Keywords: nerve regeneration, neurodegeneration, traditional Chinese medicine, Kai Xin San, insulin-degrading enzyme, amyloid-β, Alzheimer's disease, Chinese herbal compound, Aβ-degrading enzymes, neurons, Radix Ginseng, Radix Polygalae, Acorus Tatarinowii Rhizoma, neural regeneration
Abstract
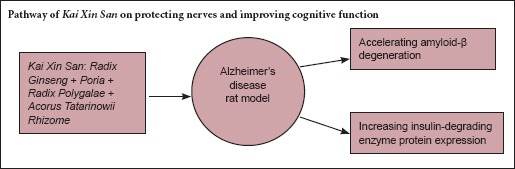
Kai Xin San is a Chinese herbal formula composed of Radix Ginseng, Poria, Radix Polygalae and Acorus Tatarinowii Rhizome. It has been used in China for many years for treating amnesia. Kai Xin San ameliorates amyloid-β (Aβ)-induced cognitive dysfunction and is neuroprotective in vivo, but its precise mechanism remains unclear. Expression of insulin-degrading enzyme (IDE), which degrades Aβ, is strongly correlated with cognitive function. Here, we injected rats with exogenous Aβ42 (200 μM, 5 μL) into the hippocampus and subsequently administered Kai Xin San (0.54 or 1.08 g/kg/d) intragastrically for 21 consecutive days. Hematoxylin-eosin and Nissl staining revealed that Kai Xin San protected neurons against Aβ-induced damage. Furthermore, enzyme-linked immunosorbent assay, western blot and polymerase chain reaction results showed that Kai Xin San decreased Aβ42 protein levels and increased expression of IDE protein, but not mRNA, in the hippocampus. Our findings reveal that Kai Xin San facilitates hippocampal Aβ degradation and increases IDE expression, which leads, at least in part, to the alleviation of hippocampal neuron injury in rats.
Introduction
Amyloid-β (Aβ), a key biomolecule in senile plaques, plays a central role in the pathology of Alzheimer's disease (AD) (Amici et al., 2016; Chai et al., 2016; Zhang et al., 2016). In the brain, under physiological conditions, Aβ is constantly generated from amyloid precursor protein and cleaved by β- and γ-secretases (Li et al., 2016; Niu et al., 2016). It is widely accepted that abnormal accumulation of Aβ, containing Aβ42 and Aβ40, participates in the pathology of AD (Awasthi et al., 2016; Wang, 2016). Aβ42, which is hydrophobic and prone to aggregation, seems to be more neurotoxic than Aβ40 in AD. It is mostly degraded by Aβ-degrading enzymes, but an imbalance between its generation and degradation leads to pathological Aβ accumulation in AD (Saido, 2013). However, anabolic increases in Aβ are rarely observed in sporadic AD cases, which constitute 95% of all AD cases (Campion et al., 1999). Several mechanisms can affect Aβ clearance and degradation, involving Aβ proteases, low-density lipoprotein receptor-related protein 1, and apolipoprotein E. Aβ degradation is correlated with Aβ-degrading enzyme dysfunction, which may explain the accumulation of Aβ in sporadic AD cases (Leissring, 2016). In addition, insulin-degrading enzyme (IDE) plays a key role in Aβ degradation, and its dysfunction contributes to the pathology of AD (Zhao et al., 2007).
The Chinese herbal formula Kai Xin San (KXS), has been used for treating amnesia for many years. Experimentally, KXS has been shown to improve learning and memory in several paradigms, including models of AD (Hu et al., 2013; Chu et al., 2016). KXS ameliorates the cognitive dysfunction induced by Aβ in vivo (Li et al., 2013) and seems to be involved in the protective effects targeting the neuropathological cascade after Aβ deposition. However, it is not known whether KXS directly mediates the catabolic metabolism of Aβ to reduce Aβ accumulation, though research on the catabolic metabolism of Aβ is ongoing. Identifying whether KXS can affect the expression of Aβ-degrading enzymes will provide important insight into treatment options for AD. To this end, we investigated the efficacy of KXS on Aβ degradation, and explored its mechanism in light of the catabolic role of IDE in Aβ metabolism. We demonstrate that KXS accelerates Aβ degradation by increasing IDE expression in the brain.
Materials and Methods
Animals
230 specific-pathogen-free Wistar rats (115 males, 115 females), weighing 200–240 g, were provided by the Laboratory Animal Center, Heilongjiang University of Chinese Medicine, China (license No. SYXK (Hei) 2013-012). Rats were housed in an animal laboratory at 22 ± 2°C and 60 ± 3% humidity, under a 12-hour dark/light cycle. The rats received a standard diet and free access to water. The animal facilities and protocols were approved by the Institutional Animal Care and Use Committee, Heilongjiang University of Chinese Medicine (No. 2014HZYLL-018). All procedures conformed with the National Institutes of Health Guide for the Care and Use of Laboratory Animals (NIH Publications No. 8023, revised, 1978).
Drugs and KXS extraction
Radix Ginseng (Renshen), Poria (Fuling), Radix Polygalae (Yuanzhi) and Acorus Tatarinowii Rhizoma (Shichangpu) were purchased from Harbin Tongrentang Drug Company (Harbin, China). From these components, we prepared KXS as described previously (Liu et al., 2014). In brief, the four dried raw herbs were mixed together in a weight ratio of 3:3:2:2 (Radix Ginseng 60 g, Poria 60 g, Radix Polygalae 40 g, Acorus Tatarinowii Rhizoma 40 g), and decocted/extracted by refluxing for 1.5 hours in 2,000 mL boiling 60% ethanol (1:10, w/v). The extracts were passed through filter paper, dried under a vacuum and stored at −80°C. The extracts were mainly characterized by 75 chemical constituents, identified by ultra-performance liquid chromatography tandem mass spectrometry.
Aβ and KXS administration
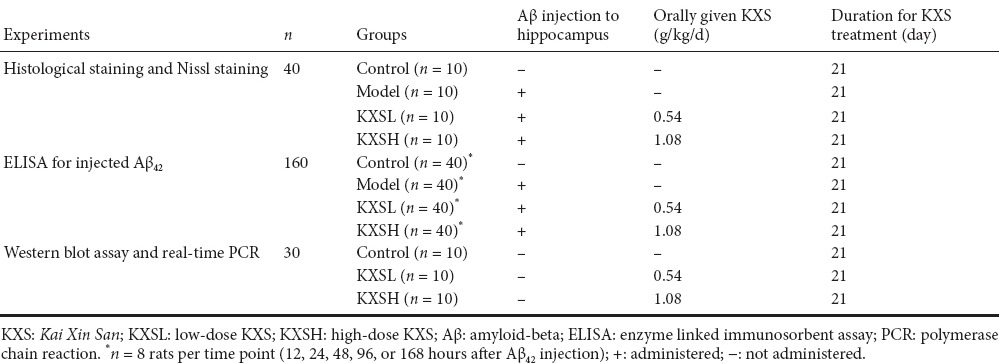
200 rats were equally and randomly allocated to four groups: control, model, low-dose KXS (KXSL) and high-dose KXS (KXSH; Table 1). Aβ42 (Wako Pure Chemical Industries, Ltd., Japan) was dissolved in dimethyl sulfoxide and diluted with 0.9% saline. For intrahippocampal Aβ42 injection (all groups except control), rats were anesthetized with 3% pentobarbital sodium (50 mg/kg) and fixed in a stereotaxic instrument as previously described (Asle-Rousta et al., 2013). Aβ42 solution was injected into the hippocampus bilaterally at the following coordinates: 3.6 mm posterior to the bregma, 2.2 mm lateral to the midline and 4.0 mm below the top of the skull. A total of 10 μL per rat (5 μL each side, 200 μM) was injected over 10 minutes. The needle was slowly withdrawn from the brain, the surgical incision was sutured, and penicillin sodium was administered to prevent infection. Control rats underwent the same procedure, but same volume of saline was injected instead of of Aβ. Subsequently, rats received KXS (KXSL group, 0.54 g/kg/d; KXSH group, 1.08 g/kg/d) or an equivalent volume of saline (control and model groups) intragastrically once a day for 21 consecutive days. Forty rats were used for histology and Nissl staining (n = 10 per group) and 160 for the enzyme-linked immunosorbent assay (ELISA) (n = 40 per group).
Table 1.
Group assignment and drug treatment
Histological staining
Rats were anesthetized with 10% (3.5 mL) chloral hydrate and decapitated after perfusion with 10% formalin and the brain was removed immediately. Brains from the rats in each group (Table 1) were fixed with 10% formalin for hematoxylin-eosin and Nissl staining. The tissue containing the hippocampus was dehydrated through a graded series of alcohol (75%, 85%, 95%, and 100%), embedded in paraffin, and sliced into 4-μm-thick sections. These sections were deparaffinized and subjected to hematoxylin-eosin and Nissl staining (Ooigawa et al., 2006; Zhou et al., 2012). Under a microscope (Olympus, Tokyo, Japan), a 300 × 100 μm2 field of view was selected at random to count the number of injured cells (showing nuclear shrinkage or disappearance) and total cells in the hippocampal dentate gyrus, and the percentage of injured cells was calculated.
ELISA assay for injected Aβ42
At five time points after injection (12, 24, 48, 96 and 168 hours; n = 8 rats per time point), brain Aβ42 concentration was measured. Rats were anesthetized with 10% (3.5 mL) chloral hydrate and decapitated. The brains were quickly dissected on ice at the allocated time-point. For ELISA, brains were homogenized in 70% formic acid buffer (1:10 w/v) for 1 hour. The lysates were centrifuged at 12,000 × g for 30 minutes. The supernatants were collected and neutralized with 1 M Tris-base solution. The solution containing Aβ42 was measured using a specific and sensitive sandwich ELISA kit (Wako Pure Chemical Industries, Ltd., Japan). Total protein concentration was measured using the BCA method.
Western blot assay
Thirty rats were equally and randomly allocated to three groups (Table 1). Rats were anesthetized with 10% (3.5 mL) chloral hydrate and decapitated. Brains were removed on ice and hippocampi were dissected out for western blot and real time-polymerase chain reaction (PCR) assays. To measure IDE content, hippocampal tissue was lysed in radioimmunoprecipitation assay buffer containing protease inhibitor. Protein content was determined using the Bradford method. Samples were mixed with an equal volume of 2× loading buffer and proteins were denatured by boiling at 100°C for 5 minutes. Protein samples (2 μg/μL) were resolved using sodium dodecyl sulfate polyacrylamide gel electrophoresis with a 10% gel and transferred to polyvinylidene fluoride membranes. Blots were blocked in 5% non-fat milk for 2 hours at room temperature, then incubated with primary polyclonal antibody against human IDE (1:400; Santa Cruz Biotechnology, Santa Cruz, CA, USA) overnight at 4°C. Immunoreactive bands were probed with a horseradish peroxidase-linked secondary polyclonal antibody (1:800; Beyotime Institute of Biotechnology, Haimen, China) for 2 hours. Enhanced chemiluminescence detection (Beyotime Institute of Biotechnology, Shanghai, China) was used to quantify the optical densities of immunoreactive bands. All protein bands were converted to gray values and compared against the internal reference protein (β-actin, Beyotime Institute of Biotechnology, Shanghai, China). Protein expression was quantified using Gel-Pro Analyzer 3.0 software (Media Cybernetics, Inc., Rockville, MD, USA).
Real time-PCR for detecting IDE mRNA
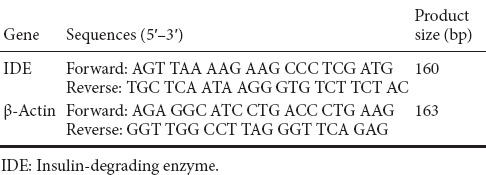
Total hippocampal RNA was extracted using TRIzol Reagent (TaKaRa Biotechnology (Dalian) Co., Ltd., Dalian, China) according to the manufacturer's protocol. RNA concentration was then measured using ultraviolet spectroscopy. Total RNA was used to prepare cDNA by reverse transcription using a PrimeScript RT reagent kit (TaKaRa Biotechnology (Dalian) Co., Ltd.). For detection of IDE mRNA, primers were designed using Primer 5 software (downloaded from https://www.genscript.com/ssl-bin/app/primer) (Table 2). cDNA was amplified using a SYBR Premix Ex Taq kit (TaKaRa Biotechnology (Dalian) Co., Ltd.). The PCR protocol consisted of denaturation at 95°C for 30 seconds followed by 40 denaturation and annealing cycles (95°C for 5 seconds, 60°C for 34 seconds). β-Actin was used as an internal reference. IDE mRNA expression was quantified using the 2−ΔΔCt method.
Table 2.
Sequences of primers used for real-time polymerase chain reaction
Statistical analysis
All data, expressed as the mean ± SD, were analyzed using SPSS 13.0 software (SPSS, Chicago, IL, USA). One-way analysis of variance was used for comparisons of multiple groups and the least significant difference test was used between two groups. P < 0.05 was considered significant.
Results
Effect of KXS on neuronal injury induced by hippocampal Aβ42 injection
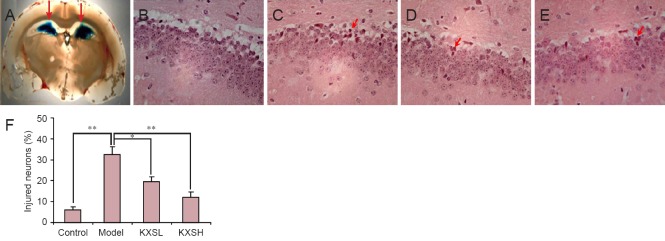
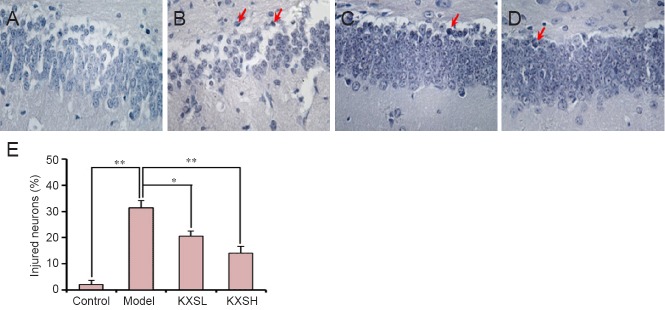
Hematoxylin-eosin and Nissl staining were performed to investigate the neuroprotective effect of KXS. As expected, Aβ42 injection induced hippocampal neuronal injury, which was prevented by KXS. Hematoxylin-eosin staining revealed irregular neurons and neuronal death around the injection site (Figure 1B). However, neuronal injuries were markedly less severe after treatment with KXS (Figure 1C, D), and the number of injured neurons in the KXS group was significantly lower than in the model group (F = 23.5; P = 0.03 and P < 0.005 Figure 1E). Accordingly, Nissl staining also revealed typical neuronal pathological changes, including neuronal loss and nucleus shrinkage or disappearance, in the hippocampal dentate gyrus in the model group (Figure 2B), and were reversed significantly after treatment with KXS (Figure 2C, D). The proportion of injured neurons was significantly lower in the KXSL group (F = 20.6; P = 0.04) and KXSH group (P = 0.007) than in the model group (Figure 2E).
Figure 1.
Effect of KXS on neuronal injury in the hippocampal dentate gyrus of rats after Aβ42 injection.
(A) Site of Aβ injection (arrows) (× 10). (B–E) Hematoxylin-eosin staining (× 200), showing neuronal injury in hippocampal dentate gyrus of control group (no Aβ or KXS) (B), model group (Aβ without KXS) (C), KXSL group (Aβ + 0.54 g/kg/d KXS, i.g.) (D), and KXSH group (Aβ + 1.08 g/kg/d KXS, i.g.) (E). Red arrows: Injured cells. (F) A 300 × 100 μm2 field was selected at random to count the number of injured (nuclear shrinkage or disappearance) and total cells in the area around the Aβ injection site. The proportion of damaged cells was significantly greater in the model group than in the control group, and significantly lower in the KXSH and KXSL groups than in the model group. Data are expressed as the mean ± SD (*P < 0.05, **P < 0.01; n = 10; one-way analysis of variance followed by the least significant difference post hoc test). KXS: Kai Xin San; KXSL: low-dose KXS; KXSH: high-dose KXS; Aβ: amyloid-beta; d: day.
Figure 2.
Hippocampal dentate gyrus histopathology.
(A–D) Representative Nissl stained sections (× 200) from animals in the control group (no Aβ or KXS) (A), model group (Aβ without KXS) (B), KXSL group (Aβ + 0.54 g/kg/d KXS, i.g.) (C), and KXSH group (Aβ + 1.08 g/kg/d KXS, i.g.) (D). Model group shows neuronal loss and nuclear shrinkage or disappearance (red arrows). KXSL and KXSH groups show fewer histopathological changes. (E) Quantification of injured cells. There was a significantly higher percentage of damaged cells in the model group than in the control group, and a significantly lower percentage in the KXSH and KXSL groups than in the model group. Data are expressed as the mean ± SD (*P < 0.05, **P < 0.01; n = 10; one-way analysis of variance followed by the least significant difference post hoc test). KXS: Kai Xin San; KXSL: low-dose KXS; KXSH: high-dose KXS; Aβ: amyloid-beta; d: day.
KXS effect on degradation of Aβ42
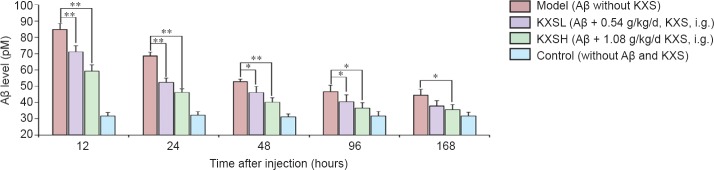
We explored why KXS reduces Aβ42-induced histological injury. Aβ42 levels in all groups declined between 6 and 96 hours, indicating that some Aβ42 clearance occurred without treatment. However, at 12, 24, 48, 96 and 168 hours, Aβ42 levels were significantly lower in the KXS groups than in the model group, and lower in the KXSH group than in the KXSL group (Figure 3).
Figure 3.
KXS accelerated the degradation of injected Aβ42.
Aβ level decreased over time in all groups. However, at 12, 24, 48, 96 and 168 hours, Aβ levels in the KXSH group and KXSL group were lower than in the model group. Data are expressed as the mean ± SD (*P < 0.05, **P < 0.01; n = 10; one-way analysis of variance followed by the least significant difference post hoc test). KXS: Kai Xin San; KXSL: low-dose KXS; KXSH: high-dose KXS; Aβ: amyloid-beta; d: day.
Effect of KXS on IDE mRNA and protein expression
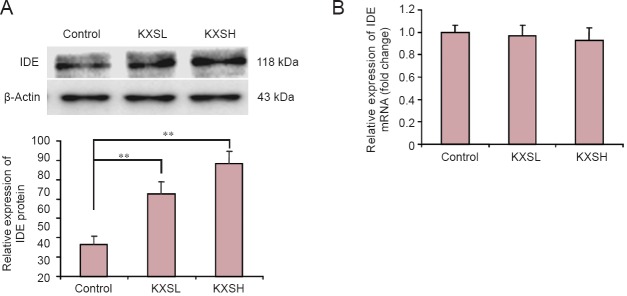
To further explore the KXS-induced reduction in Aβ42 levels, hippocampal IDE protein expression was investigated by western blot assay. Rats in the KXS groups showed significant upregulation of IDE protein after 168 hours compared with the model group (P < 0.01). IDE protein expression increased more in the KXSH group than in the KXSL group (Figure 4A). However, no significant difference in IDE mRNA expression was found between the KXS groups and the model group (Figure 4B). These results indicate that KXS accelerated Aβ degradation by upregulating IDE expression in the brain.
Figure 4.
KXS upregulates IDE protein expression but has no effect on IDE mRNA expression.
(A) Relative protein expression of IDE (gray values) in the KXSH (Aβ + 1.08 g/kg/d KXS, i.g.) and KXSL (Aβ + 0.54 g/kg/d KXS, i.g.) groups was higher than in the control group (no Aβ or KXS). (B) After 168 hours of Aβ42-induced hippocampal neuronal injury and treatment with KXS, no significant changes were observed in IDE mRNA expression (2−ΔΔCt) between the KXS groups and the model (Aβ without KXS) group. Data are expressed as the mean ± SD. **P < 0.01; n = 10; one-way analysis of variance followed by the least significant difference post hoc test. KXS: Kai Xin San; KXSL: low-dose KXS; KXSH: high-dose KXS; IDE: insulin-degrading enzyme; Aβ: amyloid-beta; d: day.
Discussion
KXS treatment improves the cognitive dysfunction induced by intrahippocampal Aβ injection (Li et al., 2013). Moreover, rat plasma containing KXS alleviated Aβ42-induced damage in PC12 cells (Wen et al., 2012). These results tightly link the pharmacological targets of KXS to Aβ42. The present study showed that KXS prevented neuronal injury induced by Aβ42 and suggested that the therapeutic efficacy of KXS was related to Aβ in treating cognitive dysfunction.
KXS improves learning and memory in several paradigms, including experimental AD (Hu et al., 2013). However, whether KXS can affect Aβ degradation and clearance is not clear. The present results show that Aβ42 levels were significantly reduced after treatment with KXS. Because the excessive accumulation of Aβ is regarded as an essential upstream process in the AD pathogenesis cascade, we speculated that KXS might promote Aβ degradation.
Although insulin was long considered the main substrate for IDE, it was recently shown that IDE also degrades Aβ into peptides in the brain (Baranello et al., 2015). In addition, the process of substrate degradation by IDE has been elucidated: a proteolytic chamber, formed by the N and C terminal units of IDE, cleaves only peptides of up to 70 amino acids (Shen et al., 2006). In the brain, IDE is secreted from neurons and microglia, and Aβ degradation takes places extracellularly as well as in the cell membrane (Qiu et al., 1998; Zhang et al., 2013). Some studies reported an association of polymorphisms in the IDE locus with the risk of AD and plasma Aβ levels (Vekrellis et al., 2000; Carrasquillo et al., 2010; Cheng et al., 2015). Most clinical studies confirm that IDE activity or expression is lower in AD and in individuals at high risk of AD (Perez et al., 2000; Del Campo et al., 2015). Moreover, an increase of cerebral endogenous Aβ was found after IDE inhibition or knockout (Shen et al., 2006). These studies suggest that IDE participates in the degradation of Aβ, and that its dysfunction contributes to the increase of Aβ in the brain. Therefore, in our study, we explored whether KXS would alter IDE expression.
Indeed, there was a significant increase in IDE protein expression after treatment with KXS, suggesting that this is an important reason for the decrease in Aβ42. Consistent with this, some active components from KXS have the ability to increase the protein level and activity of IDE. For instance, ginsenoside Rg1, one of the major active ingredients of KXS, can increase IDE expression by upregulating PPARγ, leading to lower levels of Aβ42 in the hippocampus (Quan et al., 2013). Ginsenosides Rg3 and Rb1, which act on the insulin signal transduction pathway, also can upregulate IDE protein expression, implying that the insulin pathway may be involved in regulating IDE expression (Gu et al., 2014; Jang et al., 2015). Together, these findings suggest that KXS and its active constituents can increase IDE expression. However, according to our PCR results, the mRNA level of IDE did not change. There are a number of possible reasons for these inconsistent changes. One is that rapid transcription coupled with rapid degradation will result in no change in RNA survival and availability for translation, and therefore cannot account for increased protein production. Conversely, KXS may regulate post-transcriptional changes, such as increased translation efficiency or reduced protein degradation, without affecting the mRNA level of IDE (Rodnina, 2016). These complicated molecular mechanisms represent an even greater challenge and should be addressed in subsequent studies.
In summary, the results of the present study demonstrate for the first time that KXS accelerates Aβ degradation by upregulating IDE protein expression and improving cognitive dysfunction. KXS is a promising agent for the treatment of AD.
Footnotes
Funding: This research was supported by the National Natural Science Foundation of China, No. 81303248, 81603321; the Natural Science Foundation of Heilongjiang Province of China, No. H2015028; a grant from the Nursing Program for Young Scholars of Heilongjiang Province of China, No. UNPYSCT-2016116; the Scientific Research Fund for Doctors of Qiqihar Medical University in China, No. QY2016B-09.
Conflicts of interest: None declared.
Plagiarism check: This paper was screened twice using CrossCheck to verify originality before publication.
Peer review: This paper was double-blinded and stringently reviewed by international expert reviewers.
Copyedited by Slone-Murphy J, Wang J, Li CH, Qiu Y, Song LP, Zhao M
References
- Amici S, Iannizzi P, Di Pucchio A, Abraha I, Montedori A, Chattat R, Vanacore N. Can early counselling and support for Alzheimer's disease caregivers reduce burden? Study protocol for a multicenter randomized controlled trial. Clin Trials Degener Dis. 2016;1:99–106. [Google Scholar]
- Asle-Rousta M, Kolahdooz Z, Oryan S, Ahmadiani A, Dargahi L. FTY720 (fingolimod) attenuates beta-amyloid peptide (Aβ42)-induced impairment of spatial learning and memory in rats. J Mol Neurosci. 2013;50:524–532. doi: 10.1007/s12031-013-9979-6. [DOI] [PubMed] [Google Scholar]
- Awasthi M, Singh S, Pandey VP, Dwivedi UN. Alzheimer's disease: an overview of amyloid beta dependent pathogenesis and its therapeutic implications along with in silico approaches emphasizing the role of natural products. J Neurol Sci. 2016;361:256–271. doi: 10.1016/j.jns.2016.01.008. [DOI] [PubMed] [Google Scholar]
- Baranello RJ, Bharani KL, Padmaraju V, Chopra N, Lahiri DK, Greig NH, Pappolla MA, Sambamurti K. Amyloid-beta protein clearance and degradation (ABCD) pathways and their role in Alzheimer's disease. Curr Alzheimer Res. 2015;12:32–46. doi: 10.2174/1567205012666141218140953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campion D, Dumanchin C, Hannequin D, Dubois B, Belliard S, Puel M, Thomas-Anterion C, Michon A, Martin C, Charbonnier F, Raux G, Camuzat A, Penet C, Mesnage V, Martinez M, Clerget-Darpoux F, Brice A, Frebourg T. Early-onset autosomal dominant Alzheimer disease: prevalence, genetic heterogeneity, and mutation spectrum. Am J Hum Genet. 1999;65:664–670. doi: 10.1086/302553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrasquillo MM, Belbin O, Zou F, Allen M, Ertekin-Taner N, Ansari M, Wilcox SL, Kashino MR, Ma L, Younkin LH, Younkin SG, Younkin CS, Dincman TA, Howard ME, Howell CC, Stanton CM, Watson CM, Crump M, Vitart V, Hayward C, et al. Concordant association of insulin degrading enzyme gene (IDE) variants with IDE mRNA, Abeta, and Alzheimer's disease. PLoS One. 2010;5:e8764. doi: 10.1371/journal.pone.0008764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chai GS, Wang YY, Yasheng A, Zhao P. Beta 2-adrenergic receptor activation enhances neurogenesis in Alzheimer's disease mice. Neural Regen Res. 2016;11:1617–1624. doi: 10.4103/1673-5374.193241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng H, Wang L, Shi T, Shang Y, Jiang L. Association of insulin degrading enzyme gene polymorphisms with Alzheimer's disease: a meta-analysis. Int J Neurosci. 2015;125:328–335. doi: 10.3109/00207454.2014.941440. [DOI] [PubMed] [Google Scholar]
- Chu H, Zhang A, Han Y, Lu S, Kong L, Han J, Liu Z, Sun H, Wang X. Metabolomics approach to explore the effects of Kai-Xin-San on Alzheimer's disease using UPLC/ESI-Q-TOF mass spectrometry. J Chromatogr B Analyt Technol Biomed Life Sci. 2016;1015-1016:50–61. doi: 10.1016/j.jchromb.2016.02.007. [DOI] [PubMed] [Google Scholar]
- Del Campo M, Stargardt A, Veerhuis R, Reits E, Teunissen CE. Accumulation of BRI2-BRICHOS ectodomain correlates with a decreased clearance of Abeta by insulin degrading enzyme (IDE) in Alzheimer's disease. Neurosci Lett. 2015;589:47–51. doi: 10.1016/j.neulet.2015.01.036. [DOI] [PubMed] [Google Scholar]
- Gu WJ, Liu D, Zhang MR, Zhang H. Effect of ginsenoside Rb1 on insulin signal transduction pathway in hippocampal neurons of high-glucose-fed rats. Zhongguo Zhongyao Zazhi. 2014;39:1064–1068. [PubMed] [Google Scholar]
- Hu Y, Liu M, Liu P, Yan JJ, Liu MY, Zhang GQ, Zhou XJ, Yu BY. Effect of Kai Xin San on learning and memory in a rat model of paradoxical sleep deprivation. J Med Food. 2013;16:280–287. doi: 10.1089/jmf.2012.2486. [DOI] [PubMed] [Google Scholar]
- Jang SK, Yu JM, Kim ST, Kim GH, Park da W, Lee DI, Joo SS. An Aβ42 uptake and degradation via Rg3 requires an activation of caveolin, clathrin and Aβ-degrading enzymes in microglia. Eur J Pharmacol. 2015;758:1–10. doi: 10.1016/j.ejphar.2015.03.071. [DOI] [PubMed] [Google Scholar]
- Leissring MA. Abeta-degrading proteases: therapeutic potential in alzheimer disease. CNS drugs. 2016;30:667–675. doi: 10.1007/s40263-016-0364-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Luo J, Chen D, Tong JB, Zeng LP, Cao YQ, Xiang J, Luo XG, Shi JM, Wang H, Huang JF. BACE1 in the retina: a sensitive biomarker for monitoring early pathological changes in Alzheimer's disease. Neural Regen Res. 2016;11:447–453. doi: 10.4103/1673-5374.179057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Liu B, Liu XW, Huang SM. Mechanism research of Kai Xin Granule on Aβ-induced cognitive impairment in rats. Zhongyiyao Xinxi. 2013;33:34–38. [Google Scholar]
- Liu XW, Liu S, Huang SM. Plasma pharmacoc hemistry study of effective extracivet from kai-xinsan on Alzhemer's disease. Zhongguo Shiyan Fangjixue Zazhi. 2016;20:179–183. [Google Scholar]
- Niu Jw, Zhang B, Chen H. Safety and efficacy of human umbilical cord-derived mesenchymal stem cells in patients with Alzheimer's disease: study protocol for an open-label self-control trial. Clin Trials Degener Dis. 2016;1:1–8. [Google Scholar]
- Ooigawa H, Nawashiro H, Fukui S, Otani N, Osumi A, Toyooka T, Shima K. The fate of Nissl-stained dark neurons following traumatic brain injury in rats: difference between neocortex and hippocampus regarding survival rate. Acta Neuropathol. 2006;112:471–481. doi: 10.1007/s00401-006-0108-2. [DOI] [PubMed] [Google Scholar]
- Perez A, Morelli L, Cresto JC, Castano EM. Degradation of soluble amyloid beta-peptides 1-40, 1-42, and the Dutch variant 1-40Q by insulin degrading enzyme from Alzheimer disease and control brains. Neurochem Res. 2000;25:247–255. doi: 10.1023/a:1007527721160. [DOI] [PubMed] [Google Scholar]
- Qiu WQ, Walsh DM, Ye Z, Vekrellis K, Zhang J, Podlisny MB, Rosner MR, Safavi A, Hersh LB, Selkoe DJ. Insulin-degrading enzyme regulates extracellular levels of amyloid beta-protein by degradation. J Biol Chem. 1998;273:32730–32738. doi: 10.1074/jbc.273.49.32730. [DOI] [PubMed] [Google Scholar]
- Quan Q, Wang J, Li X, Wang Y. Ginsenoside Rg1 decreases Aβ(1-42) level by upregulating PPARγ and IDE expression in the hippocampus of a rat model of Alzheimer's disease. PLoS One. 2013;8:e59155. doi: 10.1371/journal.pone.0059155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodnina MV. The ribosome in action: Tuning of translational efficiency and protein folding. Protein Sci. 2016;25:1390–1406. doi: 10.1002/pro.2950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saido TC. Metabolism of amyloid beta peptide and pathogenesis of Alzheimer's disease. Proc Jpn Acad Ser B Phys Biol Sci. 2013;89:321–339. doi: 10.2183/pjab.89.321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Y, Joachimiak A, Rosner MR, Tang WJ. Structures of human insulin-degrading enzyme reveal a new substrate recognition mechanism. Nature. 2006;443:870–874. doi: 10.1038/nature05143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vekrellis K, Ye Z, Qiu WQ, Walsh D, Hartley D, Chesneau V, Rosner MR, Selkoe DJ. Neurons regulate extracellular levels of amyloid beta-protein via proteolysis by insulin-degrading enzyme. J Neurosci. 2000;20:1657–1665. doi: 10.1523/JNEUROSCI.20-05-01657.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang GF. Effects of Bushen Yizhi Decoction on Alzheimer's disease model rats induced by D-galactose combined with amyloid-beta 25-35 and the underlying mechanism. Zhongguo Zuzhi Gongcheng Yanjiu. 2016;20:7307–7313. [Google Scholar]
- Wen W, Zhang C, Liu M, Huang SM. The rat plasma containing Kai Xin San protected PC12 from Aβ-induced toxicity. Zhongyiyao Xinxi. 2012;29:80–81. [Google Scholar]
- Zhang SG, Wang XS, Zhang YD, Di Q, Shi JP, Qian M, Xu LG, Lin XJ, Lu J. Indirubin-3’-monoxime suppresses amyloid-beta-induced apoptosis by inhibiting tau hyperphosphorylation. Neural Regen Res. 2016;11:988–993. doi: 10.4103/1673-5374.184500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Wang B, Wan H, Zhou Q, Li T. Meta-analysis of the insulin degrading enzyme polymorphisms and susceptibility to Alzheimer's disease. Neurosci Lett. 2013;541:132–137. doi: 10.1016/j.neulet.2013.01.051. [DOI] [PubMed] [Google Scholar]
- Zhao Z, Xiang Z, Haroutunian V, Buxbaum JD, Stetka B, Pasinetti GM. Insulin degrading enzyme activity selectively decreases in the hippocampal formation of cases at high risk to develop Alzheimer's disease. Neurobiol Aging. 2007;28:824–830. doi: 10.1016/j.neurobiolaging.2006.05.001. [DOI] [PubMed] [Google Scholar]
- Zhou X, Song FH, He W, Yang XY, Zhou ZB, Feng X, Zhou LH. Neonatal exposure to sevoflurane causes apoptosis and reduces nNOS protein expression in rat hippocampus. Mol Med Rep. 2012;6:543–546. doi: 10.3892/mmr.2012.976. [DOI] [PubMed] [Google Scholar]