Abstract
The anticancer effect of sclareol has long been reported, however, the exact mechanisms underlying the antitumorigenic effect of sclareol in cervical carcinoma remain to be fully elucidated. The present study analyzed cell proliferation and cell apoptosis by MTT and FITC-Annexin V assays. The protein levels of caveolin-1 (Cav-1) and copper-zinc superoxide dismutase (SOD)1 were determined by western blotting, and the interaction of Cav1 and HSC70 was investigated by co-immunoprecipitation experiments. The present study found that sclareol inhibited cell proliferation and induced apoptosis in HeLa cells. Two cancer-associated proteins, Cav1 and SOD1 were identified as potential targets of sclareol in HeLa cells. The expression of Cav1 increased when the cells were treated with sclareol, and the protein level of SOD1 was negatively correlated with Cav1. The overexpression of Cav1 enhanced the sensitivity of the HeLa cells to sclareol treatment and downregulated the protein level of SOD1, which exhibited potential associations between Cav1 and SOD1. In addition, sclareol significantly sensitized several cancer cells to the anticancer effect of bortezomib by targeting Cav1 and SOD1. Taken together, the results of the present study demonstrated that sclareol inhibited tumor cell growth through the upregulation of Cav1, and provides a potential therapeutic target for human cancer.
Keywords: sclareol, caveolin-1, copper-zinc superoxide dismutase, cervical cancer
Introduction
Cervical cancer is the third most common type of cancer and the fourth leading cause of cancer-associated mortality in women worldwide (1). In developing countries, it is the most common cause of cancer-associated mortality, and the five-year survival rate for patients with advanced cervical cancer is low (1). Thus, it is important to identify and examine effective drugs in cervical cancer treatment. It has long been reported that sclareol, a labdane-type diterpene isolated from the Salvia sclarea plant can inhibit the proliferation and induce the apoptosis of several cancer cell lines (2–6). However, the exact mechanisms underlying the antitumorigenic effect of sclareol in cervical carcinoma remain to be elucidated.
Caveolin-1 (Cav1) is an important component of caveolae and is known to function as a scaffolding protein in the regulation of several signaling pathways (7–9). The loss of Cav1 has been demonstrated to be involved in the tumorigenesis of several types of cancer, and the overexpression of Cav1 has been shown to inhibit cell and tumor growth (10–12). Thus Cav1 is regarded as a potential tumor suppressor. Although numerous studies have been performed investigating the function of Cav1 in several types of cancer, the exact role of Cav1 in cervical carcinoma remains to be elucidated (10–12).
Copper-zinc superoxide dismutase (SOD1) is an essential element of the superoxide radical eliminating system. It is the most abundant anti-oxidant enzyme and is predominantly localized in the cytoplasm, although its localization in the mitochondrial intermembrane space and nucleus has also been reported (13,14). Previous studies have revealed that SOD1 is overexpressed in various types of cancer, and that the downregulation of SOD1 leads to cancer cell death (15,16), which indicates that SOD1 is important in tumorigenesis.
In the present study, the anticancer effect of sclareol was investigated, and the expression levels of Cav1 and SOD1 in a cervical cancer cell line were investigated, in order to elucidate the potential mechanisms involved in the anticancer effect of sclareol.
Materials and methods
Cell culture
HeLa cells were obtained from the American Type Culture Collection (Rockville, MD, USA). The SW480, SW620, HepG2 and MCF-7 cancer cell lines were obtained from the Cell Resource Center of the Chinese Academy of Sciences (Shanghai, China). All cell lines were grown in Dulbecco's modified Eagle's medium (DMEM; HyClone; GE Healthcare Life Sciences, Logan, UT, USA) supplemented with 10% fetal bovine serum (HyClone; GE Healthcare Life Sciences). All cell lines were incubated in a humidified atmosphere containing 5% CO2 at 37°C.
Reagents and antibodies
Sclareol, bortezomib, E64 and pepstatin A were obtained from Sigma-Aldrich; Thermo Fisher Scientific, Inc. (Waltham, MA, USA). The Cav1, SOD1, β-tubulin and p62 antibodies were obtained from ProteinTech Group, Inc., Chicago, IL, USA), the LC3 antibody was obtained from Sigma-Aldrich; Thermo Fisher Scientific, Inc.), the Flag antibody was obtained from Prospec-Tany TechnoGene, Ltd., (East Brunswick, NJ, USA), the HA tag antibody was obtained from Cell Signaling Technology, Inc. (Beverly, MA, USA).
MTT and cell apoptosis assays
The cells were plated into 96-well tissue culture plates and seeded at a density of 10,000 cells per well. Viability of HeLa cells was determined using an MTT assay following treatment with vehicle or 10 µg/ml sclareol for 24, 48 or 72 h at 37°C. Cell apoptosis assay of HeLa cells treated with 5 µg/ml sclareol was performed for 24, 48 or 72 h at 37°C. The final volume of culture medium in each well was 100 µl. A 10 µl volume of MTT solution (5 mg/ml) was added to the 100 µl medium in each well. The plates were incubated at 37°C for 4 h, following which the supernatant was removed and 100 µl DMSO was added to each well. The absorbance signals were measured on a spectrophotometer at 490 nm. The cell death-inducing effects of drug treatment were measured using a CF488A-Annexin V and Propidium Iodide (PI) Apoptosis Assay kit (Biotium, Inc., Hayward, CA, USA). The samples and assays were prepared according to the manufacturer's protocol and then mounted onto slides. Images were captured with a Nikon fluorescence microscope (Nikon, Tokyo, Japan).
Western blot analysis
Samples were collected with 1X SDS sample buffer and were separated by 10–12% SDS-PAGE (20 µg/lane). The proteins were transferred onto an Immobilon-FL PVDF membrane (EMD Millipore, Bedford, MA, USA). Protein quantification was performed using a BCA assay kit (Thermo Fisher Scientific, Inc.). The membrane was first blocked with 5% milk for 1 h, and then incubated overnight at 4°C with the following primary antibodies: Cav 1 (cat. no. 16447-1-AP; 1:1,000; ProteinTech Group, Inc., Chicago, IL, USA), SOD1 (cat. no. 10269-1-AP; 1:1,000; ProteinTech Group, Inc.), β-tubilin (cat. no. 66240-1; 1:2,000; ProteinTech Group, Inc.), p62 (cat. no. 18420-1-AP; 1:1,000; ProteinTech Group, Inc.), LC3 (cat. no. L8918; 1:1,000; Sigma-Aldrich; Merck Millipore, Darmstadt, Germany), Flag (cat. no. ANT-222; 1:1,000; Prospec-Tany TechnoGene, Ltd., East Brunswick, NJ, USA) and HA tag (cat. no. C29F4; 1:1,000; Cell Signaling Technology, Inc., Danvers, MA, USA) diluted in the same blocking buffer. Following three washes with Tris-buffered saline with Tween 20 (TBST) containing 50 mM Tris-HCl (pH 7.5), 150 mM NaCl and 0.2% Tween 20, the blot was incubated with Dylight 680/800-conjugated secondary antibodies (cat. nos. 042-06-15-06 and 042-06-18-0; 1:10,000; KPL, Inc. Gaithersburg, MD, USA) in the dark for 2 h at room temperature. The blot was then washed again with TBST and images were captured using a Li-Cor Odyssey Clx infrared imaging system (LI-COR BioSciences, Inc., Lincoln, NE, USA).
Establishment of stable cell line
For the stable expression of Cav-1, the HeLa cells were transduced with Cav1-Flag lentiviral particles using ViraPower Lentiviral Expression system following the manufacturer's protocol (Invitrogen; Thermo Fisher Scientific, Inc.) following the manufacturer's protocol, and stable cell lines were selected using blasticidin (Invitrogen; Thermo Fisher Scientific, Inc.).
Co-immunoprecipitation assay
At 24 h post-transduction, the cells were washed with phosphate-buffered saline (PBS) and lysed in 1% NP40 lysis buffer for 30 min on ice. The cell lysates were collected in an EP tube and then centrifuged at >18,000 × g for 10 min at 4°C, the supernatants were immunoprecipitated with the Flag (1:1,000) and HA tag (1:1,000) antibodies overnight at 4°C. Prewashed protein A/G-agarose beads (Thermo Fisher Scientific, Inc.) were then added to the supernatant and incubated for another 2 h at 4°C. The immune complexes were washed with lysis buffer three times, resuspended in sample buffer and then subjected to western blot analysis.
Immunofluorescence assay
The cells were plated on coverslips at a density of 1.0×105 cells and transfected with the indicated plasmid using Lipofectamine 2000. HeLa cells and HeLa cells stably transfected with Cav1-Flag (Cav1) were transiently transfected with a plasmid expressing lysosome-associated protein 2A (Lamp-2A)-EGFP. At 24 h post-transfection, the cells were then fixed with 4% paraformaldehyde for 15 min at room temperature, rinsed with PBS and permeabilized with 0.25% Triton-x-100 for 10 min at room temperature. Following rinsing with PBS three times, the cells were blocked with 3% bovine serum albumin (Sigma-Aldrich; Merck Millipore) overnight at 4°C. The EGFP (cat. no. 50430-2; 1:1,000; Proteintech Group, Inc.), Flag (1:1,000) antibodies were added and incubated overnight at 4°C and rinsed with PBS three times The secondary antibody was added for 1 hour at room temperature, following which the cells were rinsed with PBS three times, and then incubated with DAPI for 10 min at room temperature. The cells were then rinsed, mounted and sealed. Images were captured using an Olympus confocal microscope (Olympus, Tokyo, Japan).
Plasmid construction and transfection
The full-length Cav1 open reading frame (ORF) was amplified by polymerase chain reaction (PCR) with a BamHI site-containing 5′ primer and an XbaI site-containing 3′ primer, and cloned into the pcDNA3.1(+)-flag vector (Invitrogen; Thermo Fisher Scientific, Inc.) between the BamHI and XbaI sites to generate the Cav1-Flag expression construct. The full-length heat-shock protein cognate 70 (HSC70) ORF was amplified by PCR with a BamHI site-containing 5′ primer and an XbaI site-containing 3′ primer, and cloned between the BamHI and XbaI sites of the pcDNA3.1(+)-HA vector to generate the HSC70-HA expression construct. Full-length LAMP-2A ORF was amplified by PCR with a BamHI site-containing 5′ primer and an XbaI site-containing 3′ primer, and cloned between the BamHI and XbaI sites of the pcDNA3.1(+)-EGFP vector to generate the LAMP-2A-EGFP expression construct. The human cDNA preserved in the laboratory was utilized as the template. Flag tag, HA tag and EGFP tag were cloned into the pcDNA3.1(+) vector (Life Technologies; Thermo Fisher Scientific, Inc.) between XbaI and ApaI to generate the pcDNA3.1(+)-tagged vector. For the generation of Cav1Flag lentiviral particles, the Cav1-Flag cDNA fragment was inserted into a pLenti6 lentiviral vector (Invitrogen Life Technologies; Thermo Fisher Scientific, Inc.), and the resultant plasmid was co-transfected with ViraPower Lentiviral Packaging mix (Invitrogen Life Technologies; Thermo Fisher Scientific, Inc.) into HEK293FT cells with Lipofectamine 2000. The virus-containing cell culture medium was harvested 40 h later and used to transduce the HeLa cells. The sequences of the primers are listed in Table I.
Table I.
Sequences of primers for plasmid construction.
| Primer name | Sequence (5′-3′) |
|---|---|
| Cav1-F | gatcggatccgccaccatgtctgggggcaaatacgtag |
| Cav1-R | gtagttgaacgtctttctttatagatctctag |
| HSC70-F | gatcggatccgccaccatgtccaagggacctgcagttg |
| HSC70-R | gggtggtaacttctccaactaagatctctag |
| LAMP-2A-F | gatcggatccgccaccatggtgtgcttccgcctcttc |
| LAMP-2A-R | gtacgacctatactcgttaaaagatctctag |
Restriction sites are underlined. F, forward; R, reverse; Cav1, caveolin-1; HSC70, heat shock protein cognate 70; LAMP-2A, lysosome-associated protein 2A.
Statistical analysis
Experimental data are expressed as the mean ± standard error of the mean. Data analyses were performed using ImageJ software (National Institutes of Health, Bethesda, MD, USA). Differences between groups were compared using one-way analysis of variance followed by Tukey's post-hoc test. P<0.05 was considered to indicate a statistically significant difference.
Results
Sclareol inhibits cell proliferation and induces apoptosis in HeLa cells
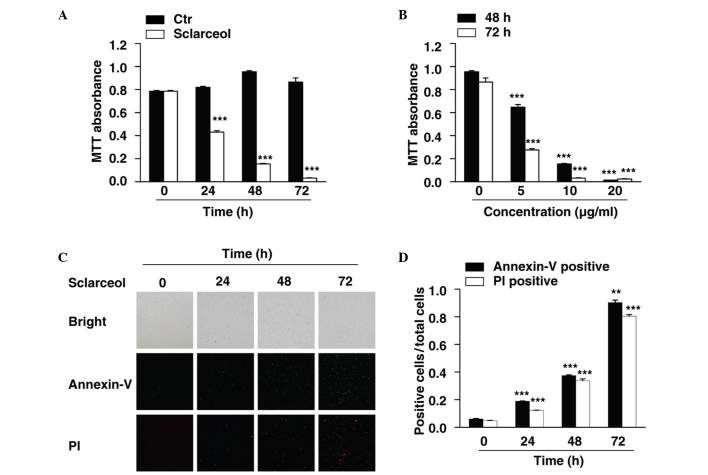
In the present study, HeLa cells were used to investigate the anticancer effects of sclareol in cervical carcinoma. An MTT assay was first performed to measure the viability of the HeLa cells. The cells were treated with sclareol for 24, 48 and 72 h, and cell viability was measured at each time point. The results showed that sclareol significantly inhibited the proliferation of the HeLa cells, and the inhibitory effect was time-dependent (Fig. 1A). In addition, when the cells were treated with different concentrations of sclareol, the proliferation of the HeLa cells was inhibited in a dose-dependent manner (Fig. 1B).
Figure 1.
Sclareol inhibits cell proliferation and induces cell apoptosis in HeLa cells. (A) Cell viability of HeLa cells was measured using an MTT assay following treatment with vehicle or 10 µg/ml sclareol for 24, 48 and 72 h. (B) Cell viability of HeLa cells was measured using an MTT assay following treatment with vehicle or sclareol at the indicated concentration for 48 and 72 h. (C) Cell apoptosis assay of HeLa cells treated with 5 µg/ml sclareol for the indicated durations. (D) Quantification of the ratio of positive cells to total cells. The number of positive cells were counted using Image J software. Data are presented as the mean + standard error of the mean of three independent experiments. *P<0.05, **P<0.01 and ***P<0.005, compared with the vehicle group. Ctr, control; PI, propidium iodide.
Treatment with sclareol also induced the apoptosis of the HeLa cells. An Annexin V-fluorescein isothiocyanate (FITC)/PI staining assay was used to assess the proportion of apoptotic cells when HeLa cells were treated with sclareol for indicated durations. The results indicated that the Annexin V-FITC-positive and PI-positive cells were significantly increased in a time-dependent manner when treated with sclareol (Fig. 1C and D).
Sclareol induces the expression of Cav1
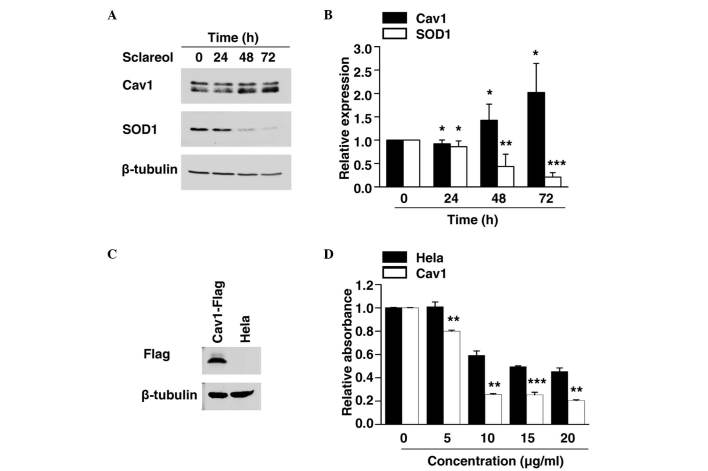
As the exact mechanism involved in the anticancer effect of sclareol remains to be fully elucidated, the present study examined the expression of various cancer-associated proteins to identify the potential target of sclareol. The cells were exposed to sclareol for 24, 48 and 72 h, and were then subjected to western blot analysis. The results showed that sclareol induced the protein levels of Cav1 in a time-dependent manner, compared with the control group (Fig. 2A and B).
Figure 2.
Sclareol affects the protein levels of Cav1 and SOD1 in HeLa cells, and overexpression of Cav1 enhances sensitivity to slcareol treatment. (A) Upregulation of the expression of Cav1 and downregulation of the expression of SOD1 were observed following treatment with 5 µg/ml sclareol. HeLa cells were treated with sclareol for the indicated duration and then harvested for western blot analysis. (B) Quantification of the protein levels of Cav1 and SOD1 in HeLa cells. Data are presented as the mean + standard error of the mean of three independent experiments. *P<0.05, **P<0.01 and ***P<0.005, compared with the vehicle group. (C) Western blot analysis of HeLa cells stably expressing Flag-tagged Cav1 protein. (D) Viability of HeLa cells and HeLa cells stably expressing Flag-tagged Cav1 protein were measured using an MTT assay following treatment with vehicle or sclareol at the indicated concentration for 24 h. Data are presented as the mean + standard error of the mean of three independent experiments. *P<0.05, **P<0.01 and ***P<0.005, compared with the control group. Cav1, caveolin-1; SOD1, superoxide dismutase 1.
Expression of SOD1 is negatively correlated with Cav1
SOD1 has previously been identified as a novel cancer gene (13,14). The results of the present study showed that the protein level of SOD1 was significantly decreased in a time-dependent manner when treated with sclareol (Fig. 2A and B), and the decreased expression of SOD1 was negatively correlated with the increased expression of Cav1.
Cells overexpressing Cav1 are more sensitive to sclareol treatment
The present study established a HeLa cell line stably expressing Cav1. The stable Cav1-expressing cell line and normal HeLa cells were treated with vehicle or sclareol at the indicated concentration for 24 h, following which the cell viability was measured using an MTT assay. It was found that stable expression of Cav1 enhanced the sensitivity of the cells to sclareol treatment (Fig. 2C and D). These results indicated that the upregulation of Cav1 was responsible for the anticancer effect of sclareol.
Cav1 promotes the downregulation of SOD1
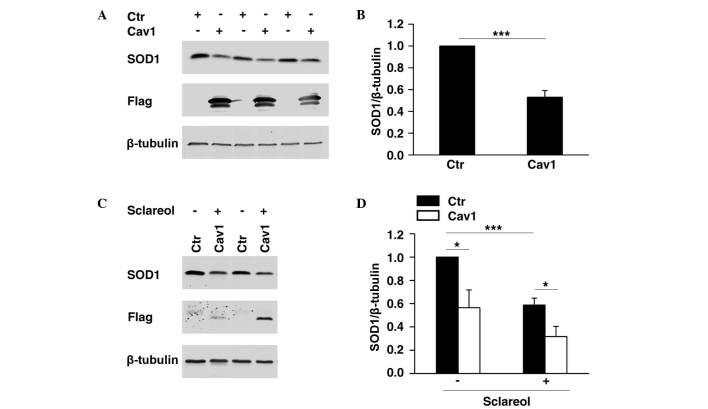
It was previously demonstrated that SOD1 is important in cancer development (10,11), whereas Cav1 has been reported to be a cancer suppressor. As shown in Fig. 2, the present study found that SOD1 was negatively correlated with Cav1. To determine whether the expression of SOD1 was directly regulated by Cav1, HeLa cells overexpressing Flag-tagged Cav1 were established, and western blot analysis was performed to determine the protein level of SOD1. The results showed that the overexpression of Cav1 significantly promoted the downregulation of SOD1 (Fig. 3A and B). In the HeLa cell line stably expressing Cav1, it was found that stable expression of Cav1 enhanced the downregulation of SOD1 when treated with sclareol (Fig. 3C and D).
Figure 3.
Overexpression of Cav1 enhances the downregulation of the protein level of SOD1 (A) Transient transfection of ectopic Cav1 in HeLa cells led to the downregulation of SOD1. HeLa cells were transfected with Flag-tagged Cav1 expression plasmid (Cav1) or empty vector (Ctr) respectively. The cells were harvested 24 h following transfection and then subjected to western blot analysis. (B) Quantification of the protein levels of SOD1 in HeLa cells. Data are presented as the mean + standard error of the mean of three independent experiments. *P<0.05, **P<0.01 and ***P<0.005, compared with the control group. (C) Stable expression of ectopic Cav1 enhanced the downregulation of the protein level of SOD1when treated with sclareol. HeLa cells (Ctr) and HeLa cells stably expressing Flag-tagged Cav1 (Cav1) were treated with vehicle and sclareol (5 µg/ml) for 48 h, and then subjected to western blot analysis. (D) Quantification of the protein levels of SOD1 in the Ctr and Cav1 cells. Data are presented as the mean + standard error of the mean of three independent experiments. *P<0.05, **P<0.01 and ***P<0.005, compared with the control group. Ctr, control; Cav1, caveolin-1; SOD1, superoxide dismutase 1.
Sclareol sensitizes cells to the antiproliferative effect of bortezomib by targeting Cav1 and SOD1
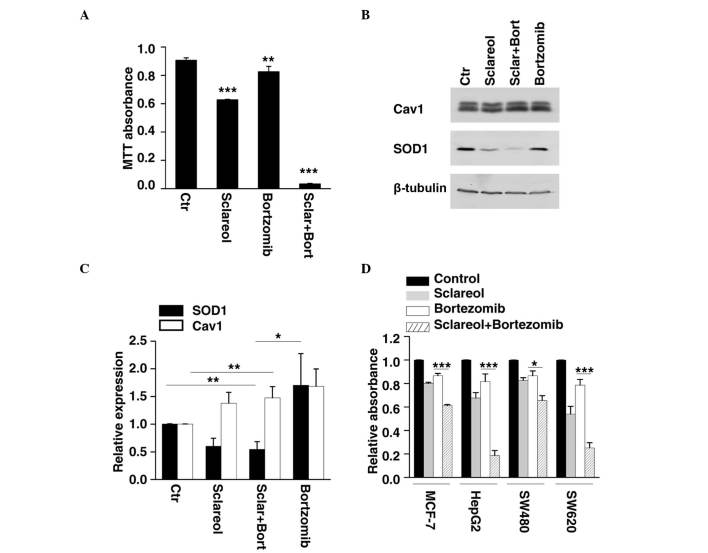
Previous investigations have revealed that Cav1 is degraded through the ubiquitin-proteasome pathway (17,18). As the results of the present study showed that sclareol inhibited cancer cell proliferation through the upregulation of Cav1, whether bortezomib, an anticancer drug and a proteasome inhibitor, enhances the anticancer effect of sclareol was investigated. The HeLa cells were treated with sclareol only, bortezomib only or sclareol and bortezomib together, and subjected to western blot analysis and an MTT assay. By measuring the cell viability of each group, it was found that sclareol significantly enhanced the antiproliferative effect of bortezomib, compared with the cells treated with either drug separately (Fig. 4A). The results of the western blot analysis showed that the protein level of Cav1 increased following treatment with bortezomib (Fig. 4B and C). In addition, the HeLa cells treated with sclareol and bortezomib together exhibited a lower level of SOD1 and higher level of Cav1, compared with the vehicle group (Fig. 4B and C). Similar results were observed when other cancer cells were treated with the drug combination of bortezomib and sclareol (Fig. 4D).
Figure 4.
Sclareol synergizes with bortezomib to inhibit cancer cell growth. (A) HeLa cells were treated with sclareol (5 µg/ml) only, bortezomib (5 nM) only, or sclareol (5 µg/ml) with bortezomib (5 nM) for 48 h, and subjected to MTT analysis. Data are presented as the mean + standard error of the mean of three independent experiments. *P<0.05, **P<0.01 and ***P<0.005, compared with the vehicle group. (B) Treated HeLa cells were also subjected to western blot analysis. (C) Quantification of the relative protein levels of SOD1 and Cav1 in HeLa cells treated with vehicle (Ctr) or indicated drugs. Data are presented as the mean + standard error of the mean of three independent experiments. *P<0.05, **P<0.01 and ***P<0.005, compared with the control group. (D) Sclareol sensitized cells to the anti-proliferative effect of bortezomib in several cancer cell lines. (A) MCF-7, HepG2, SW480 and SW620 cells were also subjected to MTT analysis. Data are presented as the mean + standard error of the mean of three independent experiments. *P<0.05, **P<0.01 and ***P<0.005 compared with control group. Ctr, control; Cav1, caveolin-1; SOD1, superoxide dismutase 1; Sclar, sclareol; Bort, bortzomib.
Cav1 may be involved in the lysosome-mediated degradation of SOD1
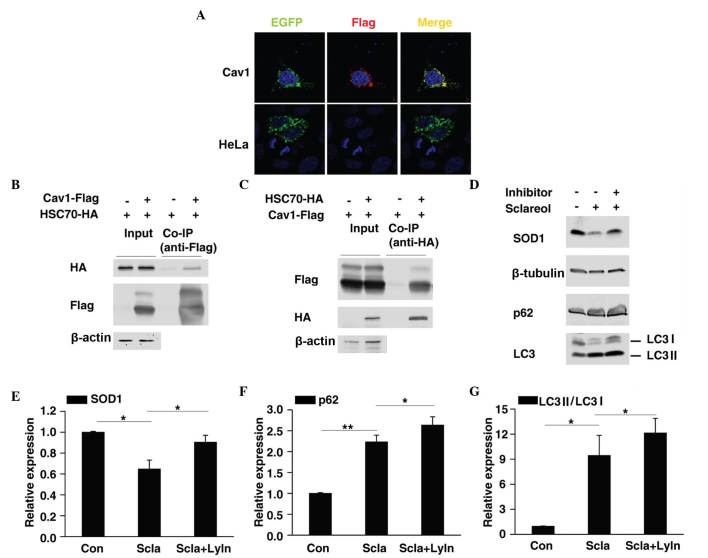
Previous reports have shown that Cav1 mediates the degradation of several proteins (19–21), thus it is possible that Cav1 promotes the degradation of SOD1. HSC70 and LAMP-2A are two important components of chaperone-mediated autophagy (CMA) (22). The present study found that Cav1 not only colocalized with LAMP-2A (Fig. 5A) but also interacted with HSC70 (Fig. 5B and C), thus it is possible that Cav1 may be involved in CMA-mediated protein degradation. CMA substrates are usually delivered to the lysosome for degradation (23). In the present study, HeLa cells were treated with sclareol only or sclareol together with lysosome inhibitors and then subjected to western blot analysis. Of note, as with p62/SQSTM1 and LC3 II, which are two autophagy-associated proteins previously reported to be degraded in lysosomes (24,25), inhibition of lysosome activity suppressed the downregulation of SOD1 caused by sclareol treatment (Fig. 5D-G), which suggested that sclareol treatment may promote the downregulation of SOD1 through lysosome mediated degradation via Cav1.
Figure 5.
Cav1 may be involved in the lysosome-mediated degradation of SOD1. (A) Colocalization of Cav1 and lysosome-associated protein 2A. Interaction of (B) Cav1 and (C) HSC70. (D) Lysosome inhibitors suppressed the downregulation in the protein level of SOD1 when treated with sclareol. HeLa cells were treated with sclareol (5 µg/ml) only or sclareol (5 µg/ml) with lysosome inhibitors (10 µM E64 and 20 µM pepstatin A) for 48 h and then subjected to western blot analysis. Quantification of the relative protein levels of (E) SOD1, (F) p62 and (G) LC3 II in HeLa cells treated with vehicle or the indicated drugs. Data are presented as the mean + standard error of the mean of three independent experiments. *P<0.05, **P<0.01 and ***P<0.005, compared with the control group. Con, vehicle control; Scla, sclareol; LyIn, lysosome inhibitor; HSC70, heat shock protein cognate 70.
Discussion
Sclareol exhibits antiproliferative and cytotoxic activity against various human cancer cell lines (2–6), and inhibits tumor growth in vivo (3,6). The anticancer effect of sclareol has long been reported, however, the exact role of sclareol and its molecular mechanisms in cervical carcinoma remain to be elucidated. The present study demonstrated that sclareol inhibited proliferation and induced apoptosis in HeLa cells, and the combination of sclareol with another anticancer drug, bortezomib, significantly inhibited tumor cell proliferation in several types of cancer cell. It was also shown that sclareol treatment induced the expression of Cav1 in HeLa cells.
Although the role of Cav1 in tumorigenesis is controversial, increasing evidence indicates that Cav1 functions as a tumor suppressor in various types of cancer. The downregulation of Cav1 has been implicated in the development and progression of cutaneous squamous cell carcinoma (26), lung carcinoma (12), alveolar rhabdomyosarcomas (10), colon cancer (11), sporadic vestibular schwannomas (27) and breast cancer (28). The overexpression of Cav1 decreases tumor cell growth (10–12) and enhances the sensitivity of cancer cells to anticancer drugs (21). However, the exact function of Cav1 in cervical carcinoma remains to be elucidated. In the present study, the upregulation of Cav1 correlated with decreased cell viability under sclareol treatment, which indicated that Cav1 may function as a tumor suppressor in HeLa cells.
Cav1 is ubiquitinated and targeted to lysosomes for degradation (17,18), and previous studies have demonstrated that Cav-1 is also a molecular target of bortezomib, which is involved in antitumorigenesis and proteasome inhibition (29), and is degraded through the proteasome pathway (30). The data obtained in the present study showed that sclareol increased the protein level of Cav1, and the combined treatment with bortezomib further enhanced the expression of Cav1. Based on these findings, it is possible that the combination of sclareol with bortezomib may have novel synergistic antitumor effects via upregulation of the cancer suppressor, Cav1. In line with this hypothesis, the present study found that bortezomib synergized with sclareol to reduce the viability of various types of cancer cell, including HeLa, HepG2, MCF-7, SW480 and SW620 cells, whereas treatment with bortezomib or sclareol alone at the same concentration had markedly lower inhibitory effects. Thus, the combination of bortezomib with sclareol may be a novel and promising approach in cancer therapy.
Previous reports have revealed the potential mechanism involved in the anticancer effect of Cav1. Cav-1 has been shown to prevent cancer growth via inhibition of the mitogen-activated protein kinase pathway (10) and to enhance the degradation of epidermal growth factor receptor (EGFR) (21). In the present study, the increased protein level of Cav1 was negatively correlated with the expression of SOD1, and the overexpression of Cav1 significantly suppressed the protein level of SOD1. These findings indicated a potential association between Cav1 and SOD1.
The expression of SOD1 has been linked to the resistance of cancer cells to pro-oxidant drugs and the anticancer drug. bortezomib (31), which raises the possibility that an SOD1 inhibitor can be used, not only as an antitumorigenic drug, but also as a molecule to challenge the drug resistance of cancer cells. The data obtained in the present study showed that the treatment of HeLa cells with sclareol led to downregulation of the protein level of SOD1, whereas the overexpression of Cav1 enhanced the suppressed expression of SOD1. These findings suggested that sclareol may function as an SOD1 inhibitor via the upregulation of Cav1. As the first anticancer drug to function as a proteasome inhibitor, bortezomib has been approved for the clinical treatment of patients with multiple myeloma (32), and has also been demonstrated to be an effective cell growth inhibitor in various cancer cell lines (33–36). However, the emergence of drug resistance has seriously affected the therapeutic effect of bortezomib. The overexpression of SOD1 has been shown to increase the resistance of cancer cells to bortezomib (31), and the results of the present study showed that combined sclareol and bortezomib treatment resulted in markedly lower protein levels of SOD1, compared with bortezomib treatment alone, which suggested that combined sclareol and bortezomib treatment may be a promising therapy in overcoming the challenge of bortezomib resistance.
Cav1 is important in Derlin-1- and p97-mediated cyclooxygenase-2 ubiquitination and degradation (19,20), and previous reports have shown that Cav1 enhances the ubiquitination and degradation of EGFR via endocytosis (21). However, how Cav1 promotes the suppressed expression of SOD1 remains to be fully elucidated. The present study found that inhibition of the lysosome rescued the decreased protein level of SOD1, which was induced by sclareol, thus Cav1 may interact with SOD1 to facilitate its degradation inside the lysosome. However, the protein level of SOD1 cannot be fully rescued by lysosome inhibition, which indicates Cav1 is not the only molecule to promote the degradation of SOD1 and that SOD1 is not only degraded by the lysosome-mediated pathway. Further detailed investigation is required to elucidate the exact mechanism involved.
In conclusion, the present study demonstrated the inhibitory effect of sclareol on the proliferation of HeLa cells. In addition, it was found that the anticancer effect of sclareol was enhanced when combined with bortezomib, which was demonstrated in various cancer cells. Further investigations revealed that the tumor suppressor, Cav1, enhanced the decreased expression of SOD1, which may be responsible for the anticancer effect of sclareol. These findings not only elucidated the potential mechanisms underlying the anticancer effect of sclareol, but also provided a novel and promising approach for cancer therapy and resolving drug resistance in cancer cells.
Acknowledgements
This study was supported by research grants from the Natural Science Foundation of Chengdu University (grant no. 2011XJZ14) and the Natural Science Foundation of China (grant no. 51402027).
References
- 1.Jemal A, Center MM, DeSantis C, Ward EM. Global patterns of cancer incidence and mortality rates and trends. Cancer Epidemiol Biomarkers Prev. 2010;19:1893–1907. doi: 10.1158/1055-9965.EPI-10-0437. [DOI] [PubMed] [Google Scholar]
- 2.Noori S, Hassan ZM, Salehian O. Sclareol reduces CD4+CD25+FoxP3+Treg cells in a breast cancer model in vivo. Iran J Immunol. 2013;10:10–21. [PubMed] [Google Scholar]
- 3.Noori S, Hassan ZM, Mohammadi M, Habibi Z, Sohrabi N, Bayanolhagh S. Sclareol modulates the Treg intra-tumoral infiltrated cell and inhibits tumor growth in vivo. Cellular Immunol. 2010;263:148–153. doi: 10.1016/j.cellimm.2010.02.009. [DOI] [PubMed] [Google Scholar]
- 4.Wang L, He HS, Yu HL, Zeng Y, Han H, He N, Liu ZG, Wang ZY, Xu SJ, Xiong M. Sclareol, a plant diterpene, exhibits potent antiproliferative effects via the induction of apoptosis and mitochondrial membrane potential loss in osteosarcoma cancer cells. Mol Med Rep. 2015;11:4273–4278. doi: 10.3892/mmr.2015.3325. [DOI] [PubMed] [Google Scholar]
- 5.Sashidhara KV, Rosaiah JN, Kumar A, Bid HK, Konwar R, Chattopadhyay N. Cell growth inhibitory action of an unusual labdane diterpene, 13-epi-sclareol in breast and uterine cancers in vitro. Phytotherapy Res. 2007;21:1105–1108. doi: 10.1002/ptr.2205. [DOI] [PubMed] [Google Scholar]
- 6.Dimas K, Hatziantoniou S, Tseleni S, Khan H, Georgopoulos A, Alevizopoulos K, Wyche JH, Pantazis P, Demetzos C. Sclareol induces apoptosis in human HCT116 colon cancer cells in vitro and suppression of HCT116 tumor growth in immunodeficient mice. Apoptosis. 2007;12:685–694. doi: 10.1007/s10495-006-0026-8. [DOI] [PubMed] [Google Scholar]
- 7.Simmons GE, Taylor HE, Hildreth JEK. Caveolin-1 suppresses human immunodeficiency virus-1 replication by inhibiting acetylation of NF-κB. Virology. 2012;432:110–119. doi: 10.1016/j.virol.2012.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Garrean S, Gao XP, Brovkovych V, Shimizu J, Zhao YY, Vogel SM, Malik AB. Caveolin-1 regulates NF-kappaB activation and lung inflammatory response to sepsis induced by lipopolysaccharide. J Immunol. 2006;177:4853–4860. doi: 10.4049/jimmunol.177.7.4853. [DOI] [PubMed] [Google Scholar]
- 9.Wang XM, Kim HP, Song R, Choi AM. Caveolin-1 confers antiinflammatory effects in murine macrophages via the MKK3/p38 MAPK pathway. Am J Respir Cell Mol Biol. 2006;34:434–442. doi: 10.1165/rcmb.2005-0376OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Huertas-Martinez J, Rello-Varona S, Herrero-Martin D, Barrau I, García-Monclús S, Sáinz-Jaspeado M, Lagares-Tena L, Núñez-Álvarez Y, Mateo-Lozano S, Mora J, et al. Caveolin-1 is down-regulated in alveolar rhabdomyosarcomas and negatively regulates tumor growth. Oncotarget. 2014;5:9744–9755. doi: 10.18632/oncotarget.2403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bender FC, Reymond MA, Bron C, Quest AF. Caveolin-1 levels are down-regulated in human colon tumors and ectopic expression of caveolin-1 in colon carcinoma cell lines reduces cell tumorigenicity. Cancer Res. 2000;60:5870–5878. [PubMed] [Google Scholar]
- 12.Belanger MM, Roussel E, Couet J. Caveolin-1 is down-regulated in human lung carcinoma and acts as a candidate tumor suppressor gene. Chest. 2004;125(Supply 5):S106. doi: 10.1378/chest.125.5_suppl.106S. [DOI] [PubMed] [Google Scholar]
- 13.Fischer LR, Igoudjil A, Magrané J, Li Y, Hansen JM, Manfredi G, Glass JD. SOD1 targeted to the mitochondrial intermembrane space prevents motor neuropathy in the Sod1 knockout mouse. Brain. 2011;134(Pt 1):196–209. doi: 10.1093/brain/awq314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Igoudjil A, Magrané J, Fischer LR, Kim HJ, Hervias I, Dumont M, Cortez C, Glass JD, Starkov AA, Manfredi G. In vivo pathogenic role of mutant SOD1 localized in the mitochondrial intermembrane space. J Neurosci. 2011;31:15826–15837. doi: 10.1523/JNEUROSCI.1965-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Somwar R, Erdjument-Bromage H, Larsson E, Shum D, Lockwood WW, Yang G, Sander C, Ouerfelli O, Tempst PJ, Djaballah H, Varmus HE. Superoxide dismutase 1 (SOD1) is a target for a small molecule identified in a screen for inhibitors of the growth of lung adenocarcinoma cell lines; Proc Natl Acad Sci USA; 2011; pp. 16375–16380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Glasauer A, Sena LA, Diebold LP, Mazar AP, Chandel NS. Targeting SOD1 reduces experimental non-small-cell lung cancer. J Clin Invest. 2014;124:117–128. doi: 10.1172/JCI71714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kirchner P, Bug M, Meyer H. Ubiquitination of the N-terminal region of caveolin-1 regulates endosomal sorting by the VCP/p97 AAA-ATPase. J Biol Chem. 2013;288:7363–7372. doi: 10.1074/jbc.M112.429076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hayer A, Stoeber M, Ritz D, Engel S, Meyer HH, Helenius A. Caveolin-1 is ubiquitinated and targeted to intralumenal vesicles in endolysosomes for degradation. J Cell Biol. 2010;191:615–629. doi: 10.1083/jcb.201003086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen SF, Wu CH, Lee YM, Tam K, Tsai YC, Liou JY, Shyue SK. Caveolin-1 interacts with Derlin-1 and promotes ubiquitination and degradation of cyclooxygenase-2 via collaboration with p97 complex. J Biol Chem. 2013;288:33462–33469. doi: 10.1074/jbc.M113.521799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen SF, Liou JY, Huang TY, Lin YS, Yeh AL, Tam K, Tsai TH, Wu KK, Shyue SK. Caveolin-1 facilitates cyclooxygenase-2 protein degradation. J Cell Biochem. 2010;109:356–362. doi: 10.1002/jcb.22407. [DOI] [PubMed] [Google Scholar]
- 21.Feldman R, Martinez JD. Growth suppression by ursodeoxycholic acid involves caveolin-1 enhanced degradation of EGFR. Biochim Biophys Acta. 2009;1793:1387–1394. doi: 10.1016/j.bbamcr.2009.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Qi L, Zhang XD, Wu JC, Lin F, Wang J, DiFiglia M, Qin ZH. The role of chaperone-mediated autophagy in huntingtin degradation. PloS One. 2012;7:e46834. doi: 10.1371/journal.pone.0046834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cuervo AM, Dice JF, Knecht E. A population of rat liver lysosomes responsible for the selective uptake and degradation of cytosolic proteins. J Biol Chem. 1997;272:5606–5615. doi: 10.1074/jbc.272.9.5606. [DOI] [PubMed] [Google Scholar]
- 24.Liang X, Wei SQ, Lee S, Fung JK, Zhang M, Tanaka A, Choi AM, Jin Y. p62 sequestosome 1/light chain 3b complex confers cytoprotection on lung epithelial cells after hyperoxia. Am J Respir Cell Mol Biol. 2013;48:489–496. doi: 10.1165/rcmb.2012-0017OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gómez-Sánchez R, Yakhine-Diop SM, Rodríguez-Arribas M, Bravo-San Pedro JM, Martínez-Chacón G, Uribe-Carretero E, de Castro DC Pinheiro, Pizarro-Estrella E, Fuentes JM, González-Polo RA. mRNA and protein dataset of autophagy markers (LC3 and p62) in several cell lines. Data Brief. 2016;7:641–647. doi: 10.1016/j.dib.2016.02.085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Trimmer C, Bonuccelli G, Katiyar S, Sotgia F, Pestell RG, Lisanti MP, Capozza F. Cav1 suppresses tumor growth and metastasis in a murine model of cutaneous SCC through modulation of MAPK/AP-1 activation. Am J Pathol. 2013;182:992–1004. doi: 10.1016/j.ajpath.2012.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Aarhus M, Bruland O, Saetran HA, Mork SJ, Lund-Johansen M, Knappskog PM. Global gene expression profiling and tissue microarray reveal novel candidate genes and down-regulation of the tumor suppressor gene CAV1 in sporadic vestibular schwannomas. Neurosurgery. 2010;67:998–1019. doi: 10.1227/NEU.0b013e3181ec7b71. [DOI] [PubMed] [Google Scholar]
- 28.Shi Y, Tan SH, Ng S, Zhou J, Yang ND, Koo GB, McMahon KA, Parton RG, Hill MM, Del Pozo MA, et al. Critical role of CAV1/caveolin-1 in cell stress responses in human breast cancer cells via modulation of lysosomal function and autophagy. Autophagy. 2015;11:769–784. doi: 10.1080/15548627.2015.1034411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Podar K, Shringarpure R, Tai YT, Simoncini M, Sattler M, Ishitsuka K, Richardson PG, Hideshima T, Chauhan D, Anderson KC. Caveolin-1 is required for vascular endothelial growth factor-triggered multiple myeloma cell migration and is targeted by bortezomib. Cancer Res. 2004;64:7500–7506. doi: 10.1158/0008-5472.CAN-04-0124. [DOI] [PubMed] [Google Scholar]
- 30.Mao M, Sudhahar V, Fukai T. P54: Nitroglycerin-induced loss of caveolin-1 results in eNOS dysfunction, peroxynitrite production and nitrate tolerance. Nitric Oxide 31, Supplement. 2013;1:S36. doi: 10.1016/j.niox.2013.02.056. [DOI] [Google Scholar]
- 31.Salem K, McCormick ML, Wendlandt E, Zhan F, Goel A. Copper-zinc superoxide dismutase-mediated redox regulation of bortezomib resistance in multiple myeloma. Redox Biol. 2015;4:23–33. doi: 10.1016/j.redox.2014.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen D, Frezza M, Schmitt S, Kanwar J, Dou QP. Bortezomib as the first proteasome inhibitor anticancer drug: Current status and future perspectives. Curr Cancer Drug Targets. 2011;11:239–253. doi: 10.2174/156800911794519752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kao C, Chao A, Tsai CL, Chuang WC, Huang WP, Chen GC, Lin CY, Wang TH, Wang HS, Lai CH. Bortezomib enhances cancer cell death by blocking the autophagic flux through stimulating ERK phosphorylation. Cell Death Dis. 2014;5:e1510. doi: 10.1038/cddis.2014.468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kretowski R, Stypulkowska A, Cechowska-Pasko M. Efficient apoptosis and necrosis induction by proteasome inhibitor: Bortezomib in the DLD-1 human colon cancer cell line. Mol Cell Biochem. 2015;398:165–173. doi: 10.1007/s11010-014-2216-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Miyamoto Y, Nakagawa S, Wada-Hiraike O, Seiki T, Tanikawa M, Hiraike H, Sone K, Nagasaka K, Oda K, Kawana K, et al. Sequential effects of the proteasome inhibitor bortezomib and chemotherapeutic agents in uterine cervical cancer cell lines. Oncol Rep. 2013;29:51–57. doi: 10.3892/or.2012.2072. [DOI] [PubMed] [Google Scholar]
- 36.Ando M, Hoyos V, Yagyu S, Tao W, Ramos CA, Dotti G, Brenner MK, Bouchier-Hayes L. Bortezomib sensitizes non-small cell lung cancer to mesenchymal stromal cell-delivered inducible caspase-9-mediated cytotoxicity. Cancer Gene Ther. 2014;21:472–482. doi: 10.1038/cgt.2014.53. [DOI] [PMC free article] [PubMed] [Google Scholar]