Abstract
We try to explore the value of aberrant DNA methylation of several cancer-related genes in plasma as non-invasive biomarkers for gastric cancer (GC) and precancerous lesions. By using methylation-specific polymerase chain reaction assay we determined the methylation status of three selected genes ZIC1, HOXD10 and RUNX3 in blood samples from patients with GC and precancerous lesions. We discovered that the methylation rate of ZIC1, HOXD10 and RUNX3 increased significantly in the progression of gastric carcinogenesis. Methylation of ZIC1 was associated with positive serum CA19-9, while that of HOXD10 was related to H. pylori status, serum CA19-9 and CEA levels and tumor invasion depth. The Odds ratios (ORs) of ZIC1, HOXD10 and RUNX3 methylation for predicting GC were 4.285 (95%CI: 2.435-7.542), 3.133 (95%CI: 1.700-5.775) and 2.674 (95%CI: 1.441-4.960), while for predicting “gastric cancer and intraepithelial neoplasia” (GnI), the ORs were 12.011 (95%CI: 0.050-28.564), 9.174 (95%CI: 3.220-26.135) and 12.794 (95%CI: 4.115-39.778), respectively. In terms of combined detection of these three genes, the sensitivity was 91.6% for GC and 89.8% for GnI, with the highest Youden index in both GC and GnI determination. Conclusively, combined detection of ZIC1, HOXD10 and RUNX3 promoter hypermethylation might be a promising strategy for early detection of GC and precancerous lesions.
Keywords: Gastric cancer, Zinc finger of the cerebellum 1, Homeobox D10, Runt-related transcription factor 3, DNA methylation
Introduction
Gastric cancer (GC) is one of most common malignancies and ranks the third leading cause of cancer-related mortality worldwide 1. The cascade of GC progress develops through chronic gastritis, atrophy, intestinal metaplasia (IM) and intraepithelial neoplasia (IN) before eventually evolving to GC 2. Gastric IM and IN are precancerous lesions that trigger at least 10-fold increase in the risk of developing GC 3, 4. Despite great improvements made in the prognosis of early gastric cancer (EGC), the survival outcomes of most GC patients were unsatisfactory since they were diagnosed at an advanced stage with a relatively poor survival outcome 5, 6. Therefore, earlier detection of GC and precancerous lesions would promote preferable management of the disease. Currently, gastroscopy with biopsy sampling is still the most effective approach for confirming GC. However, it is an invasive procedure and causes discomfort to many subjects. Frequently, patients are reluctant to undergo gastroscopy, which leads to delayed treatment. Conventional serological tumor biomarkers, such as carbohydrate antigen 19-9 (CA19-9) and carcinoembryonic antigen (CEA), may help but lack specificity and are not sensitive for identifying GC in its early stage or with precancerous lesions. Thus, new diagnostic methods with high sensitivity to detect early-stage malignancies in a non-invasive and cost-effective manner are definitely in need.
Both genetic and epigenetic alterations contribute to gastric carcinogenesis. The transcriptional silencing of tumor-suppressor genes (TSGs) through aberrant DNA methylation is thought to be a key epigenetic event in the origin of various cancers 7, 8. Accumulating data strongly suggested that methylated DNA in body fluids is detectable ahead of cancer diagnosis 9, 10. Techniques based on cell free DNA (cfDNA) circulating in body fluid of patients represent a promising alternative for GC screening. Our previous studies have discovered a panel of TSGs, including zinc finger of the cerebellum 1 (ZIC1), Klotho, Homeobox D10 (HOXD10) genes that were silenced epigenetically in GC as well as precancerous lesions 11-13. Meanwhile, we detected aberrant DNA methylation of ZIC1 in the blood samples from patients with GC and IN, indicating its potential role for early detection of GC and precancerous lesions in a non-invasive way 14. However, the sensitivity of a single gene methylation is still unsatisfying, combined detection of several genes might be a decent solution.
To our knowledge, few studies investigated aberrant DNA methylation in blood samples from GCs, in parallel with samples from patients with precancerous lesions and from healthy individuals. In this paper, we embark upon identifying potential biomarkers for the early diagnosis of GC and precancerous lesions. We firstly conducted a selection study and chose ZIC1, HOXD10 and runt-related transcription factor 3 (RUNX3) for further validation, then we examined the methylation status of the three genes in plasma from patients with advanced gastric cancer (AGC), EGC, IN, IM and normal controls (NCs). The correlation between promoter hypermethylation of these genes and clinical data was analyzed. Besides, the association of genes methylation status with histopathologic characteristics in GCs was also estimated. Finally, we categorized the subjects with GC and IN in a gastric cancer and intraepithelial neoplasia (GnI) group and compared the diagnostic and predictive ability of single detection or combination of plasma ZIC1, HOXD10 and RUNX3 promoter hypermethylation in both GC and GnI groups.
Materials and methods
Patients and blood samples
Blood samples were collected from GC, IN and IM patients diagnosed at Sir Run Run Shaw Hospital from June 2013 to December 2015. All subjects underwent gastroendoscopy and the exclusion criteria were shown as follows: patients who had gastrointestinal disorders with other causes; those with other organic diseases or gastric metastases from other malignancies; patients who had already received surgical treatment or preoperative chemotherapy or radiation therapy. We also recruited samples from healthy volunteers as NCs. Blood was prepared with Ethylenediaminetetraacetic acid (EDTA) and plasma was separated immediately by centrifuging blood at 2500 × rpm for 15 minutes. Then, samples were grouped based on the histopathological diagnosis. When multiple pathological changes occur in one case, the diagnosis is subject to the lesion with higher grade. The tumor stage of GCs was classified according to the American Joint Committee on Cancer (AJCC) staging manual 15.
All participants provided written informed consent before enrolment. The study protocol was approved by the Clinical Research Ethics Committee of the Institute of Gastroenterology of Zhejiang University.
DNA Isolation and bisulfite modification
DNA was extracted from plasma sample (400 μL) according to the instruction of the QIAamp DNA Blood Mini Kit (Qiagen, Hilden, Germany). The extracted DNA was dissolved in 50 μL of elution buffer and stored at -20°C. Bisulfite conversion was subsequently performed using the EZ DNA Methylation-Gold Kit (Zymo Research, Orange, CA, USA). The modified DNA was resuspended in 10 μL of elution buffer and stored at -20°C until use.
Methylation-specific polymerase chain reaction (MSP)
The promoter hypermethylation of ZIC1, HOXD10 and RUNX3 was detected by MSP assays utilizing the above-mentioned bisulfite-modified DNA as templates. DNA from BGC-823 (gastric cancer cell line) was used as a positive control and nuclease-free water was used as a negative control. The methylation-specific primer sequences and corresponding annealing temperatures are listed in Table S1. The PCR protocol was provided previously 14.
Evaluation of serum CEA, CA19-9 and H. pylori status
Serum CEA and CA19-9 levels were evaluated by Enzyme-linked immunosorbent assay. CEA and CA19-9 were considered positive as ≥5 ng/mL and ≥40 U/mL, respectively. H. pylori infection was diagnosed when there were positive results for both RUT and 13C-urea breath test (UBT) or histological analysis.
Statistical Analyses
All statistical analyses were performed by SPSS 20.0 software (IBM, SPSS, Chicago, IL, USA). Continuous variables were shown as mean±SD and compared by Student's t test, while categorical data were checked by Chi-square test or Fisher's exact test where appropriate. The correlation between the clinical features and DNA hypermethylation was calculated by the Chi-square test. The sensitivity and specificity with 95% confidence intervals (CIs) of DNA methylation assays of specific genes were analyzed. For predicting the risk of GC or precancerous lesions, we used odds ratios (ORs) with 95%CIs as a measure of association. Youden index was calculated to assess the accuracy of plasma DNA assays. All P values were two-sided and P<0.05 was considered statistically significant.
Results
Patient characteristics
20 samples from 10 GCs and 10 NCs were used for the selection study. Then, a total of 251 cases were enrolled in the validation study, including 131 GCs (91 AGC and 40 EGC), 56 IN patients, 30 IM patients and 34 NCs. The mean age of the GC, IN, IM and NC group was 61.1, 59.6, 57.8 and 56.5 (years), respectively. The gender ratio (male: female) was 88:43 in GC, 37:19 in IN, 3:2 in IM and 11:6 in NC group. No significant differences were found in age and sex distinction between the four groups (Table 1).
Table 1.
The general characteristics of the study subjects.
| GC (n=131) | IN (n=56) | IM (n=30) | NC (n=34) | P value | ||
|---|---|---|---|---|---|---|
| Age | 61.1±10.77 | 59.6±10.07 | 57.8±9.55 | 56.5±10.55 | 0.086 | |
| Gender | Male | 88 | 37 | 18 | 22 | 0.902 |
| Female | 43 | 19 | 12 | 12 |
GC, early gastric cancer; IN, intraepithelial neoplasia; IM, intestinal metaplasia; NC, normal control.
Selection of candidate genes
Seven tumor-related genes, including p16 (also known as CDKN2A), death-associated protein kinase (DAPK), ZIC1, klotho, HOXD10, RUNX3 and Ras association domain family 1A (RASSF1A) were reported to have frequent promoter hypermethylation in GC 11-13, 16-19. Firstly, we examined the methylation status of all 7 genes in the plasma of 10 GCs and 10 NCs. p16 and klotho showed frequent hypermethylation of 40% (4/10) and 30% (3/10) in GC cases but were also aberrantly methylated in NCs, which was regarded as an exclusion criterion. None of the plasma samples tested showed methylation in RASSF1A. Therefore, p16, klotho and RASSF1A were excluded from the selection. ZIC1 had the highest frequency of methylation (70%, 7/10) in the plasma of GCs with a lack of methylation in NCs. The methylation frequency of DAPK, HOXD10, RUNX3 in GCs was 20% (2/10), 50% (5/10) and 40% (4/10), respectively, and none of them showed methylation in NCs. From the perspective of predictive power for GC, we selected ZIC1, HOXD10 and RUNX3 as the potential markers for detection of GC and conducted a validation study with larger sample sizes for further investigation.
Frequencies of aberrant ZIC1, HOXD10 and RUNX3 promoter methylation in plasma DNA
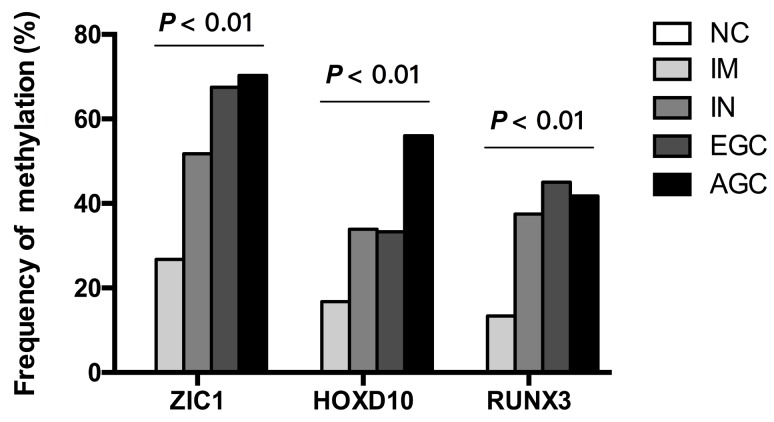
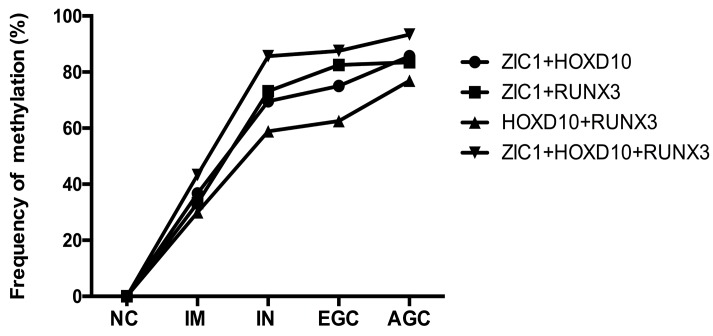
Then, we examined the methylation status of ZIC1, HOXD10 and RUNX3 in expanded 251 cases. Methylation rate (MR) of ZIC1, HOXD10 and RUNX3 in each group was shown in Figure 1. MRs increased significantly in the progression of gastric carcinogenesis from NC to IM to IN to GC samples for all 3 genes (all P<0.01 for IM, IN, EGC and AGC vs. NC). For ZIC1 and HOXD10, higher MR was found in AGC compared to EGC samples, while the MR of RUNX3 in plasma was higher in EGCs, rather than AGCs. For the NC group, none of the 3 genes were methylated in plasma DNA. In other words, the specificity of each individual biomarker was 100% for differentiating the GC and precancerous lesions from the NCs. Also, the frequencies of aberrant methylation in combined detection of multi-genes were analyzed (Figure 2, all P<0.01 for IM, IN, EGC and AGC vs. NC).
Figure 1.
Frequency of detecting methylated DNA in the plasma of AGC, EGC, IN, IM patients and NCs. ZIC1, Zinc finger of the cerebellum 1; HOXD10, Homeobox D10; RUNX3, runt-related transcription factor 3; NC, normal control; IM, intestinal metaplasia; IN, intraepithelial neoplasia; EGC, early gastric cancer; AGC, advanced gastric cancer.
Figure 2.
The methylation rates of multi-genes detection in plasma samples according to gastric neoplastic progression. ZIC1, Zinc finger of the cerebellum 1; HOXD10, Homeobox D10; RUNX3, runt-related transcription factor 3; NC, normal control; IM, intestinal metaplasia; IN, intraepithelial neoplasia; EGC, early gastric cancer; AGC, advanced gastric cancer; +, parallel testing of multi-genes.
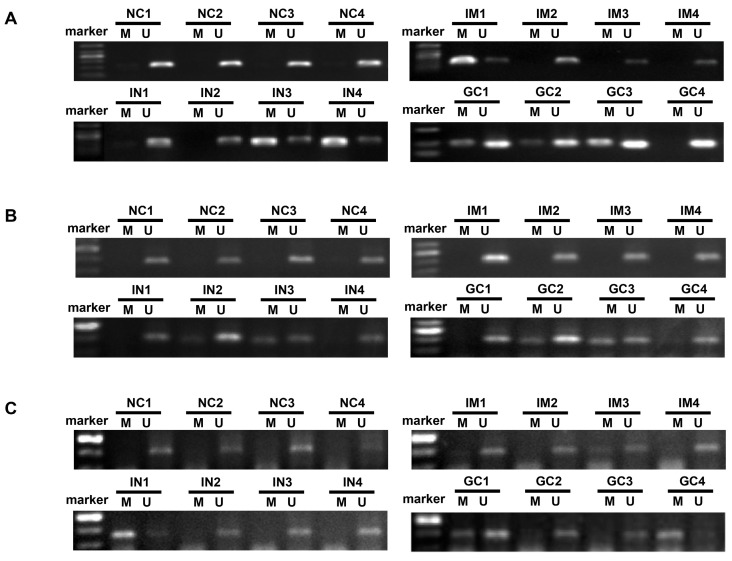
The mean number of genes methylated in IM, IN, EGC, and GC was 0.6, 1.2, 1.4 and 1.6, respectively. Concurrent methylation in 3 genes was found in none of IM, 7.1% of IN, 10% of EGC, and 13.7% of GC. Representative agarose gel electrophoresis results of the MSP for the three genes are shown in Figure 3.
Figure 3.
Representative agarose gel electrophoresis results of the methylation-specific polymerase chain reaction assay for ZIC1, HOXD10 and RUNX3 in plasma DNA (A, ZIC1; B, HOXD10; C, RUNX3). ZIC1, Zinc finger of the cerebellum 1; HOXD10, Homeobox D10; RUNX3, runt-related transcription factor 3; NC, normal control; IM, intestinal metaplasia; IN, intraepithelial neoplasia; GC, gastric cancer; M: Methylated-specific primers; U: Unmethylated-specific primers.
Relationship between DNA methylation and clinicopathological characteristics
We analyzed the correlation of DNA methylation of each gene with clinical data (age, sex, H. pylori infection and serum CEA/ CA19-9 level) of all cases (Table 2). There was no association between promoter hypermethylation of the 3 genes and age or gender. Methylation of ZIC1 was associated with positive serum CA19-9 (P<0.05). Methylation of HOXD10 was more frequent in positive H. pylori infection cases (P<0.05), in serum CA19-9 positive cases (P<0.01) and in serum CEA positive cases (P<0.01). However, no correlation was found between RUNX3 hypermethylation and H. pylori status or serum CA19-9 and CEA levels.
Table 2.
Correlation between methylation in plasma ZIC1, HOXD10 and RUNX3 and clinical parameters of all cases.
| Parameters | ZIC1 | HOXD10 | RUNX3 | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| M | U | P value | M | U | P value | M | U | P value | ||
| Age | 60.21±10.33 | 59.26±10.75 | 0.476 | 60.83±10.03 | 59.17±10.77 | 0.236 | 59.98±10.48 | 59.64±10.58 | 0.811 | |
| Gender | Male | 85 | 80 | 0.820 | 60 | 105 | 0.432 | 54 | 111 | 0.830 |
| Female | 43 | 43 | 27 | 59 | 27 | 59 | ||||
| H.pylori | Yes | 32 | 22 | 0.206 | 26 | 28 | 0.011 | 16 | 38 | 0.461 |
| No | 86 | 88 | 51 | 123 | 61 | 113 | ||||
| Serum CA19-9 | Positive | 13 | 4 | 0.026 | 11 | 6 | 0.007 | 8 | 9 | 0.177 |
| Negative | 104 | 111 | 70 | 145 | 67 | 148 | ||||
| Serum CEA | Positive | 22 | 12 | 0.072 | 20 | 14 | 0.002 | 12 | 22 | 0.689 |
| Negative | 95 | 103 | 61 | 137 | 63 | 135 | ||||
ZIC1, Zinc finger of the cerebellum 1; HOXD10, Homeobox D10; RUNX3, runt-related transcription factor 3; M = Methylated; U = Unmethylated.
We further investigated the association of the histopathologic findings with individual gene methylation in the plasma DNA from GCs (Table S2). The results revealed no marked associations between the methylation status of ZIC1, RUNX3 promoter and the histopathologic features, including tumor location, size, grade, invasion depth, lymph node metastasis, distant metastasis and TNM staging. Interestingly, we discovered that the methylation status of HOXD10 promoter was positively correlated with tumor invasion depth (P<0.05); no correlations were found with other characteristics.
Effect of the combined detection of plasma ZIC1, HOXD10 and RUNX3 methylation for GC and GnI determination
Multivariate regression analyses were performed to assess the predictive power of DNA methylation detection of plasma ZIC1, HOXD10 and RUNX3 for GC and GnI. The ORs of plasma ZIC1, HOXD10 and RUNX3 methylation for predicting the presence of GC were 4.285 (95%CI: 2.435-7.542; P<0.01), 3.133 (95%CI: 1.700-5.775; P<0.01) and 2.674(95%CI: 1.441-4.960; P<0.01), respectively. However, the OR for predicting GnI was higher for RUNX3 methylation (OR=12.794; 95%CI: 4.115-39.778; P<0.01) than for ZIC1 (OR=12.011; 95%CI: 5.050-28.564; P<0.01) or HOXD10 methylation (OR=9.174; 95%CI: 3.220-26.135; P<0.01).
In order to investigate whether combined detection of the three genes was superior for early GC determination, we analyzed the sensitivity and specificity of the methylation detection with the use of singe or multi-gene panels; Youden index was calculated to estimate the accuracy of the plasma methylation assays (Table 3). Though the specificity of combined detection was lower than that of single gene assays, the sensitivity for GC and GnI increased when 2 or 3 genes were detected together. Youden index of combined detection of 3 genes (parallel testing) was highest in both GC and GnI determination.
Table 3.
Comparison of the diagnostic and predictive ability of plasma ZIC1, HOXD10 and RUNX3 methylation, when used alone or combined, in gastric cancer and intraepithelial neoplasia.
| Sensitivity (95% CI) | Specificity (95%CI) | Youden index | |
|---|---|---|---|
| GC | |||
| ZIC1 | 69.5% (60.7%-77.0%) | 69.2% (60.0%-77.1%) | 0.386 |
| HOXD10 | 48.1% (39.3%-57.0%) | 80.0% (71.5%-86.5%) | 0.281 |
| RUNX3 | 42.7% (34.2%-51.7%) | 79.2% (70.6%-85.8%) | 0.219 |
| ZIC1+HOXD10 | 82.4% (74.6%-88.3%) | 58.3% (49.0%-67.2%) | 0.408 |
| ZIC1+RUNX3 | 83.2% (75.5%-89.0%) | 57.5% (48.1%-66.4%) | 0.407 |
| HOXD10+RUNX3 | 72.5% (63.9%-79.8%) | 65.0% (55.7%-73.3%) | 0.375 |
| ZIC1+HOXD10+RUNX3 | 91.6% (85.1%-95.5%) | 50.0% (40.8%-59.2%) | 0.416 |
| GnI | |||
| ZIC1 | 64.2% (56.8%-70.9%) | 87.5% (76.3%-94.1%) | 0.517 |
| HOXD10 | 43.9% (36.7%-51.3%) | 92.2% (82.0%-97.1%) | 0.360 |
| RUNX3 | 41.2% (34.1%-48.6%) | 93.8% (84.0%-98.0%) | 0.349 |
| ZIC1+HOXD10 | 78.6% (71.9%-84.1%) | 82.8% (70.9%-90.7%) | 0.614 |
| ZIC1+RUNX3 | 80.2% (73.6%-85.5%) | 84.4% (72.7%-91.9%) | 0.646 |
| HOXD10+RUNX3 | 68.4% (61.2%-74.9%) | 85.9% (74.5%-93.0%) | 0.544 |
| ZIC1+HOXD10+RUNX3 | 89.8% (84.4%-93.6%) | 79.7% (67.4%-88.3%) | 0.695 |
ZIC1, Zinc finger of the cerebellum 1; HOXD10, Homeobox D10; RUNX3, runt-related transcription factor 3; GC, gastric cancer; GnI, gastric cancer and intraepithelial neoplasia.
Discussions
Aberrant promoter methylation of TSGs, which leads to silencing of genes, occurs early in the course of gastric carcinogenesis. Early detection yields the opportunity for improved survival rate and less invasive treatment. The notion of utilizing molecular approaches to discover epigenetic aberrations in circulating DNA have been considered as simple and non-invasive ways for GC diagnosis in large-scale populations. Recently, several genes such as DAPK, E-cadherin, GSTP1, p16 and XAF1 have been found methylated in the tumor and blood of GC patients 18, 20. In our previous investigation of ZIC1 promoter hypermethylation in the plasma, we found that the MR of ZIC1 was 54.0% in the IN group and 60.6% in the GC group, indicating ZIC1 a potential biomarker in early diagnosis of GC and precancerous lesions. However, the sensitivity of individual gene is limited. The combination of several biomarkers achieves greater sensitivity, and thus has been an emerging field in diagnosis.
Firstly, we selected three genes, including ZIC1, HOXD10 and RUNX3, from seven frequently methylated TSGs (p16, DAPK, ZIC1, klotho, HOXD10, RUNX3, and RASSF1A) for further investigation. ZIC1, a vital transcription factor with zinc finger domains, is known to be a tumor suppressor in gastrointestinal cancers and modulate GC cell-cycle distributions and cell migration 21. HOXD10, one of the homeobox (Hox) superfamily genes, plays critical roles in the differentiation of embryonic cells and progression of GC, and suppresses GC cell invasion by targeting IGFBP3 22. RUNX3, a member of the human runt-related transcription factors, is an important target of transforming growth factor-β (TGF-β) superfamily signaling and play crucial roles not only in normal development, but also in carcinogenesis, especially stomach cancers 23, 24. We and others have previously identified ZIC1, HOXD10 and RUNX3 as novel TSGs silenced through promoter hypermethylation in GC tissues, with the methylation frequencies of 94.6% (35/37), 83.5% (81/97) and 96% (43/45), respectively 12, 13, 16. The combined application of ZIC1, HOXD10 and RUNX3 in blood DNA for GC and precancerous lesions detection is largely known.
In the present study, we found the MRs of ZIC1, HOXD10 and RUNX3 were 69.5%, 48.1% and 42.7% respectively in plasma from GCs. The fact that the MRs in plasma samples were lower than those in tissue samples may be illustrated by the explanation that tumor DNA gets into systemic circulation and then the methylation status of plasma DNA was detectable 25. However, the exact mechanism about the origin of cfDNA in circulation remains unclear for the moment. Meanwhile, the absence of methylation occurred in all 3 selected genes from plasma DNA in NC cases, which is similar to the findings of other previous studies conducted in tissue samples of NCs 12-14. Moreover, the MRs of all 3 genes increased with the histological progression from NCs, to IM, IN and GC, revealing that aberrant promoter methylation were gradually accumulated in GC development and detectable in the early stages of disease. We then analyzed the association of plasma DNA methylation with clinicopathological features. ZIC1 and HoxD10 methylation status in plasma was related to serum CA19-9/ CEA levels, which suggested that combination of promoter methylation of ZIC1 or HOXD10 with conventional serological tumor markers might be of great significance for further study. We found no marked associations between promoter methylation of ZIC1, RUNX3 and the histopathologic features of gastric tumor. Surprisingly, the presence of methylated HOXD10 in plasma was found to be associated with deeper tumor invasion, which is consistent to previous findings that ectopic expression of HOXD10 impaired GC cell invasion and HOXD10 promoter hypermethylation detected in GC tissues was associated with the TNM staging and indicated poor prognosis 12. Also, HOXD10 may be a marker in advanced stages, showing high levels of methylation in AGC but much lower levels in EGC.
Combined measurement of ZIC1, HOXD10 and RUNX3 methylation in plasma showed improved sensitivity and accuracy for early screening GC and precancerous lesions. Patients with IN lesions are more inclined to develop GC and advanced IN was considered as a warning sign. However, a very small proportion of biomarkers were validated for IN determination. Our results showed that ZIC1, HOXD10 and RUNX3 individually exhibited high risk associated with GC or GnI, and when detected together the sensitivity was improved to about 90% for GC and GnI, with the highest testing accuracy. It indicated that the combination of plasma ZIC1, HOXD10 and RUNX3 hypermethylation served as the most effective predictor of IN and GC. Since it takes a long time for the progression from precancerous lesions to GC, there may be a window of opportunity for discovering and curing advanced IN or EGC. With the help of combined methylation assays, it would be promising for early intervention in the stage of precancerous lesions and monitoring of gastric carcinogenesis. Further studies with other tools for mapping methylation aberrations are still required to estimate ZIC1, HOXD10, and RUNX3 as useful biomarkers for early detection of GC and precancerous lesions.
In conclusion, aberrant promoter methylation of TSGs in body fluids has been considered to be a decent solution for the diagnosis of various cancers. Our findings demonstrated that the promoter MRs of ZIC1, HOXD10 and RUNX3 in plasma DNA was increased with the progression from precancerous lesions to GC. Combined assays of detecting plasma ZIC1, HOXD10 and RUNX3 methylation may be of great assistance in early detection of GC and in risk evaluation of high-risk populations.
Supplementary Material
Supplementary tables.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (81372623); the Zhejiang province key science and technology innovation team (2013TD13); and the Zhejiang Provincial Medical and health research plan (2015126452; 201476310; 2014KYB121).
Author contributions
ZHL, SJC and JMS conceived of and designed the study. ZHL, MZL and XQC performed the analyses. ZHL wrote the main manuscript. ZHL, XKH, QY and SCL prepared all figures and tables. All authors reviewed the manuscript.
Abbreviations
- TSG
Tumor-suppressor gene
- GC
Gastric cancer
- IN
Intraepithelial neoplasia
- IM
Intestinal metaplasia
- NC
Normal control
- MSP
Methylation-specific polymerase chain reaction
- MR
Methylation rate
- AGC
Advanced gastric cancer
- EGC
Early gastric cancer
- GnI
gastric cancer and intraepithelial neoplasia
- CEA
Carcinoembryonic antigen
- CA19-9
Carbohydrate antigen 19-9
- cfDNA
cell free DNA
- ZIC1
Zinc finger of the cerebellum 1
- HOXD10
Homeobox D10
- RUNX3
Runt-related transcription factor 3
- RASSF1A
Ras association domain family 1A
- OR
Odds ratio
- 95%CI
95% confidence interval
- DAPK
Death-associated protein kinase
- TGF-β
Transforming growth factor-β
- RUT
Rapid Urease Test
- UBT
13C-urea breath test.
References
- 1.Ferlay J, Soerjomataram I, Dikshit R, Eser S, Mathers C, Rebelo M. et al. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. International journal of cancer Journal international du cancer. 2015;136:E359–86. doi: 10.1002/ijc.29210. [DOI] [PubMed] [Google Scholar]
- 2.Correa P. Human gastric carcinogenesis: a multistep and multifactorial process-First American Cancer Society Award Lecture on Cancer Epidemiology and Prevention. Cancer research. 1992;52:6735–40. [PubMed] [Google Scholar]
- 3.Filipe MI, Munoz N, Matko I, Kato I, Pompe-Kirn V, Jutersek A. et al. Intestinal metaplasia types and the risk of gastric cancer: a cohort study in Slovenia. International journal of cancer Journal international du cancer. 1994;57:324–9. doi: 10.1002/ijc.2910570306. [DOI] [PubMed] [Google Scholar]
- 4.You WC LJ, Blot WJ, Chang YS, Jin ML, Gail MH, Zhang L, Liu WD, Ma JL, Hu YR, Mark SD, Correa P, Joseph F. Fraumeni, JR. and Xu GW. Evolution of precancerous lesions in a rural Chinese population at high risk of gastric cancer. International journal of cancer Journal international du cancer. 1999;83:615–9. doi: 10.1002/(sici)1097-0215(19991126)83:5<615::aid-ijc8>3.0.co;2-l. [DOI] [PubMed] [Google Scholar]
- 5.Soetikno R, Kaltenbach T, Yeh R, Gotoda T. Endoscopic mucosal resection for early cancers of the upper gastrointestinal tract. J Clin Oncol. 2005;23:4490–8. doi: 10.1200/JCO.2005.19.935. [DOI] [PubMed] [Google Scholar]
- 6.Hartgrink HH, Jansen EP, van Grieken NC, van de Velde CJ. Gastric cancer. Lancet. 2009;374:477–90. doi: 10.1016/S0140-6736(09)60617-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Suva ML, Riggi N, Bernstein BE. Epigenetic reprogramming in cancer. Science. 2013;339:1567–70. doi: 10.1126/science.1230184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Qu Y, Dang S, Hou P. Gene methylation in gastric cancer. Clin Chim Acta. 2013;424:53–65. doi: 10.1016/j.cca.2013.05.002. [DOI] [PubMed] [Google Scholar]
- 9.Danese E, Minicozzi AM, Benati M, Montagnana M, Paviati E, Salvagno GL. et al. Epigenetic alteration: new insights moving from tissue to plasma - the example of PCDH10 promoter methylation in colorectal cancer. British journal of cancer. 2013;109:807–13. doi: 10.1038/bjc.2013.351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang X, Song YF, Lu HN, Wang DP, Zhang XS, Huang SL. et al. Combined detection of plasma GATA5 and SFRP2 methylation is a valid noninvasive biomarker for colorectal cancer and adenomas. World journal of gastroenterology: WJG. 2015;21:2629–37. doi: 10.3748/wjg.v21.i9.2629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang LJ, Wang X, Wang XJ, Jie P, Lu HQ, Zhang SJ. et al. Klotho is silenced through promoter hypermethylation in gastric cancer. American Journal of Cancer Research. 2011;1:111–9. [PMC free article] [PubMed] [Google Scholar]
- 12.Wang L, Chen S, Xue M, Zhong J, Wang X, Gan L. et al. Homeobox D10 gene, a candidate tumor suppressor, is downregulated through promoter hypermethylation and associated with gastric carcinogenesis. Molecular medicine. 2012;18:389–400. doi: 10.2119/molmed.2011.00172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang LJ, Jin HC, Wang X, Lam EKY, Bin Zhang J, Liu X. et al. ZIC1 is downregulated through promoter hypermethylation in gastric cancer. Biochem Bioph Res Co. 2009;379:959–63. doi: 10.1016/j.bbrc.2008.12.180. [DOI] [PubMed] [Google Scholar]
- 14.Chen X, Lin Z, Xue M, Si J, Chen S. Zic1 Promoter Hypermethylation in Plasma DNA Is a Potential Biomarker for Gastric Cancer and Intraepithelial Neoplasia. PloS one. 2015;10:e0133906. doi: 10.1371/journal.pone.0133906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Edge SB, Compton CC. The American Joint Committee on Cancer: the 7th edition of the AJCC cancer staging manual and the future of TNM. Annals of surgical oncology. 2010;17:1471–4. doi: 10.1245/s10434-010-0985-4. [DOI] [PubMed] [Google Scholar]
- 16.Homma N. Spreading of methylation within RUNX3 CpG island in gastric cancer. Cancer Sci; 2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fan XY, Hu XL, Han TM, Wang NN, Zhu YM, Hu W. et al. Association between RUNX3 promoter methylation and gastric cancer: a meta-analysis. BMC gastroenterology. 2011;11:92. doi: 10.1186/1471-230X-11-92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee TL, Leung WK, Chan MW, Ng EK, Tong JH, Lo KW. et al. Detection of Gene Promoter Hypermethylation in the Tumor and Serum of Patients with Gastric Carcinoma. Clinical cancer research: an official journal of the American Association for Cancer Research. 2002;8:1761–6. [PubMed] [Google Scholar]
- 19.Ye M, Xia B, Guo Q, Zhou F, Zhang X. Association of diminished expression of RASSF1A with promoter methylation in primary gastric cancer from patients of central China. Bmc Cancer. 2007;7:120. doi: 10.1186/1471-2407-7-120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ling ZQ, Lv P, Lu XX, Yu JL, Han J, Ying LS. et al. Circulating Methylated XAF1 DNA Indicates Poor Prognosis for Gastric Cancer. PloS one. 2013;8:e67195. doi: 10.1371/journal.pone.0067195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhong J, Chen SJ, Xue M, Du Q, Cai JT, Jin HC, ZIC1 modulates cell-cycle distributions and cell migration through regulation of sonic hedgehog, PI3K and MAPK signaling pathways in gastric cancer. Bmc Cancer; 2012. p. 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xue M, Fang Y, Sun G, Zhuo W, Zhong J, Qian C. et al. IGFBP3, a transcriptional target of homeobox D10, is correlated with the prognosis of gastric cancer. PloS one. 2013;8:e81423. doi: 10.1371/journal.pone.0081423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li QL, Ito K, Sakakura C, Fukamachi H, Inoue K, Chi XZ. et al. Causal relationship between the loss of RUNX3 expression and gastric cancer. Cell. 2002;109:113–24. doi: 10.1016/s0092-8674(02)00690-6. [DOI] [PubMed] [Google Scholar]
- 24.Levanon D, Bettoun D, Harris-Cerruti C, Woolf E, Negreanu V, Eilam R. et al. The Runx3 transcription factor regulates development and survival of TrkC dorsal root ganglia neurons. Embo J. 2002;21:3454–63. doi: 10.1093/emboj/cdf370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gormally E, Caboux E, Vineis P, Hainaut P. Circulating free DNA in plasma or serum as biomarker of carcinogenesis: practical aspects and biological significance. Mutation research. 2007;635:105–17. doi: 10.1016/j.mrrev.2006.11.002. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary tables.