Keywords: nerve regeneration, three-dimensional printing, traumatic brain injury, tissue engineering, scaffolds, magnetic resonance imaging, collagen, chitosan, mimics, neural regeneration
Abstract
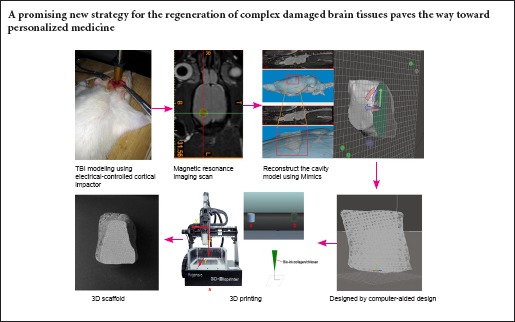
Conventional fabrication methods lack the ability to control both macro- and micro-structures of generated scaffolds. Three-dimensional printing is a solid free-form fabrication method that provides novel ways to create customized scaffolds with high precision and accuracy. In this study, an electrically controlled cortical impactor was used to induce randomized brain tissue defects. The overall shape of scaffolds was designed using rat-specific anatomical data obtained from magnetic resonance imaging, and the internal structure was created by computer-aided design. As the result of limitations arising from insufficient resolution of the manufacturing process, we magnified the size of the cavity model prototype five-fold to successfully fabricate customized collagen-chitosan scaffolds using three-dimensional printing. Results demonstrated that scaffolds have three-dimensional porous structures, high porosity, highly specific surface areas, pore connectivity and good internal characteristics. Neural stem cells co-cultured with scaffolds showed good viability, indicating good biocompatibility and biodegradability. This technique may be a promising new strategy for regenerating complex damaged brain tissues, and helps pave the way toward personalized medicine.
Introduction
Traumatic brain injury (TBI) is a major source of morbidity and mortality that significantly impacts the overall quality of life of patients around the world (Haghbayan et al., 2016; Villapol, 2016). The pathophysiology of TBI is complex, in part because of nerve tissue loss resulting from primary and secondary injuries (Ray et al., 2002; Bramlett and Dietrich, 2015). As key factors for nerve regeneration, biological scaffolds can provide physical support to bridge the gap between tissues, and promote new tissue formation (Johnson et al., 2015; Lee et al., 2017). While the favorable effect of traditional scaffolds in repairing nerve injuries has been proven (Guan et al., 2013; Tian et al., 2015; Duan et al., 2016), intrinsic limitations remain including precise control of the external shape and internal architecture of scaffolds.
Therefore, newly developed three-dimensional printing (3DP) techniques may offer a novel strategy for future therapeutic brain tissue regeneration (Suri et al., 2011; Hsieh et al., 2015). In recent years, 3DP technologies have been used in various medical fields (Chimene et al., 2016; Patra and Young, 2016). Additional studies have reported relatively accurate printing of standardized scaffolds using 3DP (Huang et al., 2012; Shi et al., 2012; Álvarez et al., 2014; Yang and Li, 2016). Cylindrical scaffolds with axial microgrooves and orthogonally intersecting channels from a 3D printer can benefit brain tissue regeneration, as channels and microgrooves can be oriented toward the desired direction of cellular migration and neuronal alignment (Wong et al., 2008). A subsequent systematic investigation revealed that the 3D constructs with anisotropic structures could be organized to produce the best possible outcome (Wüst et al., 2015). Although, it should be noted that these experimental models reflect a relatively uncommon injury in the clinical setting (Turtzo et al., 2013; Bramlett and Dietrich, 2015). Fortunately, technological developments combining imaging and 3DP make it possible to create complex, anatomically customized, high-fidelity scaffolds (Mitsouras et al., 2015; Marro et al., 2016). The key is the ability to precisely control structural and material properties in 3D based on high-definition imaging data, such as computed tomography, magnetic resonance imaging (MRI), and/or various ultrasonic apparatuses providing patient-specific anatomical information (Zhang and Zhang, 2015; Crozier et al., 2016). For use in tissue regeneration, one-stop manufacturing from medical imaging to 3DP technology allows for fabrication of customized scaffolds for brain tissue engineering (Radenkovic et al., 2016).
Two primary limitations remain for state-of-the-art scaffolds fabricated for brain tissue repair. First, standardized TBI modeling, by conventional methods such as blunt cutting (Huang et al., 2012) or frostbite (Yasuda et al., 2009), often result in regular acute cavities, which exhibit vastly different pathophysiological processes than injury in TBI patients. Second, unification of scaffold manufacturing as, traditionally, mold scaffolds have had a fixed shape before transplantation (Huang et al., 2014), making it difficult to adapt to individual cavity differences in patients with TBI. As such, this study addresses the necessity for a convenient and rapid method to obtain irregular cavity models after TBI, including improved 3DP joint imaging technology to produce complex 3D scaffolds and testing of scaffold bioactivity/regenerative potential for future therapeutic strategies.
Materials and Methods
Ethics statement and animals
All animal procedures were approved by the Animal Ethics Committee of the Medical College of Chinese Armed Police Forces (approval No. 20160420) and conducted in accordance with the National Institutes of Health Guide for the Care and Use of Laboratory Animals. Precautions were taken to minimize suffering and number of animals used in each experiment.
Adult male Sprague-Dawley rats (n = 22; mean body weight 300 ± 10 g, specific-pathogen free) aged 3–6 months and pregnant (day 14) rats (n = 2) were obtained from the Laboratory Animal Center of The Academy of Military Medical Sciences (animal license No. SCXK-2012-0004, Beijing, China). All animals were housed under a 12-hour light/dark cycle at 22–25°C in cages, and allowed free access to food and water. Ten male rats were subjected to TBI modeling and MRI scans (model group), with five used for histological observation (slice group). Twelve male rats were used for scaffold degradation experiments (degradation group). Two pregnant rats were used for neural stem cell (NSC) extraction.
TBI model establishment
Adult rats (n = 10) were anesthetized with isoflurane (4%) in oxygen (0.8 L/min), placed in a stereotaxic frame, and prepped in a sterile fashion. After scalp incision, a 6-mm craniotomy was performed over the right motor cortex (2.0 mm posterior of bregma and the lateral edge adjacent to the right lateral side) (Kharatishvili et al., 2009) using a micromotor drill with a dental bit (SUESHIN, Seoul, Korea). TBI was produced by the electrically controlled cortical impactor (Custom & Design Company, Richmond, VA, USA), as previously described (Dixon et al., 1991), impacting the right (ipsilateral) cortex with an aluminum impactor tip (5 mm) at an impact velocity of 5 m/s, a depth of 3 mm, and a dwell time of 200 m/s. After carefully probing the damaged area of the brain, the hematoma was totally removed. During the surgical procedure, core body temperature of the rats was monitored continuously and maintained at 37°C.
MRI imaging data acquisition
In vivo MRI of the brain was performed using a 3.0 T MRI scanner (Siemens Trio Tim, Berlin and Munich, Germany) 48 hours after TBI. Model rats were anesthetized by isoflurane and kept warm with a circulating water pad. Conventional T-1 3D images were acquired with the following parameters: repetition time = 1,600 ms, echo time = 3.52 ms, slice thickness = 3 mm, matrix: 192 × 186. Conventional T-2 3D images were acquired with the following parameters: repetition time = 1,000 ms, echo time = 146 ms, slice thickness = 2 mm, matrix: 256 × 128. Data were recorded in the digital imaging and communications in medicine (DICOM) format.
MRI-based 3D data processing and customized cavity-scaffold design
The cavity-scaffold design considers a combination of specific anatomical shape and optimized internal architecture. Design of the cavity anatomical shape commences with import of the DICOM MRI file into Mimics 16.0 (Materialise, Leuven, Belgium) for digital processing. The scope of the cavity is first determined by positioning and the navigation functions unit. Cavity structures are then segmented using a combination of automatic and semiautomatic intensity thresholding, a growing dynamic region, and a crop mask tool. To reconstruct the entire cavity structure, the left (normal) side of the brain is mirrored to the right (lesion) side as a template for digital editing. Next, the fine details of segmentation are addressed by one-by-one manual contouring of the tri-planer views (i.e., axial, coronal, and sagittal). The segmented cavity is then converted to a meshed, smoothed and remeshed model using Meshmixer v10.9 software (Autodesk, San Rafael, USA) and saved as a Stereo Lithography file for prototyping. Finally, Solidworks 2014 software (Dassault Systemes, Willis, France) was used to create orthogonally intersecting channels inside the scaffold model.
Comparison of histological samples and reconstruction model
For histological studies, the day after TBI establishment, five TBI model rats were intraperitoneally anesthetized with 4% chloral hydrate and perfused transcardially with normal saline followed by 4% paraformaldehyde. Following complete perfusion, brains were extracted and fixed in fresh 4% paraformaldehyde for 12 hours. Each whole brain specimen was photographed and compared with the reconstructed model of the brain. The dentate gyrus was selected for coronal serial sections, stained for toluidine blue, and observed by bright-field microscopy using an Olympus BX51 microscope (Olympus Corp, Tokyo, Japan).
Scaffold fabrication
A 3D-Bioplotter™ system (Regenovo, Hangzhou, China) was employed for printing scaffolds. The system integrates a personal computer, an x–y–z motion nozzle, and platform with temperature controllers (Figure 1A). The computer was used for loading of the scaffold model, planning of manufacturing paths, and motion control of the nozzle. For biocompatibility and biodegradability, collagen-chitosan was selected as the fabrication material. Chitosan (448877-250G medium molecular weight; Lot#STBF3507V, Cas Number: 9012-76-4; Mw = 1,526.454 g/mol; 200–800 cP, DDA 75–85%; Sigma-Aldrich, St. Louis, MO, USA) was dissolved in a 1% (v/v) ethylic acid solution via agitation, and collagen (Tianjin ShiJi KangTai Biomedical Engineering Co., Ltd., Tianjin, China) was swelled in acetic acid solution, and then mixed with chitosan solution (Zeng et al., 2014). In this work, collagen was blended at 3:1, 2:1, and 1:1 ratios with chitosan to explore optimal 3DP molding parameters. Finally, we chose a ratio of 1:1, which is sufficient to match the printing conditions. The computer automatically sends commands to the nozzle for pressure and temperature control units so that each layer of the sieved gelatin powder was spread, and the binder was printed selectively to form a 2D pattern. This process was repeated layer by layer to form the designed shape and inner structures of the scaffold. As the scaffold was too small, when the filament was superimposed, the material did not have sufficient time to mold to the frame structure and fused together immediately. Upon attempting to integrate the model into a larger cube to increase cooling time, we encountered great difficult in separating the scaffolds from the cube after formation. Thus, we magnified the size of the cavity model from the prototype by five-fold for 3DP. Next, scaffolds were placed into Petri dishes, vacuum freeze-dried for 24 hours, and soaked in a 1% NaOH solution for 24 hours. Finally, scaffolds were repeatedly rinsed in deionized water and sterilized by cobalt-60 irradiation.
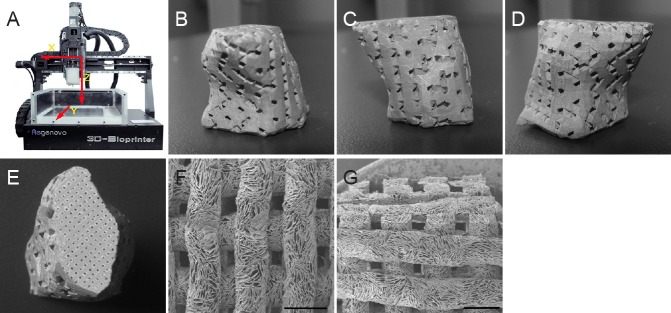
Figure 1.
Regenovo 3D-Bioplotter™ system and three dimensional printing (3DP) scaffold.
(A) Regenovo 3D-Bioplotter™ system. (B–D) 3DP scaffold with an external shape matching the brain cavity design (front view, left view and right view) and predefined internal architecture (E). Scanning electron microscopy images of the scaffold showing top (F) and cross-sectional (G) views. Scale bars: 1 mm.
Performance of collagen-chitosan scaffolds
The compression modulus and strengths of the scaffolds were evaluated using an Instron 5865 (Instron, Norwood, MA, USA). The load was 0.1 kN, the sinusoidal waveform was 5 Hz, and the maximum compressive strain was 10% (Monsell et al., 2014).
NSCs from 14-day-old embryonic Sprague-Dawley rat brains were isolated as previously described (Jiao et al., 2011). We examined the growth and morphology of NSCs on the collagen-chitosan scaffold with an inverted phase contrast microscope (Nikon, Tokyo, Japan). The microcosmic morphology of cell-scaffold constructs was observed by scanning electron microscopy (QUANTA 200/FEI, Hillsboro, OR, USA).
Cell proliferation on scaffolds was assessed using a Cell Counting Kit-8 assay (CCK-8) (Guan et al., 2015). Briefly, NSCs were seeded onto scaffolds (scaffold group) and complete DMEM (control group) at a density of 3 × 103 cells per well. Twentymicroliters of CCK-8 reagents (Dojindo, Kumamoto, Japan) were added to each well and incubated at 37°C for 3 hours at 1, 3, 5, 7, and 9 days, respectively. The absorbance at 450 nm was determined using a microplate reader (Thermo Fisher, MA, USA) for intergroup comparison.
In addition, to evaluate the degradation rate of scaffolds in vivo, scaffolds (3 mm × 3 mm) were implanted into the brain tissues of Sprague-Dawley rats (n = 12), as previously described (Wong et al., 2008). Scaffolds were harvested from three rats at 1, 3, 6, 9 and 12 weeks after implantation. The weight of each scaffold was recorded before (m1) and after (m2) implantation, and the degradation rate was calculated using the following formula: (m1 – m2)/m1 × 100%.
Statistical analysis
Data, expressed as the mean ± SD, were analyzed using SPSS 21.0 (IBM, Armonk, NY, USA) software. Statistical analysis of results was performed using one-way analysis of variance. Tests were conducted with a confidence interval of 95% (P < 0.05).
Results
Macroarchitecture of cavity scaffolds designed using rat-specific anatomical data
One day after surgery, a cortically localized region of low signal intensity was observed on coronal T2-weighted rat brain MRIs (Figure 2a–c). This region was consistent with the TBI injury site. Therefore, MRI scans could yield extremely detailed images of damaged brain areas in vivo and provide data for 3D reconstruction. Next, we modeled these digital images by utilizing Mimics software to create a brain cavity anatomical shape. Figure 2a–c exhibits a three-axial MRI image of a rat head in the Mimics software interface and the cavity model constructed by additional software processing. Following removal of the surrounding bone tissue, information for all soft tissue of the selected brain was obtained by adjusting the gray-value threshold to the minimum. As a result, information about the cavity body was completely preserved without extra information or background (Figure 2d).
Figure 2.
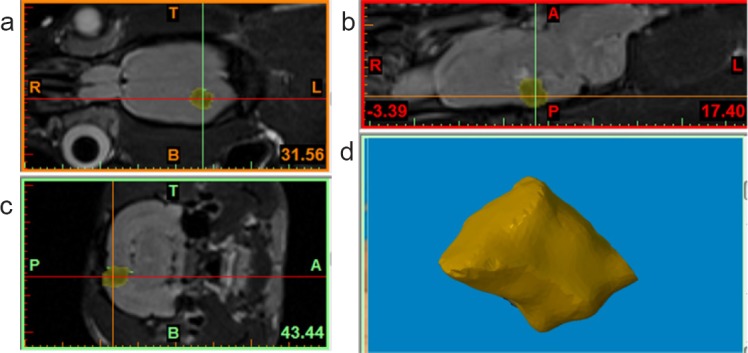
Macroarchitecture of cavity scaffolds designed using rat-specific anatomical data.
(a–c) Three-axial (i.e., axial, coronal, and sagittal) magnetic resonance imaging images of a rat head in the Mimics software interface. Yellow area indicates the cavity body. A: Anterior; P: posterior; T: top; B: bottom; R: right; L: left. (d) The cavity body was completely preserved without extra information or background.
Congruence between models obtained from histological and 3D reconstruction methods
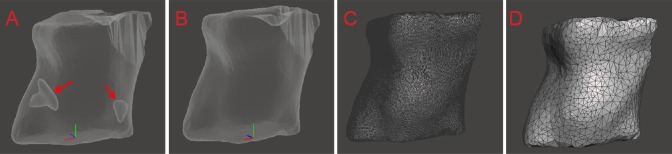
Figure 3A shows brain tissue with traditional pathology methods (gross morphology). Figure 3B shows the same brain tissue with 3D reconstruction by Mimics (reconstruction model). The merged image shown in Figure 3C demonstrates striking consistencies between these two models. Furthermore, a cross-section of the cavity model (Figure 3D, E) compared favorably with brain tissue sections stained with toluidine blue (Figure 3F), indicating the accuracy and reliability of our method.
Figure 3.
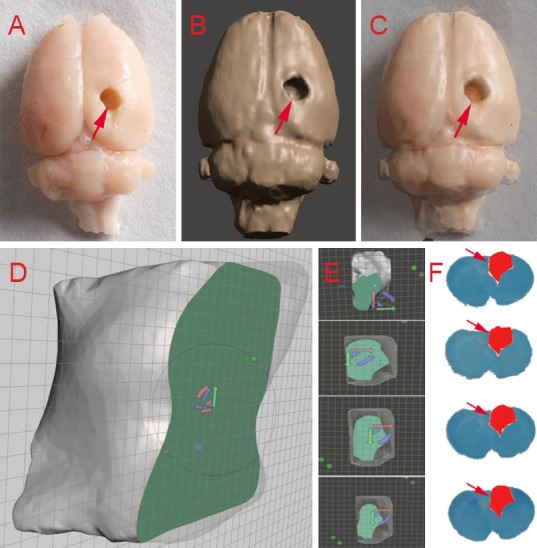
Congruence between models obtained from histological and three-dimensional reconstruction methods.
Brain tissue shown with traditional pathology methods (A) three-dimensional reconstruction via Mimics (B) for the same rat brain; merged image of the two brain models (C). Comparison of several cross sections of the brain cavity by Meshmixer software (D–E) and toluidine blue staining (F). Arrows indicate damaged areas.
Optimization and microarchitecture of the cavity-scaffold design
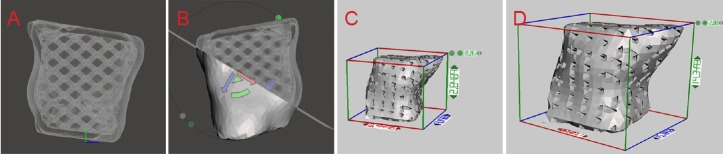
To optimize the model, state-of-the-art software (Meshmixer) was used to fit triangle meshes. Figure 4A shows the raw model prior to cleaning has inclusions. Figure 4B shows a smoothed model following cleaning and polishing. Additionally, the model is shown before (Figure 4C) and after (Figure 4D) remeshing. Finally, an individual cavity-scaffold model with a 3D orthogonal periodic porous architecture was designed by computer-aided design (CAD) (Figure 5A). The cavity model was then rotated 360° in a transparent mode to observe and generate a 3D incision (Figure 5B). The size of the cavity scaffold was scaled up from the prototype (Figure 5C) by five-fold (Figure 5D) for 3DP.
Figure 4.
Split-based method of cavity model imaging by Meshmixer software.
(A) Raw model prior to cleaning; (B) smoothed model following cleaning and polishing; cavity model before (C) and after remeshing (D).
Figure 5.
3D orthogonal periodic porous architecture of the cavity scaffold model designed by CAD.
(A) An individual cavity scaffold model with 3D orthogonal periodic porous architecture characterized by CAD. (B) The cavity model was observed in 360° and 3D incision was generated. The size of the cavity scaffold was scaled up from the prototype (C) to five-fold (D) for 3D printing. CAD: Computer-aided design; 3D: three-dimensional.
Collagen-chitosan scaffold fabricated by 3DP
The 3DP scaffold revealed an external shape matching the brain cavity design (Figure 1B–E; front view, left view, right view and back view), with a predefined internal architecture (Figure 1F, G). Scanning electron microscopy images of scaffolds revealed that the morphology of each layer was well preserved, and pores were clearly identifiable.
Performance of the collagen-chitosan scaffold
Mechanical parameters of the collagen-chitosan scaffold indicated compression displacement of 0.29316 ± 0.00573 mm, compression stress of 0.00476 ± 0.00023 MPa, and a modulus of elasticity of 15.27274 ± 3.20038 kPa.
Co-culture of NSCs and scaffold was observed by optical microscopy. NSC clusters dispersed within the scaffold layer and formed a multi-level connection (Figure 6A). Neurites extending from cell bodies into the scaffold could be observed at different layers (Figure 6B). Proliferation of NSCs on scaffolds and in culture medium (control), as measured in terms of absorbance, is shown in Figure 6C. There was no significant difference between the two groups (P > 0.05). Scanning electron microscopy images of NSCs cultured for 7 days on the surface of a scaffold are presented in Figure 6D, E. NSC bodies appear to be fully embedded within the scaffolds, where they intimately interact with the surrounding scaffold by forming neurites (Figure 6D). Scaffolds undergo faster degradation during the initial stage, after which the rate of degradation slowed (Figure 6F). After 12 weeks of degradation within the brain, 90% of the original weight had been lost.
Figure 6.
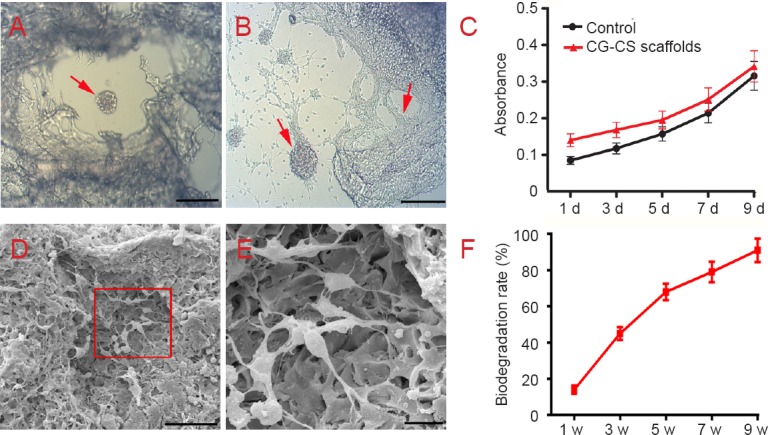
Biocompatibility of collagen-chitosan (CG-CS) composite scaffolds.
(A) Co-culture of neural stem cells (NSCs) and scaffolds was observed by optical microscopy. (B) Neurites extending from cell bodies to the scaffold can be observed. Arrows in A and B indicate NSCs. (C) Proliferation of NSCs on collagen-chitosan composite scaffolds and culture medium (control) over time as detected by Cell Counting Kit-8 staining. (D) Electron microscopy shows NSCs adhered to the scaffold surface and infiltrated into inner pores. (E) Higher magnification of the rectangular area in D. Scale bars: 200 μm in A and B, 50 μm in D, 10 μm in E. (F) Percentage of scaffold biodegradation increased gradually with time. d: Day(s); w: week(s).
Discussion
In this study, we simulated real TBI using an electrically controlled cortical impactor approach to induce a randomized brain tissue defect model. We successfully merged digital scanning technology with a 3DP technique to fabricate customized scaffolds mimicking a rat brain cavity. Briefly, we modeled digital MRI images using Mimics software to create the anatomical shape of the brain cavity. Along with shape, the optimized internal architecture of the model was also designed using CAD. The biocompatibility of scaffolds was demonstrated using NSCs, the most commonly used cell source for nerve tissue engineering. As a result, we demonstrated the potential of this technique as a novel strategy to regenerate damaged complex brain tissues, paving the way toward personalized medicine.
A large mass lesion forming within the highly complex nervous system after severe TBI is a frequent and devastating medical problem (Chimene et al., 2016). For tissue engineering, scaffolds are critical to provide both physical connections for injured tissue and biochemical cues to direct cell behavior (Leung et al., 2012). Perhaps the best example includes use of polymer scaffolds alone to treat brain defects, with results demonstrating beneficial effects on the progression of defect size in a rat model of TBI (Wong et al., 2007).
Scaffolds have been developed to reconstitute damaged brain areas using 3DP techniques. During their processes, in general, defect size was standardized to the same size as the implant using identical trephines and forceps (Wüst et al., 2015). However, the complex reality of anatomical defects observed during brain trauma results in pathophysiological heterogeneity and variability in the location, size, and shape of injuries (Wong et al., 2008; Bramlett and Dietrich, 2015). Only an accurately fabricated scaffold fits into the defect appropriately and reduces the probability of subsequent movement, dislodgement and extrusion (Maravelakis et al., 2008). Even a small malposition during transplantation of scaffolds into the brain can have potentially devastating consequences (Klein et al., 2013). Therefore, 3DP is the optimal solution to solve this critical issue for thick and complex tissue fabrication, as 3D tissues with complex structures can be printed layer-by-layer based on scanned computed tomography or MRI images (Mitsouras et al., 2015; Crozier et al., 2016; Marro et al., 2016). This additive process prints the object one layer at a time by extruding materials through a nozzle, while moving around a build plate, which provides unique ways to manufacture accurate scaffolds compared with conventional fabrication techniques (Ventola, 2014; Chia and Wu, 2015).
Our study demonstrated the feasibility of fabricating customized scaffolds based on imaging data and 3DP. During scaffold fabrication, we overcame two major challenges. First, materials used to support the cavity scaffold should be biocompatible and show plasticity. Collagen is widely used for wound healing and tissue regeneration, but its mechanical properties are poor (Mahmoud and Salama, 2016). The incorporation of chitosan into a collagen scaffold improves its biomechanical properties (Yan et al., 2015). We confirmed a mass ratio of 50/50 collagen/chitosan for balancing degradation and printing characteristics in this study. The second problem is how to fabricate these small scaffolds with complex geometries and interconnected porous structures. We tested an indirect stereolithography approach wherein a positive replica of the desired shape was printed using a photopolymer, and the final scaffold was directly produced from the printed mold. Although this method can form the outer contour of small scaffolds, it cannot control internal scaffold structure. Thus, we attempted to directly print scaffolds by sequential fiber deposition using a 3D-Bioplotter™ system. Liquid material pushed from the nozzle to the platform needs some time to freeze the molding. Moreover, this method requires 2 days to manufacture scaffolds and additional time to accommodate lag between trauma and scaffold implantation. To reduce scaffold manufacturing time in a feasible manner, direct-write bioprinting appears to a promising method (Ahu et al., 2016; Mandrycky et al., 2016; Park et al., 2017). With this technology, it is possible to instantly print and transplant on the day of surgery to eliminate lag-time between trauma and scaffold implantation.
In this study, we successfully designed and fabricated customized cavity scaffolds using a technique integrating 3DP and imaging. To a certain extent, this emerging technology may overcome the shortcomings of former brain tissue scaffolds. Design of bionic brain scaffolds has the following characteristics. First, radiographic images can be converted to 3DP files to create complex, customized anatomical structures. Recently, several methods have been utilized to generate 3D object renderings, including computed tomography, laser scanning, and MRI, to provide representation of what is inside the body (Kikinis and Pieper, 2011; Xu et al., 2014). Additionally, this allows for quick production of custom scaffolds to repair lesions with various shapes (Lee et al., 2013; Jardini et al., 2014; Otto et al., 2015). Such scaffolds can be accurately matched with each defect and possess many advantages compared with standard scaffolds containing several regular channels used in earlier research. However, as previously mentioned, TBI-induced brain tissue defects/cavities change over time, and the most appropriate model design and timing of scaffold implantation require further study. Furthermore, the neurosurgeon could analyze each patient's defect, as well as other characteristics, such as age, race, or gender, to determine an optimal scaffold form. A neurosurgeon or tissue engineer could then print personalized implantable scaffolds via an automated 3DP system.
Another beneficial feature offered by this technology is the ability to accurately control the internal structure of scaffolds. In this study, orthogonally interconnected channels, which have been proven to enhance cell viability and proliferation (Domingos et al., 2013), were created in the cavity model using CAD software. Porosity and pore size, as well as open porous and interconnected networks, have direct implications on cell biological behavior (Wüst et al., 2015). The use of CAD is an attempt to closely mimic the complexities of the extracellular matrix by providing infiltrating or embedded cells with an environment similar to that of native brain tissue (Moroni et al., 2008). In view of this, orthogonally interconnected channels alone are not sufficient; more complex structures need incorporation into bionic scaffolds to highly simulate the extracellular matrix.
An additional strength of 3DP technology is the ability to freely select an appropriate “biological ink” (i.e., material). Currently, the state of 3DP technology for tissue regeneration is severely limited by the imbalance between printable materials and high-precision printing. While industrial 3DP methods, such as stereolithography, have achieved high resolution in the last few years, there are a limited number of biodegradable, biocompatible materials (Patra and Young, 2016). In contrast, key advantages of biological 3DP include material flexibility and room temperature processing, but the primary disadvantage is low resolution (Patra and Young, 2016). In this study, a 3D-Bioplotter™ system, based on extrusion of a viscous liquid material from a pressurized syringe onto a platform, was employed in printing scaffolds. Based on the previous experimental work of our laboratory, we decided to magnify the size of the cavity model from the prototype five-fold for 3DP. In addition to size, performance of the material for printing is important. It is a core principle of 3DP to identify materials capable of supporting the scaffold that also demonstrate biocompatibility and plasticity. Although materials encountered by neural cells in their native niche (such as laminin or hyaluronic acid) play a key role in replacing extracellular matrix (Ajioka et al., 2014; Duan et al., 2016), they are not suitable for printing. Meanwhile, natural biological materials, such as chitosan (Zhang et al., 2006) and collagen (Eagle et al., 1995) have been shown to possess excellent biocompatibility, low antigenicity, stable physical and chemical characteristics, and abundance in mammals. However, the primary limitation of collagen is its low mechanical strength and the rapid rate at which it degrades. Combining collagen with chitosan yields scaffolds with increased mechanical stability and appropriate degradation rates because of an ionic complex formed between positively charged chitosan and negatively charged collagen (Lee et al., 2004). In this work, collagen was blended at a 1:1 ratio with chitosan to produce a material strong enough to accommodate scaffold-printing conditions and overcome challenges related to printing precision and compatibility of materials.
Although the elastic modulus of collagen-chitosan (1:1) scaffolds used in this experiment is greater than brain tissue (Engler et al., 2006), the majority of NSCs adhered to scaffolds, spread, and exhibited survival and differentiation ability. More work examining mechanisms related to this phenomenon is clearly needed. It is important to also note that mechanical match between brain tissue and implants can positively impact the ability of the implant to enhance neural regeneration (Ventola, 2014). According to this and previous reports, reducing stiffness can result from changing ratios of materials and/or print parameters. With the upgrade of 3DP, we expected to be able to use a reduced proportion of chitosan (primary contributor to stiffness). Domingos et al. (2013) first investigated the influence of pore size and shape on mechanical and biological performance. Results of mechanical performance tests suggested weak scaffold stiffness with increasing porosity and number of deposition angles (from 0°/90° to 0°/45°/90°/135°) (Domingos et al., 2013). This research clearly demonstrated that mimicking brain elasticity could benefit from specifically designed architectures with tailored mechanical properties. Thus, the reality of an ideal printed neural restoration scaffold is still some way off. Although this magnified scaffold was not implanted into the rat brain defect to verify its effect on brain tissue regeneration, in vitro experiments confirmed good biocompatibility. To examine the function of brain tissue reconstruction in vivo, this technique could be applied to prototype scaffold implants for larger animal models. In addition, further study of materials utilized for 3DP must consider the biodegradability, porosity, strength, and location of brain tissue application to increase nerve regeneration.
This study elucidated techniques for the fabrication of custom scaffolds with specific external contours and orthogonally interconnected channels using a technique combining 3DP technology and MRI imaging. Generated scaffolds were found to be cytocompatible and biocompatible, thus offering great potential as bridges for brain tissue repair. We were unable to achieve scaffolds prototyping because the brain of the rodent model was too small and the bioprinting machine was not accurate enough. However, we provided a proof-of-concept illustration that an imaging-coupled 3D printing process can facilitate customized biomimetic scaffolds for neuroregeneration in previously inaccessible ways. Further investigation of this type of scaffold is needed, including identification of the most suitable materials and internal architecture for inducing differentiation of nerve cells in vitro and promoting functional tissue regeneration in vivo. In addition, the best time-point for scaffold design after TBI and appropriate clinical applications must be investigated. Regardless, it is conceivable that this technique may be a promising new strategy for regeneration of damaged complex brain tissues, thus paving the way toward personalized medicine.
Footnotes
Funding: This work was supported by the National Natural Science Foundation of China, No. 81301050, 81401067, 81271392, 81471275, 81541034; the Natural Science Foundation of Tianjin City of China, No. 14JCQNJC10200, 15JCQNJC11100, 16JCYBJC27600.
Conflicts of interest: None declared.
Plagiarism check: This paper was screened twice using CrossCheck to verify originality before publication.
Peer review: This paper was double-blinded and stringently reviewed by international expert reviewers.
Copyedited by Deusen AV, Maxwell R, Yu J, Li CH, Qiu Y, Song LP, Zhao M
References
- Ahu AY, Rami El A, Pu C, Sinan G, Fatih I, Utkan D. Towards artificial tissue models: past, present, and future of 3D bioprinting. Biofabrication. 2016;8:014103. doi: 10.1088/1758-5090/8/1/014103. [DOI] [PubMed] [Google Scholar]
- Ajioka I, Jinnou H, Okada K, Sawada M, Saitoh S, Sawamoto K. Enhancement of neuroblast migration into the injured cerebral cortex using laminin-containing porous sponge. Tissue Eng Part A. 2014;21:193–201. doi: 10.1089/ten.TEA.2014.0080. [DOI] [PubMed] [Google Scholar]
- Álvarez Z, Castaño O, Castells AA, Mateos-Timoneda MA, Planell JA, Engel E, Alcántara S. Neurogenesis and vascularization of the damaged brain using a lactate-releasing biomimetic scaffold. Biomaterials. 2014;35:4769–4781. doi: 10.1016/j.biomaterials.2014.02.051. [DOI] [PubMed] [Google Scholar]
- Bramlett HM, Dietrich WD. Long-term consequences of traumatic brain injury: current status of potential mechanisms of injury and neurological outcomes. J Neurotrauma. 2015;32:1834–1848. doi: 10.1089/neu.2014.3352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chia HN, Wu BM. Recent advances in 3D printing of biomaterials. J Biol Eng. 2015;9:4. doi: 10.1186/s13036-015-0001-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chimene D, Lennox KK, Kaunas RR, Gaharwar AK. Advanced bioinks for 3D printing: a materials science perspective. Ann Biomed Eng. 2016;44:2090–2102. doi: 10.1007/s10439-016-1638-y. [DOI] [PubMed] [Google Scholar]
- Crozier A, Augustin CM, Neic A, Prassl AJ, Holler M, Fastl TE, Hennemuth A, Bredies K, Kuehne T, Bishop MJ, Niederer SA, Plank G. Image-based personalization of cardiac anatomy for coupled electromechanical modeling. Ann Biomed Eng. 2016;44:58–70. doi: 10.1007/s10439-015-1474-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon CE, Clifton GL, Lighthall JW, Yaghmai AA, Hayes RL. A controlled cortical impact model of traumatic brain injury in the rat. J Neurosci Methods. 1991;39:253–262. doi: 10.1016/0165-0270(91)90104-8. [DOI] [PubMed] [Google Scholar]
- Domingos M, Intranuovo F, Russo T, Santis RD, Gloria A, Ambrosio L, Ciurana J, Bartolo P. The first systematic analysis of 3D rapid prototyped poly(ɛ-caprolactone) scaffolds manufactured through BioCell printing: the effect of pore size and geometry on compressive mechanical behaviour and in vitro hMSC viability. Biofabrication. 2013;5:045004. doi: 10.1088/1758-5082/5/4/045004. [DOI] [PubMed] [Google Scholar]
- Duan H, Li X, Wang C, Hao P, Song W, Li M, Zhao W, Gao Y, Yang Z. Functional hyaluronate collagen scaffolds induce NSCs differentiation into functional neurons in repairing the traumatic brain injury. Acta Biomater. 2016;45:182–195. doi: 10.1016/j.actbio.2016.08.043. [DOI] [PubMed] [Google Scholar]
- Eagle KS, Chalmers GR, Clary DO, Gage FH. Axonal regeneration and limited functional recovery following hippocampal deafferentation. J Comp Neurol. 1995;363:377–388. doi: 10.1002/cne.903630304. [DOI] [PubMed] [Google Scholar]
- Engler AJ, Sen S, Sweeney HL, Discher DE. Matrix elasticity directs stem cell lineage specification. Cell. 2006;126:677–689. doi: 10.1016/j.cell.2006.06.044. [DOI] [PubMed] [Google Scholar]
- Guan J, Zhu Z, Zhao RC, Xiao Z, Wu C, Han Q, Chen L, Tong W, Zhang J, Han Q, Gao J, Feng M, Bao X, Dai J, Wang R. Transplantation of human mesenchymal stem cells loaded on collagen scaffolds for the treatment of traumatic brain injury in rats. Biomaterials. 2013;34:5937–5946. doi: 10.1016/j.biomaterials.2013.04.047. [DOI] [PubMed] [Google Scholar]
- Guan Y, Yang F, Yao Q, Shi J, Wang G, Gu Z, Zhou F, Shen J. Impacts of phosphatase and tensin homology deleted on chromosome ten (PTEN)-inhibiting chitosan scaffold on growth and differentiation of neural stem cells. Int J Clin Exp Med. 2015;8:14308–14315. [PMC free article] [PubMed] [Google Scholar]
- Haghbayan H, Boutin A, Laflamme M, Lauzier F, Shemilt M, Moore L, Zarychanski R, Fergusson D, Turgeon AF. The prognostic value of magnetic resonance imaging in moderate and severe traumatic brain injury: a systematic review and meta-analysis protocol. Syst Rev. 2016;5:10. doi: 10.1186/s13643-016-0184-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh FY, Lin HH, Hsu SH. 3D bioprinting of neural stem cell-laden thermoresponsive biodegradable polyurethane hydrogel and potential in central nervous system repair. Biomaterials. 2015;71:48–57. doi: 10.1016/j.biomaterials.2015.08.028. [DOI] [PubMed] [Google Scholar]
- Huang KF, Hsu WC, Chiu WT, Wang JY. Functional improvement and neurogenesis after collagen-GAG matrix implantation into surgical brain trauma. Biomaterials. 2012;33:2067–2075. doi: 10.1016/j.biomaterials.2011.11.040. [DOI] [PubMed] [Google Scholar]
- Huang KF, Hsu WC, Hsiao JK, Chen GS, Wang JY. Collagen-glycosaminoglycan matrix implantation promotes angiogenesis following surgical brain trauma. Biomed Res Int 2014. 2014 doi: 10.1155/2014/672409. 672409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jardini AL, Larosa MA, Filho RM, Zavaglia CA, Bernardes LF, Lambert CS, Calderoni DR, Kharmandayan P. Cranial reconstruction: 3D biomodel and custom-built implant created using additive manufacturing. J Craniomaxillofac Surg. 2014;42:1877–1884. doi: 10.1016/j.jcms.2014.07.006. [DOI] [PubMed] [Google Scholar]
- Jiao Q, Zhang HX, Lv HX, Liu Y, Li JL. Organotypic slice culture of neonatal rat cortex and induced neural stem cell differentiation. Nan Fang Yi Ke Da Xue Xue Bao. 2011;31:1318–1322. [PubMed] [Google Scholar]
- Johnson BN, Lancaster KZ, Zhen G, He J, Gupta MK, Kong YL, Engel EA, Krick KD, Ju A, Meng F, Enquist LW, Jia X, McAlpine MC. 3D Printed Anatomical Nerve Regeneration Pathways. Adv Funct Mater. 2015;25:6205–6217. doi: 10.1002/adfm.201501760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kharatishvili I, Sierra A, Immonen RJ, Gröhn OHJ, Pitkänen A. Quantitative T2 mapping as a potential marker for the initial assessment of the severity of damage after traumatic brain injury in rat. Exp Neurol. 2009;217:154–164. doi: 10.1016/j.expneurol.2009.01.026. [DOI] [PubMed] [Google Scholar]
- Kikinis R, Pieper S. 3D Slicer as a tool for interactive brain tumor segmentation. Conf Proc IEEE Eng Med Biol Soc. 2011;2011:6982–6984. doi: 10.1109/IEMBS.2011.6091765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein GT, Lu Y, Wang MY. 3D printing and neurosurgery--ready for prime time? World Neurosurg. 2013;80:233–235. doi: 10.1016/j.wneu.2013.07.009. [DOI] [PubMed] [Google Scholar]
- Lee JE, Kim KE, Kwon IC, Ahn HJ, Lee SH, Cho H, Kim HJ, Seong SC, Lee MC. Effects of the controlled-released TGF-β1 from chitosan microspheres on chondrocytes cultured in a collagen/chitosan/glycosaminoglycan scaffold. Biomaterials. 2004;25:4163–4173. doi: 10.1016/j.biomaterials.2003.10.057. [DOI] [PubMed] [Google Scholar]
- Lee JY, Choi B, Wu B, Lee M. Customized biomimetic scaffolds created by indirect three-dimensional printing for tissue engineering. Biofabrication. 2013;5:045003. doi: 10.1088/1758-5082/5/4/045003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SJ, Zhu W, Heyburn L, Nowicki M, Harris B, Zhang LG. Development of novel 3-D printed scaffolds with core-shell nanoparticles for nerve regeneration. IEEE Trans Biomed Eng. 2017;64:408–418. doi: 10.1109/TBME.2016.2558493. [DOI] [PubMed] [Google Scholar]
- Leung GKK, Wang YC, Wu W. Peptide nanofiber scaffold for brain tissue reconstruction. Methods Enzymol. 2012;508:177–190. doi: 10.1016/B978-0-12-391860-4.00009-4. [DOI] [PubMed] [Google Scholar]
- Mahmoud AA, Salama AH. Norfloxacin-loaded collagen/chitosan scaffolds for skin reconstruction: Preparation, evaluation and in-vivo wound healing assessment. Eur J Pharm Sci. 2016;83:155–165. doi: 10.1016/j.ejps.2015.12.026. [DOI] [PubMed] [Google Scholar]
- Mandrycky C, Wang Z, Kim K, Kim DH. 3D bioprinting for engineering complex tissues. Biotechnol Adv. 2016;34:422–434. doi: 10.1016/j.biotechadv.2015.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maravelakis E, David K, Antoniadis A, Manios A, Bilalis N, Papaharilaou Y. Reverse engineering techniques for cranioplasty: a case study. J Med Eng Technol. 2008;32:115–121. doi: 10.1080/03091900600700749. [DOI] [PubMed] [Google Scholar]
- Marro A, Bandukwala T, Mak W. Three-dimensional printing and medical imaging: a review of the methods and applications. Curr Probl Diagn Radiol. 2016;45:2–9. doi: 10.1067/j.cpradiol.2015.07.009. [DOI] [PubMed] [Google Scholar]
- Mitsouras D, Liacouras P, Imanzadeh A, Giannopoulos AA, Cai T, Kumamaru KK, George E, Wake N, Caterson EJ, Pomahac B, Ho VB, Grant GT, Rybicki FJ. Medical 3D printing for the radiologist. Radiographics. 2015;35:1965–1988. doi: 10.1148/rg.2015140320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monsell F, Hughes AW, Turner J, Bellemore MC, Bilston L. Can the material properties of regenerate bone be predicted with non-invasive methods of assessment? Exploring the correlation between dual X-ray absorptiometry and compression testing to failure in an animal model of distraction osteogenesis. Strategies Trauma Limb Reconstr. 2014;9:45–51. doi: 10.1007/s11751-014-0188-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moroni L, de Wijn JR, van Blitterswijk CA. Integrating novel technologies to fabricate smart scaffolds. J Biomater Sci Polym Ed. 2008;19:543–572. doi: 10.1163/156856208784089571. [DOI] [PubMed] [Google Scholar]
- Otto IA, Melchels FP, Zhao X, Randolph MA, Kon M, Breugem CC, Malda J. Auricular reconstruction using biofabrication-based tissue engineering strategies. Biofabrication. 2015;7:032001. doi: 10.1088/1758-5090/7/3/032001. [DOI] [PubMed] [Google Scholar]
- Park JH, Jang J, Lee JS, Cho DW. Three-dimensional printing of tissue/organ analogues containing living cells. Ann Biomed Eng. 2017;45:180–194. doi: 10.1007/s10439-016-1611-9. [DOI] [PubMed] [Google Scholar]
- Patra S, Young V. A review of 3D printing techniques and the future in biofabrication of bioprinted tissue. Cell Biochem Biophys. 2016;74:93–98. doi: 10.1007/s12013-016-0730-0. [DOI] [PubMed] [Google Scholar]
- Radenkovic D, Solouk A, Seifalian A. Personalized development of human organs using 3D printing technology. Med Hypotheses. 2016;87:30–33. doi: 10.1016/j.mehy.2015.12.017. [DOI] [PubMed] [Google Scholar]
- Ray SK, Dixon CE, Banik NL. Molecular mechanisms in the pathogenesis of traumatic brain injury. Histol Histopathol. 2002;17:1137–1152. doi: 10.14670/HH-17.1137. [DOI] [PubMed] [Google Scholar]
- Shi W, Nie D, Jin G, Chen W, Xia L, Wu X, Su X, Xu X, Ni L, Zhang X, Zhang X, Chen J. BDNF blended chitosan scaffolds for human umbilical cord MSC transplants in traumatic brain injury therapy. Biomaterials. 2012;33:3119–3126. doi: 10.1016/j.biomaterials.2012.01.009. [DOI] [PubMed] [Google Scholar]
- Später T, Frueh FS, Metzger W, Menger MD, Laschke MW. In vivo biocompatibility, vascularization, and incorporation of Integra® dermal regenerative template and flowable wound matrix. J Biomed Mater Res B Appl Biomater. 2016 doi: 10.1002/jbm.b.33813. doi: 101002/jbmb33813:n/a-n/a. [DOI] [PubMed] [Google Scholar]
- Suri S, Han LH, Zhang W, Singh A, Chen S, Schmidt CE. Solid freeform fabrication of designer scaffolds of hyaluronic acid for nerve tissue engineering. Biomed Microdevices. 2011;13:983–993. doi: 10.1007/s10544-011-9568-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian L, Prabhakaran MP, Ramakrishna S. Strategies for regeneration of components of nervous system: scaffolds, cells and biomolecules. Regen Biomater. 2015;2:31–45. doi: 10.1093/rb/rbu017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turtzo LC, Budde MD, Gold EM, Lewis BK, Janes L, Yarnell A, Grunberg NE, Watson W, Frank JA. The evolution of traumatic brain injury in a rat focal contusion model. NMR Biomed. 2013;26:468–479. doi: 10.1002/nbm.2886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ventola CL. Medical applications for 3D printing: current and projected uses. P T. 2014;39:704–711. [PMC free article] [PubMed] [Google Scholar]
- Villapol S. Consequences of hepatic damage after traumatic brain injury: current outlook and potential therapeutic targets. Neural Regen Res. 2016;11:226–227. doi: 10.4103/1673-5374.177720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wüst S, Müller R, Hofmann S. 3D Bioprinting of complex channels—Effects of material, orientation, geometry, and cell embedding. J Biomed Mater Res A. 2015;103:2558–2570. doi: 10.1002/jbm.a.35393. [DOI] [PubMed] [Google Scholar]
- Wong DY, Krebsbach PH, Hollister SJ. Brain cortex regeneration affected by scaffold architectures. J Neurosurg. 2008;109:715–722. doi: 10.3171/JNS/2008/109/10/0715. [DOI] [PubMed] [Google Scholar]
- Wong DY, Hollister SJ, Krebsbach PH, Nosrat C. Poly(epsilon-caprolactone) and poly (L-lactic-co-glycolic acid) degradable polymer sponges attenuate astrocyte response and lesion growth in acute traumatic brain injury. Tissue Eng. 2007;13:2515–2523. doi: 10.1089/ten.2006.0440. [DOI] [PubMed] [Google Scholar]
- Xu X, Chen X, Zhang J, Zheng Y, Sun G, Yu X, Xu B. Comparison of the Tada formula with software slicer: precise and low-cost method for volume assessment of intracerebral hematoma. Stroke. 2014;45:3433–3435. doi: 10.1161/STROKEAHA.114.007095. [DOI] [PubMed] [Google Scholar]
- Yan F, Yue W, Zhang YL, Mao GC, Gao K, Zuo ZX, Zhang YJ, Lu H. Chitosan-collagen porous scaffold and bone marrow mesenchymal stem cell transplantation for ischemic stroke. Neural Regen Res. 2015;10:1421–1426. doi: 10.4103/1673-5374.163466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang P, Li CD. Construction of a polycaprolactone/bone extracellular matrix scaffold with three-dimensional printing technology and its osteoinductivity in vitro. Zhongguo Zuzhi Gongcheng Yanjiu. 2016;20:7773–7780. [Google Scholar]
- Yasuda H, Kuroda S, Shichinohe H, Kamei S, Kawamura R, Iwasaki Y. Effect of biodegradable fibrin scaffold on survival, migration, and differentiation of transplanted bone marrow stromal cells after cortical injury in rats. J Neurosurg. 2009;112:336–344. doi: 10.3171/2009.2.JNS08495. [DOI] [PubMed] [Google Scholar]
- Zeng W, Rong M, Hu X, Xiao W, Qi F, Huang J, Luo Z. Incorporation of chitosan microspheres into collagen-chitosan scaffolds for the controlled release of nerve growth factor. PLoS One. 2014;9:e101300. doi: 10.1371/journal.pone.0101300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang WG, Lü DC, Fu CY, Qü W. Favorable effect of chitosan sustained-release FK506 incorporated conduits on axonal regeneration in rat sciatic nerve. Zhonghua Yi Xue Za Zhi. 2006;86:1065–1068. [PubMed] [Google Scholar]
- Zhang X, Zhang Y. Tissue Engineering Applications of Three-Dimensional Bioprinting. Cell Biochem Biophys. 2015;72:777–782. doi: 10.1007/s12013-015-0531-x. [DOI] [PubMed] [Google Scholar]