Abstract
AMP-activated protein kinase (AMPK) and sterol regulatory element binding protein (SREBP)-1c are major therapeutic targets in the treatment of metabolic diseases. In the present study, the fat-reducing mechanisms of berberine (BBR), a natural isoquinoline, was investigated by examining the AMPK-mediated modulation of SREBP-1c in 3T3-L1 adipocytes. BBR activated AMPK in a dose- and time-dependent manner, and increased the phosphorylation of the 125-kDa precursor form of SREBP-1c, which suppressed its proteolytic processing into the mature 68-kDa form and its subsequent nuclear translocation. The binding of nuclear SREBP-1c to its E-box motif-containing target DNA sequence was decreased following treatment with BBR, which led to a decrease in the expression of lipogenic genes and subsequently reduced intracellular fat accumulation. Transfection with AMPKα1 siRNA, and not control siRNA, inhibited BBR-induced phosphorylation of the 125-kDa SREBP-1c, which confirmed that AMPK was responsible for phosphorylating SREBP-1c. AMPKα1 siRNA transfection rescued the proteolytic processing, nuclear translocation and target DNA binding of SREBP-1c that had been suppressed by BBR. In addition, BBR-induced suppression of lipogenic gene expression and intracellular fat accumulation were rescued by AMPKα1 siRNA transfection. In conclusion, the results of the present study demonstrate that BBR activates AMPK to induce phosphorylation of SREBP-1c, thereby suppressing proteolytic processing, nuclear translocation and target DNA binding of SREBP-1c, which leads to a reduction in lipogenic gene expression and intracellular fat accumulation. The results of the present study indicate that BBR may be a potential candidate for the development of drugs to treat obesity.
Keywords: AMP-activated protein kinases, sterol regulatory element binding protein 1, berberine, adipogenesis, 3T3-L1
Introduction
AMP-activated protein kinase (AMPK), a sensor of cellular energy status, is one of the major therapeutic targets in the treatment of metabolic diseases. AMPK is composed of a catalytic α subunit and regulatory β and γ subunits (1). The AMPK α subunit consists of two isoforms, α1 and α2; of which α1 is the predominant isoform expressed in adipocytes (2). Phosphorylation of Thr172 in the α subunit activates AMPK >100-fold, and 5′-adenosine monophosphate maintains AMPK in an activated state by enhancing phosphorylation of the α subunit, as well as by allosteric activation (3).
Berberine (BBR) is a natural isoquinoline alkaloid, which has been reported to exert therapeutic effects in a number of metabolic diseases, such as diabetes and hypercholesterolemia (4). The beneficial effects of BBR are attributable to its effect as an AMPK activator. BBR has been demonstrated to activate AMPK in adipocytes and muscle cells, as well as in animal models of diabetes and obesity, thus resulting in reduced fat accumulation and improved insulin sensitivity (5). In addition, BBR has been reported to inhibit mitochondrial respiratory chain complex I, thereby activating AMPK (6). However, its downstream mechanisms have not yet been fully elucidated.
The sterol regulatory element-binding protein (SREBP) is the major transcription factor involved in regulating lipogenic gene expression (7). Previous studies have suggested that BBR reduces fat accumulation in white adipose tissue and adipocytes via downregulation of SREBPs (8,9). In the present study, the detailed molecular mechanisms of the action of BBR were elucidated, whereby a signaling pathway involving BBR-mediated AMPK activation and SREBP-1c downregulation in 3T3-L1 adipocytes was identified.
Materials and methods
Chemicals and reagents
Cell culture reagents, including Dulbecco's modified Eagle's medium (DMEM), fetal bovine serum, calf serum and antibiotics were obtained from Thermo Fisher Scientific, Inc. (Waltham, MA, USA). AMPKα1 small interfering (si)RNA and control siRNA were purchased from Santa Cruz Biotechnology, Inc. (Dallas, TX, USA). Lipofectamine RNAiMAX transfection reagent was purchased from Invitrogen; Thermo Fisher Scientific, Inc. BBR, insulin, dexamethasone and 3-isobutyl-1-methylxanthine were obtained from Sigma-Aldrich; Merck KGaA (Darmstadt, Germany).
Cell culture and differentiation
3T3-L1 pre-adipocytes were purchased from the American Type Culture Collection (Manassas, VA, USA) and seeded in 12-well plates at a density of 1.5×105 cells/well with DMEM containing 10% calf serum at 37°C in 5% CO2 atmosphere with relative humidity of 85–95%. When the cells were 100% confluent following 3 days of culture (taken as day 0), the cells were induced to differentiate by incubation in a differentiation induction medium consisting of DMEM, 10% fetal bovine serum, 1 µg/ml insulin, 0.25 µM dexamethasone and 0.5 mM 3-isobutyl-1-methylxanthine for 2 days. The culture medium was then replaced with differentiation maintenance medium, consisting of DMEM, 1 µg/ml insulin and 10% fetal bovine serum on days 2 and 4, and the cells were harvested on day 7. To examine the effects of BBR on cell differentiation, BBR was dissolved in dimethylsulfoxide and diluted >1,000-fold in differentiation maintenance medium. In the present study, BBR was administered during the late phase of adipogenic differentiation (days 2–7) in order to analyze the effects of BBR on fat accumulation and avoid its effects on mitotic clonal expansion, as BBR was previously reported to inhibit the mitotic clonal expansion of 3T3-L1 cells (10). In dose-response experiments, cells were treated with BBR (0, 5, 10, 15 and 20 µM) at day 2 and day 4, when media were replaced, and harvested at day 7. In time-response experiments, cells were treated with 0 or 20 µM BBR at day 2 and day 4, when media were replaced, and harvested at days 3, 5 and 7. Cells harvested at day 3 were treated with 20 µM BBR at day 2 only. Control cells were treated with an equal volume of dimethylsulfoxide diluted in differentiation maintenance medium. Intracellular fat accumulation in 1.2×106 cells was measured by 0.3% Oil red O staining solution at room temperature for 30 min followed by light microscopy with a magnification of ×400. Oil red O dye in intracellular fat droplets was extracted by adding 700 µl 60% isopropanol per well and shaking at 100 rpm for 2 h at room temperature. The extracted dye was measured by optical density at 510 nm using the methods described previously (11). Cell viability was determined according to the manufacturer's protocol using a CellCountEZ™ Cell Survival assay kit (Rockland Immunochemicals, Inc., Pottstown, PA, USA), which is based on the ability of viable mammalian cells to convert hydroxyethyl disulfide into mercaptoethanol (12). Cell number was determined using a hemocytometer following the harvesting of cells by trypsinization.
Reverse transcription-quantitative polymerase chain reaction (RT-qPCR) analysis
RT-qPCR experiments were performed as described previously (13). Cells (1.2×106) were treated with BBR and harvested as described above. Total RNA was extracted using the RNeasy Mini kit (Qiagen GmbH, Hilden, Germany), according to the manufacturer's protocol. RNA (1 µg) was reverse-transcribed at 37°C using the High-Capacity cDNA Reverse Transcription kit (Applied Biosystems; Thermo Fisher Scientific, Inc.), according to the manufacturer's protocol. qPCR was performed using a StepOne Real-Time PCR System (Applied Biosystems; Thermo Fisher Scientific, Inc.), in triplicate, in a final volume of 20 µl, which consisted of 10 µl TaqMan Gene Expression Master Mix (Applied Biosystems; Thermo Fisher Scientific, Inc.) containing Taq polymerase and dNTPs, 1 µl TaqMan Gene Expression assay (Applied Biosystems; Thermo Fisher Scientific, Inc.) containing 20X TaqMan probe and primers, cDNA (1/10th that produced by RT) and distilled water. The reaction mixtures were preheated at 95°C for 10 min to activate the enzyme, followed by 40 cycles of melting at 95°C for 15 sec and annealing/extension at 60°C for 1 min. TaqMan Gene Expression assays containing TagMan probe and primers were used to evaluate the mRNA levels of the following genes: peroxisome proliferator-activated receptor-γ (PPARγ; assay ID, Mm00440945_m1), CCAAT-enhancer-binding protein α (C/EBPα; assay ID, Mm01265914_s1), SREBP-1c (assay ID, Mm00550338_m1), lipoprotein lipase (LPL; assay ID, Mm00434764_m1), fatty acid synthase (FAS; assay ID, Mm01253292_m1) and acetyl-CoA carboxylase-1 (ACC-1; assay ID, Mm01304257_m1) and 18S ribosomal (r) RNA (assay ID, Hs99999901_s1). The 18S rRNA was used as an internal control, as previously described (11). For each sample, target gene mRNA levels were normalized against the level of 18S rRNA, and the ratio of normalized mRNA in each sample to that of the control sample was determined using the comparative Cq method (14).
Western blotting
Western blotting was performed using the methods described previously (13). Cells (1.2×106) were treated with BBR and harvested as described above. Cells were lysed for 1 h in ice-cold radioimmunoprecipitation assay buffer containing 25 mM Tris-HCl (pH 7.6) 150 mM NaCl, 1% Nonidet P-40, 1% sodium deoxycholate, 0.1% SDS and a protease inhibitor cocktail (Sigma-Aldrich; Merck KGaA). The cell lysates were centrifuged at 19,000 × g for 20 min at 4°C to remove insoluble materials. The protein concentrations were determined using a BCA protein assay kit (Pierce; Thermo Fisher Scientific, Inc.). Protein extracts (50 µg) were resolved by 10% SDS-PAGE and electro-transferred to nitrocellulose membranes at 150 mA for 1 h. The membranes were then blocked for 2 h at room temperature with PBS containing 5% skim milk and 0.1% Tween-20 and, after washing for 10 min three times with PBS containing 0.1% Tween 20, were incubated with primary antibodies (diluted 1:1,000 in the blocking solution) overnight at 4°C and subsequently with horseradish peroxidase-conjugated anti-rabbit or anti-mouse secondary antibodies (diluted 1:1,000 in the blocking solution) for 1 h at room temperature. Protein bands were visualized using Pierce ECL Western Blotting Substrate, according to the manufacturer's protocol (Thermo Fisher Scientific, Inc.). An anti-β-Actin antibody (diluted 1:1,000 in the blocking solution) was used as an endogenous control to confirm that equal amounts of proteins were loaded. Anti-phosphorylated (p)-AMPKα (cat. no. 2535), anti-AMPKα (cat. no. 2603), anti-p-SREBP-1c (cat. no. 9874) and anti-PPARγ (cat. no. 2430) primary antibodies, as well as anti-mouse IgG (cat. no. 7076S) and anti-rabbit IgG (cat. no. 7074) secondary antibodies were purchased from Cell Signaling Technology, Inc. (Danvers, MA, USA). The anti-SREBP-1c (cat. no. 557036) antibody was purchased from BD Biosciences (Franklin Lakes, NJ, USA). Anti-C/EBPα (cat. no. sc-61), anti-TATA box-binding protein (TBP; cat. no. sc-204) and anti-β-actin (cat. no. sc-47778) primary antibodies were purchased from Santa Cruz Biotechnology, Inc.
Analysis of nuclear SREBP-1c levels
Cells (1.2×106) were treated with BBR and harvested as described above. Cells were harvested using cell scrapers, and nuclear lysates were prepared using a Nuclear Extract kit (Active Motif, Inc. Carlsbad, CA, USA) according to the manufacturer's protocol. The protein concentration of nuclear lysates was determined using a BCA protein assay kit. To determine nuclear SREBP-1c protein expression levels, 10 µg nuclear protein was separated by 10% SDS-PAGE, followed by western blotting using anti-SREBP-1c antibody followed by secondary antibody. The procedures and antibodies used for western blotting were identical to those described above.
Determination of SREBP-1c binding to target DNA
Binding of nuclear SREBP-1c to the target DNA sequence, 5′-TCACCTGA-3′, which contains an E-box motif, was measured using a SREBP-1 Transcription Factor ELISA kit (cat. no. 10010854; Cayman Chemical Company, Ann Arbor, MI, USA). Briefly, 10 µg protein from the nuclear lysates was added to wells of a 96-well plate that were provided as part of the ELISA kit. The wells included in the kit were already coated with the 5′-TCACCTGA-3′ oligonucleotide. Following incubation for 1 h at room temperature, unbound nuclear proteins were removed by washing five times with a wash buffer provided in the kit. Each well was incubated with 100 µl anti-SREBP-1 (1:100) primary antibody for 1 h at room temperature followed by 100 µl horseradish peroxidase-conjugated secondary antibody (1:100) for 1 h at room temperature. Optical density at 450 nm was measured following the addition of 100 µl developing solution followed by 100 µl stop solution provided in the kit.
AMPK activity assay
The kinase activity of AMPK was measured by ELISA using an AMPK assay kit (cat. no. CY-1182; Cyclex Inc., Columbia, MD, USA). Cells (1.2×106) were lysed in 0.2 ml lysis buffer containing 20 mM Tris HCl (pH 7.5) 250 mM NaCl, 10% glycerol, 0.5% Nonidet P-40, 0.2 mM PMSF, 1 µg/ml pepstatin, 0.5 µg/ml leupeptin, 5 mM NaF, 2 mM Na3VO4, 2 mM β-glycerophosphate and 1 mM dithiothreitol for 90 min at 4°C, followed by centrifugation at 22,000 × g for 10 min at 4°C. Cell lysates (10 µl) and 90 µl kinase reaction buffer with 50 µM ATP (included in the kit) were added to wells of a 96-well plate coated with peptides consisting of the sequence surrounding Ser789 of insulin receptor substrate (IRS)-1, which is efficiently phosphorylated by AMPK, and incubated at 30°C for 30 min. The quantity of phosphorylated substrate was measured by incubating with 100 µl anti-p-IRS-1 antibody at room temperature for 30 min followed by incubation a with 100 µl horseradish peroxidase-conjugated secondary antibody at room temperature for 30 min. The primary and secondary antibodies included in the ELISA kit were prediluted. Subsequently, 100 µl substrate reagent was added at room temperature for 10 min followed by 100 µl stop solution included in the kit. Optical density was measured at 450 nm.
Transfection of siRNA
At one day prior to reaching confluence (taken as day-1), 3T3-L1 cells were incubated in serum-free medium for 1 h and then transfected with 12.5, 25, 50, or 75 nM AMPKα1 siRNA, or 50 or 75 nM control siRNA using Lipofectamine RNAiMAX transfection reagent according to the manufacturer's protocol. The following day (on day 0), the transfected cells were induced to differentiate by replacing the medium with differentiation-induction medium. Following 7 days of differentiation into adipocytes, cells were harvested for Oil red O staining, western blotting, determination of SREBP-1c binding to target DNA or RT-qPCR analysis using the aforementioned methods.
Statistical analysis
Data are presented as the mean ± standard deviation of four replicate experiments. Differences among multiple groups were determined using one-way analysis of variance followed by post hoc analysis using Duncan's multiple range test, and comparisons between two groups were analyzed using an unpaired Student's t-test. Statistical tests were performed using SPSS software (version, 14.0; SPSS, Inc., Chicago, IL, USA).
Results
Dose-dependent effect of BBR on AMPK, SREBP-1c and lipogenesis-associated gene expression
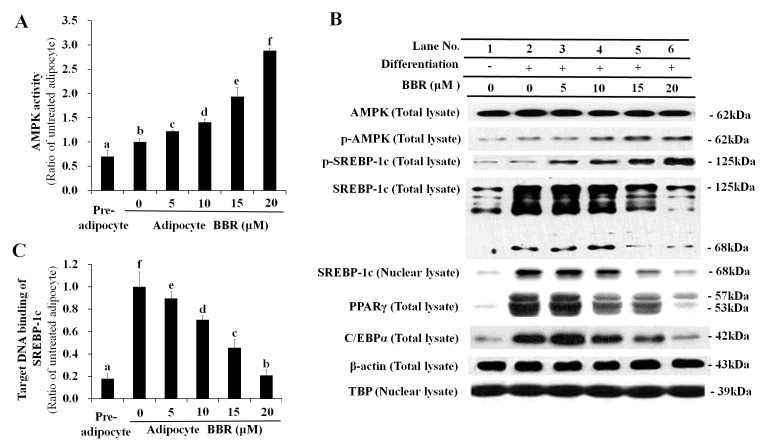
SREBP-1c is synthesized as a 125-kDa precursor, which is processed by proteolytic cleavage into a mature 68-kDa form that translocates into the nucleus. Phosphorylation of the SREBP-1c precursor suppresses its proteolytic maturation (15). In the present study BBR activated AMPK in a dose-dependent manner (Fig. 1A), which was associated with elevation of p-AMPK and p-SREBP-1c levels (Fig. 1B). The increase in p-SREBP-1c levels was accompanied by reduced proteolytic processing of the 125-kDa SREBP-1c precursor into the 68-kDa mature form (Fig. 1B). Western blotting of total SREBP-1c revealed gradual proteolytic degradation of the 125-kDa precursor form into the 68-kDa mature form (Fig. 1B). The level of the mature 68-kDa SREBP-1c isoform was decreased in the total cell lysate and nuclear lysate in a dose-dependent manner following treatment with BBR, as proteolytic maturation is required for the nuclear translocation of SREBP-1c (Fig. 1B). Similarly, the binding of nuclear SREBP-1c to its E-box motif-containing target DNA sequence was reduced in a dose-dependent manner following BBR treatment (Fig. 1C).
Figure 1.
Dose-dependent effects of BBR on AMPK, SREBP-1c and lipogenesis-associated protein expression levels. 3T3-L1 adipocytes were treated with 0, 5, 10, 15 or 20 µM BBR from days 2–7 of differentiation, and cells were harvested on day 7 for analysis. (A) AMPK activity as determined by its capability to phosphorylate Ser789 on its substrate, IRS-1. This was measured in cell lysates by ELISA. (B) The level of p-AMPK and p-SREBP-1c, as well as the 125-kDa precursor form and mature 68-kDa form, were measured in total cell lysates by western blotting. Levels of the 68-kDa mature SREBP-1c form were analyzed in nuclear lysates to determine its nuclear translocation. Levels of the major adipogenic transcription factors, PPARγ and C/EBPα, were analyzed in total cell lysates. β-actin and TBP were used as endogenous controls for protein expression in total cell lysates and nuclear lysates, respectively. (C) The binding of nuclear SREBP-1c to its E-box motif-containing target DNA sequence, 5′-TCACCTGA-3′, as determined by ELISA. Different letters indicate significant differences between groups (P<0.05). BBR, berberine; AMPK, AMP-activated protein kinase; SREBP-1c, sterol regulatory element binding protein-1c; p-AMPK, phosphorylated AMPK; IRS-1, insulin receptor substrate-1; p-SREBP-1c, phosphorylated SREBP-1c; PPARγ, peroxisome proliferator-activated receptor-γ; C/EBPα, CCAAT/enhancer-binding protein α; TBP, TATA box-binding protein.
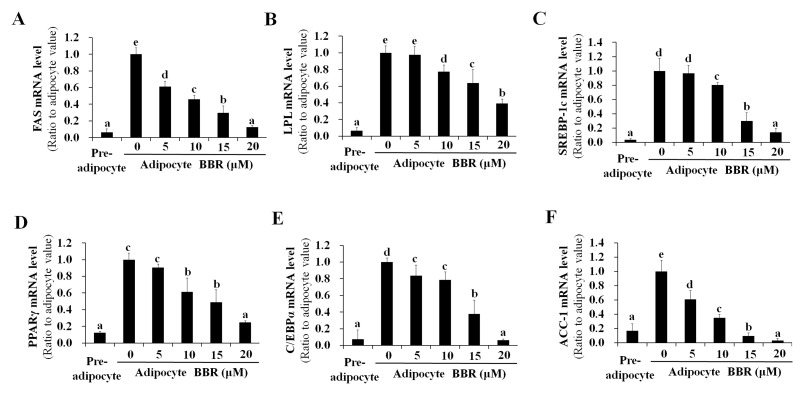
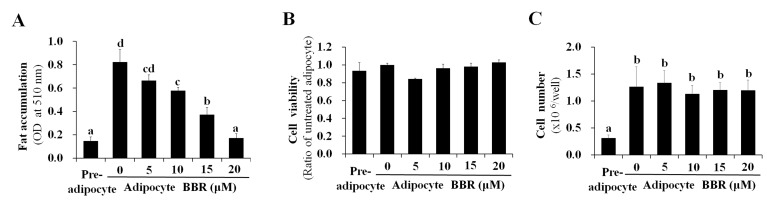
SREBP-1c is a transcription factor that binds to the promoter region of numerous lipogenesis-associated genes (16–21). As a result of decreased binding of nuclear SREBP-1c to target DNA, the mRNA expression of lipogenic genes, including FAS, LPL, SREBP-1c, PPARγ, C/EBPα and ACC-1 were reduced by BBR treatment in a dose-dependent manner (Fig. 2). This is consistent with the dose-dependent reduction in PPARγ and C/EBPα protein expression levels demonstrated in Fig. 1B. Intracellular fat accumulation, which is mediated by the collective actions of lipogenic genes, was reduced by BBR treatment in a dose-dependent manner (Fig. 3A). BBR exhibited no significant effect on cell viability, which was maintained at >90% of untreated adipocytes (Fig. 3B). This indicates that the BBR-induced suppression of nuclear SREBP-1c binding to its target DNA, as well as the reduction in lipogenic gene expression and intracellular fat accumulation, were not caused by increased cytotoxicity. Previously, BBR was reported to inhibit the mitotic clonal expansion of 3T3-L1 cells when treated during the early phase of differentiation (10) and the present study aimed to elucidate the molecular mechanism of BBR for reducing fat accumulation without its effects on mitotic clonal expansion. In the present study, BBR was administered during the late phase of adipogenic differentiation (from days 2–7) in order to avoid its effects on mitotic clonal expansion, which occurs during the early phase of differentiation. As demonstrated in Fig. 3C, increasing concentrations of BBR demonstrated no significant effects on the number of cells, demonstrating that mitotic clonal expansion was not affected in this experimental condition (Fig. 3C).
Figure 2.
Dose-dependent effects of BBR on the expression of lipogenesis-associated genes. 3T3-L1 adipocytes were treated with 0, 5, 10, 15 or 20 µM BBR from days 2–7 of differentiation, and cells were harvested on day 7 for analysis. The mRNA levels of (A) FAS, (B) LPL, (C) SREBP-1c, (D) PPARγ, (E) C/EBPα and (F) ACC-1 were measured by reverse transcription-quantitative polymerase chain reaction. Different letters indicate significant differences between groups (P<0.05). BBR, berberine; FAS, fatty acid synthase; LPL, lipoprotein lipase; SREBP1-c, sterol regulatory element binding protein-1c; PPARγ, peroxisome proliferator-activated receptor-γ; C/EBPα, CCAAT/enhancer-binding protein α; ACC-1, acetyl-CoA carboxylase-1.
Figure 3.
Dose-dependent effects of BBR on intracellular fat accumulation, cell viability and cell number. 3T3-L1 adipocytes were treated with 0, 5, 10, 15 or 20 µM BBR from days 2–7 of differentiation, and cells were harvested on day 7 for analysis. (A) Oil red O stain in intracellular fat droplets was extracted using isopropyl alcohol and quantified by measuring the OD at 510 nm. (B) Analysis of the viability of 3T3-L1 adipocytes following treatment with increasing concentrations of BBR. (C) The cell number was determined using a hemocytometer following trypsinization of the cells. Different letters indicate significant differences between groups (P<0.05). BBR, berberine; OD, optical density.
Effect of BBR on AMPK, SREBP-1c and the expression of lipogenesis-associated genes over time
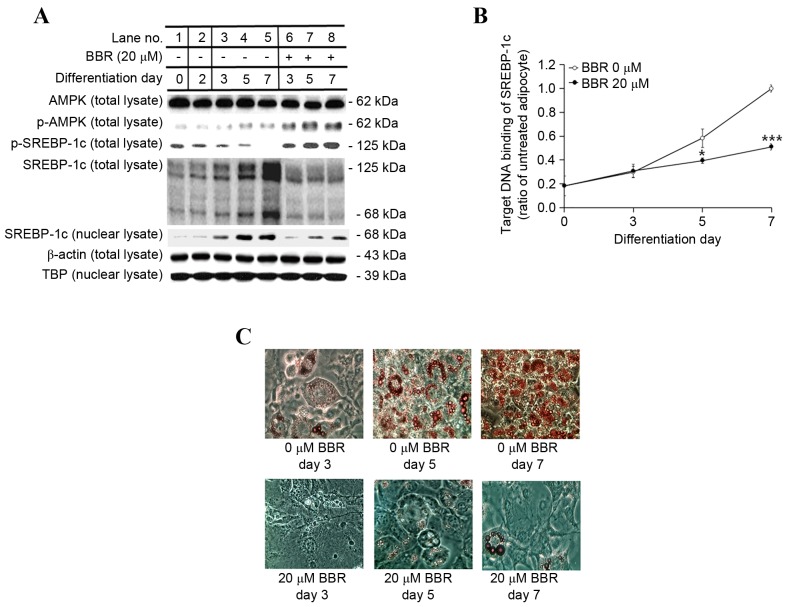
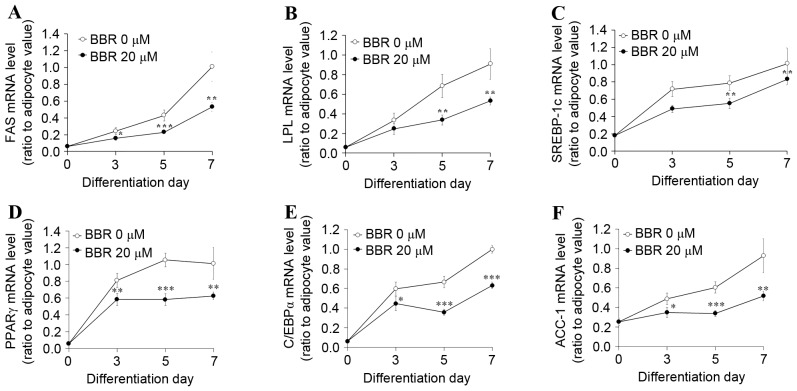
In order to examine the effect of BBR exposure over time, 3T3-L1 adipocytes were treated with 0 or 20 µM BBR from days 2–7, and were analyzed on days 3, 5 and 7 of adipogenic differentiation. Under these conditions, the protein expression levels of p-AMPK and p-SREBP-1c were increased in BBR-treated cells relative to the untreated cells (Fig. 4A). The protein expression levels of the 125-kDa SREBP-1c precursor and its processed 68-kDa mature form were reduced in BBR-treated cells when compared with in untreated cells (Fig. 4A). In addition, the nuclear levels of mature 68-kDa SREBP-1c were reduced in BBR-treated cells when compared with untreated cells, as proteolysis is required for nuclear translocation (Fig. 4A). The binding of nuclear SREBP-1c to its target DNA sequence was elevated in a time-dependent manner in untreated cells, which was significantly suppressed in BBR-treated cells, in a similar manner to nuclear SREBP-1c levels (Fig. 4B). Intracellular fat accumulation was reduced in BBR-treated cells when compared with the untreated cells (Fig. 4C). Consistent with these observations, the mRNA levels of lipogenesis-associated genes, including SREBP-1c, were significantly suppressed in BBR-treated cells when compared with the untreated cells (Fig. 5).
Figure 4.
Effect of BBR on AMPK and SREBP-1c protein expression and intracellular fat accumulation over time. 3T3-L1 adipocytes were treated with 0 or 20 µM BBR commencing at day 2, and cells were harvested at days 3, 5 and 7 of differentiation for time course analyses. (A) The protein levels of p-AMPK and p-SREBP-1c, as well as the 125-kDa precursor form of SREBP-1c and the mature 68-kDa form were analyzed in total cell lysates by western blotting. The level of the mature 68-kDa SREBP-1c isoform was analyzed in nuclear lysates. β-actin and TBP were used as endogenous controls for the analysis of protein expression in total cell lysates and nuclear lysates, respectively. (B) The binding of nuclear SREBP-1c to its E-box motif-containing target DNA sequence, 5′-TCACCTGA-3′, was measured by ELISA. (C) Intracellular fat droplets were stained with Oil red O dye and examined by light microscopy. Magnification, ×400. *P<0.05 and ***P<0.001 vs. untreated adipocytes at the same day of differentiation. BBR, berberine; AMPK, AMP-activated protein kinase; SREBP-1c, sterol regulatory element binding protein-1c; p-AMPK, phosphorylated AMPK; p-SREBP-1c, phosphorylated-SREBP-1c; TBP, TATA box-binding protein.
Figure 5.
Effect of BBR on the expression of lipogenesis-associated genes over time. 3T3-L1 adipocytes were treated with 0 or 20 µM BBR commencing at day 2 until cells were harvested at days 3, 5 or 7 of differentiation. The mRNA levels of (A) FAS, (B) LPL, (C) SREBP-1c, (D) PPARγ, (E) C/EBPα and (F) ACC-1 were measured by reverse transcription-quantitative polymerase chain reaction. *P<0.05, **P<0.01 and ***P<0.001 vs. untreated adipocytes at same day of differentiation. BBR, berberine; FAS, fatty acid synthase; LPL, lipoprotein lipase; SREBP1-c, sterol regulatory element binding protein-1c; PPARγ, peroxisome proliferator-activated receptor-γ; C/EBPα, CCAAT/enhancer-binding protein α; ACC-1, acetyl-CoA carboxylase-1.
Effect of AMPK knockdown on SREBP-1c processing, lipogenesis-associated gene expression and intracellular fat accumulation
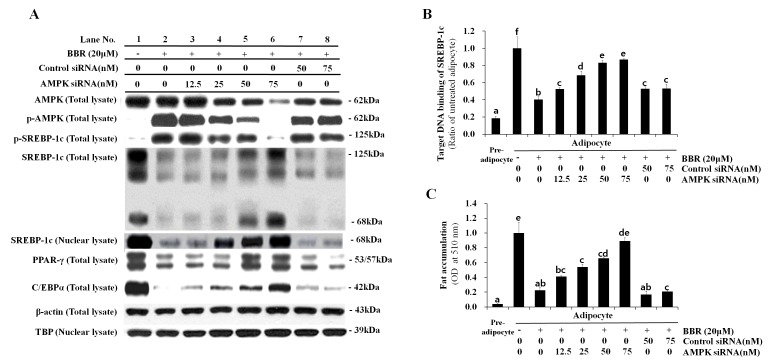
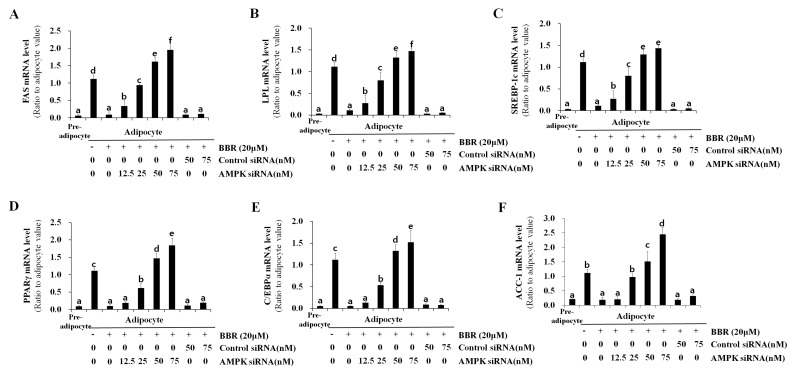
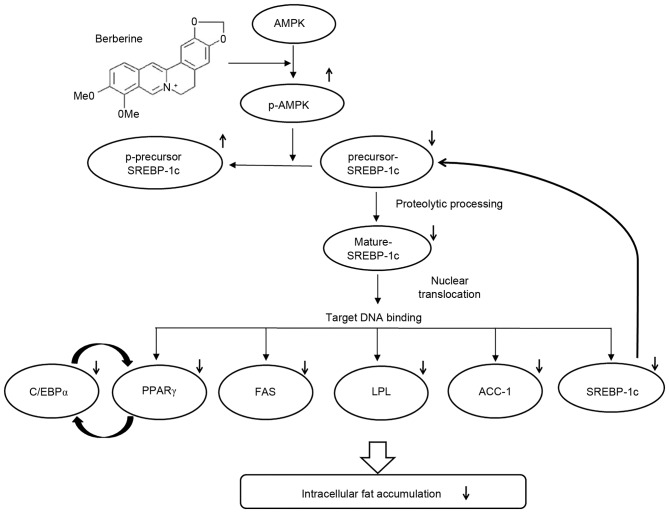
In order to confirm the essential role of AMPK in BBR-induced SREBP-1c phosphorylation and processing, 3T3-L1 cells were transfected with different concentrations of control siRNA or AMPKα1 siRNA. siRNA sequences targeting the α1 isoform of AMPK, rather than α2, were employed for knockdown experiments in the present study, as α1 is the predominant isoform expressed in adipocytes (2). Levels of p-AMPK and p-SREBP-1c were elevated in adipocytes treated with BBR alone when compared with untreated adipocytes (Fig. 6A). Levels of p-SREBP-1c were reduced in a dose-dependent manner following transfection with increasing concentrations of AMPKα1 siRNA, whereas, this effect was not observed following transfection with control siRNA (Fig. 6A). This confirmed that SREBP-1c is a substrate of AMPK. Conversely, the level of total SREBP-1c, including the 125-kDa precursor form and the mature 68-kDa form, were reduced in BBR-treated cells when compared with in untreated cells (Fig. 6A). These levels increased in a dose-dependent manner in AMPKα1 siRNA-transfected cells, however, not in control siRNA-transfected cells (Fig. 6A). The nuclear expression levels of the 68-kDa SREBP-1c form demonstrated a similar pattern to that observed in the total cell lysate. The binding of nuclear SREBP-1c to its target DNA sequence, which was suppressed by BBR, was rescued by AMPKα1 siRNA transfection in a dose-dependent manner (Fig. 6B). This effect was not observed following transfection with control siRNA. Intracellular fat accumulation, which was suppressed by BBR exposure, was rescued by AMPKα1 siRNA transfection, whereas transfection with control siRNA demonstrated no effect (Fig. 6C). In addition, the mRNA levels of lipogenesis-associated genes, including FAS, LPL, SREBP-1c, PPARγ, C/EBPα and ACC-1, which were suppressed by BBR treatment, were rescued AMPKα1 siRNA transfection in a dose-dependent manner, whereas the control siRNA sequences demonstrated no significant effect (Fig. 7). Similar to the protein expression levels of PPARγ and C/EBPα (Fig. 6A), the mRNA expression levels of these factors were rescued by AMPKα1 siRNA transfection in a dose-dependent manner (Fig. 7). Therefore, the results of the present study demonstrate that AMPK serves an essential role in the BBR-induced phosphorylation of SREBP-1c and the suppression of proteolytic processing, nuclear translocation and target DNA binding of SREBP-1c, which leads to the downregulation of lipogenesis-associated gene expression and decreased intracellular fat accumulation (Fig. 8).
Figure 6.
Effect of AMPK knockdown on SREBP-1c expression and intracellular fat accumulation following BBR treatment. 3T3-L1 cells were transfected with 12.5, 25, 50, or 75 nM AMPKα1 siRNA, or 50 or 75 nM control siRNA at one day prior to induction of differentiation. Cells were treated with 20 µM BBR from days 2–7 of differentiation, and were harvested on day 7 for analysis. (A) The protein expression levels of p-AMPK and p-SREBP-1c, as well as the 125-kDa precursor SREBP-1c isoform and the mature 68-kDa isoform, were analyzed in total cell lysates by western blotting. The levels of the mature 68-kDa SREBP-1c isoform were analyzed in nuclear lysates in order to examine its nuclear translocation. Levels of the major adipogenic transcription factors, PPARγ and C/EBPα, were analyzed in total cell lysates. β-actin and TBP were used as endogenous controls for the analysis of protein expression in total cell lysates and nuclear lysates, respectively. (B) The binding of nuclear SREBP-1c to its E-box motif-containing target DNA sequence, 5′-TCACCTGA-3′, was measured by ELISA. (C) Oil red O stain in intracellular fat droplets was extracted using isopropyl alcohol and quantified by measuring the OD at 510 nm. Different letters indicate significant differences between groups (P<0.05). AMPK, AMP-activated protein kinase; SREBP-1c, sterol regulatory element binding protein-1c; BBR, berberine; siRNA, small-interfering RNA; p-AMPK, phosphorylated-AMPK; p-SREBP-1c, phosphorylated-SREBP-1c; PPARγ, peroxisome proliferator-activated receptor-γ; C/EBPα, CCAAT/enhancer-binding protein α; TBP, TATA box-binding protein; OD, optical density.
Figure 7.
Effect of AMPK knockdown on the expression of lipogenesis-associated genes. 3T3-L1 cells were transfected with 12.5, 25, 50, or 75 nM AMPKα1 siRNA, or 50 or 75 nM control siRNA at one day prior to induction of differentiation. Cells were treated with 20 µM BBR from days 2–7 of differentiation, and were harvested on day 7 for analysis. The mRNA level of (A) FAS, (B) LPL, (C) SREBP-1c, (D) PPARγ, (E) C/EBPα and (F) ACC-1 were measured by reverse transcription-quantitative polymerase chain reaction. Different letters indicate significant differences between groups (P<0.05). AMPK, AMP-activated protein kinase; siRNA, small-interfering RNA; BBR, berberine; FAS, fatty acid synthase; LPL, lipoprotein lipase; SREBP1-c, sterol regulatory element binding protein-1c; PPARγ, peroxisome proliferator-activated receptor-γ; C/EBPα, CCAAT/enhancer-binding protein α; ACC-1, acetyl-CoA carboxylase-1.
Figure 8.
A schematic depicting the molecular mechanisms underlying the anti-adipogenic activity of berberine. AMPK, AMP-activated protein kinase; p-AMPK, phosphorylated-AMPK; SREBP1-c, sterol regulatory element binding protein-1c; C/EBPα, CCAAT/enhancer-binding protein α; PPARγ, peroxisome proliferator-activated receptor-γ; FAS, fatty acid synthase; LPL, lipoprotein lipase; ACC-1, acetyl-CoA carboxylase-1.
Discussion
SREBPs are helix-loop-helix-leucine zipper transcription factors that regulate lipid homeostasis by controlling the expression of genes required for the synthesis of fatty acids, triacylglycerol and cholesterol (7). The three SREBP isoforms, SREBP-1a, SREBP-1c and SREBP-2, serve different roles in lipid metabolism. SREBP-1c and SREBP-1a arise from the SREBP-1 gene through the use of alternative first exons, and SREBP-1c is the predominant isoform in the majority of tissues, including adipose tissue, liver and skeletal muscle. SREBP-1c is involved in fatty acid synthesis and lipogenesis, whereas SREBP-2 is primarily involved in cholesterol synthesis. The transcription of SREBP-1c is induced by insulin, and the SREBP-1c protein is synthesized as a 125-kDa precursor containing an N-terminal transcription factor domain and a C-terminal regulatory domain. Precursor SREBP-1c remains in the membrane of the endoplasmic reticulum until it is transported to the Golgi apparatus. In the Golgi apparatus, the membrane-bound 125-kDa precursor SREBP-1c undergoes proteolytic processing to generate the soluble 68-kDa mature SREBP-1c, which contains a transcription factor domain and a nuclear localization signal. Once soluble, the 68-kDa mature SREBP-1c enters the nucleus to bind its target DNA sequence within the promoters of a number of lipogenic genes (7,22,23).
In the nucleus, SREBP-1c, also termed adipocyte determination- and differentiation-dependent factor 1, binds to its binding sequence, E-box or a sterol regulatory element (SRE) in the promoter region of many lipogenesis-associated genes, including that of SREBP-1. In this way, mature SREBP-1c upregulates its own mRNA expression by binding to the promoter of the SREBP-1 gene (16). The promoter of the PPARγ gene contains an E-box motif that mediates its regulation by SREBP-1 (17). PPARγ immediately induces C/EBPα to construct a mutual activation loop (24). SREBP-1c has been reported to transactivate the promoter of the LPL gene in 3T3-L1 cells (18). The promoter of the ACC gene contains an E-box motif for SREBP-1c binding, together with binding sites for additional transcription factors, such as upstream stimulatory factor (USF) and nuclear factor (NF)-Y (19). FAS gene transcription is controlled by USF and SREBP-1c, which bind at the SRE located in its proximal promoter region (20).
AMPK is a serine/threonine kinase that regulates metabolic processes by upregulating catabolism and downregulating anabolism, and its therapeutic importance for metabolic diseases has led many researchers to search for its downstream target proteins as well as upstream activators (1). Recently, it was demonstrated that AMPK phosphorylates precursor SREBP-1c at its Ser372 residue, which suppresses the proteolytic processing of the precursor 125-kDa SREBP-1c into the mature 68-kDa SREBP-1c in HepG2 hepatocellular carcinoma cells and liver tissues of low-density lipoprotein receptor-deficient diabetic mice (15). However, this AMPK-SREBP-1c signaling pathway has not been reported as a mechanism of action of any anti-adipogenic agent in adipocytes.
In the present study, BBR, an AMPK activator of natural origin, induced the phosphorylation of precursor SREBP-1c in a time- and dose-dependent manner, suppressing its proteolytic processing into mature SREBP-1c, its nuclear translocation and target DNA binding. The ability of nuclear SREBP-1c to bind to its target DNA sequence was similar to the observed pattern of mRNA expression of lipogenesis-associated genes in BBR-treated cells. Knockdown of AMPK was associated with inhibition of BBR-induced phosphorylation of precursor SREBP-1c, thus confirming that AMPK is the kinase responsible for phosphorylating SREBP-1c in adipocytes. AMPKα1 siRNA rescued the proteolytic processing, nuclear translocation and target DNA binding of SREBP-1c, which was suppressed by BBR. Accordingly, the expression of lipogenesis-associated genes, which were suppressed by BBR, was rescued by AMPKα1 siRNA. SREBP-1c is one of the target genes of which transcriptions were activated by the binding of mature SREBP-1c, and levels of SREBP-1c mRNA and SREBP-1c precursor protein were rescued by AMPKα1 siRNA. The mRNA levels of a number of lipogenic genes demonstrated similar expression patterns; however, they did not correspond completely with the ability of nuclear SREBP-1c to bind to target DNA. The may be due to the possibility that additional transcription factors, including USF, NF and C/EBPα, may additionally be involved in these processes in cooperation with SREBP-1c (19,20).
Acknowledgements
The present study was supported by the Korean Health Technology Research and Development Project of the Ministry of Health and Welfare (grant nos. HI15C0075 and HI14C2687).
Glossary
Abbreviations
- ACC
acetyl-CoA carboxylase
- AMPK
AMP-activated protein kinase
- BBR
berberine
- C/EBP
CCAAT/enhancer-binding protein
- FAS
fatty acid synthase
- IRS
insulin receptor substrate
- LPL
lipoprotein lipase
- NF
nuclear factor
- PPAR
peroxisome proliferator-activated receptor
- SRE
sterol regulatory element
- SREBP
sterol regulatory element binding protein
- TBP
TATA box-binding protein
- USF
upstream stimulatory factor
References
- 1.Steinberg GR, Kemp BE. AMPK in health and disease. Physiol Rev. 2009;89:1025–1078. doi: 10.1152/physrev.00011.2008. [DOI] [PubMed] [Google Scholar]
- 2.Daval M, Diot-Dupuy F, Bazin R, Hainault I, Viollet B, Vaulont S, Hajduch E, Ferré P, Foufelle F. Anti-lipolytic action of AMP-activated protein kinase in rodent adipocytes. J Biol Chem. 2005;280:25250–25257. doi: 10.1074/jbc.M414222200. [DOI] [PubMed] [Google Scholar]
- 3.Gowans GJ, Hawley SA, Ross FA, Hardie DG. AMP is a true physiological regulator of AMP-activated protein kinase by both allosteric activation and enhancing net phosphorylation. Cell Metab. 2013;18:556–566. doi: 10.1016/j.cmet.2013.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vuddanda PR, Chakraborty S, Singh S. Berberine: A potential phytochemical with multispectrum therapeutic activities. Expert Opin Investig Drugs. 2010;19:1297–1307. doi: 10.1517/13543784.2010.517745. [DOI] [PubMed] [Google Scholar]
- 5.Lee YS, Kim WS, Kim KH, Yoon MJ, Cho HJ, Shen Y, Ye JM, Lee CH, Oh WK, Kim CT, et al. Berberine, a natural plant product, activates AMP-activated protein kinase with beneficial metabolic effects in diabetic and insulin-resistant states. Diabetes. 2006;55:2256–2264. doi: 10.2337/db06-0006. [DOI] [PubMed] [Google Scholar]
- 6.Turner N, Li JY, Gosby A, To SW, Cheng Z, Miyoshi H, Taketo MM, Cooney GJ, Kraegen EW, James DE, et al. Berberine and its more biologically available derivative, dihydroberberine, inhibit mitochondrial respiratory complex I: A mechanism for the action of berberine to activate AMP-activated protein kinase and improve insulin action. Diabetes. 2008;57:1414–1418. doi: 10.2337/db07-1552. [DOI] [PubMed] [Google Scholar]
- 7.Jeon TI, Osborne TF. SREBPs: Metabolic integrators in physiology and metabolism. Trends Endocrinol Metab. 2012;23:65–72. doi: 10.1016/j.tem.2011.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li GS, Liu XH, Zhu H, Huang L, Liu YL, Ma CM, Qin C. Berberine-improved visceral white adipose tissue insulin resistance associated with altered sterol regulatory element-binding proteins, liver × receptors and peroxisome proliferator-activated receptors transcriptional programs in diabetic hamsters. Biol Pharm Bull. 2011;34:644–654. doi: 10.1248/bpb.34.644. [DOI] [PubMed] [Google Scholar]
- 9.Hu Y, Kutscher E, Davies GE. Berberine inhibits SREBP-1-related clozapine and risperidone induced adipogenesis in 3T3-L1 cells. Phytother Res. 2010;24:1831–1838. doi: 10.1002/ptr.3204. [DOI] [PubMed] [Google Scholar]
- 10.Huang C, Zhang Y, Gong Z, Sheng X, Li Z, Zhang W, Qin Y. Berberine inhibits 3T3-L1 adipocyte differentiation through the PPARgamma pathway. Biochem Biophys Res Commun. 2006;348:571–578. doi: 10.1016/j.bbrc.2006.07.095. [DOI] [PubMed] [Google Scholar]
- 11.Gustafson B, Smith U. Cytokines promote Wnt signaling and inflammation and impair the normal differentiation and lipid accumulation in 3T3-L1 preadipocytes. J Biol Chem. 2006;281:9507–9516. doi: 10.1074/jbc.M512077200. [DOI] [PubMed] [Google Scholar]
- 12.Li J, Zhang D, Ward KM, Prendergast GC, Ayene IS. Hydroxyethyl disulfide as an efficient metabolic assay for cell viability in vitro. Toxicology in Vitro. 2012;26:603–612. doi: 10.1016/j.tiv.2012.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee H, Bae S, Yoon Y. The anti-adipogenic effects of (−)epigallocatechin gallate are dependent on the WNT/β-catenin pathway. J Nutr Biochem. 2013;24:1232–1240. doi: 10.1016/j.jnutbio.2012.09.007. [DOI] [PubMed] [Google Scholar]
- 14.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 15.Li Y, Xu S, Mihaylova MM, Zheng B, Hou X, Jiang B, Park O, Luo Z, Lefai E, Shyy JY, et al. AMPK phosphorylates and inhibits SREBP activity to attenuate hepatic steatosis and atherosclerosis in diet-induced insulin-resistant mice. Cell Metab. 2011;13:376–388. doi: 10.1016/j.cmet.2011.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Amemiya-Kudo M, Shimano H, Yoshikawa T, Yahagi N, Hasty AH, Okazaki H, Tamura Y, Shionoiri F, Iizuka Y, Ohashi K, et al. Promoter analysis of the mouse sterol regulatory element-binding protein-1c gene. J Biol Chem. 2000;275:31078–31085. doi: 10.1074/jbc.M005353200. [DOI] [PubMed] [Google Scholar]
- 17.Fajas L, Schoonjans K, Gelman L, Kim JB, Najib J, Martin G, Fruchart JC, Briggs M, Spiegelman BM, Auwerx J. Regulation of peroxisome proliferator-activated receptor gamma expression by adipocyte differentiation and determination factor 1/sterol regulatory element binding protein 1: Implications for adipocyte differentiation and metabolism. Mol Cell Biol. 1999;19:5495–5503. doi: 10.1128/MCB.19.8.5495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schoonjans K, Gelman L, Haby C, Briggs M, Auwerx J. Induction of LPL gene expression by sterols is mediated by a sterol regulatory element and is independent of the presence of multiple E boxes. J Mol Biol. 2000;304:323–334. doi: 10.1006/jmbi.2000.4218. [DOI] [PubMed] [Google Scholar]
- 19.Barber MC, Vallance AJ, Kennedy HT, Travers MT. Induction of transcripts derived from promoter III of the acetyl-CoA carboxylase-alpha gene in mammary gland is associated with recruitment of SREBP-1 to a region of the proximal promoter defined by a DNase I hypersensitive site. Biochem J. 2003;375:489–501. doi: 10.1042/bj20030480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Griffin MJ, Sul HS. Insulin regulation of fatty acid synthase gene transcription: Roles of USF and SREBP-1c. IUBMB Life. 2004;56:595–600. doi: 10.1080/15216540400022474. [DOI] [PubMed] [Google Scholar]
- 21.Bene H, Lasky D, Ntambi JM. Cloning and characterization of the human stearoyl-CoA desaturase gene promoter: Transcriptional activation by sterol regulatory element binding protein and repression by polyunsaturated fatty acids and cholesterol. Biochem Biophys Res Commun. 2001;284:1194–1198. doi: 10.1006/bbrc.2001.5102. [DOI] [PubMed] [Google Scholar]
- 22.Wang X, Sato R, Brown MS, Hua X, Goldstein JL. SREBP-1, a membrane-bound transcription factor released by sterol-regulated proteolysis. Cell. 1994;77:53–62. doi: 10.1016/0092-8674(94)90234-8. [DOI] [PubMed] [Google Scholar]
- 23.Eberle D, Hegarty B, Bossard P, Ferré P, Foufelle F. SREBP transcription factors: Master regulators of lipid homeostasis. Biochimie. 2004;86:839–848. doi: 10.1016/j.biochi.2004.09.018. [DOI] [PubMed] [Google Scholar]
- 24.Zhao QH, Wang SG, Liu SX, Li JP, Zhang YX, Sun ZY, Fan QM, Tian JW. PPARγ forms a bridge between DNA methylation and histone acetylation at the C/EBPα gene promoter to regulate the balance between osteogenesis and adipogenesis of bone marrow stromal cells. FEBS J. 2013;280:5801–5814. doi: 10.1111/febs.12500. [DOI] [PubMed] [Google Scholar]