Abstract
An agonist that acts through a single receptor can activate numerous signaling pathways. Recent studies have suggested that different ligands can differentially activate these pathways by stabilizing a limited range of receptor conformations, which in turn preferentially drive different downstream signaling cascades. This concept, termed “biased signaling” represents an exciting therapeutic opportunity to target specific pathways that elicit only desired effects, while avoiding undesired effects mediated by different signaling cascades. The cannabinoid receptors CB1 and CB2 each activate multiple pathways, and evidence is emerging for bias within these pathways. This review will summarize the current evidence for biased signaling through cannabinoid receptor subtypes CB1 and CB2.
Keywords: : agonist bias, cannabinoid receptors, functional selectivity, G protein-coupled receptor
Introduction
Identifying and characterizing the molecular determinants of agonist efficacy in signaling pathway activation are a vital requisite of contemporary drug design. One of the determinants of agonist efficacy is the molecular structure of the agonist, and thus the receptor conformation that it induces. However, receptor conformation is also affected by interactions with various intracellular signaling proteins.1–3 For example, the conformation of the β2 adrenergic receptor has been demonstrated to be quite distinct in the presence of the second messenger protein Gs.2,3 Receptor activation will therefore be both ligand and tissue specific, as the assortment and abundance of intracellular signaling constituents vary between cell types. Biased signaling is the concept that different ligands acting on the same G protein-coupled receptor (GPCR), in the same tissue, can give rise to markedly different cellular responses (Fig. 1), and this is likely due to each ligand stabilizing different receptor conformations. This concept has been given many different names—“stimulus trafficking,” “functional selectivity,” and more recently, “agonist bias” or “biased signaling.” It is important to note that differential signaling pathway activation by different agonists can probably also arise as a consequence of kinetics; if there are significant differences in agonist binding kinetics, the more slowly dissociating ligands may allow receptor conformations that favor low-affinity interactions for a particular receptor/signaling molecule pair to persist long enough for productive coupling.
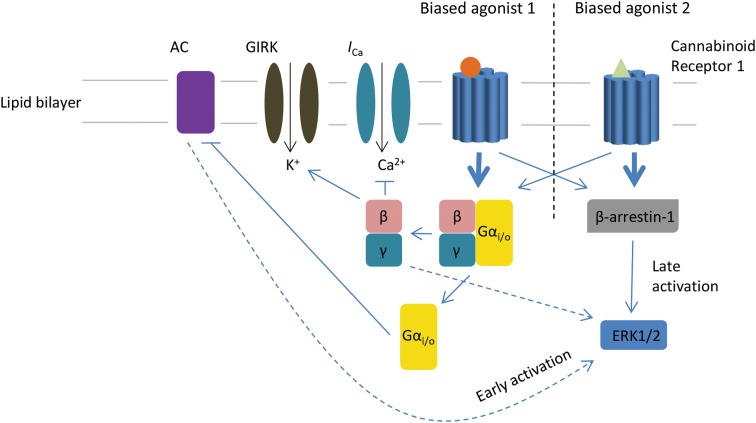
FIG. 1.
Biased agonist 1 or 2 binds to the seven-transmembrane cannabinoid receptor 1. Structurally different ligands will induce diverse conformations of the receptor, which may then favor one of the possible signaling pathways over others. In this diagram, agonist 1 is biased toward the activation of the Gαi/o heterotrimer over β-arrestin-1, while agonist 2 favorably activates β-arrestin-1. Activation of Gαi/o prompts the release of the Gβγ subunit, which inhibits voltage-dependent calcium channels (ICa) and activates GIRK. The Gαi/o subunit inhibits AC, which stimulates the phosphorylation and early activation of ERK1/2. The activation of β-arrestin-1 conversely induces late activation of ERK1/2. AC, adenylyl cyclase; ERK1/2, extracellular signal-regulated kinase 1 or 2; GIRK, G protein-gated inwardly rectifying potassium channels.
Traditional approaches to demonstrating bias have focused on comparisons of EC50 or Emax values within different pathways, but such methods may not account for inherent differences between pathway stoichiometry. For example, some pathways may achieve maximum response at lower receptor occupancy, resulting in higher potency for all agonists within this pathway (pathway bias). Quantifying biased signaling involves determining the effects of two or more agonists on two or more cellular responses, and comparing the agonist profiles for each pathway. More recently, the operational model of bias has been utilized4; this approach compares all ligands within each pathway against a reference ligand and then makes comparisons of the relative shifts between pathways relative to the reference.
Therapeutically, it is hoped that the study of biased signaling of GPCR-directed therapeutics will enable engagement of desired pathways over those not involved in the therapeutic effect. Ideally, this would eliminate on-target but unwanted or adverse effects; agonists or allosteric modulators that induce signaling in a biased manner could potentially revolutionize future therapeutic drugs. The μ-opioid receptor presents a prime example where biased signaling might be exploited at the translational level. Although opioid receptors are useful targets of analgesics, μ-opioid receptor activation also causes respiratory depression, which is suggested to be a product of β-arrestin2 recruitment.5 Hence, the development of an agonist that preserves the analgesic (G protein-mediated) properties of activated μ-opioid receptors without any respiratory side effects would be beneficial to avoid adverse effects, and such compounds have recently been developed in light of this hypothesis.6,7
The cannabinoid receptor family consists of two GPCRs, cannabinoid receptor 1 (CB1) and cannabinoid receptor 2 (CB2), and they will be the focus of this review. CB1 plays a role in regulating neurotransmission in many brain regions. When activated, CB2 regulates immune responses and inflammatory pathways. Mice lacking CB2 often demonstrate an exacerbated inflammatory phenotype8 and besides roles in the periphery expression in brain microglia suggest a role for CB2 in neuroinflammation.9,10 Recently, CB2 has been suggested to contribute to neuronal plasticity in mouse hippocampal neurons11 potentially expanding the role of this receptor in the brain. The endogenous lipids anandamide and 2-arachidonoylglycerol (2-AG) are the physiological cannabinoid receptor agonists and are hence known as endocannabinoids.12–14 A great deal of interest has centered on the potential role of CB1 in targeting a range of central nervous system (CNS) disorders such as pain,15 anxiety,16 multiple sclerosis,17 obesity,18 nicotine addiction,19 Huntington disease,20 and Parkinson's disease.21 In more recent times CB2 has also become a focus in peripheral inflammatory disorders such as nephrotoxicity.8,22
In addition to the two well-established cannabinoid receptors, several other GPCRs have been reported to be activated by cannabinoid drugs or endocannabinoids and related molecules, including GPR55,23 GPR18,24 and GPR119.25 Furthermore, endogenous and synthetic cannabinoids can activate and potentiate transient receptor potential (TRP) channels26 and glycine receptors,27 respectively. The potential contribution of these receptors to the therapeutic effects of cannabinoids or the physiological effects of endocannabinoids are only beginning to be explored, and much less is known about how ligands regulate their signaling.
The widespread distribution of CB1 in the CNS provides a strong rationale for developing ligands with biased profiles to potentially avoid the consequences of activating multiple signaling pathways in many different brain regions. Biased ligands could also potentially have context-dependent effects, providing effective modulation of pathways dysregulated by disease in restricted subsets of neurons. Modulating selected downstream pathways would result in a more targeted pharmacological response, but considerable research is still required to understand which of the characterized pathways are therapeutically desirable, and under which disease condition.
Signaling of Cannabinoid Receptors
G protein coupling of cannabinoid receptors
First cloned in the early 1990s,28,29 both CB1 and CB2 were initially described as exhibiting pertussis toxin-sensitive signaling through Gαi/o-type G proteins,30 however, differences in signaling were reported from the outset. When expressed in AtT-20 cells, CB1 mediated an inhibition of P/Q-type calcium channels and an activation of G protein-gated inwardly rectifying potassium channels (GIRKs) in addition to inhibition of adenylyl cyclase.30 By contrast, CB2 was not reported to modulate the activity of either channel in AtT-20 cells but did inhibit adenylyl cyclase.30 This poor coupling may be an example of functional selectivity as it was recently found that mouse CB2 (mCB2) can inhibit voltage-gated calcium channels, in a manner strongly dependent on the CB2 ligand used, with CP55,940, but not WIN55,212-2, able to inhibit the channels via a Gβγ pathway.31 It is unclear how far this apparent biased signaling extends, however, as both WIN55,212-2 and CP55,940 hyperpolarize AtT-20 cells expressing human CB2 (hCB2), and this response almost certainly reflects Gβγ subunit-mediated activation of GIRKs.32
In studies using purified G proteins, Glass and Northup33 demonstrated that while CB1 and CB2 had similar affinity for Gαi-type G proteins, CB2 had significantly lower affinity for Gαo than Gαi. CB1 is promiscuous in its G protein coupling, with activation of Gαs- and Gαq-dependent signaling under some conditions, or in some cells. Gαs-like coupling has been suggested under circumstances where Gαi activation is limited, such as following pertussis toxin treatment, or simultaneous activation of CB1 with other Gαi linked receptors such as D234–36; indeed it appears that in some cell types the Gαs linkage is dominant.37,38 Studies have suggested that the second intracellular loop of CB1 mediates both Gαs and Gαi coupling specificity,39 but a detailed understanding of what regulates the specific G protein coupling is still lacking. Gαq coupling of CB1 has been suggested for only a few ligands40 and will be discussed in more detail below. Finally, in vivo studies have also suggested coupling of CB1 to the Gαi/o-related G protein Gαz.41 The ability to activate such a diverse range of G proteins strongly suggests that biased signaling between different G protein pathways could be achieved, and examples of this are emerging and described below. Apart from the interactions with Gαi and Gαo less is known about additional G proteins interacting with CB2.
The consequences of activating Gαi, Gαo, Gαs, and Gαq heterotrimers have been described for many GPCRs, and this signaling seems similar for the most part for CB1 and CB2. Gαi/o subunits inhibit adenylyl cyclase or couple to the mitogen-activated protein kinase (MAPK) pathway. Gαs stimulates adenylyl cyclase (and subsequent phosphorylation of cAMP response element-binding protein [CREB]). Gαq couples to phospholipase C and promotes release of intracellular calcium ([Ca]i). Gβγ subunits derived from Gαi/o activate GIRKs (comprising KIR 3.X heteromers), activate phosphatidylinositide-3-kinase, and inhibit voltage-dependent calcium channels (ICa).42–44
Originally described as “G protein-coupled” proteins based on their modulation of classical signaling pathways via heterotrimeric G proteins, GPCRs also recruit other proteins for signaling, most prominently β-arrestin-1 and -2. Arrestins serve multiple functions as regulators of G protein coupling and receptor localization, and are essential elements of multiple GPCR signaling cascades involving kinases, phosphatases, and ubiquitin ligases.45 Arrestin signaling has been generally thought to occur in the absence of bound G protein, and although this has been recently challenged,46 arrestin-mediated signaling is likely to be mediated by a different ligand/receptor conformation from those involved in Gα interactions.
Activation of MAPK cascades
MAPK family members have been found to regulate diverse biological functions by phosphorylation of specific target molecules (such as transcription factors) and thereby participate in the regulation of a variety of cellular processes, including cell proliferation, differentiation, and apoptosis.47
As for most GPCRs, there are multiple pathways by which CB1 activation can lead to phosphorylation of extracellular signal-regulated kinase 1 or 2 (ERK1/2). In Chinese hamster ovary (CHO), U373 MG, and PC-3 cells, phosphatidylinositide 3-kinase activation is required for CB1 activation of ERK1/2, and the Gβγ subunit, rather than Gα, may transduce the CB1 signal.48,49 In contrast to this, ERK1/2 activation in N1E-115 mouse neuroblastoma cells, or mouse hippocampal slices, is reported to be downstream of inhibition of cyclic adenosine monophosphate (cAMP)/protein kinase A.50,51
CB1 may also transactivate members of the receptor tyrosine kinase family leading to subsequent activation of ERK1/2. Several tyrosine kinase receptors have been implicated in this pathway, including the vascular endothelial growth factor receptor52 and epidermal growth factor receptor.53 The Src-family kinase, Fyn, is also likely to be involved in the activation of ERK1/2 by the CB1-transactivated tyrosine kinase receptors, as Fyn knockout (KO) mice show no elevation of phosphorylated ERK1/2 (pERK1/2) in response to cannabinoid administration.51
Regardless of the precise pathway mediating pERK1/2 downstream of CB1 activation, all of the above studies found the activation to be downstream of Gαi/o protein signaling (and therefore pertussis toxin sensitive), however, pertussis toxin-insensitive ERK1/2 activation has also been demonstrated in rat CB (rCB1)-transfected human embryonic kidney 293 (HEK293) cells.54 The Gαi/o protein-independent pERK1/2 activation might arise from the recruitment of β-arrestin-1 as mutation of phosphorylation sites in the CB1 C-terminal (S426A/S430A) has been suggested to result in β-arrestin-1-mediated pERK activation,55 which in general occurs later than the G protein-mediated activation.56 Similarly, as discussed below, ORG27569, an allosteric modulator of CB1, has been suggested by some studies57,58 but not others59,60 to activate β-arrestin-1-mediated pERK1/2 signaling through CB1.
Activation of other MAPKs downstream of CB1 has been reported, although there is considerable divergence between tissues and ligands used among the studies, which makes reconciling contrasting results difficult. In rat hippocampal slices, anandamide, CP55,940, WIN55,212-2, and Δ9-tetrahydrocannabinol (THC) activate p38 MAPK, but not c-Jun N-terminal kinase (JNK).61 In cultured cortical neurons, JNK was activated when stimulating with THC in a pertussis toxin-sensitive manner,62 however, HU-210 failed to stimulate ERK1/2, p38 MAPK, or JNK in a subsequent study on cultured hippocampal neurons.63 In Neuro-2a cells endogenously expressing mCB1, HU-210 activated only ERK1/2, but not JNK or p38 MAPK pathways,64 although a different study found that HU-210 stimulated JNK activation in these cells.65 Rueda et al.66 showed that THC-mediated activation of JNK was dependent on Gαi/o proteins, phosphatidylinositide 3-kinase, and Ras, and involved platelet-derived growth factor (PDGF) receptor transactivation in CHO cells. In contrast, in the same cells, THC activation of p38 MAPK was not dependent on PDGF receptor activation,66 suggesting the possibility of multiple CB1-stimulated pathways for kinase activation in the same cells.
There are relatively few studies on CB2 activation of MAPK pathways. While several studies have investigated the ability of cannabinoid agonists to modulate ERK1/2 activation in response to inflammatory stimuli,67,68 only one study has examined the pathway by which CB2 may link to ERK activation. In this study, hCB2 coupling to ERK1/2 was shown to be pertussis toxin sensitive but independent of cAMP.69 Some of the variability observed in the pathways activating kinases downstream of both CB1 and CB2 may reflect different levels of receptor expression between cells as previous studies have demonstrated that the ability to activate some kinases (most notably pAkt) required high receptor expression, whereas ERK1/2 activation occurred at all levels of expression.70
Desensitization, arrestin recruitment, and signaling
A common pathway for uncoupling GPCRs from G protein-dependent signaling is receptor phosphorylation followed by binding of an arrestin.71 In this sequence of events, G protein receptor kinases (GRKs) have a prominent role phosphorylating the GPCR after agonist binding, and this phosphorylation increases the affinity of arrestin for the receptor. Arrestin binds to domains of the receptor thought to interact with G proteins, preventing receptor activation of these effectors. It is also possible that phosphorylation of a receptor before arrestin binding disrupts receptor/G protein interactions. The ultimate consequences of arrestin binding are complex; for many receptors, arrestin binding is a step in a pathway that leads to removal of the receptor from the plasma membrane, however, arrestin also acts as a scaffolding molecule for non-G protein-mediated signaling pathways such as activation of ERK.
In a classic series of studies, Howlett et al. delineated CB1-mediated inhibition of agonist-stimulated adenylyl cyclase activity in N18TG2 cells.72 This work included showing that desacetyllevonantradol (CP54,939) inhibition of adenylyl cyclase maximally desensitized within 30 min of drug exposure.72 The desensitization of CB1 responses was homologous, as it did not affect the muscarinic receptor-mediated inhibition of adenylyl cyclase and did not affect the maximum accumulation of cAMP produced by the stimulatory agonist, secretin.73 THC also produced desensitization in this assay, but it was slower and less complete than that produced by CP54,939. Similar effects of CP55,940 on hCB1 expressed in CHO cells indicate that rapid desensitization of CB1 coupling to adenylyl cyclase is common.74 While these assays provide a good indication that receptor desensitization is rapid, interpretation of the quantitative elements of the studies is difficult as the assays utilized 20 min or more of agonist exposure to define control responses, and this period almost certainly encompasses many significant regulatory events.
By contrast to traditional single-point assays of cAMP accumulation or ERK phosphorylation, GPCR modulation of ion channels provides a continuous and relatively direct readout of receptor activity, particularly when Gβγ inhibition of ICa or activation of GIRK is measured.75 Unfortunately, there are very few cell lines where native CB1 receptors coexist with voltage-gated ICa or GIRK. NG-108-15 and N18 cells, with endogenous CB1, were used for the very earliest descriptions of CB1 receptor inhibition of ICa,76–78 however, receptor regulation was not examined. Recombinant CB1 receptors expressed in murine AtT-20 cells couple to both inhibition of ICa and activation of GIRK,79 and desensitization of GIRK activation has been reported.80 AtT-20 cells and Xenopus oocytes have been used to provide some insight into CB1 regulation of ion channels.80,81 In oocytes, desensitization of rCB1-mediated activation of GIRK was shown to be stimulated by coexpression of both GRK3 and β-arrestin-2, but not affected by either protein alone.80 The efficiency of other members of the GRK family, or of β-arrestin-1, was not addressed in the oocyte studies, nor were the effects of coexpression of these molecules on basal coupling of CB1 to GIRK.
Mutation of two of six available serine/threonine residues in the C-terminal tail (S426/S430) of rCB1 was sufficient to block GRK3/β-arrestin-2-mediated desensitization of coupling to GIRK in oocytes.80 When CB1 missing the last 55 intracellular residues was expressed in AtT-20 cells, WIN55,212-2 desensitization was abolished suggesting a role for the putative GRK phosphorylation sites contained in the missing domain in desensitization. Unfortunately, coupling of the S426A/S430A mutant to native GIRK in AtT-20 cells was not measured, and the role of GRK3 (or other GRK family members) in regulation of CB1 in these cells has not been directly addressed.
Direct recruitment of β-arrestin-2 to activated CB1 has also been demonstrated in both AtT20 and HEK293 cells.54,82 In a detailed study of C-terminal mutants, Daigle et al.82 found that all internalization-competent CB1 mutants could recruit β-arrestin-2, although some mutants appeared to recruit β-arrestin-2 at a reduced rate, while mutation of all six serine/threonine residues in the rCB1 C-terminus (460–473) prevented internalization and also failed to recruit β-arrestin-2. A recent detailed bioluminescence resonance energy transfer (BRET)-based study of CB1 arrestin interactions, suggested a low-affinity, transient interaction between CB1 and β-arrestin-2, with no interaction in late endosomes consistent with a family-A interaction, while recruitment of β-arrestin-1 by orthosteric ligands was not observed.83 Structural studies have suggested an interaction between β-arrestin-1 with a synthesized CB1 C-terminus,84,85 and this has recently been observed in a whole cell.86
β-arrestin-2 KO mice revealed a role of arrestin in CB1-mediated signaling. THC produced both greater antinociception and greater decreases in body temperature in β-arrestin-2 KO mice compared with wild-type mice, consistent with a role for arrestins in blunting receptor signaling, however, the action of a range of synthetic ligands was normal.87 Tolerance to THC antinociceptive effects was also reduced in KO mice and decreased downregulation of CB1 was observed.88 These studies could suggest substantial agonist differences in arrestin recruitment for different assays, which requires further study. Alternatively, as THC is a partial agonist at CB1, it may be more sensitive to subtle changes in receptor availability as presumably it requires full occupancy to exert maximum effect. Finally, as with all studies of global KO animals, it is possible that the signaling of many GPCRs in the circuits that mediate cannabinoid effects is altered, and has been since the start of the animals' life. Single-cell studies examining CB1 receptor function and regulation have not be done using cells from arrestin KO animals. Studies on Gαi/o-coupled μ-opioid receptors in single neurons from KO animals show that there is not always a clear correlation between changes in agonist actions at these neurons and with changes in behavior.89,90
Surprisingly, there has been little detailed study of CB2-mediated arrestin recruitment. McGuinness et al.91 and Dhopeshwarkar and Mackie92 utilized the PathHunter DiscoveRx assay to investigate the ability of a range of cannabinoid ligands to recruit β-arrestin-2 at hCB2 and mCB2, respectively. This assay utilizes enzyme complementation to detect recruitment of tagged arrestin to the receptor. McGuinness et al. observed robust and potent recruitment of arrestin to a range of cannabinoid ligands, including CP55,940, JWH015, and WIN55,212-2.91 Dhopeshwarkar and Mackie found that nonclassic ligands (CP55,940) recruited arrestin, whereas classic cannabinoid ligands (THC, JWH133, KM233, etc.) did not, suggesting a strong functional bias with structure.92 Recently, Soethoudt et al.32 also investigated the ability of a wide range of ligands to activate arrestin utilizing this assay and found that all ligands modulated arrestin recruitment to some extent. CP55,940 was the most potent ligand in this assay, followed by WIN55,212-2. Only CP55,940 acted as full agonist, while WIN55,212-2, JWH133, JWH015, HU-308, HU-910 were partial (Emax 50–70%) and the endocannabinoids (Emax 40–80%) only partially recruited arrestin.32 Interestingly, JWH133, HU-308, and HU-910 were all significantly less potent on mCB2 than hCB2 in arrestin recruitment, strongly suggesting species differences. A potential confounding issue with all these assays is the protein modules added to the receptor and arrestin constructs to enable visualization of the interactions using optical techniques. The signaling capacity of these constructs is not usually assessed, and so it is not obvious that the conformational changes being reported faithfully reflect those of the native receptor and effector. In summary, while there is some evidence to suggest that arrestin-biased signaling is observed between cannabinoid ligands, the current lack of consistency between studies needs to be resolved to enable the design of genuinely biased ligands.
Post-endocytic regulation of cannabinoid receptors
CB1 undergoes rapid internalization following agonist stimulation.93,94 While early studies suggested that CB1 may recycle following internalization,94,95 subsequent detailed studies have suggested that the internalized receptor enters degradative pathways96 and is rapidly degraded, with resensitization requiring the delivery of newly synthesized receptors.93
Internalization of CB1 has generally been shown to be clathrin mediated,94,97 although in some systems, caveolae-mediated internalization has been observed.97 Mutation of a highly conserved aspartate residue in the second intracellular loop (D164 in rCB1) resulted in a loss of CB1 internalization in AtT20 cells.81 Intriguingly, receptors with this mutation demonstrated preserved binding, cAMP inhibition, and inhibition of Ca2+ currents but did not activate GIRK.81 This mutation has also been suggested to decrease the constitutive activity of CB1.98 Further studies identified the extreme carboxy-terminal tail of the rCB1 as a central mediator of agonist-induced internalization of the receptor in AtT20 cells.94 Subsequently, truncation of this region was also shown to prevent arrestin recruitment to CB1 in AtT20 cells.82 However, the truncated receptor internalized normally when expressed in HEK293 cells.82 Mutation of all of the serine/threonine residues in this region resulted in failure to recruit arrestin and a decreased extent of internalization, although the initial rates were normal.82 Changes in the extent of internalization are challenging to interpret, as higher expression levels frequently result in a decreased extent of internalization of receptors in transfected cell lines, likely due to saturation of the internalization machinery. Thus, the precise mechanisms controlling internalization kinetics are far from clear.
Studies on CB1 trafficking have been complicated by the presence of a large intracellular pool of receptors observed in both primary and recombinant cell lines, and native expressing tissues.93 While it was predicted that these receptors represented a pool of receptors from which CB1 could be rapidly mobilized to the cell surface,95 mobilization has not yet been observed, although the receptors do undergo constitutive synthesis and degradation.93 Recent studies have suggested that these intracellular receptors may be involved in signaling,99,100 a suggestion made more feasible by the high lipid solubility of the endocannabinoid ligands. Intracellular CB1 has recently been suggested to be present in mitochondria in neurons and astrocytes, suggested to signal through mitochondrial G proteins leading to inhibition of cAMP and mitochondrial respiration.101,102
Studies detailing the trafficking properties of CB2 are limited. Early studies utilizing an antibody that failed to detect hCB2 when phosphorylated at serine 352 suggested that CB2 undergoes agonist-mediated phosphorylation in response to CP55,940. The phosphorylation was accompanied by decreased signaling in both CHO and HL60 cells.103,104 Agonist-mediated internalization has also been observed in response to CP55,940103,105 and 2-AG.103,106 Early studies suggested that CB2 was repeatedly phosphorylated and dephosphorylated by alternate stimulation with agonist and inverse agonist implying that CB2 recycling may occur,69 and this was more recently demonstrated to be the case, and to be Rab11 dependent.107 The effect of the inverse agonist SR144528 on CB2 internalization is unclear as some find a decrease in cell surface CB2,103 while others find no change.106 One recent study has suggested that chronic treatment with a cannabinoid agonist leads to CB2-mediated upregulation of GRK5 in rat prefrontal cortex via a β-arrestin-2-dependent pathway,108 but further molecular studies are required.
For many receptors, bias has been observed in the ability of ligands to induce internalization.109 The consequences of such bias are not straightforward to interpret. We can speculate that if a receptor remained on the cell surface longer in the presence of a particular ligand, it might exhibit prolonged signaling, however, equally, if the receptors are desensitized but remain on the cell surface, feedback mechanisms governing the recycling of the receptor (for CB2) or delivery of new receptor to the cell surface (for CB1) might be altered leading to an overall reduction in signaling.
Biased signaling of cannabinoid receptors
The diverse chemical structures of commonly used cannabinoid ligands increase the likelihood of identifying biased signaling, and evidence of agonist bias has emerged from a number of studies.
The first evidence for biased signaling through CB1 came from assays in which membranes of CB1 expressing Sf9 cells, stripped of their endogenous G proteins, were reconstituted with purified G proteins.33 These studies demonstrated that the relative activation of Gαi and Gαo is dependent on the agonist. HU-210, WIN55,212-2, and anandamide all elicited maximal Gαi activation, whereas THC caused only partial Gαi activation. In contrast, only HU-210 effected maximal CB1 stimulation of Gαo, with anandamide, WIN55,212-2, and THC all partially stimulating.33 This work was further extended by coimmunoprecipitation of activated G proteins in N18TG2 cells, which demonstrated that WIN55,212-2 behaved as a full agonist for all three Gαi subtypes, while methanandamide appeared to be an agonist at Gαi3 and an inverse agonist at Gαi1 and Gαi2.110 In addition, relative signaling efficacies and potencies of CB1 ligands differ in different brain regions,111–113 which may represent the different G protein compositions of different regions. Intriguingly, plasmon-waveguide resonance (PWR) spectroscopy has demonstrated that CP55,940 and WIN55,212-2 produced distinct spectral changes (PWR shifts in opposite directions) on binding to the hCB1 indicating that the two agonists produce qualitatively distinct active conformations of the receptor, which have differing affinity for Gαi.114 Differential signaling by WIN55,212-2 and CP55,940 is consistent with the suggestion that these ligands have overlapping but distinct binding sites,115,116 a finding supported by molecular docking in the recently described crystal structure of CB1.117,118
Laprairie et al.20 investigated the biased signaling of WIN55,212-2, CP55,940, 2-AG, anandamide, THC, cannabidiol, and the combination THC+cannabidiol on several signaling pathways. The agonists were used on in vitro medium spiny projection neurons having a wild-type (STHdhQ7/Q7) or Huntington disease (STHdhQ111/Q111) background. The effect on a range of cannabinoid-dependent signaling pathways was measured via pERK1/2 (Gαi/o mediated), β-arrestin-1 recruitment to CB1 (by BRET), phosphorylation of CREB (pCREB; suggested to be Gαs mediated), phosphorylation of phospholipase C (pPLCβ3; suggested to be Gαq mediated), and phosphorylation of Akt (pAkt) (Gβγ mediated). The signaling bias was calculated relative to WIN55,212-2 signaling. This study found that CP55,940 induced signaling biased toward Gαs and β-arrestin-1 compared to Gαi/o, while Gαi/o signaling was biased compared to Gαq and Gβγ in both cell types (i.e., Gαs > β-arrestin-1 > Gαi/o > Gαq >Gβγ). 2-AG elicited signaling bias toward Gβγ compared to Gαi/o (in STHdhQ7/Q7 cells), and Gαi/o-biased signaling compared to β-arrestin-1 (in STHdhQ7/Q7 cells) and Gαq (predominantly in STHdhQ111/Q111) (i.e., Gβγ > Gαi/o > β-arrestin-1 > Gαq). Similar to 2-AG, anandamide signaling was biased toward Gβγ compared to Gαi/o (in STHdhQ7/Q7 cells), while Gαi/o signaling was biased compared to β-arrestin-1 and Gαq (mostly in STHdhQ111/Q111) (i.e., Gβγ >Gαi/o > β-arrestin1 > Gαq). THC produced signaling biased toward β-arrestin-1, Gαq, and Gβγ compared to Gαi/o in both cell types (i.e., β-arrestin-1 >Gαq=Gβγ > Gαi/o). Since cannabidiol treatment only evoked significant Gαs-mediated pCREB, bias values could not be calculated for this ligand. The combination of THC+cannabidiol induced signaling biased toward Gαs compared with Gαi/o, while signaling was biased toward Gαi/o compared with β-arrestin-1, Gαq, and Gβγ (predominantly in STHdhQ7/Q7 cells) (i.e., Gαs > Gαi/o > β-arrestin1=Gαq=Gβγ).
Khajehali et al.60 investigated biased signaling and allosteric modulation between cAMP and ERK1/2 activation using CHO cells stably expressing CB1 treated with CP55,940, HU-210, WIN55,212-2, THC, methanandamide, anandamide, and 2-AG. pERK1/2 and cAMP levels were measured in response to the treatments. The signaling bias induced by each ligand was calculated relative to 2-AG. CP55,940, HU-210, WIN55,212-2, THC, methanandamide, and anandamide all showed a preference toward cAMP inhibition compared to pERK1/2. HU-210 and methanandamide displayed strong bias toward cAMP inhibition, whereas CP55,940, THC, and anandamide showed nonsignificant bias toward cAMP inhibition. As both these pathways are pertussis toxin sensitive in these cells, this may indicate bias between α- and βγ-mediated pathways (although the exact pathway for pERK1/2 activation was not defined). The mechanism by which α and βγ bias could be mediated is currently unclear.
The putative Gαs coupling of CB1 has also shown potential for agonist bias. While equivalent rank order of potencies was observed for inhibiting or stimulating cAMP in CHO-hCB1 cells, anandamide and CP55,940 were significantly less efficacious in stimulating the accumulation of cAMP than in inhibiting its formation.119 Forskolin acts synergistically with CB1-activated Gαs at adenylyl cyclase.35 Cannabinoid receptor-mediated stimulation of cAMP also revealed differences among agonists in as much as forskolin enhanced the potency of HU-210 and CP55,940 by ∼100-fold but had no effect on the potency of WIN55,212–2 or anandamide.119
Perhaps the most extreme example of agonist bias for one G protein over another through CB1 is that observed in the proposed coupling of CB1 to Gαq. Lauckner et al.40 demonstrated that high concentrations of WIN55,212-2, but not THC, HU-210, 2-AG, or methanandamide resulted in increased release of [Ca]i through a Gαq pathway. More recently, rat hippocampal autaptic long-term potentiation has been suggested to be mediated by 2-AG-induced CB1 activation of Gαq, suggesting that biased signaling may not hold between different cell types.120 N-arachidonoyl dopamine (NADA) represents an interesting case for a putative highly biased CB1 agonist as it is currently only known to affect very select pathways.121 NADA is an endocannabinoid agonist of both CB1 and TRPV1.122,123 NADA binds orthosterically to CB1 with high nanomolar affinity but has no substantial effect on GIRK-mediated hyperpolarization, cAMP levels, pERK, or adenylyl cyclase activity. At concentrations above 30 μM, NADA elevates [Ca]i levels from intracellular stores in cell cultures and causes a slow internalization of CB1 from the cell surface. The data strongly suggest that NADA signals via a Gαq-mediated pathway. Although NADA did not potently induce [Ca]i, it has been demonstrated to be more potent under different in vitro assay conditions122 and in brain slices.124,125 In summary, significant bias in activating Gαq-mediated responses occurs, with only a small subset of ligands reporting this (WIN55,212-2, NADA, and 2-AG), and then only at high concentrations and in a tissue-dependent manner.
For most GPCRs, interest in biased signaling has focused predominantly on the ability of agonists to differentially modulate G protein and arrestin pathways. The greatest promise for this to date through CB1 has come from allosteric modulators. While ORG27569 has been suggested to generally inhibit agonist-mediated G protein activation on its own,112 it has been suggested to exhibit biased signaling toward pERK1/2 pathways via β-arrestin-1.57,58 This finding is not consistent across all reports, however, as others59,60 did not find that treatment with ORG27569 alone induced pERK1/2. The different findings could arise from differences in assay design or differences in the relative receptor expression, and signaling systems between the different cells utilized.57,58,60,126 In particular, time-dependent effects on CB1 signaling have been reported in several studies of ORG27569 and functionally related molecules,127,128 which may account for some variability.
Pregnenolone, a putative endogenous allosteric modulator of CB1 function, modifies agonist signaling in a biased or selective manner, inhibiting pERK1/2 signaling while not modifying cAMP signaling.129 Interestingly, recent studies have suggested a similar profile for Src homology 3-domain growth factor receptor-bound 2-like (endophilin) interacting protein 1 (SGIP1), a CB1 interacting protein expressed in HEK293 cells.130 These findings emphasize the challenges in interpreting pERK1/2 studies when the pathway regulating it (G protein versus arrestin mediated) is generally not well defined. An understanding of the conformation of the receptors driven by the presence of allosteric modulators may help guide the development of highly biased agonists as it is likely that a greater range of conformations may be achieved by targeting regions outside of the relatively constrained orthosteric binding site.
Similarly, receptor mutation studies may enhance our understanding of the molecular drivers for bias. A few examples of this are currently emerging, for example, mutations in potential phosphorylation sites in rCB1 (S426A/S430A) drive preferential signaling via β-arrestin-1,55 while different mutations in the DRY motif drive conformations to preferentially signal through either G proteins or β-arrestins.131
Comparatively, there is little evidence for agonist bias through G proteins for CB2, although some signaling biases have been described. Shoemaker et al.105 compared CP55,940, 2-AG, and 2-arachidonoylglyceryl-ether using MAPK activation, stimulation of calcium transients, and inhibition of adenylyl cyclase as the signaling pathways. Each ligand differed in its rank order of potency in the three assays despite similar efficacies. Schuehly et al.132 found that AM630 displayed inverse agonist/antagonist actions on CB2-mediated inhibition of cAMP production and was silent in its effects on [Ca]i transients. On the contrary, a novel CB2 ligand, 4-O-methylhonokiol, was an inverse agonist/antagonist with regard to cAMP production but potentiated the effects of 2-AG on calcium transients. Atwood et al.31 have further demonstrated that CP55,940, but not WIN55,212-2, leads to inhibition of voltage-gated calcium channels through CB2, in addition to different actions on receptor trafficking.
Recent studies have suggested agonist bias in CB2 internalization. Atwood et al.31 found that while CP55,940 induced robust internalization of rCB2, WIN55,212-2 failed to promote receptor internalization, despite both agonists activating pERK, and arrestin recruitment. Extending these studies, Dhopeshwarkar and Mackie92 examined a range of ligands in inhibiting adenylyl cyclase, internalization, and arrestin recruitment to mCB2. Of the most commonly utilized ligands, CP55,940 and JWH015 were the most balanced compounds evaluated although JWH015 had lower efficacy. The majority of the other compounds screened were G protein biased, while a few less commonly utilized ligands (STS135; UR144; 4-O-methylhonokiol; and GW833972A) were more arrestin biased. It will be interesting to see if this difference results in different tolerance profiles to these agonists in CB2-mediated effects.
In a highly detailed analysis of functional selectivity of CB2, Soethoudt et al.32 recently analyzed the ability of a wide range of ligands to signal through hCB2 and analyzed the data with operational analyses. This analysis suggested that THC showed bias toward pERK signaling compared to arrestin and GTPγS, and intriguingly, THC did not activate GIRK, indicative of high bias against this pathway. (R,S)-AM1241 was biased toward arrestin coupling and pERK signaling compared to GIRK channel activation. JWH133 was moderately biased toward arrestin compared to GIRK, whereas both WIN55,212-2 and JWH015 showed preference for GIRK compared to cAMP signaling. Anandamide showed preference for pERK and GIRK signaling compared to cAMP, whereas 2-AG was significantly biased toward GIRK compared to cAMP and G protein signaling. On comparison between arrestin coupling and cAMP signaling, all ligands appear to be significantly biased, however, this may reflect the choice of CP55,940 as the reference ligand, as this ligand appears itself biased toward cAMP signaling. The authors of this study concluded that HU-910 and HU-308 were well-balanced ligands without significant bias toward any signal transduction pathway on hCB2, but this study also highlighted species differences, as at mCB2, HU-910 and HU-308 were significantly biased toward G protein signaling over arrestin coupling. A limitation of this study is that the different assays were conducted in different cells lines, and thus, some differences may represent different expressions of signaling molecules (e.g., different βγ-subunit expression), which requires further investigation. The study concludes that THC, 2-AG, and (R,S)-AM1241 are highly biased CB2 agonists and underscores that biased signaling at CB2 is subject to species variation.
Conclusions
The concept of agonist bias provides an exciting new direction for developing therapeutics with less adverse effects. However, for the cannabinoid receptors, this field is still in its infancy. The influence of different cell types, with different receptor numbers and second-messenger expressions in the signaling pathways measured, remains to be fully elucidated. This review highlights that CB1- and CB2-biased signaling can be different between tissues and species, which might prove an issue for translating results from in vitro studies to in vivo. As a large body of the work describing biased signaling of the cannabinoid receptors is performed using heterologously expressed receptors, future work should include endogenously expressed receptors to determine if previous observations are relevant.
Importantly, almost all of the studies described here measured single time points for each signaling assay, rather than clearly defining and comparing the kinetics of each assay and it is clear that detection of bias can change across time.133 Finally, and most importantly, we do not yet know which pathways are mediating desired therapeutic effects, and it remains an open question whether or not these can be clearly defined or if the therapeutic effects are mediated through a combination of signaling pathways.
Abbreviations Used
- 2-AG
2-arachidonoylglycerol
- CB1
cannabinoid receptor 1
- CB2
cannabinoid receptor 2
- CHO
Chinese hamster ovary
- CNS
central nervous system
- ERK1/2
extracellular signal-regulated kinase 1 or 2
- GIRKs
G protein-gated inwardly rectifying potassium channels
- GPCR
G protein-coupled receptor
- GRKs
G protein receptor kinases
- hCB2
human CB2
- JNK
c-Jun-n terminal kinase
- KO
knockout
- MAP
mitogen-activated protein
- mCB2
mouse CB2
- NADA
N-arachidonoyl dopamine
- PDGF
platelet-derived growth factor
- pERK1/2
phosphorylated ERK1/2
- PWR
plasmon-waveguide resonance
- rCB1
rat CB
- THC
tetrahydrocannabinol
- TRP
transient receptor potential
Author Disclosure Statement
No competing financial interests exist.
References
- 1.Kumari P, Srivastava A, Banerjee R, et al. Functional competence of a partially engaged GPCR-beta-arrestin complex. Nat Commun. 2016;7:13416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rasmussen SG, Choi HJ, Fung JJ, et al. Structure of a nanobody-stabilized active state of the beta(2) adrenoceptor. Nature. 2011;469:175–180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rasmussen SG, DeVree BT, Zou Y, et al. Crystal structure of the beta2 adrenergic receptor-Gs protein complex. Nature. 2011;477:549–555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kenakin T, Christopoulos A. Signalling bias in new drug discovery: detection, quantification and therapeutic impact. Nat Rev Drug Discov. 2013;12:205–216 [DOI] [PubMed] [Google Scholar]
- 5.Raehal KM, Walker JK, Bohn LM. Morphine side effects in beta-arrestin 2 knockout mice. J Pharmacol Exp Ther. 2005;314:1195–1201 [DOI] [PubMed] [Google Scholar]
- 6.DeWire SM, Yamashita DS, Rominger DH, et al. A G protein-biased ligand at the mu-opioid receptor is potently analgesic with reduced gastrointestinal and respiratory dysfunction compared with morphine. J Pharmacol Exp Ther. 2013;344:708–717 [DOI] [PubMed] [Google Scholar]
- 7.Manglik A, Lin H, Aryal DK, et al. Structure-based discovery of opioid analgesics with reduced side effects. Nature. 2016;537:185–190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Turcotte C, Blanchet MR, Laviolette M, et al. The CB2 receptor and its role as a regulator of inflammation. Cell Mol Life Sci. 2016;73:4449–4470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ashton JC, Glass M. The cannabinoid CB2 receptor as a target for inflammation-dependent neurodegeneration. Curr Neuropharmacol. 2007;5:73–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Palazuelos J, Aguado T, Pazos MR, et al. Microglial CB2 cannabinoid receptors are neuroprotective in Huntington's disease excitotoxicity. Brain. 2009;132:3152–3164 [DOI] [PubMed] [Google Scholar]
- 11.Stempel AV, Stumpf A, Zhang HY, et al. Cannabinoid type 2 receptors mediate a cell type-specific plasticity in the hippocampus. Neuron. 2016;90:795–809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Devane WA, Hanus L, Breuer A, et al. Isolation and structure of a brain constituent that binds to the cannabinoid receptor. Science. 1992;258:1946–1949 [DOI] [PubMed] [Google Scholar]
- 13.Mechoulam R, Ben-Shabat S, Hanus L, et al. Identification of an endogenous 2-monoglyceride, present in canine gut, that binds to cannabinoid receptors. Biochem Pharmacol. 1995;50:83–90 [DOI] [PubMed] [Google Scholar]
- 14.Sugiura T, Kondo S, Sukagawa A, et al. 2-Arachidonoylglycerol: a possible endogenous cannabinoid receptor ligand in brain. Biochem Biophys Res Commun. 1995;215:89–97 [DOI] [PubMed] [Google Scholar]
- 15.Iversen L, Chapman V. Cannabinoids: a real prospect for pain relief? Curr Opin Pharmacol. 2002;2:50–55 [DOI] [PubMed] [Google Scholar]
- 16.Rubino T, Zamberletti E, Parolaro D. Endocannabinoids and mental disorders. Handb Exp Pharmacol. 2015;231:261–283 [DOI] [PubMed] [Google Scholar]
- 17.Pertwee RG. Cannabinoids and multiple sclerosis. Pharmacol Ther. 2002;95:165–174 [DOI] [PubMed] [Google Scholar]
- 18.Horvath TL. Endocannabinoids and the regulation of body fat: the smoke is clearing. J Clin Invest. 2003;112:323–326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Le Foll B, Goldberg SR. Cannabinoid CB1 receptor antagonists as promising new medications for drug dependence. J Pharmacol Exp Ther. 2005;312:875–883 [DOI] [PubMed] [Google Scholar]
- 20.Laprairie RB, Bagher AM, Kelly ME, et al. Biased type 1 cannabinoid receptor signaling influences neuronal viability in a cell culture model of huntington disease. Mol Pharmacol. 2016;89:364–375 [DOI] [PubMed] [Google Scholar]
- 21.Segovia G, Mora F, Crossman AR, et al. Effects of CB1 cannabinoid receptor modulating compounds on the hyperkinesia induced by high-dose levodopa in the reserpine-treated rat model of Parkinson's disease. Mov Disord. 2003;18:138–149 [DOI] [PubMed] [Google Scholar]
- 22.Mukhopadhyay P, Baggelaar M, Erdelyi K, et al. The novel, orally available and peripherally restricted selective cannabinoid CB2 receptor agonist LEI-101 prevents cisplatin-induced nephrotoxicity. Br J Pharmacol. 2016;173:446–458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pertwee RG. Receptors and channels targeted by synthetic cannabinoid receptor agonists and antagonists. Curr Med Chem. 2010;17:1360–1381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McHugh D, Page J, Dunn E, et al. Delta(9)-tetrahydrocannabinol and N-arachidonyl glycine are full agonists at GPR18 receptors and induce migration in human endometrial HEC-1B cells. Br J Pharmacol. 2012;165:2414–2424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chu ZL, Carroll C, Chen R, et al. N-oleoyldopamine enhances glucose homeostasis through the activation of GPR119. Mol Endocrinol. 2010;24:161–170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Di Marzo V, De Petrocellis L. Endocannabinoids as regulators of transient receptor potential (TRP) channels: a further opportunity to develop new endocannabinoid-based therapeutic drugs. Curr Med Chem. 2010;17:1430–1449 [DOI] [PubMed] [Google Scholar]
- 27.Hejazi N, Zhou C, Oz M, et al. Delta9-tetrahydrocannabinol and endogenous cannabinoid anandamide directly potentiate the function of glycine receptors. Mol Pharmacol. 2006;69:991–997 [DOI] [PubMed] [Google Scholar]
- 28.Matsuda LA, Lolait SJ, Brownstein MJ, et al. Structure of a cannabinoid receptor and functional expression of the cloned cDNA. Nature. 1990;346:561–564 [DOI] [PubMed] [Google Scholar]
- 29.Munro S, Thomas KL, Abu-Shaar M. Molecular characterization of a peripheral receptor for cannabinoids. Nature. 1993;365:61–65 [DOI] [PubMed] [Google Scholar]
- 30.Felder CC, Joyce KE, Briley EM, et al. Comparison of the pharmacology and signal transduction of the human cannabinoid CB1 and CB2 receptors. Mol Pharmacol. 1995;48:443–450 [PubMed] [Google Scholar]
- 31.Atwood BK, Wager-Miller J, Haskins C, et al. Functional selectivity in CB(2) cannabinoid receptor signaling and regulation: implications for the therapeutic potential of CB(2) ligands. Mol Pharmacol. 2012;81:250–263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Soethoudt M, Grether U, Fingerle J, et al. Cannabinoid CB2 receptor ligand profiling reveals biased signaling and off-target activity: implications for drug discovery. Nat Commun. 2017;8:13958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Glass M, Northup JK. Agonist selective regulation of G proteins by cannabinoid CB(1) and CB(2) receptors. Mol Pharmacol. 1999;56:1362–1369 [DOI] [PubMed] [Google Scholar]
- 34.Calandra B, Portier M, Kerneis A, et al. Dual intracellular signaling pathways mediated by the human cannabinoid CB1 receptor. Eur J Pharmacol. 1999;374:445–455 [DOI] [PubMed] [Google Scholar]
- 35.Glass M, Felder CC. Concurrent stimulation of cannabinoid CB1 and dopamine D2 receptors augments cAMP accumulation in striatal neurons: evidence for a Gs linkage to the CB1 receptor. J Neurosci. 1997;17:5327–5333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gonzalez B, Paz F, Floran L, et al. Cannabinoid agonists stimulate [3H]GABA release in the globus pallidus of the rat when G(i) protein-receptor coupling is restricted: role of dopamine D2 receptors. J Pharmacol Exp Ther. 2009;328:822–828 [DOI] [PubMed] [Google Scholar]
- 37.Bash R, Rubovitch V, Gafni M, et al. The stimulatory effect of cannabinoids on calcium uptake is mediated by Gs GTP-binding proteins and cAMP formation. Neurosignals. 2003;12:39–44 [DOI] [PubMed] [Google Scholar]
- 38.Stamer WD, Golightly SF, Hosohata Y, et al. Cannabinoid CB(1) receptor expression, activation and detection of endogenous ligand in trabecular meshwork and ciliary process tissues. Eur J Pharmacol. 2001;431:277–286 [DOI] [PubMed] [Google Scholar]
- 39.Chen XP, Yang W, Fan Y, et al. Structural determinants in the second intracellular loop of the human cannabinoid CB1 receptor mediate selective coupling to G(s) and G(i). Br J Pharmacol. 2010;161:1817–1834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lauckner JE, Hille B, Mackie K. The cannabinoid agonist WIN55,212-2 increases intracellular calcium via CB1 receptor coupling to Gq/11 G proteins. Proc Natl Acad Sci U S A. 2005;102:19144–19149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Garzon J, de la Torre-Madrid E, Rodriguez-Munoz M, et al. Gz mediates the long-lasting desensitization of brain CB1 receptors and is essential for cross-tolerance with morphine. Mol Pain. 2009;5:11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kamato D, Mitra P, Davis F, et al. Gaq proteins: molecular pharmacology and therapeutic potential. Cell Mol Life Sci. 2016; DOI: 10.1007/s00018-016-2405-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Khan SM, Sung JY, Hebert TE. Gbetagamma subunits-Different spaces, different faces. Pharmacol Res. 2016;111:434–441 [DOI] [PubMed] [Google Scholar]
- 44.Neves SR, Ram PT, Iyengar R. G protein pathways. Science. 2002;296:1636–1639 [DOI] [PubMed] [Google Scholar]
- 45.Shukla AK, Xiao K, Lefkowitz RJ. Emerging paradigms of beta-arrestin-dependent seven transmembrane receptor signaling. Trends Biochem Sci. 2011;36:457–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Thomsen AR, Plouffe B, Cahill TJ 3rd, et al. GPCR-G protein-beta-arrestin super-complex mediates sustained G protein signaling. Cell. 2016;166:907–919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Keshet Y, Seger R. The MAP kinase signaling cascades: a system of hundreds of components regulates a diverse array of physiological functions. Methods Mol Biol. 2010;661:3–38 [DOI] [PubMed] [Google Scholar]
- 48.Galve-Roperh I, Rueda D, Gomez del Pulgar T, et al. Mechanism of extracellular signal-regulated kinase activation by the CB(1) cannabinoid receptor. Mol Pharmacol. 2002;62:1385–1392 [DOI] [PubMed] [Google Scholar]
- 49.Sanchez MG, Ruiz-Llorente L, Sanchez AM, et al. Activation of phosphoinositide 3-kinase/PKB pathway by CB(1) and CB(2) cannabinoid receptors expressed in prostate PC-3 cells. Involvement in Raf-1 stimulation and NGF induction. Cell Signal. 2003;15:851–859 [DOI] [PubMed] [Google Scholar]
- 50.Davis MI, Ronesi J, Lovinger DM. A predominant role for inhibition of the adenylate cyclase/protein kinase A pathway in ERK activation by cannabinoid receptor 1 in N1E-115 neuroblastoma cells. J Biol Chem. 2003;278:48973–48980 [DOI] [PubMed] [Google Scholar]
- 51.Derkinderen P, Valjent E, Toutant M, et al. Regulation of extracellular signal-regulated kinase by cannabinoids in hippocampus. J Neurosci. 2003;23:2371–2382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rubovitch V, Gafni M, Sarne Y. The involvement of VEGF receptors and MAPK in the cannabinoid potentiation of Ca2+ flux into N18TG2 neuroblastoma cells. Brain Res Mol Brain Res. 2004;120:138–144 [DOI] [PubMed] [Google Scholar]
- 53.Hart S, Fischer OM, Ullrich A. Cannabinoids induce cancer cell proliferation via tumor necrosis factor alpha-converting enzyme (TACE/ADAM17)-mediated transactivation of the epidermal growth factor receptor. Cancer Res. 2004;64:1943–1950 [DOI] [PubMed] [Google Scholar]
- 54.Daigle TL, Kearn CS, Mackie K. Rapid CB1 cannabinoid receptor desensitization defines the time course of ERK1/2 MAP kinase signaling. Neuropharmacology. 2008;54:36–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Delgado-Peraza F, Ahn KH, Nogueras-Ortiz C, et al. Mechanisms of biased beta-arrestin-mediated signaling downstream from the cannabinoid 1 receptor. Mol Pharmacol. 2016;89:618–629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nogueras-Ortiz C, Yudowski GA. The multiple waves of cannabinoid 1 receptor signaling. Mol Pharmacol. 2016;90:620–626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ahn KH, Mahmoud MM, Kendall DA. Allosteric modulator ORG27569 induces CB1 cannabinoid receptor high affinity agonist binding state, receptor internalization, and Gi protein-independent ERK1/2 kinase activation. J Biol Chem. 2012;287:12070–12082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ahn KH, Mahmoud MM, Shim JY, et al. Distinct roles of beta-arrestin 1 and beta-arrestin 2 in ORG27569-induced biased signaling and internalization of the cannabinoid receptor 1 (CB1). J Biol Chem. 2013;288:9790–9800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gamage TF, Anderson JC, Abood ME. CB1 allosteric modulator Org27569 is an antagonist/inverse agonist of ERK1/2 signaling. Cannabis Cannabinoid Res. 2016;1:272–280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Khajehali E, Malone DT, Glass M, et al. Biased agonism and biased allosteric modulation at the CB1 cannabinoid receptor. Mol Pharmacol. 2015;88:368–379 [DOI] [PubMed] [Google Scholar]
- 61.Derkinderen P, Ledent C, Parmentier M, et al. Cannabinoids activate p38 mitogen-activated protein kinases through CB1 receptors in hippocampus. J Neurochem. 2001;77:957–960 [DOI] [PubMed] [Google Scholar]
- 62.Downer EJ, Fogarty MP, Campbell VA. Tetrahydrocannabinol-induced neurotoxicity depends on CB1 receptor-mediated c-Jun N-terminal kinase activation in cultured cortical neurons. Br J Pharmacol. 2003;140:547–557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Molina-Holgado F, Pinteaux E, Heenan L, et al. Neuroprotective effects of the synthetic cannabinoid HU-210 in primary cortical neurons are mediated by phosphatidylinositol 3-kinase/AKT signaling. Mol Cell Neurosci. 2005;28:189–194 [DOI] [PubMed] [Google Scholar]
- 64.Graham ES, Ball N, Scotter EL, et al. Induction of Krox-24 by endogenous cannabinoid type 1 receptors in Neuro2A cells is mediated by the MEK-ERK MAPK pathway and is suppressed by the phosphatidylinositol 3-kinase pathway. J Biol Chem. 2006;281:29085–29095 [DOI] [PubMed] [Google Scholar]
- 65.He JC, Gomes I, Nguyen T, et al. The G alpha(o/i)-coupled cannabinoid receptor-mediated neurite outgrowth involves Rap regulation of Src and Stat3. J Biol Chem. 2005;280:33426–33434 [DOI] [PubMed] [Google Scholar]
- 66.Rueda D, Galve-Roperh I, Haro A, et al. The CB(1) cannabinoid receptor is coupled to the activation of c-Jun N-terminal kinase. Mol Pharmacol. 2000;58:814–820 [DOI] [PubMed] [Google Scholar]
- 67.Adhikary S, Kocieda VP, Yen JH, et al. Signaling through cannabinoid receptor 2 suppresses murine dendritic cell migration by inhibiting matrix metalloproteinase 9 expression. Blood. 2012;120:3741–3749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Merighi S, Gessi S, Varani K, et al. Cannabinoid CB(2) receptors modulate ERK-1/2 kinase signalling and NO release in microglial cells stimulated with bacterial lipopolysaccharide. Br J Pharmacol. 2012;165:1773–1788 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 69.Bouaboula M, Poinot-Chazel C, Marchand J, et al. Signaling pathway associated with stimulation of CB2 peripheral cannabinoid receptor. Involvement of both mitogen-activated protein kinase and induction of Krox-24 expression. Eur J Biochem. 1996;237:704–711 [DOI] [PubMed] [Google Scholar]
- 70.Cudaback E, Marrs W, Moeller T, et al. The expression level of CB1 and CB2 receptors determines their efficacy at inducing apoptosis in astrocytomas. PLoS One. 2010;5:e8702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Moore CA, Milano SK, Benovic JL. Regulation of receptor trafficking by GRKs and arrestins. Annu Rev Physiol. 2007;69:451–482 [DOI] [PubMed] [Google Scholar]
- 72.Howlett AC, Champion-Dorow TM, McMahon LL, et al. The cannabinoid receptor: biochemical and cellular properties in neuroblastoma cells. Pharmacol Biochem Behav. 1991;40:565–569 [DOI] [PubMed] [Google Scholar]
- 73.Dill JA, Howlett AC. Regulation of adenylate cyclase by chronic exposure to cannabimimetic drugs. J Pharmacol Exp Ther. 1988;244:1157–1163 [PubMed] [Google Scholar]
- 74.Rinaldi-Carmona M, Le Duigou A, Oustric D, et al. Modulation of CB1 cannabinoid receptor functions after a long-term exposure to agonist or inverse agonist in the Chinese hamster ovary cell expression system. J Pharmacol Exp Ther. 1998;287:1038–1047 [PubMed] [Google Scholar]
- 75.Connor M, Osborne PB, Christie MJ. Mu-opioid receptor desensitization: is morphine different? Br J Pharmacol. 2004;143:685–696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Caulfield MP, Brown DA. Cannabinoid receptor agonists inhibit Ca current in NG108-15 neuroblastoma cells via a pertussis toxin-sensitive mechanism. Br J Pharmacol. 1992;106:231–232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Mackie K, Devane WA, Hille B. Anandamide, an endogenous cannabinoid, inhibits calcium currents as a partial agonist in N18 neuroblastoma cells. Mol Pharmacol. 1993;44:498–503 [PubMed] [Google Scholar]
- 78.Mackie K, Hille B. Cannabinoids inhibit N-type calcium channels in neuroblastoma-glioma cells. Proc Natl Acad Sci U S A. 1992;89:3825–3829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Mackie K, Lai Y, Westenbroek R, et al. Cannabinoids activate an inwardly rectifying potassium conductance and inhibit Q-type calcium currents in AtT20 cells transfected with rat brain cannabinoid receptor. J Neurosci. 1995;15:6552–6561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Jin W, Brown S, Roche JP, et al. Distinct domains of the CB1 cannabinoid receptor mediate desensitization and internalization. J Neurosci. 1999;19:3773–3780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Roche JP, Bounds S, Brown S, et al. A mutation in the second transmembrane region of the CB1 receptor selectively disrupts G protein signaling and prevents receptor internalization. Mol Pharmacol. 1999;56:611–618 [DOI] [PubMed] [Google Scholar]
- 82.Daigle TL, Kwok ML, Mackie K. Regulation of CB1 cannabinoid receptor internalization by a promiscuous phosphorylation-dependent mechanism. J Neurochem. 2008;106:70–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Gyombolai P, Boros E, Hunyady L, et al. Differential beta-arrestin2 requirements for constitutive and agonist-induced internalization of the CB1 cannabinoid receptor. Mol Cell Endocrinol. 2013;372:116–127 [DOI] [PubMed] [Google Scholar]
- 84.Bakshi K, Mercier RW, Pavlopoulos S. Interaction of a fragment of the cannabinoid CB1 receptor C-terminus with arrestin-2. FEBS Lett. 2007;581:5009–5016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Singh SN, Bakshi K, Mercier RW, et al. Binding between a distal C-terminus fragment of cannabinoid receptor 1 and arrestin-2. Biochemistry. 2011;50:2223–2234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Laprairie RB, Bagher AM, Kelly ME, et al. Type 1 cannabinoid receptor ligands display functional selectivity in a cell culture model of striatal medium spiny projection neurons. J Biol Chem. 2014;289:24845–24862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Breivogel CS, Lambert JM, Gerfin S, et al. Sensitivity to delta9-tetrahydrocannabinol is selectively enhanced in beta-arrestin2-/- mice. Behav Pharmacol. 2008;19:298–307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Nguyen PT, Schmid CL, Raehal KM, et al. beta-arrestin2 regulates cannabinoid CB1 receptor signaling and adaptation in a central nervous system region-dependent manner. Biol Psychiatry. 2012;71:714–724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Connor M, Bagley EE, Chieng BC, et al. Beta-arrestin-2 knockout prevents development of cellular mu-opioid receptor tolerance but does not affect opioid-withdrawal-related adaptations in single PAG neurons. Br J Pharmacol. 2015;172:492–500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Dang VC, Chieng B, Azriel Y, et al. Cellular morphine tolerance produced by betaarrestin-2-dependent impairment of mu-opioid receptor resensitization. J Neurosci. 2011;31:7122–7130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.McGuinness D, Malikzay A, Visconti R, et al. Characterizing cannabinoid CB2 receptor ligands using DiscoveRx PathHunter beta-arrestin assay. J Biomol Screen. 2009;14:49–58 [DOI] [PubMed] [Google Scholar]
- 92.Dhopeshwarkar A, Mackie K. Functional selectivity of CB2 cannabinoid receptor ligands at a canonical and noncanonical pathway. J Pharmacol Exp Ther. 2016;358:342–351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Grimsey NL, Graham ES, Dragunow M, et al. Cannabinoid receptor 1 trafficking and the role of the intracellular pool: implications for therapeutics. Biochem Pharmacol. 2010;80:1050–1062 [DOI] [PubMed] [Google Scholar]
- 94.Hsieh C, Brown S, Derleth C, et al. Internalization and recycling of the CB1 cannabinoid receptor. J Neurochem. 1999;73:493–501 [DOI] [PubMed] [Google Scholar]
- 95.Leterrier C, Bonnard D, Carrel D, et al. Constitutive endocytic cycle of the CB1 cannabinoid receptor. J Biol Chem. 2004;279:36013–36021 [DOI] [PubMed] [Google Scholar]
- 96.Martini L, Waldhoer M, Pusch M, et al. Ligand-induced down-regulation of the cannabinoid 1 receptor is mediated by the G-protein-coupled receptor-associated sorting protein GASP1. FASEB J. 2007;21:802–811 [DOI] [PubMed] [Google Scholar]
- 97.Keren O, Sarne Y. Multiple mechanisms of CB1 cannabinoid receptors regulation. Brain Res. 2003;980:197–205 [DOI] [PubMed] [Google Scholar]
- 98.Nie J, Lewis DL. Structural domains of the CB1 cannabinoid receptor that contribute to constitutive activity and G-protein sequestration. J Neurosci. 2001;21:8758–8764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Brailoiu GC, Oprea TI, Zhao P, et al. Intracellular cannabinoid type 1 (CB1) receptors are activated by anandamide. J Biol Chem. 2011;286:29166–29174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Rozenfeld R, Devi LA. Regulation of CB1 cannabinoid receptor trafficking by the adaptor protein AP-3. FASEB J. 2008;22:2311–2322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Benard G, Massa F, Puente N, et al. Mitochondrial CB(1) receptors regulate neuronal energy metabolism. Nat Neurosci. 2012;15:558–564 [DOI] [PubMed] [Google Scholar]
- 102.Hebert-Chatelain E, Desprez T, Serrat R, et al. A cannabinoid link between mitochondria and memory. Nature. 2016;539:555–559 [DOI] [PubMed] [Google Scholar]
- 103.Bouaboula M, Dussossoy D, Casellas P. Regulation of peripheral cannabinoid receptor CB2 phosphorylation by the inverse agonist SR 144528. Implications for receptor biological responses. J Biol Chem. 1999;274:20397–20405 [DOI] [PubMed] [Google Scholar]
- 104.Derocq JM, Jbilo O, Bouaboula M, et al. Genomic and functional changes induced by the activation of the peripheral cannabinoid receptor CB2 in the promyelocytic cells HL-60. Possible involvement of the CB2 receptor in cell differentiation. J Biol Chem. 2000;275:15621–15628 [DOI] [PubMed] [Google Scholar]
- 105.Shoemaker JL, Ruckle MB, Mayeux PR, et al. Agonist-directed trafficking of response by endocannabinoids acting at CB2 receptors. J Pharmacol Exp Ther. 2005;315:828–838 [DOI] [PubMed] [Google Scholar]
- 106.Carrier EJ, Kearn CS, Barkmeier AJ, et al. Cultured rat microglial cells synthesize the endocannabinoid 2-arachidonylglycerol, which increases proliferation via a CB2 receptor-dependent mechanism. Mol Pharmacol. 2004;65:999–1007 [DOI] [PubMed] [Google Scholar]
- 107.Grimsey NL, Goodfellow CE, Dragunow M, et al. Cannabinoid receptor 2 undergoes Rab5-mediated internalization and recycles via a Rab11-dependent pathway. Biochim Biophys Acta. 2011;1813:1554–1560 [DOI] [PubMed] [Google Scholar]
- 108.Franklin JM, Carrasco GA. G-protein receptor kinase 5 regulates the cannabinoid receptor 2-induced up-regulation of serotonin 2A receptors. J Biol Chem. 2013;288:15712–15724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Raehal KM, Schmid CL, Groer CE, et al. Functional selectivity at the mu-opioid receptor: implications for understanding opioid analgesia and tolerance. Pharmacol Rev. 2011;63:1001–1019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Mukhopadhyay S, Howlett AC. Chemically distinct ligands promote differential CB1 cannabinoid receptor-Gi protein interactions. Mol Pharmacol. 2005;67:2016–2024 [DOI] [PubMed] [Google Scholar]
- 111.Breivogel CS, Childers SR. Cannabinoid agonist signal transduction in rat brain: comparison of cannabinoid agonists in receptor binding, G-protein activation, and adenylyl cyclase inhibition. J Pharmacol Exp Ther. 2000;295:328–336 [PubMed] [Google Scholar]
- 112.Breivogel CS, Sim LJ, Childers SR. Regional differences in cannabinoid receptor/G-protein coupling in rat brain. J Pharmacol Exp Ther. 1997;282:1632–1642 [PubMed] [Google Scholar]
- 113.Sim LJ, Selley DE, Childers SR. In vitro autoradiography of receptor-activated G proteins in rat brain by agonist-stimulated guanylyl 5′-[gamma-[35S]thio]-triphosphate binding. Proc Natl Acad Sci U S A. 1995;92:7242–7246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Georgieva T, Devanathan S, Stropova D, et al. Unique agonist-bound cannabinoid CB1 receptor conformations indicate agonist specificity in signaling. Eur J Pharmacol. 2008;581:19–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.McAllister SD, Rizvi G, Anavi-Goffer S, et al. An aromatic microdomain at the cannabinoid CB(1) receptor constitutes an agonist/inverse agonist binding region. J Med Chem. 2003;46:5139–5152 [DOI] [PubMed] [Google Scholar]
- 116.Song ZH, Bonner TI. A lysine residue of the cannabinoid receptor is critical for receptor recognition by several agonists but not WIN55212-2. Mol Pharmacol. 1996;49:891–896 [PubMed] [Google Scholar]
- 117.Hua T, Vemuri K, Pu M, et al. Crystal structure of the human cannabinoid receptor CB1. Cell. 2016;167:750.e714–762.e714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Shao Z, Yin J, Chapman K, et al. High-resolution crystal structure of the human CB1 cannabinoid receptor. Nature. 2016;540:602–606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Bonhaus DW, Chang LK, Kwan J, et al. Dual activation and inhibition of adenylyl cyclase by cannabinoid receptor agonists: evidence for agonist-specific trafficking of intracellular responses. J Pharmacol Exp Ther. 1998;287:884–888 [PubMed] [Google Scholar]
- 120.Kellogg R, Mackie K, Straiker A. Cannabinoid CB1 receptor-dependent long-term depression in autaptic excitatory neurons. J Neurophysiol. 2009;102:1160–1171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Redmond WJ, Cawston EE, Grimsey NL, et al. Identification of N-arachidonoyl dopamine as a highly biased ligand at cannabinoid CB1 receptors. Br J Pharmacol. 2016;173:115–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Bisogno T, Melck D, Bobrov M, et al. N-acyl-dopamines: novel synthetic CB(1) cannabinoid-receptor ligands and inhibitors of anandamide inactivation with cannabimimetic activity in vitro and in vivo. Biochem J. 2000;351:817–824 [PMC free article] [PubMed] [Google Scholar]
- 123.Huang SM, Bisogno T, Trevisani M, et al. An endogenous capsaicin-like substance with high potency at recombinant and native vanilloid VR1 receptors. Proc Natl Acad Sci U S A. 2002;99:8400–8405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Fawley JA, Hofmann ME, Andresen MC. Cannabinoid 1 and transient receptor potential vanilloid 1 receptors discretely modulate evoked glutamate separately from spontaneous glutamate transmission. J Neurosci. 2014;34:8324–8332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Marinelli S, Di Marzo V, Florenzano F, et al. N-arachidonoyl-dopamine tunes synaptic transmission onto dopaminergic neurons by activating both cannabinoid and vanilloid receptors. Neuropsychopharmacology. 2007;32:298–308 [DOI] [PubMed] [Google Scholar]
- 126.Baillie GL, Horswill JG, Anavi-Goffer S, et al. CB(1) receptor allosteric modulators display both agonist and signaling pathway specificity. Mol Pharmacol. 2013;83:322–338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Cawston EE, Connor M, Di Marzo V, et al. Distinct temporal fingerprint for cyclic adenosine monophosphate (cAMP) signaling of indole-2-carboxamides as allosteric modulators of the cannabinoid receptors. J Med Chem. 2015;58:5979–5988 [DOI] [PubMed] [Google Scholar]
- 128.Cawston EE, Redmond WJ, Breen CM, et al. Real-time characterization of cannabinoid receptor 1 (CB1) allosteric modulators reveals novel mechanism of action. Br J Pharmacol. 2013;170:893–907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Vallee M, Vitiello S, Bellocchio L, et al. Pregnenolone can protect the brain from cannabis intoxication. Science. 2014;343:94–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Hajkova A, Techlovska S, Dvorakova M, et al. SGIP1 alters internalization and modulates signaling of activated cannabinoid receptor 1 in a biased manner. Neuropharmacology. 2016;107:201–214 [DOI] [PubMed] [Google Scholar]
- 131.Gyombolai P, Toth AD, Timar D, et al. Mutations in the ‘DRY’ motif of the CB1 cannabinoid receptor result in biased receptor variants. J Mol Endocrinol. 2015;54:75–89 [DOI] [PubMed] [Google Scholar]
- 132.Schuehly W, Paredes JM, Kleyer J, et al. Mechanisms of osteoclastogenesis inhibition by a novel class of biphenyl-type cannabinoid CB(2) receptor inverse agonists. Chem Biol. 2011;18:1053–1064 [DOI] [PubMed] [Google Scholar]
- 133.Klein Herenbrink C, Sykes DA, Donthamsetti P, et al. The role of kinetic context in apparent biased agonism at GPCRs. Nat Commun. 2016;7:10842. [DOI] [PMC free article] [PubMed] [Google Scholar]
References
Cite this article as: Ibsen MS, Connor M, Glass M (2017) Cannabinoid CB1 and CB2 receptor signaling and bias, Cannabis and Cannabinoid Research 2:1, 48–60, DOI: 10.1089/can.2016.0037.