Abstract
Partial deletions on the long arm of chromosome 13 lead to a number of different phenotypes depending on the size and position of the deleted region. The present study investigated 2 patients with 13q terminal (13qter) deletion syndrome, which manifested as anal atresia with rectoperineal fistula, complex type congenital heart disease, esophageal hiatus hernia with gastroesophageal reflux, facial anomalies and developmental and mental retardation. Array comparative genomic hybridization identified 2 regions of deletion on chromosome 13q31-qter; 20.38 Mb in 13q31.3-qter and 12.99 Mb in 13q33.1-qter in patients 1 and 2, respectively. Comparisons between the results observed in the present study and those obtained from patients in previous studies indicate that the gene encoding ephrin B2 (EFNB2) located in the 13q33.3-q34 region, and the gene coding for endothelin receptor type B, in the 13q22.1–31.3 region, may be suitable candidate genes for the observed urogenital/anorectal anomalies. In addition, the microRNA-17-92a-1 cluster host gene and the glypican 6 gene in the 13q31.3 region, as well as EFNB2 and the collagen type IV a1 chain (COL4A1) and COL4A2 genes in the 13q33.1-q34 region may together contribute to cardiovascular disease development. It is therefore possible that these genes may be involved in the pathogenesis of complex type congenital heart disease in patients with 13q deletion syndrome.
Keywords: chromosome 13 deletion syndrome, array comparative genomic hybridization, genotype-phenotype, urogenital/anorectal anomalies, congenital heart disease, gastrointestinal tract anomalies
Introduction
Chromosome 13q deletion syndrome, a rare genetic disorder, is characterized by partial deletions of one of the long arms of chromosome 13, which leads to a number of human birth defects. The clinical symptoms vary widely among patients and may include, developmental delays (mental and growth retardation), facial anomalies (microcephaly, hypertelorism, flattened nasal bridge and micrognathia), and severe malformations in the distal limbs, central nervous system (posterior encephalocele, holoprosencephaly and neural tube defects), eyes (micro-ophthalmia and retinoblastoma), heart (congenital heart defects), lungs, kidneys, gastrointestinal tract and genitourinary tract (penoscrotal transposition and hypospadias, ambiguous genitalia, reduced anogenital distance, imperforate anus, bicornuate uterus and imperforate anus with vaginal fistula or cloaca) (1–4).
Clinical characteristics and severity depend on the size of the deleted region and the location on chromosome 13. Chromosome 13 deletion syndrome was first described in 1969 (1), and efforts since then have been made to identify the critical region involved in specific anomalies using genotype-phenotype analysis (5,6). However, due to the limited number of cases and the different levels of penetrance, the causative genes have yet to be determined. Previous studies have proposed the existence of the following 3 groups of genotype-phenotypes based on the involvement of the critical 13q32 band in the deletion (2,5): i) proximal deletions with a non-deleted 13q32 band are observed primarily in patients with mild mental retardation, growth delay and inconstant retinoblastoma; ii) a 13q32 band deletion is associated with severe congenital malformations; iii) a distal deletion without a 13q32 deletion has been observed in patients with severe mental retardation, no brain malformation or growth delay. Previous studies involving the use of array comparative genomic hybridization (CGH) coupled with fluorescence in situ hybridization, reverse transcription-quantitative polymerase chain reaction (qPCR) and multiple ligation-depended probe amplification to determine the precise breakpoint of the unbalanced chromosomal translocation have been conducted (3–5,7–11). This analysis has provided information regarding the molecular genotype-phenotype in chromosome 13 deletion syndrome.
In the present study, two patients diagnosed with chromosome 13 deletion syndrome, which harbored 13q31.3q terminal (qter) and 13q33.1qter deletions, respectively, were recruited for genotype-phenotype analysis using array-CGH and qPCR.
Case report
The present study was approved by the institutional review board of Shengjing Hospital affiliated to China Medical University (Shenyang, China). Written informed consent regarding participation and the publication of clinical information was obtained from the parents of patients 1 and 2, and all clinical investigations were conducted according to the principles expressed in the Declaration of Helsinki.
Clinical descriptions
Patient 1 (gender, female; age, 14 months) was diagnosed with anal atresia with rectoperineal fistula following birth, and was referred to Shengjing Hospital for surgery in June 2009 due to recurrent constipation. Patient 1 was born at full-term by Caesarean section, and was the first-born child to non-consanguineous parents. Following birth, the patient was blue and was diagnosed with a heart murmur as well as an imperforate anus with navicular fossa fistula. At 1 month of age, the patient suffered recurrent seizures. At 14 months-old, physical examination identified marked growth retardation, with a height of 69 cm, which was <3 standard deviations below the average for the patient's age; a body weight of 7.8 kg and a head circumference of 44 cm. In addition, facial dysmorphism was observed, including hypotelorism, blepharophimosis and a broad nasal bridge with a flat philtrum. In addition, psychomotor milestones, determined by developmental quotient evaluation, were markedly delayed, as they were equivalent to the level observed in a 5-month-old child. Angioma was observed in the lateral side of the left foot and digits. A systolic-diastolic phase heart murmur and a number of complex type congenital heart defects were identified by echocardiography including ventricular septal defect, double outlet right ventricle (DORV) defects, single atrium (SA) defects, mixed type atrial septal defect (ASD), persistent left superior vena cava (PLSVC) defects and severe pulmonary stenosis (PS). Radiography analysis of the fistula and anoplasty revealed a fistula with a 0.3-cm opening at the navicular fossa (also known as the fossa of the vaginal vestibule) located before the terminal rectum, which was 1.5 cm in length, stopping 0.5-cm from the anterior wall of the rectal blind end. Magnetic resonance imaging (MRI) of the brain revealed cortical atrophy, agenesis of the corpus callosum, cerebral ventricle dilation, a small cerebral cortex hippocampus, bilateral otitis media and mastoiditis. The patient was treated with anoplasty and the fistula was closed, however, the cardiac anomalies were not corrected.
Patient 2 was a newborn (gender, female; age, 46 h), born at full-term to a 39-year-old mother and 43-year-old father by Caesarean section due to oligohydramnios. The parents were non-consanguineous. At birth, patient 2 weighed 2,460 g (<3rd centile), was 42 cm in length (<3rd centile) and demonstrated a head circumference of 31.5 cm (<3rd centile). The patient presented with poor feeding and vomiting following birth. Facial dysmorphism was observed, including a round face with a small forehead, hypotelorism and blepharophimosis. The patient's heartbeat was strong with a regular rhythm, and no murmur was observed. A nasogastric tube was passed into the stomach with great difficulty, and an esophageal hiatus hernia and gastroesophageal reflux were identified following upper gastrointestinal contrast X-ray analysis. MRI results revealed limited damage to the cerebral white matter, which was only observed in the posterior horn of the left lateral ventricle, and suggested an optimistic prognosis. Following treatment with gastrointestinal decompression, the patient's vomiting was greatly relieved.
Cytogenetic analysis
Cytogenetic investigation using Giemsa-banding with trypsin as the proteolytic enzyme (GTG banding) was performed on metaphase spreads of peripheral blood lymphocytes using standard procedures (12). The following detailed procedures were followed: heparinized human whole blood (0.5 ml) was cultured at 37°C for 70 h in 8 ml Gibco Chromosome Medium (Thermo Fisher Scientific, Inc., Waltham, MA, USA). Cells were arrested with colchicine (10 ug/ml) for 90 min. Chromosome preparations were made by incubating the cell suspension in 0.075 mol/l potassium chloride at 37°C for 30 min, followed by a fixation step in a freshly prepared mixture of 3:1 methanol:acetic acid at −20°C for 30 min. GTG banding was performed by incubating the glass slides in a 0.05% trypsin solution at 37°C for 15 sec, followed by rinsing the slides in PBS buffer and staining in a 5% Giemsa stain for 8 min at room temperature. The slides were rinsed with water and air dried. Cytogenetic analysis was performed on GTG-banded metaphase spreads collected from the patients and their parents at a resolution of 400 bands according to standard lab procedures. A total of 15 metaphases were analyzed for each individual sample.
DNA isolation and array-CGH analysis
Genomic DNA was isolated from the peripheral blood samples using the DNeasy Blood & Tissue kit (Qiagen GmbH, Hilden, Germany). DNA quality was confirmed by gel electrophoresis with a 1.5% agarose gel, and the yield was confirmed by spectrophotomery (NanoDrop ND-1,000; NanoDrop Technologies; Thermo Fisher Scientific, Inc., Wilmington, DE, USA). Genomic DNA was labeled using the Affymetrix Cytogenetics Reagent kit (Affymetrix, Inc., Santa Clara, CA, USA) and the labeled DNA was loaded on to an Affymetrix Cytogenetics Array Cytoscan 750K Chip (containing 7.5 million copy number markers; Affymetrix, Inc.), which was performed by Gene Tech (Shanghai, China). The array was scanned and the data were analyzed using the Affymetrix Chromosome Analysis Suite (version 2.1; Affymetrix, Inc.).
qPCR validation
PCR was performed on lymphocyte DNA extracts using the 7900HT Fast Real-Time PCR System (Applied Biosystems; Thermo Fisher Scientific, Inc.). A total of 3 sequence-tagged sites on chromosome 13 (D13S797 in 13q33.2, D13S628 in 13q31.1 and D13S258 in 13q21.33) were detected. The primers used are shown in Table I. The NCBI reference sequence of D13S797 used to determine copy number alterations was NG_012694.1 (https://www.ncbi.nlm.nih.gov/nuccore/255958284) from 19563 nt to 19758 nt, the amplified product length is 196 bp. NCBI reference sequence for D13S258 was AL356754.18 (https://www.ncbi.nlm.nih.gov/nuccore/AL356754) from 969 nt to 1239 nt, the amplified product length is 271 bp. NCBI reference sequence for was AL160154.11 (https://www.ncbi.nlm.nih.gov/nucleotide/14160914); from 1004 nt to 1243 nt, the amplified product length is 240 bp. PCR reactions were prepared using the SYBR Premix Ex Taq II PCR reagent kit (Takara Biotechnology Co., Ltd., Dalian, China) according to the manufacturer's instructions. Amplification was performed in a final reaction volume of 10 µl, containing 50 ng genomic DNA, 0.4 µM PCR forward primer, 0.4 µM PCR reverse primer, ROX Reference Dye II (1X) and SYBR Premix Ex Taq (Tli RNase H Plus; 1X). The thermal cycling conditions were as follows: initial denaturation at 94°C for 30 sec, followed by 40 cycles of denaturation at 95°C for 5 sec and annealing at 60°C for 35 sec. Amplification levels were calculated using the 2−ΔΔCq method (13).
Table I.
Primer sequences of STS markers used for PCR analysis.
| STS | Forward primer | Reverse primer |
|---|---|---|
| D13S797 | 5′-GGTTTGCTGGCATCTGTATT-3′ | 5′-TGTCTGGAGGCTTTTCAGTC-3′ |
| D13S258 | 5′-ACCTGCCAAATTTTACCAGG-3′ | 5′-GACAGAGAGAGGGAATAAACC-3′ |
| D13S628 | 5′-CGCCACTTTTCTAAATGCC-3′ | 5′-GGAGTAACAAATAGCAAGGCT-3′ |
STSs, sequence tagged sites.
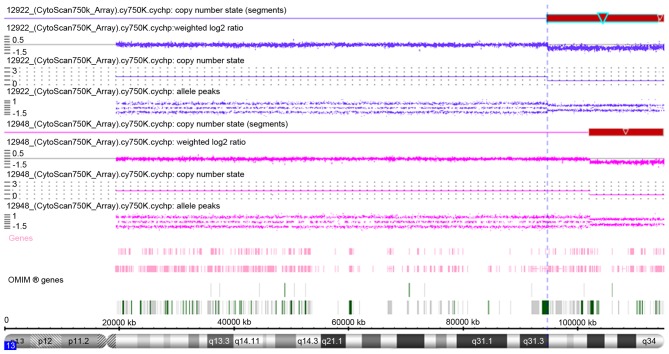
The array-CGH results are shown in Fig. 1 and Table II, and a summary of the regions on chromosome 13 identified in the present and previous studies (3,4,6–8,10,11,14–22) are reviewed in Fig. 2. Cytogenetic analysis of patient 1 revealed a karyotype of 46,XX,del(13)(pter→q31; data not shown), and the array-CGH results revealed a distal 20.38 Mb deletion in the 13q31.3-qter region, from 94,724,977 to 115,107,733 bp (terminal end). A total of 59 Refseq genes were detected in this region of deletion, as determined by analysis by Affymetrix Chromosome Analysis Suite software. The qPCR results verified the presence of a deletion at D13S797 only, but not at D13S628 and D13S258 (data not shown). The karyotype and qPCR results of the parents of patient 1 were normal, demonstrating that the deletion was not hereditary (data not shown).
Figure 1.
Array-comparative genomic hybridization results of patient 1 (12922; blue regions) and patient 2 (12948; pink regions) as indicated by copy number state using the Affymetrix Cytoscan 750K. The deleted region 13q31.3-qter of patient 1 from 94,724,977 bp to 115,107,733 bp is indicated by red box with light blue outline, with the darker blue line indicating that copy number state is 1 (normal copy number state is 2). The deleted region 13q33.1-qter of patient 2 from 102,110,842 bp to 115,107,733 bp is indicated by red box, with the pink line indicating that the copy number state is 1. OMIM, Online Mendelian Inheritance in Man.
Table II.
Amplified DNA copy numbers in the 13q31.3-q34 regions as determined using the Affymetrix Cytoscan 750K.
| A, Patient 1 | |||||||
|---|---|---|---|---|---|---|---|
| CN state | Type | Chromosome band | Size (kbp) | Marker count | Confidence | Start (bp) | End (bp) |
| LOH | 13q33.1-q33.3 | 6,293.846 | 651 | 1 | 104,002,716 | 110,296,562 | |
| 1 | Loss | 13q34-q34 | 861.336 | 320 | 0.9014088 | 114,246,397 | 115,107,733 |
| 1 | Loss | 13q31.3-q34 | 19,490.64 | 5,095 | 0.91336507 | 94,724,977 | 114,215,617 |
| B, Patient 2 | |||||||
| CN state | Type | Chromosome band | Size (kbp) | Marker count | Confidence | Min (bp) | Max (bp) |
| LOH | 13q33.1-q34 | 12,987.398 | 1,170 | 1 | 102,108,307 | 115,095,705 | |
| 1 | Loss | 13q31.3-q34 | 12,996.891 | 3,262 | 0.9176309 | 102,110,842 | 115,107,733 |
CN, copy number; LOH, loss of heterozygosity.
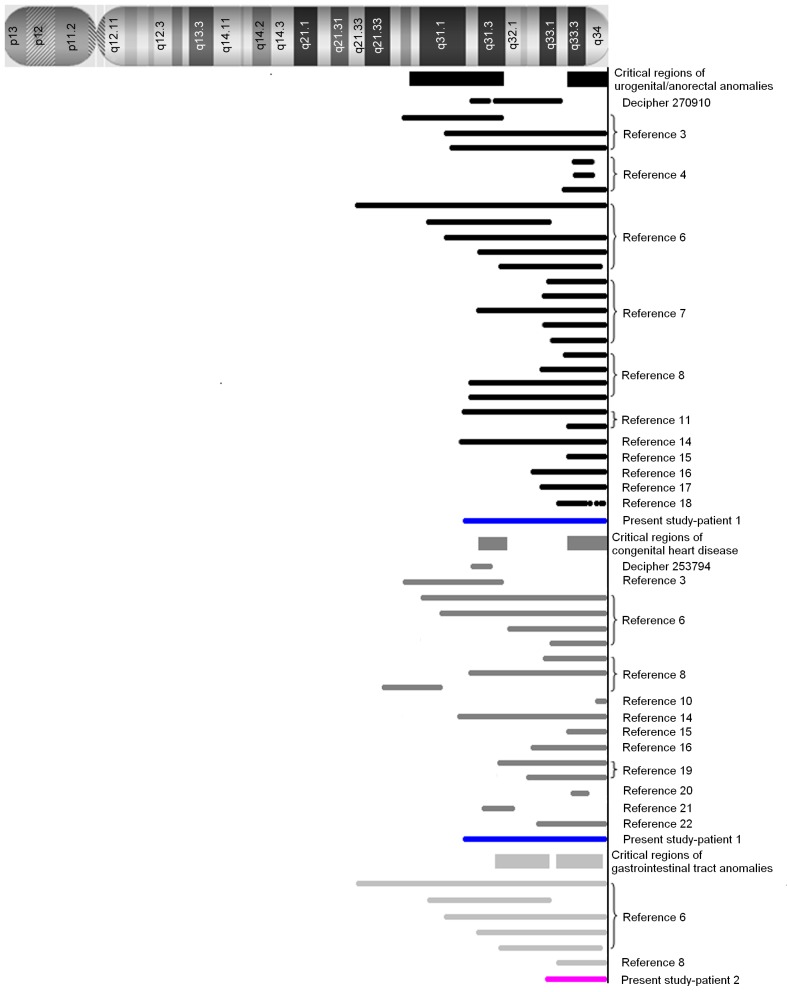
Figure 2.
The critical regions of chromosome 13 associated with anorectal/genitourinary anomalies (black rectangles), congenital heart disease (grey rectangles) and gastrointestinal anomalies (light grey rectangles), respectively. The deleted regions of chromosome 13 reported by previous studies are indicated by dark/light bars, and deleted regions in patients 1 and 2 of the present study are indicated by blue and pink bars, respectively.
GTG banding analysis revealed that the karyotype of patient 2 was 46,XX,del(13)(pter→q33:)(data not shown), and the array-CGH results revealed a distal deletion of 12.99 Mb in the 13q33.1-qter region spanning 102,110,842 to 115,107,733 bp. Analysis by Affymetrix Chromosome Analysis Suite software revealed that a total of 34 Refseq genes are located in this region of deletion. The qPCR results verified the deletion at positions D13S797 and D13S628, but not D13S258 (data not shown). The karyotype and qPCR results of the parents of patient 2 were normal (data not shown), which suggested that this deletion was not hereditary.
Discussion
Congenital heart disease, anorectal/genitourinary and gastrointestinal tract malformations are the predominant anomalies observed in 13q deletion syndrome, particularly as a part of VACTERL association, a disorder characterized by vertebral anomalies, anal atresia, cardiac defects, tracheoesophageal fistula, renal anomalies and limb defects (14,23–25). The aim of phenotype-genotype association analysis between these anomalies and deleted regions of chromosome 13, is to identify a limited number of candidate genes located in narrow regions of deletion that may provide novel targets for molecular pathogenesis research into the development of heart, anorectal/genitourinary and gastrointestinal tract diseases.
Anorectal/genitourinary anomalies in 13q deletion syndrome are generally rare and vary in manifestation and severity. Typical cases observed in male patients involve anal atresia with severe hypospadias and perineal fistula, whereas other patients may present with distal hypospadias without anorectal anomalies. By contrast, females are often diagnosed with anal atresia and vaginal fistula (4,7). Human embryological studies have demonstrated that the primitive urogenital sinus and anorectal canal originate from the primitive cloaca, and there are number of molecular events that govern the developmental processes underlying cloacal septation, closure of the perineum and scrotum, urethral tubularization and penoscrotal positioning (26–28). The results obtained from patient 1 in the present study, together with those obtained from patients with genitourinary/anorectal anomalies in 13q deletion syndrome from previous studies (3,4,6–8,11,14–18), have identified two critical regions, 13q33.3-qter and 13q22.1–31.3. In recent years, the non-morbid online Mendelian Inheritance in Man (OMIM) gene, ephrin B2 (EFNB2) located in 13q33.3, has been recognized as a strong candidate gene for hypospadias or anorectal anomalies in 13q deletion syndrome in a number of studies (4,7–9,18). Animal experiments have demonstrated that a partial loss-of-function EFNB2 mutation in heterozygous male mice induces severe hypospadias and incomplete cloacal septation, and female mice exhibit similar defects in their external genitalia (29). Molecular data has further verified that the reverse signal, EFNB2 activation by EPH receptor B2 (EPHB2) on EPHB2-expressing cells, is exhibited in a dominant-negative manner in Efnb2LacZ/+ mice with hypospadias, which display a reduction in the effect of tyrosine phosphorylation (29). The haploinsufficiency of the EFNB2 gene product may provide a causative explanation for the clinical results observed in patient 1 of the present study. However, a limited number of studies have observed no mutations in the EFNB2 gene among 331 patients with isolated anorectal malformations (11), nor in patients with persistent cloaca and associated kidney malformations (30). This suggests that mutations in the EFNB2 gene do not exclusively affect the development of the anorectal and geniourinary tract. An additional important region, 13q22.1–31.3, was identified in two patients harboring an interstitial deletion (3), Database of Chromosomal Imbalance and Phenotype in Humans using Ensembl Resources (DECIPHER) ID 270910] (31). This region contains 27 OMIM genes, including the following 5 morbid genes: the ceroid-lipofuscinosis, neuronal 5 gene, the endothelin receptor B gene (EDNRB), the SLIT and NTRK like family member 1 gene, the microRNA-17-92a-1 cluster host gene (MIR17HG) and the glypican 6 gene (GPC6). Until recently, none of these genes have been proven to be associated with urogenital/anorectal anomalies, however, previous studies have indicated they may induce a number of congenital malformations (32–39). By contrast, homozygous or heterozygous mutations in EDNRB, located in 13q22.3, are thought to give rise to 3 types of allelic disease: Hirschsprung disease, albinism, black lock, cell migration disorder syndrome (ABCD syndrome) and Waardenburg syndrome, which are attributed to a defect in the migration of neural crest cells (33–35). The occurrence of comorbid Hirschsprung disease and hypospadisa/anorectal malformations has been reported in a number of studies (40–42). We hypothesize that aganglionic alterations that occur as a result of EDNRB defects may be involved in the underlying mechanisms of urogenital/anorectal anomalies. Therefore, the role of the EDNRB gene in the development of the urogenital tract requires further investigation to elucidate the molecular and pathological effects of the EDNRB gene.
Congenital heart disease in 13q deletion syndrome is more complex than in isolated cases, and include cases of Tetralogy of Fallot combined with additional heart defects (6), at least 2 heart anomalies in one patient (14,15,19), or rare type complex heart anomalies (20,43). In the present study, five of the six heart anomalies identified in patient 1 were rare types, and included, DORV, SA defects, mixed-type ASD, PLSVC and severe PS. These complex conditions associated with cardiovascular anomalies in 13q deletion syndrome, suggest that multiple genes may be involved in its pathogenesis. To date, at least 2 critical regions, 13q31.3 and 13q33.3–13q34, have been reviewed by the literature (3,6,8,10,14–16,19–22) (DECIPHER ID 253794). In the 13q33.3–13q34 region, morbid OMIM genes collagen type IV α1 chain (COL4A1) and COL4A2, were identified as potential candidates for heart development in a patient with an interstitial deletion of 13q33.3-q34 that presented with DORV (20). Previous studies have demonstrated that the collagen IV protein encoded by COL4A1 and COL4A2 serves a vital role during early cardiac development, and specifically in the development of the atria and outflow tract in mouse and human cardiovascular progenitor cells in fetal hearts (44,45). EFNB2 located in 13q33.3, though not a morbid OMIM gene, is an excellent candidate for congenital heart disease. Efnb2−/− null homozygotes display early embryonic lethality due to severe defects in cardiovascular development (46,47). However, the majority of Efnb2lacZ/lacZ homozygotes survive embryonic development and are born live only to perish within the first day due to cardiac abnormalities (48). These data suggest that insufficient EFNB2 expression may serve an important role in the pathogenesis of cardiovascular abnormalities. In an additional region of 13q31.3, which was identified in two patients with a microdeletion (DECIPHER ID 253794) (20,29), 2 OMIM morbid genes MIR17HG and GPC6 were present. MIR17HG is associated with Feingold syndrome type 2 (37), which is an autosomal dominant disorder characterized by variable combinations of microcephaly, limb malformations, esophageal and duodenal atresia, and learning disability/mental retardation. Cardiac and renal malformations, vertebral anomalies and deafness have been described in a minority of patients. GPC6 is associated with omodysplasia 1 (38,39), a rare autosomal recessive skeletal dysplasia characterized by severe congenital micromelia with shortening and distal tapering of the humeri and femora producing a club-like appearance. Patients with GPC6 mutations may additionally present with cryptorchidism, hernias, congenital heart defects and cognitive delay (38,39). Heart development is a complex process, which involves atrial and ventricular septation, giant vascular sprouting, branching and tubularization. It has been hypothesized that multiple proteins and factors may be required, with precise timing and spatial expression patterns. Therefore, the haploid insufficiency of MIR17HG, GPC6, EFNB2, COL4A1 and COL4A2 genes may co-contribute to the complex heart anomalies observed in patient 1 with a 13q31.3-qter deletion in the present study.
Gastrointestinal tract anomalies are rare in 13q deletion syndrome, and primarily involve the esophagus and its neighboring organs. They include the development of tracheoesophageal fistula, esophageal atresia (4), pyloric stenosis (8) and esophageal hiatus hernia with gastroesophageal reflux, and gastroesophageal reflux was observed in patient 2 of the present study. Additional digestive anomalies include common mesentery, pancreas anomalies, gall bladder agenesis/hypoplasia and spleen hypoplasia/supernumerary spleen (6). Two regions spanning 13q31.3-q33.1 and 13q33.2-q34 are thought to be involved, however, it is not conclusive due to the limited number of cases. Among the morbid OMIM genes in the 13q31.3-q33.1 region, only MIR17HG in 13q31.3, which induces Feingold syndrome type 2, has been associated with tracheoesophageal fistula and esophageal atresia (37). However, in the 13q33.2-q34 region, no morbid OMIM gene may provide an explanation for these anomalies. The non-morbid gene EFNB2 in 13q33.3, is thought to be involved in intestinal epithelial architecture via the EPH receptor B2-EFNB signaling pathway (49,50), which may explain, in part, these gastrointestinal anomalies. However, this does not explain the majority of reported cases, and thus the precise regions require further investigation with more detailed cytogenetic information and molecular data on the complex symptoms of gastrointestinal anomalies.
When combining the information from patient 1 and 2 of the present study with the results of previous studies involving patients with VACTERL syndrome, it is apparent that the urogenital/anorectal anomalies, congenital heart disease and gastrointestinal tract anomalies may involve common or overlapping regions of deletion in chromosome 13q, and suggests they may share a common molecular mechanism. Increasing numbers of microdeletions are being identified in patients using the array-CGH technique, and the morbid genes identified may therefore provide a greater understanding of the molecular mechanisms underlying chromosome 13q deletion syndrome. In addition, obtaining molecular data from non-morbid OMIM genes in knockout animal models may reveal the pathological processes during development.
Acknowledgements
The authors would like to thank the patients and their parents, as well as all the associated physicians who contributed to the analysis of patient samples and collection of clinical data. The present study was funded by the Liaoning Province Natural Science Foundation (grant no. 2013021021).
References
- 1.Allderdice PW, Davis JG, Miller OJ, Kliner HP, Warburton D, Miller DA, Allen FH, Jr, Abrams CA, McGilvray E. The 13q-deletion syndrome. Am J Hum Genet. 1969;21:499–512. [PMC free article] [PubMed] [Google Scholar]
- 2.Brown S, Gersen S, Anyane-Yeboa K, Warburton D. Preliminary definition of a ‘critical region’ of chromosome 13 in q32: Report of 14 cases with 13q deletions and review of the literature. Am J Med Genet. 1993;45:52–59. doi: 10.1002/ajmg.1320450115. [DOI] [PubMed] [Google Scholar]
- 3.Ballarati L, Rossi E, Bonati MT, Gimelli S, Maraschio P, Finelli P, Giglio S, Lapi E, Bedeschi MF, Guerneri S, et al. 13q Deletion and central nervous system anomalies: Further insights from karyotype-phenotype analyses of 14 patients. J Med Genet. 2007;44:e60. doi: 10.1136/jmg.2006.043059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Walczak-Sztulpa J, Wisniewska M, Latos-Bielenska A, Linné M, Kelbova C, Belitz B, Pfeiffer L, Kalscheuer V, Erdogan F, Kuss AW, et al. Chromosome deletions in 13q33-34: Report of four patients and review of the literature. Am J Med Genet A. 2008;146A:337–342. doi: 10.1002/ajmg.a.32127. [DOI] [PubMed] [Google Scholar]
- 5.Brown S, Russo J, Chitayat D, Warburton D. The 13q-syndrome: The molecular definition of a critical deletion region in band 13q32. Am J Hum Genet. 1995;57:859–866. [PMC free article] [PubMed] [Google Scholar]
- 6.Quélin C, Bendavid C, Dubourg C, de la Rochebrochard C, Lucas J, Henry C, Jaillard S, Loget P, Loeuillet L, Lacombe D, et al. Twelve new patients with 13q deletion syndrome: Genotype-phenotype analyses in progress. Eur J Med Genet. 2009;52:41–46. doi: 10.1016/j.ejmg.2008.10.002. [DOI] [PubMed] [Google Scholar]
- 7.Garcia NM, Allgood J, Santos LJ, Lonergan D, Batanian JR, Henkemeyer M, Bartsch O, Schultz RA, Zinn AR, Baker LA. Deletion mapping of critical region for hypospadias, penoscrotal transposition and imperforate anus on human chromosome 13. J Pediatr Urol. 2006;2:233–242. doi: 10.1016/j.jpurol.2006.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kirchhoff M, Bisgaard AM, Stoeva R, Dimitrov B, Gillessen-Kaesbach G, Fryns JP, Rose H, Grozdanova L, Ivanov I, Keymolen K, et al. Phenotype and 244k array-CGH characterization of chromosome 13q deletions: An update of the phenotypic map of 13q21.1-qter. Am J Med Genet Part A. 2009;149A:894–905. doi: 10.1002/ajmg.a.32814. [DOI] [PubMed] [Google Scholar]
- 9.Shojaei A, Behjati F, Derakhshandeh-Peykar P, Razzaghy-Azar M, Otukesh H, Kariminejad R, Dowlati MA, Rashidi-Nezhad A, Tavakkoly-Bazzaz J. Partial trisomy 7q and monosomy 13q in a child with disorder of sex development: Phenotypic and genotypic findings. Gene. 2013;517:137–145. doi: 10.1016/j.gene.2012.11.013. [DOI] [PubMed] [Google Scholar]
- 10.Yang YF, Ai Q, Huang C, Chen JL, Wang J, Xie L, Zhang WZ, Yang JF, Tan ZP. A 1.1Mb deletion in distal 13q deletion syndrome region with congenital heart defect and postaxial polydactyly: Additional support for a CHD locus at distal 13q34 region. Gene. 2013;528:51–54. doi: 10.1016/j.gene.2013.03.145. [DOI] [PubMed] [Google Scholar]
- 11.Dworschak GC, Draaken M, Marcelis C, et al. De novo 13q deletions in two patients with mild anorectal malformations as part of VATER/VACTERL and VATER/VACTERL-like association and analysis of EFNB2 in patients with anorectal malformations. Am J Med Genet A. 2013;161A:3035–3041. doi: 10.1002/ajmg.a.36153. [DOI] [PubMed] [Google Scholar]
- 12.McKay RD. The mechanism of G and C banding in mammalian metaphase chromosomes. Chromosoma. 1973;44:1–14. doi: 10.1007/BF00372569. [DOI] [PubMed] [Google Scholar]
- 13.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 14.Walsh LE, Vance GH, Weaver DD. Distal 13q Deletion Syndrome and the VACTERL association: Case report, literature review, and possible implications. Am J Med Gene. 2001;98:137–144. doi: 10.1002/1096-8628(20010115)98:2<137::AID-AJMG1022>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 15.Kaylor J, Alfaro M, Ishwar A, Sailey C, Sawyer J, Zarate YA. Molecular and cytogenetic evaluation of a patient with ring chromosome 13 and discordant results. Cytogenet Genome Res. 2014;144:104–108. doi: 10.1159/000368649. [DOI] [PubMed] [Google Scholar]
- 16.Cain CC, Saul DO, Oehler E, Blakemore K, Stetten G. Prenatal detection of a subtle unbalanced chromosome rearrangement by karyotyping, FISH and array comparative genomic hybridization. Fetal Diagn Ther. 2008;24:286–290. doi: 10.1159/000158519. [DOI] [PubMed] [Google Scholar]
- 17.Kataoka A, Hirakawa S, Iwamoto M, Sakumura Y, Yoshinaga R, Ohba T. Prenatal diagnosis of a case of partial monosomy/monosomy 13 mosaicism: 46,XX,r(13)(p11q33)/45,XX,-13 suspected by nuchal fold translucency increasing. Kurume Med J. 2011;58:127–130. doi: 10.2739/kurumemedj.58.127. [DOI] [PubMed] [Google Scholar]
- 18.Andresen JH, Aftimos S, Doherty E, Love DR, Battin M. 13q33.2 deletion: A rare cause of ambiguous genitalia in a male newborn with growth restriction. Acta Paediatr. 2010;99:784–786. doi: 10.1111/j.1651-2227.2010.01683.x. [DOI] [PubMed] [Google Scholar]
- 19.Mimaki M, Shiihara T, Watanabe M, Hirakata K, Sakazume S, Ishiguro A, Shimojima K, Yamamoto T, Oka A, Mizuguchi M. Holoprosencephaly with cerebellar vermis hypoplasia in 13q deletion syndrome: Critical region for cerebellar dysgenesis within 13q32.2q34. Brain Dev. 2015;37:714–718. doi: 10.1016/j.braindev.2014.10.009. [DOI] [PubMed] [Google Scholar]
- 20.McMahon CJ, Breathnach C, Betts DR, Sharkey FH, Greally MT. De Novo interstitial deletion 13q33.3q34 in a male patient with double outlet right ventricle, microcephaly, dysmorphic craniofacial findings, and motor and developmental delay. Am J Med Genet A. 2015;167A:1134–1141. doi: 10.1002/ajmg.a.36978. [DOI] [PubMed] [Google Scholar]
- 21.Valdes-Miranda JM, Soto-Alvarez JR, Toral-Lopez J, González-Huerta L, Perez-Cabrera A, Gonzalez-Monfil G, Messina-Bass O, Cuevas-Covarrubias S. A novel microdeletion involving the 13q31.3-q32.1 region in a patient with normal intelligence. Eur J Med Genet. 2014;57:60–64. doi: 10.1016/j.ejmg.2014.01.006. [DOI] [PubMed] [Google Scholar]
- 22.Huang C, Yang YF, Yin N, Chen JL, Wang J, Zhang H, Tan ZP. Congenital heart defect and mental retardation in a patient with a 13q33.1–34 deletion. Gene. 2012;498:308–310. doi: 10.1016/j.gene.2012.01.083. [DOI] [PubMed] [Google Scholar]
- 23.Czeizel A, Ludányi I. An aetiological study of the VACTERL-association. Eur J Pediatr. 1985;144:331–337. doi: 10.1007/BF00441773. [DOI] [PubMed] [Google Scholar]
- 24.McMullen KP, Karnes PS, Moir CR, Michels VV. Familial recurrence of tracheoesophageal fistula and associated malformations. Am J Med Genet. 1996;63:525–528. doi: 10.1002/(SICI)1096-8628(19960628)63:4<525::AID-AJMG3>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- 25.Quan L, Smith DW. The VATER association. Vertebral defects, anal atresia, T-E fistula with esophageal atresia, radial and rnal dysplasia: A spectrum of associated defects. J Pediatr. 1973;82:104–107. doi: 10.1016/S0022-3476(73)80024-1. [DOI] [PubMed] [Google Scholar]
- 26.Rogers DS, Paidas CN, Morreale RF, Huthcins GM. Septation of the anorectal and genitourinary tract in the human embryo: Crucial role of the catenoidal shape of the urorectal sulcus. Teratology. 2002;66:144–152. doi: 10.1002/tera.10041. [DOI] [PubMed] [Google Scholar]
- 27.Hynes PJ, Fraher JP. The development of the male genitourinary system. I. The origin of the urorectal septum and the formation of the perineum. Br J Plast Surg. 2004;57:27–36. doi: 10.1016/j.bjps.2003.08.019. [DOI] [PubMed] [Google Scholar]
- 28.Baskin LS, Erol A, Jegatheesan P, Li Y, Liu W, Cunha GR. Urethral seam formation and hypospadias. Cell Tissue Res. 2001;305:379–387. doi: 10.1007/s004410000345. [DOI] [PubMed] [Google Scholar]
- 29.Dravis C, Yokoyama N, Chumley MJ, Cowan CA, Silvany RE, Shay J, Baker LA, Henkemeyer M. Bidirectional signaling mediated by ephrin-B2 and EphB2 controls urorectal development. Dev Biol. 2004;271:272–290. doi: 10.1016/j.ydbio.2004.03.027. [DOI] [PubMed] [Google Scholar]
- 30.Jenkins D, Bitner-Glindzicz M, Thomasson L, Malcolm S, Warne SA, Feather SA, Flanagan SE, Ellard S, Bingham C, Santos L, et al. Mutational analyses of UPIIIA, SHH, EFNB2 and HNF1beta in persistent cloaca and associated kidney malformations. J Pediatr Urol. 2007;3:2–9. doi: 10.1016/j.jpurol.2006.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Firth HV, Richards SM, Bevan AP, Clayton S, Corpas M, Rajan D, Van Vooren S, Moreau Y, Pettett RM, Carter NP. DECIPHER: Database of chromosomal imbalance and phenotype in humans using ensembl resources. Am J Hum Genet. 2009;84:524–533. doi: 10.1016/j.ajhg.2009.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xin W, Mullen TE, Kiely R, Min J, Feng X, Cao Y, O'Malley L, Shen Y, Chu-Shore C, Mole SE, et al. CLN5 mutations are frequent in juvenile and late-onset non-Finnish patients with NCL. Neurology. 2010;74:565–571. doi: 10.1212/WNL.0b013e3181cff70d. [DOI] [PubMed] [Google Scholar]
- 33.Tanaka H, Moroi K, Iwai J, Takahashi H, Ohnuma N, Hori S, Takimoto M, Nishiyama M, Masaki T, Yanagisawa M, et al. Novel mutations of the endothelin B receptor gene in patients with Hirschsprung's disease and their characterization. J Biol Chem. 1998;273:11378–11383. doi: 10.1074/jbc.273.18.11378. [DOI] [PubMed] [Google Scholar]
- 34.Puffenberger EG, Hosoda K, Washington SS, Nakao K, deWit D, Yanagisawa M, Chakravarti A. A missense mutation of the endothelin-B receptor gene in multigenic Hirschsprung's disease. Cell. 1994;79:1257–1266. doi: 10.1016/0092-8674(94)90016-7. [DOI] [PubMed] [Google Scholar]
- 35.Verheij JB, Kunze J, Osinga J, van Essen AJ, Hofstra RM. ABCD syndrome is caused by a homozygous mutation in the EDNRB gene. Am J Med Genet. 2002;108:223–225. doi: 10.1002/ajmg.10172. [DOI] [PubMed] [Google Scholar]
- 36.Abelson JF, Kwan KY, O'Roak BJ, Baek DY, Stillman AA, Morgan TM, Mathews CA, Pauls DL, Rasin MR, Gunel M, et al. Sequence variants in SLITRK1 are associated with Tourette's syndrome. Science. 2005;310:317–320. doi: 10.1126/science.1116502. [DOI] [PubMed] [Google Scholar]
- 37.de Pontual L, Yao E, Callier P, Faivre L, Drouin V, Cariou S, Van Haeringen A, Geneviève D, Goldenberg A, Oufadem M, et al. Germline deletion of the miR-17-92 cluster causes skeletal and growth defects in humans. Nat Genet. 2011;43:1026–1030. doi: 10.1038/ng.915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Albano LM, Oliveira LA, Bertola DR, Mazzu JF, Kim CA. Omodysplasia: The first reported Brazilian case. Clinics (Sao Paulo) 2007;62:531–534. doi: 10.1590/S1807-59322007000400023. [DOI] [PubMed] [Google Scholar]
- 39.Elcioglu NH, Gustavson KH, Wilkie AO, Yüksel-Apak M, Spranger JW. Recessive omodysplasia: Five new cases and review of the literature. Pediat Radiol. 2004;34:75–82. doi: 10.1007/s00247-003-1064-9. [DOI] [PubMed] [Google Scholar]
- 40.Lukong CS, Mshelbwala PM, Anumah MA, Ameh EA, Nmadu PT. Anorectal malformation coexisting with Hirschsprung's disease: A report of two patients. Afr J Paediatr Surg. 2012;9:166–168. doi: 10.4103/0189-6725.99409. [DOI] [PubMed] [Google Scholar]
- 41.Metts JC, III, Kotkin L, Kasper S, Shyr Y, Adams MC, Brock JW., III Genital malformations and coexistent urinary tract or spinal anomalies in patients with imperforate anus. J Urol. 1997;158:1298–1300. doi: 10.1016/S0022-5347(01)64460-4. [DOI] [PubMed] [Google Scholar]
- 42.Raboei EH. Patients with anorectal malformation and Hirschsprung's disease. Eur J Pediatr Surg. 2009;19:325–327. doi: 10.1055/s-0029-1224131. [DOI] [PubMed] [Google Scholar]
- 43.Jobanputra V, Wilson A, Shirazi M, Feenstra H, Levy B, Anyane-Yeboa K, Warburton D. Partial uniparental disomy with mosaic deletion 13q in an infant with multiple congenital anomalies. Am J Med Genet A. 2013;161A:2393–2395. doi: 10.1002/ajmg.a.36040. [DOI] [PubMed] [Google Scholar]
- 44.Hanson KP, Jung JP, Tran QA, Hsu SP, lida R, Ajeti V, Campagnola PJ, Eliceiri KW, Squirrell JM, Lyons GE, Ogle BM. Spatial and temporal analysis of extracellular matrix proteins in the developing murine heart: A blueprint for regeneration. Tissue Eng Part A. 2013;19:1132–1143. doi: 10.1089/ten.tea.2012.0316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schenke-Leyland K, Nsair A, Van Handel B, Angelis E, Gluck JM, Votteler M, Goldhaber JI, Mikkola HK, Kahn M, MacLellan WR. Recapitulation of the embryonic cardiovascular progenitor cell niche. Biomaterials. 2011;32:2748–2756. doi: 10.1016/j.biomaterials.2010.12.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Adams RH, Wilkinson GA, Weiss C, Diella F, Gale NW, Deutsch U, Risau W, Klein R. Roles of ephrinB ligands and EphB receptors in cardiovascular development: Demarcation of arterial/venous domains, vascular morphogenesis, and sprouting angiogenesis. Genes Dev. 1999;13:295–306. doi: 10.1101/gad.13.3.295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang HU, Chen ZF, Anderson DJ. Molecular distinction and angiogenic interaction between embryonic arteries and veins revealed by ephrin-B2 and its receptor Eph-B4. Cell. 1998;93:741–753. doi: 10.1016/S0092-8674(00)81436-1. [DOI] [PubMed] [Google Scholar]
- 48.Cowan CA, Yokoyama N, Saxena A, Chumley MJ, Silvany RE, Baker LA, Srivastava D, Henkemeyer M. Ephrin-B2 reverse signaling is required for axon pathfinding and cardiac valve formation but not early vascular development. Dev Biol. 2004;271:263–271. doi: 10.1016/j.ydbio.2004.03.026. [DOI] [PubMed] [Google Scholar]
- 49.Batlle E, Henderson JT, Beghtel H, van den Born MM, Sancho E, Huls G, Meeldijk J, Robertson J, van de Wetering M, Pawson T, Clevers H. Beta-catenin and TCF mediate cell positioning in the intestinal epithelium by controlling the expression of EphB/ephrinB. Cell. 2002;111:251–263. doi: 10.1016/S0092-8674(02)01015-2. [DOI] [PubMed] [Google Scholar]
- 50.Batlle E, Bacani J, Begthel H, Jonkheer S, Gregorieff A, van de Born M, Malats N, Sancho E, Boon E, Pawson T, et al. EphB receptor activity suppresses colorectal cancer progression. Nature. 2005;435:1126–1130. doi: 10.1038/nature03626. [DOI] [PubMed] [Google Scholar]