Abstract
The aim of the present study was to detect the expression of the key molecules, including transforming growth factor-β1 (TGF-β1), phosphatidylinositol 3-kinase (PI3K), protein kinase B (Akt) of TGF-β1/mammalian target of rapamycin (mTOR) pathway in pathological scar fibroblasts. Immunofluorescence, reverse transcription-polymerase chain reaction (RT-PCR) and western blot analysis were used to detect the expression of the key molecules TGF-β1, PI3K, Akt, mTOR in fibroblasts of normal skin tissue and pathological scar tissue. Immunofluorescence showed that the expression of TGF-β1, PI3K and Akt was significantly enhanced (P<0.05) in pathological scar fibroblasts, and mainly expressed in the cell nucleus, but not in normal skin tissue or fibroblasts. RT-PCR and western blot test results revealed that the TGF-β1, PI3K, Akt, and mTOR mRNA and protein expression in pathological scar fibroblasts were significantly higher (P<0.05) than in the normal skin tissue. Expression of the TGF-β1/mTOR signaling pathway in pathological scar fibroblasts was significantly increased. Data suggest that this expression may be an important mechanism for pathological scar formation.
Keywords: pathological scar, fibroblasts, TGF-β1/mTOR signaling pathway
Introduction
Pathological scar is the pathological product formed during the process of wound healing. As typical fiber hyperplastic disease, its pathological changes are mainly characterized by massive proliferation of fibroblasts and excessive deposition of extracellular matrix dominated by collagen (1). Since the pathogenesis of pathological scar is intricate and complex, it has yet to be elucidated. However, the excessive proliferation of fibroblasts and its increasing activity play a critical role during its formation. The proliferation and activation of pathological scar fibroblasts are regulated and controlled by various cytokines, among which transforming growth factor-β1 (TGF-β1) is the cytokine recognized to have the closest relationship with the formation of scars at present (2). The transduction process of cell signals is inseparable to the development of biological activity of TGF-β1. An increasing number of studies have been focused on the typical TGF-β1/Smads signal pathway. However, the underlying mechanism of the proliferation of pathological scar fibroblasts remains to be determined.
In recent years, it was shown that various independent Smad pathways, in addition to the typical Smad pathway, have been involved in the transduction process of TGF-β1 signals. Additionally, some of these pathways were directly or indirectly involved tumor formation and were closely related to the proliferation, invasion and transfer of tumor (3). Pathologically, pathological scar belongs to benign solid tumor. Keloid can invasively grow to surrounding normal tissues and its pathological characteristics are similar to that of tumor, with tumor-like characteristics (4). Therefore, it is of great significance to implement studies on pathological scar from signal pathway closely associated with tumor.
Previous findings have shown that mammalian target of rapamycin (mTOR) was closely related to excessive cell proliferation and the occurrence of tumor and significantly strengthened its expression in various tumor tissues (5). The mTOR pathway plays an important role in the process of TGF-β1-mediated epithelial-mesenchymal transition (EMT) (6). Therefore, the TGF-β1/mTOR signal pathway also plays an important role during the process of the proliferation of pathological scar fibroblasts. To examine this hypothesis, the present study applied immunofluorescence, reverse transcription-polymerase chain reaction (RT-PCR), western blotting and other technologies to detect the expression of the key molecules of TGF-β1/mTOR pathway in pathological scar fibroblasts to clarify the mechanism related to the proliferation of pathological scar fibroblasts.
Materials and methods
Specimen source
The specimens were pathological scar tissue after surgical resection for treatment at the Department of Dermatology of Xuzhou Hospital of Traditional Chinese Medicine (Jiangsu, China), and normal skin tissue was resected from patients with redundant prepuce at the Department of Urology Surgery of Xuzhou Hospital of Traditional Chinese Medicine.
Main reagents
D-Hank's liquid (Shanghai Kang Lang Biological Technology Co., Ltd., Shanghai, China), 0.25% trypsin (Shanghai Bioleaf Biotech Co., Ltd., Shanghai, China), fetal bovine serum (Shanghai Bioleaf Biotech Co., Ltd.), Triton X-100 (Shanghai Lianshuo Biological Technology Co., Ltd., Shanghai, China), reverse transcription kit superscript III (Invitrogen, Carlsbad, CA, USA), SDS-PAGE gel preparation kit (Shanghai Solarbio Bioscience & Technology Co., Ltd., Shanghai, China), rabbit anti-human TGF-β1 monoclonal antibody (Wuhan Boster Biological Technology Co., Ltd., Wuhan, China), rabbit anti-human monoclonal antibody (Wuhan Boster Biological Technology Co., Ltd.), rabbit anti-human protein kinase B (Akt) monoclonal antibody (Wuhan Boster Biological Technology Co., Ltd.); rabbit anti-human mTOR monoclonal antibody (Wuhan Boster Biological Technology Co., Ltd.), and goat anti-rabbit secondary antibody (Jackson ImmunoResearch Inc., West Grove, PA, USA) labeled by fluorescein isothiocyanate were used in the study.
Main instruments
Instruments used included: Optical microscope (Olympus Corp., Tokyo, Japan), CO2 incubator (Binder GmbH, Tuttlingen, Germany), laser confocal microscope with high sensitivity (Carl Zeiss, Inc., Oberkochen, Germany), PCR instrument (Thermo Fisher Scientific, Waltham, MA, USA), centrifuge with low temperature and high speed (Thermo Fisher Scientific), Image Pro-Plus image analysis system (Media Cybernetics, Inc., Rockville, MD, USA), infrared fluorescence imaging instrument (LI-COR Biosciences, Lincoln, NE, USA).
In vitro culture of fibroblasts
Under aseptic conditions, pathological scar tissue and normal skin tissue specimens were respectively cut. Ophthalmic scissors was applied to carefully remove epithelial and subcutaneous tissues and the specimens were washed three times with D-Hank's liquid. The specimens were cut into 1 mm3 tissue block, placed into a sterile bottle, filtered after digestion into floccules by 0.25% trypsin EDTA, centrifuged at 650 × g for 5 min, well mixed and precipitated with 5 ml of DMEM liquid containing 20% fetal bovine serum, and placed in a 25 ml culture flask to culture in an incubator of 5% CO2 at 37°C. DMEM liquid (20%) containing 20% fetal bovine serum was used to change liquid once every 3–4 days. The cell growth was observed under microscope (Olympus Corp.). When the cells were close to overgrowing, passage was conducted according to the proportion of 1:3 and the 5th generation cells were taken to implement the experimental study.
Immunofluorence test
The 5th generation fibroblasts were taken, 4% paraformaldehyde was added, fixed at 25°C for 30 min, washed three times with phosphate buffer solution (PBS), incubated at 0.1% Triton X-100 37°C for 20 min, washed with PBS, and sealed with non-immune goat serum for 1 h. Primary antibody was added after the complete suction of serum, and they were incubated overnight at 4°C. Rabbit anti-human TGF-β1 monoclonal antibody (dilution: 1:500; catalog no. 5154LC) and rabbit anti-human PI3K monoclonal antibody (dilution: 1:500; catalog no. 9655T), rabbit anti-human protein kinase B (Akt) monoclonal antibody (dilution: 1:500; catalog nos. 9474S) and rabbit anti-human mTOR monoclonal antibody (dilution: 1:500; catalog no. 8665T) were from Cell Signaling Technology (Danvers, MA, USA). The following day, after they were washed with PBS, secondary antibody (dilution: 1:200; catalog no. 4412S; Cell Signaling Technology) was added, incubated at 37°C for 60 min, washed with PBS, re-dyed with DAPI in the dark, and observed under fluorescence microscope (Olympus Corp.) after 10 min.
Expression of mRNA detected by RT-PCR
After the 5th generation fibroblasts were incubated for 48 h under standard environment, a TRIzol reagent total RNA extraction kit was applied to conduct total RNA extraction and the operating steps were implemented according to the manufacturer's instruction. Electrophoresis was applied to identify the integrity of RNA extraction. By measuring, the A260/A280 of all the samples (the ratio of light absorption value at 260 nm/280 nm) was >1.8, which indicated high sample purity without obvious protein pollution. Total RNA purity was uniformly adjusted to be 0.5 g/l and the specimens were stored at 70°C.
RNA (6 µl) was taken as a reverse transcription template. Reverse transcription reaction system was 20 µl, with the reaction condition of 37°C for 1 h and 95°C for 3 min. Standard curve quantitation method was used to prepare standard substance of cDNA template and it was diluted by 10-time step dilution successively to prepare five gradients of standard substances of cDNA template. The mRNA sequence of target genes was obtained in the GenBank database and specific primers were designed in CDS area by applying Primer Express version 2.0 software (Thermo Fisher Scientific, Inc., Beijing, China). Table I shows the sequence of specific primers. Each sample was amplified by genes to be detected and reference genes. The amplification system was established according to the following system: cDNA 2 µl, qPCR mix 10 µl, primer F 1 µl, primer R 1 µl and ddH2O 6 µl. The reaction was conducted on ABI 7900 qPCR instrument according to the following conditions: Pre-degeneration at 95°C for 2 min, degeneration at 94°C for 20 sec, annealing at 65°C for 20 sec, extending at 72°C for 30 sec and 40 cycles. Quantity-one gel imaging analysis system was used to analyze gray values of all bands and the relative quantity (RQ) of mRNA was shown by the ratio of the gray values of target genes and reference gene β-actin. Gene quantity was RQ=2−ΔΔCq. A higher RQ value was considered higher gene quantity.
Table I.
PI3K, Akt, mTOR gene primer sequences and amplification fragment length of the product.
| Genes | Sequence (5′→3′) | Product size (bp) |
|---|---|---|
| TGF-β1 | U: GCCCTGGACACCAACTATTG | 1,724 |
| D: CGTGTCCAGGCTCCAAATG | ||
| PI3K | U: GATTCTCAGCAGCCAGCTCTGAT | 863 |
| D: GCAGGCTGTCGTTCATTCCAT | ||
| Akt | U: GACAACCGCCATCCAGAC | 851 |
| D: CCAGGGACACCTCCATCTC | ||
| mTOR | U: ACTCGCTTCTATGACCAACTGA | 494 |
| D: TTTCCATGACAACTGGGTCATTG |
U, upstream; D, downstream; PI3K, phosphatidylinositol 3-kinase; Akt, protein kinase B; mTOR, mammalian target of rapamycin.
The calculation method of ΔΔCq was: ΔΔCq = (Ct mean of target gene of the sample to be detected - Ct mean of the housekeeping gene of the sample to be detected) - (Ct mean of target gene of teh control sample - Ct mean of the housekeeping gene of the control sample).
Expression of western blot detection protein
Cultured fibroblasts were collected and washed with pre-cooling PBS twice. Cell lysis buffer was added, and the cultured fibroblasts implemented were placed on ice for 10–20 min. After the homogenate was scrapped by cell scraper, the cells were centrifuged at 8,400 × g at 4°C for 10 min and the supernatant was taken. Lowry was applied to conduct protein quantification for supernatant. Protein sample (50 µg) was loaded to 15% SDS-polyacrylamide gel electrophoresis. After bromophenol blue entered the bottom of the gel, protein was reimprinted on the nitrocellulose membrane. Primary antibody was added into fibroblasts which were incubated overnight at 4°C. Corresponding secondary antibody marked with horseradish peroxidase was added. Following incubation at room temperature for 1 h, the membranes were washed, and Odyssey infrared fluorescence imager was used to scan membranes and conduct imaging. Images were analyzed by applying Image Pro-Plus image analysis system.
Statistical analysis
Experimental data were expressed as mean ± standard deviation. SPSS 18.0 software (SPSS, Inc., Chicago, IL, USA) was used for data analysis. Homogeneity of variance was tested using the t-test, and non-parametric was used to test whether homogeneity of variance was not met. P<0.05 was considered to indicate a statistically significant difference.
Results
Immunofluorescence detection
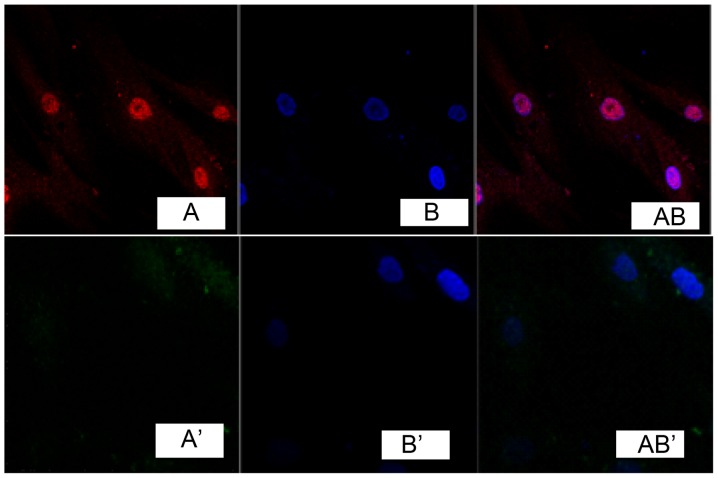
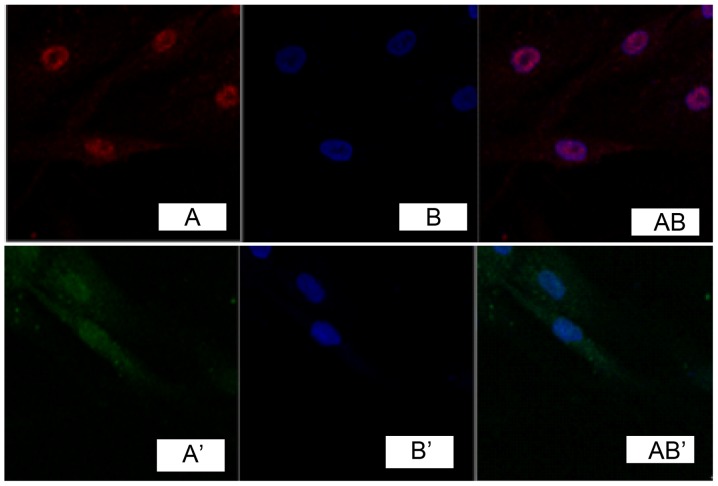
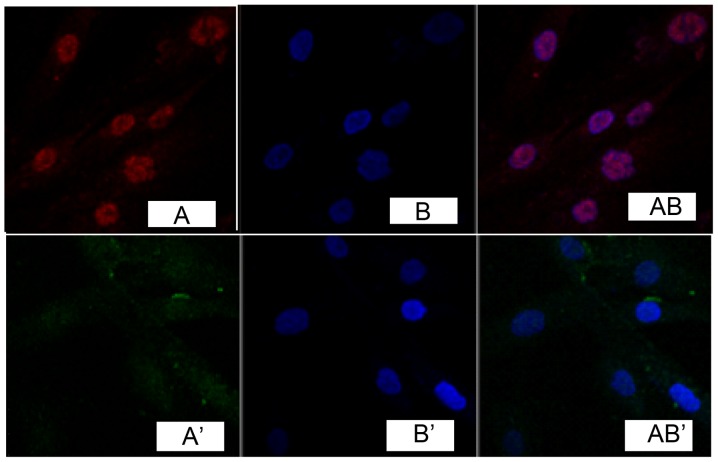
Immunofluorescence detection results of cells showed that, TGF-β1 was marked in red (Fig. 1A). DAPI marked nucleus indicated blue fluorescence (Fig. 1B). Fig. 1A and B overlapped when merging, indicating that TGF-β1 was expressed in the cell nucleus, while no obvious expression was found in fibroblasts of the normal skin tissue (Fig. 1A', B' and AB'). Immunocytochemistry detection showed that, phosphatidylinositol 3-kinase (PI3K) and Akt were expressed in the cell nucleus in pathological scar fibroblasts (Figs. 2 and 3), while no obvious expression was found in normal skin tissue (Figs. 2 and 3).
Figure 1.
Expression of transforming growth factor-β1 in fibroblast cells.
Figure 2.
Expression of phosphatidylinositol 3-kinase in fibroblast cells.
Figure 3.
Expression of protein kinase B in fibroblast cells.
RT-PRC result
Relative quantities of the expression of TGF-β1, PI3K, ATK and mTOR mRNA in pathological scar fibroblasts were significantly higher (P<0.05) than those observed in normal skin tissue (Table II).
Table II.
mRNA relative expression quantities of TGF-β1, PI3K, Akt, and mTOR in the two groups.
| Groups | TGF-β1 | PI3K | Akt | mTOR |
|---|---|---|---|---|
| Normal skin | 1.014±0.203 | 1.005±0.121 | 1.014±0.197 | 1.002±0.047 |
| Pathological scar | 3.563±0.349a | 4.598±0.558b | 3.136±0.219c | 5.799±0.227d |
Versus normal skin group, P<0.05. TGF-β1, transforming growth factor-β1, PI3K, phosphatidylinositol 3-kinase; Akt, protein kinase B; mTOR, mammalian target of rapamycin.
Western blot analysis
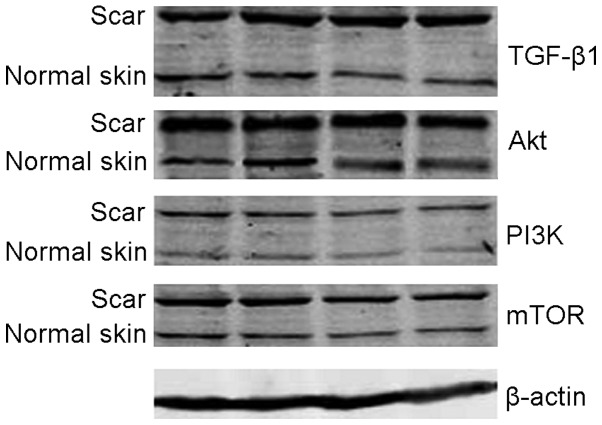
The analysis of Image Pro-Plus imaging analysis system revealed that, relative expression quantities of TGF-β1, PI2K, Akt and mTOR protein in pathological scar fibroblasts were higher than those in normal skin tissue, and there were significant differences between two groups (P<0.05) (Table III and Fig. 4).
Table III.
The protein relative expression quantities of TGF-β1, PI3K, Akt, and mTOR in the two groups.
| Groups | TGF-β1 | PI3K | Akt | mTOR |
|---|---|---|---|---|
| Normal skin | 0.717±0.074 | 0.636±0.064 | 0.552±0.054 | 0.895±0.102 |
| Pathological scar | 1.439±0.047a | 1.380±0.052b | 1.879±0.063c | 1.695±0.069d |
Versus normal skin group, P<0.05. TGF-β1, transforming growth factor-β1, PI3K, phosphatidylinositol 3-kinase; Akt, protein kinase B; mTOR, mammalian target of rapamycin.
Figure 4.
Protein expression of electrophoresis of transforming growth factor-β1 (TGF-β1), phosphatidylinositol 3-kinase (PI3K), protein kinase B (Akt), mammalian target of rapamycin (mTOR), β-actin of the two groups.
Discussion
The excessive proliferation of fibroblast and its increasing activity played a critical role in the formation process of pathological scar. After the occurrence of trauma, the fibroblast at injured sites proliferates in a large number. Its synthesis and the number of extracellular matrix, including collagen secretion, began to increase 5–6 days after injuries. The proliferation and activation of pathological scar fibroblasts were regulated and controlled by various cytokines, including (TGF-β1), connective tissue growth factor, hyaluronic acid stimulating factor, interleukin (7), insulin-like growth factor (8) and tumor necrosis factor. Under certain circumstances, these cytokines led to the proliferation of fibroblasts through several links and the increased collagen synthesis with decreased decomposition, which led to massive deposit of collagen and the formation of pathological scar (9). TGF-β1 is the cytokine most closely related to scars and is a strong chemotactic factor of fibroblasts. It can act on cells in a direct or indirect manner, and exerts independent or synergistic effects of paracrine and autocrine to generate three types of important biological effects, i.e., chemotactic migration, proliferation and differentiation as well as synthesis and secretion.
Life process is a biochemical reaction process on the basis of cell signal transduction, as are wound healing and scar formation. Therefore, during the process of the formation of pathological scar, excessive proliferation of fibroblasts was the result of integrated response of cells to the stimulation of various extracellular signals (including TGF-β1). Since this integrated response was completed in cells, it inevitably involved the process of cell signal transduction. The process of cell signal transduction was inseparable for the biological activity of TGF-β1. At present, more studies focused on typical TGF-β1/Smads signal pathway; however, this did not fully reveal the specific mechanism of the formation of keloid fibroblasts as the formation of keloid involved multiple genes and multiple signal pathways is a very complicated process. In recent years, an increasing number of studies have shown that independent Smad pathway, in addition to the typical Smad pathway, are involved in the transduction process of TGF-β1 signals. Some of the pathways were involved directly or indirectly during tumor formation and were closely related to the proliferation, invasion and transfer of tumor (3).
Tumor-related genes are involved in the process of formation of keloid (10,11). Some methods and drugs for treating tumor were also effective for keloid (12). These indicated great significance for studying pathological scar. The mTOR signaling pathway is considered important in the process of TGF-β1 signal transduction. For instance, the level of mTOR mRNA and protein expression increased during the process of TGF-β1-induced EMT related to tumorigenesis as well as migration and invasion. As the main signaling pathway that regulates and controls protein synthesis, the mTOR signaling pathway was involved in cell regulation such as proliferation and differentiation, was identified in previous studies, and gradually became a research hotspot, especially in studies on tumor. The mTOR signal pathway includes upstream PI3K, protein kinase B (PKB, also known as Akt), phosphatase and tensin homolog (PTEN) deleted on chromosome 10 and tuberous sclerosis complex 1/2 (TSC 1/2) (13), of which PTEN is a negative feedback regulating factor; downstream ribosomal protein S6 kinase (S6K), eukaryotic cell interpretation initiation factor 4E binding protein (4EBP) (14) and both of them are key regulatory factors of protein translation. After extracellular signal combines with transmembrane tyrosine kinase receptor, receptor tyrosine residues were phosphorylated to be activated. Thus, combined with regulatory subunit p85 of PI3K leads to PI3K activation. The stimulated PI3K can activate downstream Akt (15). Activated Akt can directly stimulate mTOR or mTOR signal by inhibiting the formation of TSC 1/2. mTOR is the regulator key for protein synthesis and stimulated mTOR acts on two main downstream target proteins, i.e., S6K and 4EBP. Recent studies proved that mTOR gene was closely related to abnormal cell proliferation and its expression in the occurrence of many tumors, including glioma, esophageal cancer and colorectal cancer (5), markedly increased. Pathologically, pathological scar belongs to benign solid tumor. Keloid can invasively grow to surrounding normal tissues and its pathological characteristics are very similar to that of tumor, with tumor-like characteristics. This is an indication suggesting that the mTOR signaling pathway may be important in the process of the proliferation of pathological scar fibroblasts. To prove the hypothesis, this study group implemented some explorations on the subject.
The present study applied cellular immunofluorescence to detect the expression of key molecules of TGF-β1/mTOR signaling pathway, and found the obviously strengthened expression of TGF-β1, PI3K and Akt in pathological scar fibroblasts, mainly in cell nucleus, without obvious expression in fibroblasts of normal skin tissue. The results of RT-PCR and western blot detection showed that the quantity of expression of TGF-β1, PI3K, Akt, and mTOR mRNA and protein in pathological scar fibroblasts was obviously higher than that of fibroblasts of normal skin tissue, with statistically significant differences (P<0.05). Additionally, there was abnormally activated TGF-β1/mTOR signal pathway in pathological scar fibroblasts, which may be the important mechanism in the formation of pathological scar and may achieve some breakthroughs to prevent and successfully treat pathological scar by taking the key molecules of this signal pathway as target spots. However, it is unclear which approach could be used by abnormally activated TGF-β1/mTOR signaling pathway promoting excessive proliferation and activation of pathological scar fibroblasts that require implementation of further explorations.
References
- 1.van der Veer WM, Bloemen MC, Ulrich MM, Molema G, Van Zuijlen PP, Middelkoop E, Niessen FB. Potential cellular and molecular causes of hypertrophic scar formation. Burns. 2009;35:15–29. doi: 10.1016/j.burns.2008.06.020. [DOI] [PubMed] [Google Scholar]
- 2.Rhett JM, Ghatnekar GS, Palatinus JA, O'Quinn M, Yost MJ, Gourdie RG. Novel therapies for scar reduction and regenerative healing of skin wounds. Trends Biotechnol. 2008;26:173–180. doi: 10.1016/j.tibtech.2007.12.007. [DOI] [PubMed] [Google Scholar]
- 3.Sun Q, Guo S, Wang CC, Sun X, Wang D, Xu N, Jin SF, Li KZ. Cross-talk between TGF-β/Smad pathway and Wnt/β-catenin pathway in pathological scar formation. Int J Clin Exp Pathol. 2015;8:7631–7639. [PMC free article] [PubMed] [Google Scholar]
- 4.Huang LP, Mao Z, Zhang L, Liu XX, Huang C, Jia ZS. Screening of differentially expressed genes in pathological scar tissues using expression microarray. Genet Mol Res. 2015;14:10743–10751. doi: 10.4238/2015.September.9.13. [DOI] [PubMed] [Google Scholar]
- 5.Steelman LS, Chappell WH, Abrams SL, Kempf RC, Long J, Laidler P, Mijatovic S, Maksimovic-Ivanic D, Stivala F, Mazzarino MC, et al. Roles of the Raf/MEK/ERK and PI3K/PTEN/Akt/mTOR pathways in controlling growth and sensitivity to therapy-implications for cancer and aging. Aging (Albany NY) 2011;3:192–222. doi: 10.18632/aging.100296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lamouille S, Connolly E, Smyth JW, Akhurst RJ, Derynck R. TGF-β-induced activation of mTOR complex 2 drives epithelial-mesenchymal transition and cell invasion. J Cell Sci. 2012;125:1259–1273. doi: 10.1242/jcs.095299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ghazizadeh M, Tosa M, Shimizu H, Hyakusoku H, Kawanami O. Functional implications of the IL-6 signaling pathway in keloid pathogenesis. J Invest Dermatol. 2007;127:98–105. doi: 10.1038/sj.jid.5700564. [DOI] [PubMed] [Google Scholar]
- 8.Ishihara H, Yoshimoto H, Fujioka M, Murakami R, Hirano A, Fujii T, Ohtsuru A, Namba H, Yamashita S. Keloid fibroblasts resist ceramide-induced apoptosis by overexpression of insulin-like growth factor I receptor. J Invest Dermatol. 2000;115:1065–1071. doi: 10.1046/j.1523-1747.2000.00180.x. [DOI] [PubMed] [Google Scholar]
- 9.van den Bogaerdt AJ, van der Veen VC, Van Zuijlen PP, Reijnen L, Verkerk M, Bank RA, Middelkoop E, Ulrich MM. Collagen cross-linking by adipose-derived mesenchymal stromal cells and scar-derived mesenchymal cells: Are mesenchymal stromal cells involved in scar formation? Wound Repair Regen. 2009;17:548–558. doi: 10.1111/j.1524-475X.2009.00501.x. [DOI] [PubMed] [Google Scholar]
- 10.Chen W, Fu X, Sun T, Sun X, Zhao Z, Sheng Z. Change of gene expression of transforming growth factor-beta1, Smad 2 and Smad 3 in hypertrophic scars skins. Zhonghua Wai Ke Za Zhi. 2002;40:17–19. (In Chinese) [PubMed] [Google Scholar]
- 11.Teofoli P, Barduagni S, Ribuffo M, Campanella A, De Pita' O, Puddu P. Expression of Bcl-2, p53, c-jun and c-fos protooncogenes in keloids and hypertrophic scars. J Dermatol Sci. 1999;22:31–37. doi: 10.1016/S0923-1811(99)00040-7. [DOI] [PubMed] [Google Scholar]
- 12.Kleinerman R, Kilmer SL, Chotzen VA. Mitomycin C in the treatment of keloids: A case and review. J Drugs Dermatol. 2013;12:701–703. [PubMed] [Google Scholar]
- 13.Matsuoka T, Yashiro M. The role of PI3K/Akt/mTOR signaling in gastric carcinoma. Cancers (Basel) 2014;6:1441–1463. doi: 10.3390/cancers6031441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gomez-Pinillos A, Ferrari AC. mTOR signaling pathway and mTOR inhibitors in cancer therapy. Hematol Oncol Clin North Am. 2012;26:483–505. doi: 10.1016/j.hoc.2012.02.014. [DOI] [PubMed] [Google Scholar]
- 15.Xavier S, Niranjan T, Krick S, Zhang T, Ju W, Shaw AS, Schiffer M, Böttinger EP. TbetaRI independently activates Smad- and CD2AP-dependent pathways in podocytes. J Am Soc Nephrol. 2009;20:2127–2137. doi: 10.1681/ASN.2008070806. [DOI] [PMC free article] [PubMed] [Google Scholar]