate constants, as shown in Fig. 6B. Thus, according to Eqs. 4A and 4B the reduction of the redox switch inhibits the transmission of H2O2 signals, as it is observed for the reductions of OxyR by glutaredoxin 1 [29], of Yap1 by thioredoxins 1 and 2 [30], of Pap1 by thioredoxins 1 and 3 [31], [32], of PTP1B by redoxin TRP14 and thioredoxin 1 [33], [34], of PTEN by thioredoxin 1 [34], and of the NRF2/KEAP1 system by the thioredoxin system [35].
kswitchoff values for protein phosphatases PTP1B and SHP-2 – 2×10−3 s−1 – [23] are about three orders of magnitude lower than for peroxiredoxins – 2 s−1 [36]. This large difference reflects the value of the rate constant for the reduction of phosphatases – 700 M−1s−1 for PTP1B (estimated from [37]) – being much lower than the rate constant for the reduction of peroxiredoxins by thioredoxin – 2.1×105 M−1s−1 [38], 2.2×105 M−1s−1 (estimated from [39]) and (2–8)×105 M−1s−1 (estimated from [40]) for peroxiredoxins 2, 3, and 5, respectively. Thus, the input dynamic range for peroxiredoxins is not as low as it could be expected from their high reactivity towards H2O2, and may even overlap with that of less reactive proteins. For example, input dynamic ranges for PTP1B and peroxiredoxin 5 are predicted to overlap (Table 2).
Table 2.
H2O2 dynamic range and response time for thiol proteins. Calculation of H2O2 dynamic range was done with Eq. 4B, assuming 10% and 90% of target oxidation, respectively for the lower and upper limit. Calculation of the response time to H2O2 was done with Eq. 5B, assuming 50% of target oxidation. For PTP1B and SHP-2 data for the reactivity obtained in the presence of CO2[26] is also shown.
| Redox target | ktarget+H2O2(M−1s−1) | kswitchoff(s−1) | H2O2dynamic range (µM) | Response time to H2O2(s) |
|---|---|---|---|---|
| PTP1B | 24 | 2×10−3 | 9–750 | 173 |
| +CO2 | 396 | 2×10−3 | 0.6–45 | 173 |
| SHP‐2 | 20 | 2×10−3 | 11–900 | 173 |
| +CO2 | 167 | 2×10−3 | 1.3–108 | 173 |
| Prx5 | 3×105 | 2 | 0.7–60 | 0.2 |
| Prx2 | 1×107 | 2 | 0.02–1.8 | 0.2 |
Table 2 shows the input dynamic range predicted according to published kinetic data. For H2O2 signaling concentrations lower than 1 µM, signaling is probably intermediated by a high reactive protein such as peroxiredoxin 2. For H2O2 concentrations higher than 1 µM, mediation of signaling by proteins with different reactivity towards H2O2 is feasible, but mediation by a high or by a low reactive protein is not equivalent as the response time is different (see below).
Not considered here is the hyperoxidation of peroxiredoxins, which also mediates transmission of information encoded in H2O2 [41], [42], [43]. Having different input dynamic ranges depending on modified forms of the same protein gives adaptability to signaling systems [44].
2.4. Response time to H2O2
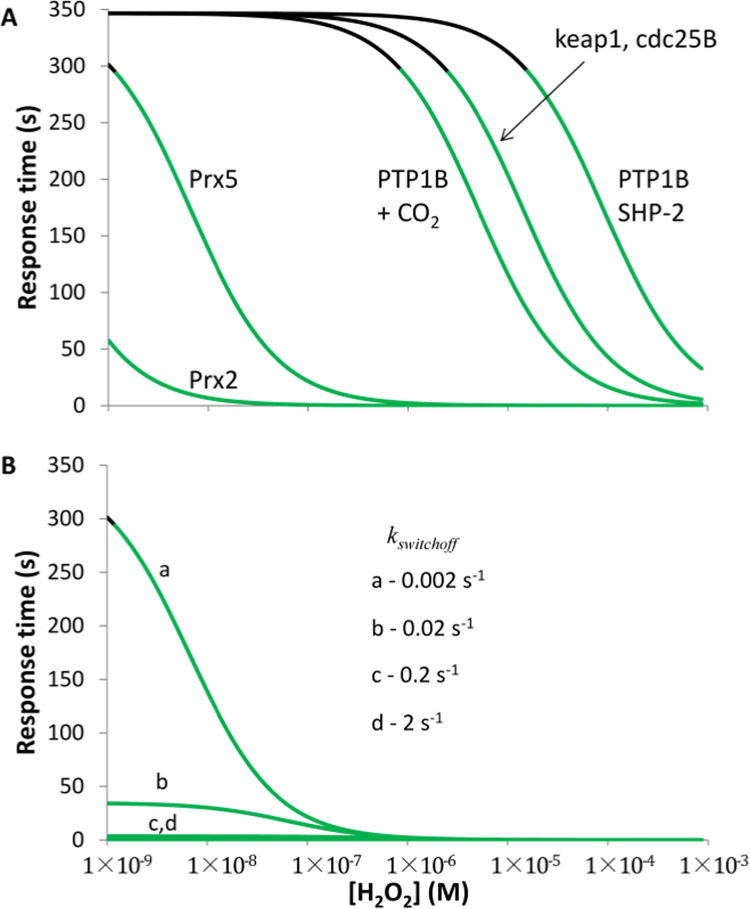
Similarly to the input range, the response time also depends on the reactivity of proteins towards H2O2 and on the value of kswitchoff (Eqs. 5A and 5B). But contrary to the input dynamic range, the response does not depend on the ratio of the rate constant values for these processes, but rather on their sum. Therefore, between two proteins with similar input dynamic ranges, the one with higher reactivity towards H2O2 displays a faster response time for a given H2O2 concentration. For example, PTP1B will respond with a time of approximately 3 min, while peroxiredoxin 5 responds in less than 1 s, even if they have similar input dynamic ranges (Table 2). The response time for PTP1B calculated here is much faster than published estimations [8], [26], [36] because previous analyses considered only the oxidation of the redox switch, neglecting the impact of its reduction in the acceleration of the response time.
Response time values shown in Table 2 were calculated with Eq. 5 B, which hides the influence of H2O2 concentration and ktarget+H2O2. For example, in spite of very different reactivity towards H2O2, peroxiredoxins 2 and 5 have a similar response time – 0.2 s. Implicit H2O2 concentrations used are, however, different for peroxiredoxins 2 and 5, being those that induce 50% of peroxiredoxin oxidation, as calculated by Eq. 4B. For the same H2O2 concentration, peroxiredoxin 2 has a much faster response time than peroxiredoxin 5, as shown in Fig. 7A.
Fig. 7.
Response time of redox switches to H2O2. Plots of Eq. 5A show the time needed to reach 50% of the overall response of sensors to H2O2. Values of rate constants are the same as those used in Fig. 6. In (A) kswitchoff value was 2×10−3 s−1, while in (B) the influence of a range of kswitchoff values is shown for peroxiredoxin 5. Input H2O2 concentrations supporting rapid information transmission by redox switches are shown in green and were defined as the input eliciting responses faster than 10 min. Nonetheless, some processes may be compatible with longer responses times to H2O2, including for example apoptosis [45] or adaptation pathways [46], [47].
2.5. Analyzing typical experiments
In addition to provide new insights on the mechanisms of H2O2 signaling, the two sets of Eqs. 4 and 5 may be applied to learn new information from typical redox signaling experiments that measure the oxidation time course of redox switches. To this end, three experimental measurements are useful: (i) the fraction of the redox target in the oxidized form, (ii) the response time, and (iii) the H2O2 input dynamic range. The oxidation levels of the redox target and the response time can be estimated from the time course of the oxidation profile of the target under analysis. The measurement of the input dynamic range is more difficult. When H2O2 is added externally, the intracellular concentration of H2O2 is lower than that applied extracellularly, and a gradient across the plasma membrane is established [48]. The magnitude of this gradient is unknown and depends on the cell type and whether peroxiredoxins are active at the external H2O2 concentration applied in the experimental set up. The presence of active peroxiredoxins increases gradients by approximately two orders of magnitude, from values under 10 [48], [49], [50] to values in the 650–1000 range [51], [52]. Thus, uncertainties in the values of intracellular H2O2 concentrations impact the determination of the input dynamic range when H2O2 is added extracellularly. Alternatively, if endogenous production of H2O2 is stimulated with a signaling molecule, like a growth factor, the intracellular H2O2 concentration is also unknown. Even if the intracellular H2O2 level is followed with a probe, the conversion of the signal measured to H2O2 concentrations values is usually not done. In spite of these caveats, useful information can still be obtained from the concentration of H2O2 applied in experiments as exemplified below.
Stat3 is inhibited by 5 µM extracellular H2O2 [53], which corresponds to an intracellular concentration in the range of 0.5–0.005 µM if gradients are considered. In spite of this wide range of possible H2O2 concentrations, the involvement of a sensor with reactivity similar to peroxiredoxin 2 can be predicted (Table 2). In addition, the response time observed experimentally is below 60 s, for an extracellular H2O2 concentration of 50 µM [53]. A peroxiredoxin-like sensor is needed to attain such rapid response according to Eqs. 5 A and 5B. In fact, peroxiredoxin 2 acts as a sensor, reacting with H2O2 and then relaying the oxidation to form disulfide links between Stat3 monomers [53]. Of note, when oxidation relays are involved response times are slower than those indicated in Table 2 as the oxidation relay step introduces an additional delay not considered in the minimal model of Fig. 4.
In another example, a response time of about 4–5 min is estimated from the PTP1B oxidation profile observed when the endogenous production of H2O2 is triggered by EGF in A431 cells [54]. This slow response time is compatible with the direct reaction of H2O2 with PTP1B (see Table 2). In addition, in this case the H2O2 signaling concentration attained in the vicinity of PTP1B can be estimated as rate constants for PTP1B are known: if the level of PTP1B oxidation measured experimentally – approximately 50% [54] – together with ktarget+H2O2 =396 M−1s−1 and kswitchoff =2×10−3 s−1, is introduced in equation 4B, a concentration of 5 µM is estimated. By a similar approach, the H2O2 concentration attained in the vicinity of SHP-2 during stimulation of Rat-1 cells by PDGF is calculated to be 6 µM, based on the observed SHP-2 oxidation profile [55]. H2O2 concentrations in the order of 5–6 µM are much higher than the bulk steady-state H2O2 concentration, estimated in the range 0.1–0.01 µM [5], but are still plausible as localized pools of H2O2 probably play an important role during signaling [13], [36], [41], [56]. The plausibility of this estimation is reinforced by noting that oxidation profiles of protein phosphatases PTP1B and SHP-2 observed with growth factors are similar to those observed with extracellular H2O2 concentrations close to 100 µM [23], [55], [57]. An extracellular 100 µM H2O2 concentration corresponds to an intracellular concentration of 5 µM if an H2O2 gradient across the plasma membrane of 20 is established, which is plausible taking into consideration that at this relatively high external H2O2 levels peroxiredoxin systems are at least partially inhibited [36], [58].
Nonetheless, H2O2 concentrations attained during signaling are most probably pathway dependent. As referred above, low H2O2 extracellular concentrations, in the order of 5 µM, are in play during the formation of disulfide-linked Stat3 oligomers [53]. On the other hand, the inhibition of protein phosphatase 1 (PP1) probably needs a high dose of H2O2 because an associated IC50 of 67 µM was measured in vitro [13]; in fact, this inhibition is probably mediated by localized production of H2O2 because colocalization of NOX4 and PP1 was observed and both proteins were identified together in a complex [13].
The previous discussion illustrates how Eqs. 4 and 5 give new insights on the mechanisms of H2O2 signaling and how new information is learned from common experimental measures. In addition, other applications for the equations are possible, including for example their fitting to experimental data to determine rate constants [23].
3. Final remarks
The main results of the quantitative biology analysis of H2O2 signaling presented here are summarized in the form of Eqs. 4 and 5 and are depicted in Fig. 8. Eqs. 4 and 5 govern the biology of H2O2 signaling and provide a quantitative framework with predictive power. In addition, common experimental measurements, like response time and oxidation profile of redox switches, may be analyzed with these equations, giving hints on the mechanisms of signal processing underlying experimental observations. Although the analysis was focused on thiol switches, these equations also apply to other types of redox switches.
Fig. 8.
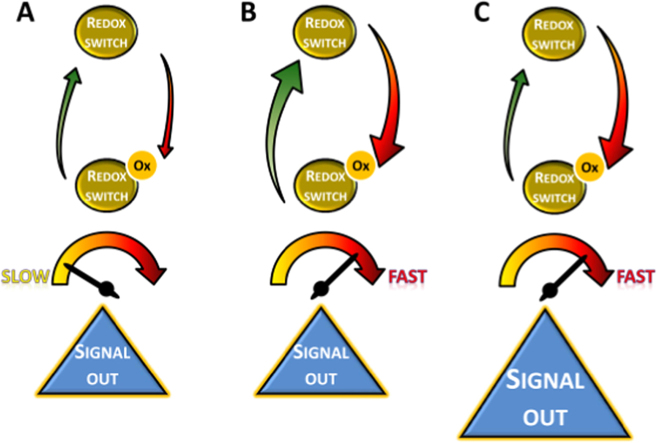
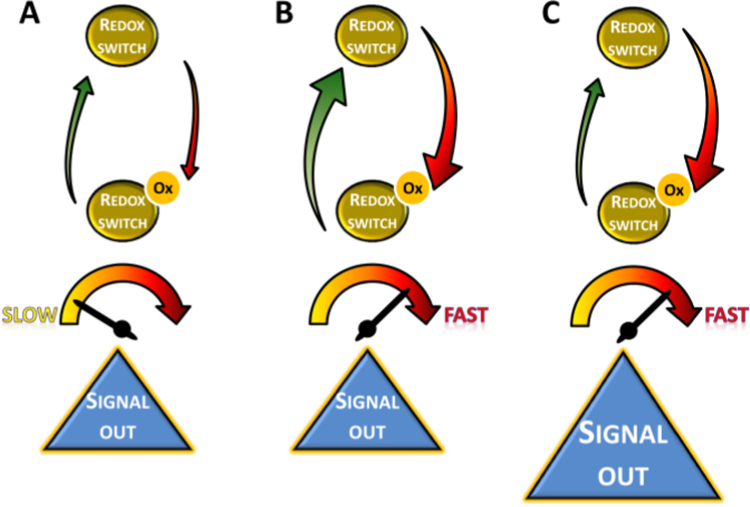
Redox signaling properties. Redox characteristics of the redox switch define the properties of the transmission of information encoded in H2O2. When the ratio between oxidation and reduction is similar the same information output is observed (panels A and B), but the redox switch with higher redox turnover has a faster response (panel B). If oxidation dominates over reduction, signal transduction is favored by displacing the redox switch to the oxidized form (Panel C).
The input dynamic range, i.e., the H2O2 concentration range to which redox switches respond, depends not only on their H2O2-induced oxidability but also on their rate of reduction (Fig. 8). Eq. 4 indicates that the ratio between the kinetics constants of these two processes defines the sensitivity of the redox switch to H2O2. Peroxiredoxins and protein phosphatases respond in ranges of H2O2 concentration that are not as far apart as it could be predicted based solely in their very large different reactivity towards H2O2, because peroxiredoxins are reduced faster than protein phosphatases (Table 2). The response time also depends on the rates of oxidation and reduction of the redox switch given by Eq. 5. In this case, it is not the ratio between oxidation and reduction that determines the response time, but their effects add up to increase the rate of response.
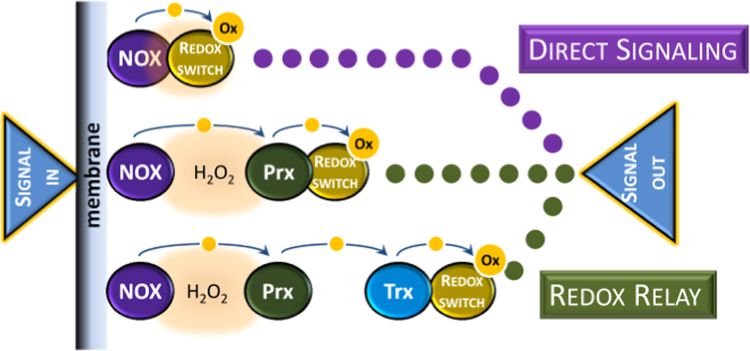
Redox switches transmit information along a signaling cascade after being oxidized by H2O2. This oxidation may be direct or, alternatively, indirect when the redox sensor is a high reactive protein, like a peroxiredoxin, that relays the oxidation to a redox switch with low reactivity towards H2O2 (Fig. 9) [4], [59], [60]. Examples of relay circuits already identified include the original discovery of the Gpx3/Yap1 [61], and subsequently Tpx1/Pap1 [32], [62], Tsa1/Sty1 [63], Prx1/Ask1 [64], and Prx2/Stat3 [53]. In the thioredoxin-peroxiredoxin model, thioredoxin, or another protein responsible for the reduction of the redox sensor, acts as a redox relay, mediating the oxidation of a downstream redox switch [60], [65].
Fig. 9.
Mechanisms of H2O2 signaling. In the direct signaling pathway, H2O2 reacts directly with a redox switch. In the redox relay mechanism, the sensor of H2O2, a high-reactive protein such as a peroxiredoxin, relays the oxidation to a redox switch. The redox relay mechanism can be made more complex by the intervention of additional intermediates, as exemplified in the thioredoxin-peroxiredoxin model in which the protein responsible for the reduction of the redox sensor acts as a redox relay.
Independently of the specific mechanism, localized interactions are probably important to attain accurate information transmission [66]. These localized interactions include (1) complexes of NOX with redox switches, favoring switch on of a specific redox sensor, and (2) the interaction of a high reactive sensor, such as a peroxiredoxin, with a target protein, sustaining a specific relay of the oxidative signal. In this second case, the peroxiredoxin will not only relay the oxidative signal downstream but also trap H2O2 [36], avoiding H2O2 diffusion outside the signal locus and, consequently, preventing either unspecific signaling messages or even some form of pathological stress.
In conclusion, the relatively weak oxidation potential of H2O2 is coupled to timely and accurate transmission of information by a combination of chemical reactions that balance oxidation and reduction of redox switches, together with specific protein interactions and localized H2O2 pools.
Acknowledgement
Supported by Fundação para a Ciência e a Tecnologia (FCT), Portugal (PEst-OE/QUI/UI0612/2013 and VIH/SAU/0020/2011)
References
- 1.Jones D.P., Sies H. The redox code. Antioxid. Redox Signal. 2015;23:734–746. doi: 10.1089/ars.2015.6247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brandes R.P., Weissmann N., Schröder K. Nox family NADPH oxidases: molecular mechanisms of activation. Free Radic. Biol. Med. 2014;76:208–226. doi: 10.1016/j.freeradbiomed.2014.07.046. [DOI] [PubMed] [Google Scholar]
- 3.Sies H. Hydrogen peroxide as a central redox signaling molecule in physiological oxidative stress: oxidative eustress. Redox Biol. 2017;11:613–619. doi: 10.1016/j.redox.2016.12.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Flohé L. The impact of thiol peroxidases on redox regulation. Free Radic. Res. 2016;50:126–142. doi: 10.3109/10715762.2015.1046858. [DOI] [PubMed] [Google Scholar]
- 5.Chance B., Sies H., Boveris A. Hydroperoxide metabolism in mammalian organs. Physiol. Rev. 1979;59:527–605. doi: 10.1152/physrev.1979.59.3.527. [DOI] [PubMed] [Google Scholar]
- 6.Urban N., Tsitsipatis D., Hausig F., Kreuzer K., Erler K., Stein V., Ristow M., Steinbrenner H., Klotz L.-O. Non-linear impact of glutathione depletion on C. elegans life span and stress resistance. Redox Biol. 2017;11:502–515. doi: 10.1016/j.redox.2016.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.C. Espinosa-Diez, V. Miguel, D. Mennerich, T. Kietzmann, P. Sánchez-Pérez, S. Cadenas, S. Lamas, Antioxidant responses and cellular adjustments to oxidative stress, Redox Biol. 6 183–197. doi:10.1016/j.redox.2015.07.008, 2015. [DOI] [PMC free article] [PubMed]
- 8.Marinho H.S., Real C., Cyrne L., Soares H., Antunes F. Hydrogen peroxide sensing, signaling and regulation of transcription factors. Redox Biol. 2014;2:535–562. doi: 10.1016/j.redox.2014.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brandes N., Schmitt S., Jakob U. Thiol-based redox switches in eukaryotic proteins. Antioxid. Redox Signal. 2009;11:997. doi: 10.1089/ars.2008.2285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brigelius-Flohé R., Flohé L. Basic principles and emerging concepts in the redox control of transcription factors. Antioxid. Redox Signal. 2011;15:2335–2381. doi: 10.1089/ars.2010.3534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Go Y.-M., Jones D.P. The redox proteome. J. Biol. Chem. 2013;288:26512–26520. doi: 10.1074/jbc.R113.464131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lee J.-W., Helmann J.D. The PerR transcription factor senses H2O2 by metal-catalysed histidine oxidation. Nature. 2006;440:363–367. doi: 10.1038/nature04537. [DOI] [PubMed] [Google Scholar]
- 13.Santos C.X., Hafstad A.D., Beretta M., Zhang M., Molenaar C., Kopec J., Fotinou D., Murray T.V., Cobb A.M., Martin D., Silva M.Z., Anilkumar N., Schröder K., Shanahan C.M., Brewer A.C., Brandes R.P., Blanc E., Parsons M., Belousov V., Cammack R., Hider R.C., Steiner R.A., Shah A.M. Targeted redox inhibition of protein phosphatase 1 by Nox4 regulates eIF2α‐mediated stress signaling. EMBO J. 2016;35:319–334. doi: 10.15252/embj.201592394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ferrer-Sueta G., Manta B., Botti H., Radi R., Trujillo M., Denicola A. Factors affecting protein thiol reactivity and specificity in peroxide reduction. Chem. Res. Toxicol. 2011;24:434–450. doi: 10.1021/tx100413v. [DOI] [PubMed] [Google Scholar]
- 15.Winterbourn C.C., Hampton M.B. Thiol chemistry and specificity in redox signaling. Free Radic. Biol. Med. 2008;45:549–561. doi: 10.1016/j.freeradbiomed.2008.05.004. [DOI] [PubMed] [Google Scholar]
- 16.Boveris A., Cadenas E. Cellular sources and steady-state levels of reactive oxygen species. In: Clerch L.B., Massaro D.J., editors. Oxyg. Gene Expr. Cell. Funct. Marcel Dekker; New York: 1997. pp. 1–25. [Google Scholar]
- 17.Buettner G.R., Wagner B.A., Rodgers V.G.J. Quantitative Redox Biology: an approach to understanding the role of reactive species in defining the cellular redox environment. Cell Biochem. Biophys. 2013;67 doi: 10.1007/s12013-011-9320-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pillay C.S., Eagling B.D., Driscoll S.R.E., Rohwer J.M. Quantitative measures for redox signaling. Free Radic. Biol. Med. 2016;96:290–303. doi: 10.1016/j.freeradbiomed.2016.04.199. [DOI] [PubMed] [Google Scholar]
- 19.Bae Y.S., Kang S.W., Seo M.S., Baines I.C., Tekle E., Chock P.B., Rhee S.G. Epidermal Growth Factor (EGF)-induced generation of hydrogen peroxide role in egf receptor-mediated tyrosine phosphorylation. J. Biol. Chem. 1997;272:217–221. [PubMed] [Google Scholar]
- 20.Forman H.J., Maiorino M., Ursini F. Signaling functions of reactive oxygen species. Biochem. (Mosc.). 2010;49:835–842. doi: 10.1021/bi9020378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Oshino N., Chance B., Sies H. The properties of the secundary catalase-peroxide complex (compund II) in the hemoglobin-free perfused rat liver. Arch. Biochem. Biophys. 1973;159:704–711. [Google Scholar]
- 22.Hanschmann E.-M., Godoy J.R., Berndt C., Hudemann C., Lillig C.H. Thioredoxins, glutaredoxins, and peroxiredoxins—molecular mechanisms and health significance: from cofactors to antioxidants to redox signaling. Antioxid. Redox Signal. 2013;19:1539–1605. doi: 10.1089/ars.2012.4599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brito P.M., Antunes F. Estimation of kinetic parameters related to biochemical interactions between hydrogen peroxide and signal transduction proteins. Cell. Biochem. 2014;2:82. doi: 10.3389/fchem.2014.00082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Maxima, Maxima, a Computer Algebra System., n.d. 〈http://maxima.sourceforge.net/〉.
- 25.LaButti J., Chowdhury G., Reilly T.J., Gates K.S. Redox regulation of protein tyrosine phosphatase 1B (PTP1B) by peroxymonophosphate (=O3POOH) J. Am. Chem. Soc. 2007;129:5320. doi: 10.1021/ja070194j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhou H., Singh H., Parsons Z.D., Lewis S.M., Bhattacharya S., Seiner D.R., LaButti J.N., Reilly T.J., Tanner J.J., Gates K.S. The biological buffer bicarbonate/CO 2 potentiates H2O2 -mediated inactivation of protein tyrosine phosphatases. J. Am. Chem. Soc. 2011;133:15803–15805. doi: 10.1021/ja2077137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Trindade D.F., Cerchiaro G., Augusto O. A role for peroxymonocarbonate in the stimulation of biothiol peroxidation by the bicarbonate/carbon dioxide pair. Chem. Res. Toxicol. 2006;19:1475–1482. doi: 10.1021/tx060146x. [DOI] [PubMed] [Google Scholar]
- 28.Zhang Z.-Y. Protein tyrosine phosphatases: structure and function, substrate specificity, and inhibitor development. Annu. Rev. Pharmacol. Toxicol. 2002;42:209–234. doi: 10.1146/annurev.pharmtox.42.083001.144616. [DOI] [PubMed] [Google Scholar]
- 29.Zheng M. Activation of the OxyR transcription factor by reversible disulfide bond formation. Science. 1998;279:1718–1722. doi: 10.1126/science.279.5357.1718. [DOI] [PubMed] [Google Scholar]
- 30.Delaunay A., Isnard A.-D., Toledano M.B. H2O2 sensing through oxidation of the Yap1 transcription factor. EMBO J. 2000;19:5157–5166. doi: 10.1093/emboj/19.19.5157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Brown J.D., Day A.M., Taylor S.R., Tomalin L.E., Morgan B.A., Veal E.A., Peroxiredoxin A. Promotes H2O2 signaling and oxidative stress resistance by oxidizing a thioredoxin family protein. Cell Rep. 2013;5:1425–1435. doi: 10.1016/j.celrep.2013.10.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Calvo I.A., Boronat S., Domènech A., García-Santamarina S., Ayté J., Hidalgo E. Dissection of a redox relay: H2O2-dependent activation of the transcription factor Pap1 through the peroxidatic Tpx1-thioredoxin cycle. Cell Rep. 2013;5:1413–1424. doi: 10.1016/j.celrep.2013.11.027. [DOI] [PubMed] [Google Scholar]
- 33.Dagnell M., Frijhoff J., Pader I., Augsten M., Boivin B., Xu J., Mandal P.K., Tonks N.K., Hellberg C., Conrad M., Arner E.S.J., Ostman A. Selective activation of oxidized PTP1B by the thioredoxin system modulates PDGF-? Receptor tyrosine kinase signaling. Proc. Natl. Acad. Sci. USA. 2013;110:13398–13403. doi: 10.1073/pnas.1302891110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schwertassek U., Haque A., Krishnan N., Greiner R., Weingarten L., Dick T.P., Tonks N.K. Reactivation of oxidized PTP1B and PTEN by thioredoxin 1. FEBS J. 2014;281:3545–3558. doi: 10.1111/febs.12898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cebula M., Schmidt E.E., Arnér E.S.J. TrxR1 as a potent regulator of the Nrf2-Keap1 response system. Antioxid. Redox Signal. 2015;23:823–853. doi: 10.1089/ars.2015.6378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Travasso R.D.M., dos Aidos F.S., Bayani A., Abranches P., Salvador A. Localized redox relays as a privileged mode of cytoplasmic hydrogen peroxide signaling. Redox Biol. 2017;12:223–245. doi: 10.1016/j.redox.2017.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Parsons Z.D., Gates K.S. Thiol-dependent recovery of catalytic activity from oxidized protein tyrosine phosphatases. Biochemistry. 2013;52:6412–6423. doi: 10.1021/bi400451m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Manta B., Hugo M., Ortiz C., Ferrer-Sueta G., Trujillo M., Denicola A. The peroxidase and peroxynitrite reductase activity of human erythrocyte peroxiredoxin 2. Arch. Biochem. Biophys. 2009;484:146–154. doi: 10.1016/j.abb.2008.11.017. [DOI] [PubMed] [Google Scholar]
- 39.Hanschmann E.-M., Lönn M.E., Schütte L.D., Funke M., Godoy J.R., Eitner S., Hudemann C., Lillig C.H. Both thioredoxin 2 and glutaredoxin 2 contribute to the reduction of the mitochondrial 2-Cys peroxiredoxin Prx3. J. Biol. Chem. 2010;285:40699–40705. doi: 10.1074/jbc.M110.185827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Seo M.S., Kang S.W., Kim K., Baines I.C., Lee T.H., Rhee S.G. Identification of a new type of mammalian peroxiredoxin that forms an intramolecular disulfide as a reaction intermediate. J. Biol. Chem. 2000;275:20346–20354. doi: 10.1074/jbc.M001943200. [DOI] [PubMed] [Google Scholar]
- 41.Karplus P.A. A primer on peroxiredoxin biochemistry. Free Radic. Biol. Med. 2015;80:183–190. doi: 10.1016/j.freeradbiomed.2014.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rhee S.G., Woo H.A., Kil I.S., Bae S.H. Peroxiredoxin functions as a peroxidase and a regulator and sensor of local peroxides. J. Biol. Chem. 2012;287:4403–4410. doi: 10.1074/jbc.R111.283432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Peskin A.V., Dickerhof N., Poynton R.A., Paton L.N., Pace P.E., Hampton M.B., Winterbourn C.C. Hyperoxidation of peroxiredoxins 2 and 3: rate constants for the reactions of the sulfenic acid of the peroxidatic cysteine. J. Biol. Chem. 2013;288:14170–14177. doi: 10.1074/jbc.M113.460881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Brenner N., Friedlander T. Adaptive response and enlargement of dynamic range. Math. Biosci. Eng. 2011;8:515–528. doi: 10.3934/mbe.2011.8.515. [DOI] [PubMed] [Google Scholar]
- 45.Antunes F., Cadenas E., Brunk U.T. Apoptosis induced by exposure to a low steady-state concentration of H2O2 is a consequence of lysosomal rupture. Biochem. J. 2001;356:549–555. doi: 10.1042/0264-6021:3560549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Davies J.M., Lowry C.V., Davies K.J. Transient adaptation to oxidative stress in yeast. Arch. Biochem. Biophys. 1995;317:1–6. doi: 10.1006/abbi.1995.1128. [DOI] [PubMed] [Google Scholar]
- 47.Wiese A.G., Pacifici R.E., Davies K.J. Transient adaptation of oxidative stress in mammalian cells. Arch. Biochem. Biophys. 1995;318:231–240. doi: 10.1006/abbi.1995.1225. [DOI] [PubMed] [Google Scholar]
- 48.Antunes F., Cadenas E. Estimation of H2O2 gradients across biomembranes. FEBS Lett. 2000;475:121–126. doi: 10.1016/s0014-5793(00)01638-0. [DOI] [PubMed] [Google Scholar]
- 49.Oliveira-Marques V., Cyrne L., Marinho H.S., Antunes F. A quantitative study of NF-{kappa}B activation by H2O2: relevance in inflammation and synergy with TNF-{alpha} J. Immunol. 2007;178:3893–3902. doi: 10.4049/jimmunol.178.6.3893. [DOI] [PubMed] [Google Scholar]
- 50.Oliveira-Marques V., Silva T., Cunha F., Covas G., Marinho H.S., Antunes F., Cyrne L. A quantitative study of the cell-type specific modulation of c-Rel by hydrogen peroxide and TNF-α. Redox Biol. 2013;1:347–352. doi: 10.1016/j.redox.2013.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Adimora N.J., Jones D.P., Kemp M.L. A model of redox kinetics implicates the thiol proteome in cellular hydrogen peroxide responses. Antioxid. Redox Signal. 2010;13:731–743. doi: 10.1089/ars.2009.2968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Huang B.K., Sikes H.D. Quantifying intracellular hydrogen peroxide perturbations in terms of concentration. Redox Biol. 2014;2:955–962. doi: 10.1016/j.redox.2014.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sobotta M.C., Liou W., Stöcker S., Talwar D., Oehler M., Ruppert T., Scharf A.N.D., Dick T.P. Peroxiredoxin-2 and STAT3 form a redox relay for H2O2 signaling. Nat. Chem. Biol. 2015;11:64–70. doi: 10.1038/nchembio.1695. [DOI] [PubMed] [Google Scholar]
- 54.Lee S.R., Kwon K.S., Kim S.R., Rhee S.G. Reversible inactivation of protein-tyrosine phosphatase 1B in A431 cells stimulated with epidermal growth factor. J. Biol. Chem. 1998;273:15366–15372. doi: 10.1074/jbc.273.25.15366. [DOI] [PubMed] [Google Scholar]
- 55.Meng T.-C., Fukada T., Tonks N.K. Reversible oxidation and inactivation of protein tyrosine phosphatases in vivo. Mol. Cell. 2002;9:387–399. doi: 10.1016/s1097-2765(02)00445-8. [DOI] [PubMed] [Google Scholar]
- 56.Heppner D.E., Janssen-Heininger Y.M.W., van der Vliet A. The role of sulfenic acids in cellular redox signaling: Reconciling chemical kinetics and molecular detection strategies. Arch. Biochem. Biophys. 2017;616:40–46. doi: 10.1016/j.abb.2017.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rinna A., Torres M., Forman H.J. Stimulation of the alveolar macrophage respiratory burst by ADP causes selective glutathionylation of protein tyrosine phosphatase 1B. Free Radic. Biol. Med. 2006;41:86–91. doi: 10.1016/j.freeradbiomed.2006.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sobotta M.C., Barata A.G., Schmidt U., Mueller S., Millonig G., Dick T.P. Exposing cells to H2O2: a quantitative comparison between continuous low-dose and one-time high-dose treatments. Free Radic. Biol. Med. 2013;60:325–335. doi: 10.1016/j.freeradbiomed.2013.02.017. [DOI] [PubMed] [Google Scholar]
- 59.Fomenko D.E., Koc A., Agisheva N., Jacobsen M., Kaya A., Malinouski M., Rutherford J.C., Siu K.-L., Jin D.-Y., Winge D.R., Gladyshev V.N. Thiol peroxidases mediate specific genome-wide regulation of gene expression in response to hydrogen peroxide. Proc. Natl. Acad. Sci. USA. 2011;108:2729–2734. doi: 10.1073/pnas.1010721108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Netto L.E.S., Antunes F. The roles of peroxiredoxin and thioredoxin in hydrogen peroxide sensing and in signal transduction. Mol. Cells. 2016;39:65–71. doi: 10.14348/molcells.2016.2349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Delaunay A., Pflieger D., Barrault M.B., Vinh J., Toledano M.B. A thiol peroxidase is an H2O2 receptor and redox-transducer in gene activation. Cell. 2002;111:471–481. doi: 10.1016/s0092-8674(02)01048-6. [DOI] [PubMed] [Google Scholar]
- 62.Vivancos A.P., Castillo E.A., Biteau B., Nicot C., Ayté J., Toledano M.B., Hidalgo E. A cysteine-sulfinic acid in peroxiredoxin regulates H2O2-sensing by the antioxidant Pap1 pathway. Proc. Natl. Acad. Sci. USA. 2005;102:8875–8880. doi: 10.1073/pnas.0503251102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Veal E.A., Findlay V.J., Day A.M., Bozonet S.M., Evans J.M., Quinn J., Morgan B.A. A 2-Cys peroxiredoxin regulates peroxide-induced oxidation and activation of a stress-activated MAP kinase. Mol. Cell. 2004;15:129–139. doi: 10.1016/j.molcel.2004.06.021. [DOI] [PubMed] [Google Scholar]
- 64.Jarvis R.M., Hughes S.M., Ledgerwood E.C. Peroxiredoxin 1 functions as a signal peroxidase to receive, transduce, and transmit peroxide signals in mammalian cells. Free Radic. Biol. Med. 2012;53:1522–1530. doi: 10.1016/j.freeradbiomed.2012.08.001. [DOI] [PubMed] [Google Scholar]
- 65.Netto L.E.S., de Oliveira M.A., Tairum C.A., da Silva Neto J.F. Conferring specificity in redox pathways by enzymatic thiol/disulfide exchange reactions. Free Radic. Res. 2016;50:206–245. doi: 10.3109/10715762.2015.1120864. [DOI] [PubMed] [Google Scholar]
- 66.Winterbourn C.C. Are free radicals involved in thiol-based redox signaling? Free Radic. Biol. Med. 2015;80:164–170. doi: 10.1016/j.freeradbiomed.2014.08.017. [DOI] [PubMed] [Google Scholar]