Abstract
Human herpesvirus 6A (HHV-6A) starts its replication cycle following the action of immediate early proteins that transactivate viral promoters. Immediate early protein 2 (IE2) of HHV-6A is a 1500 amino acid polypeptide with a C-terminal region that is conserved among beta-herpesvirus subfamily members. In this study, a structural domain in the homologous C-terminal region was subjected to secondary structure prediction, and residues 1324–1500 were subsequently designated as the C-terminal domain of IE2 (IE2-CTD). The gene fragment encoding IE2-CTD was inserted into an E. coli expression vector and expressed as a fusion protein with maltose binding protein (MBP) at the N-terminus. IE2-CTD has a theoretical isoelectric point (pI) of 9.29, and strong cation exchange column chromatography was effective for purification. Needle-shaped crystals of IE2-CTD were obtained using the sitting-drop vapour diffusion method, and larger selenomethionine-labelled crystals of space group P21 diffracted X-rays to 2.5 Å resolution using synchrotron radiation. Data were collected at the selenium absorption peak wavelength for experimental phasing by the single anomalous dispersion method. The resulting electron density map clearly shows the protein backbone, and full structural determination and refinement are in progress.
Keywords: Human Herpesvirus 6A, Immediate Early Protein 2, Transactivator, X-ray Crystallography
Human herpesvirus 6A (HHV-6A) belongs to the beta-herpesvirus subfamily, which also includes three other pathogenic human herpesviruses, namely, human herpesvirus 6B (HHV-6B), human herpesvirus 7 (HHV-7) and human cytomegalovirus (HCMV). HHV-6B is the causative agent of exanthema subitum (roseola) in children (1), and is highly similar to HHV-6A, but they can be distinguished by several characteristic differences (2–9). The pathogenicity of HHV-6A is still unclear, although it has been linked to multiple sclerosis (10,11).
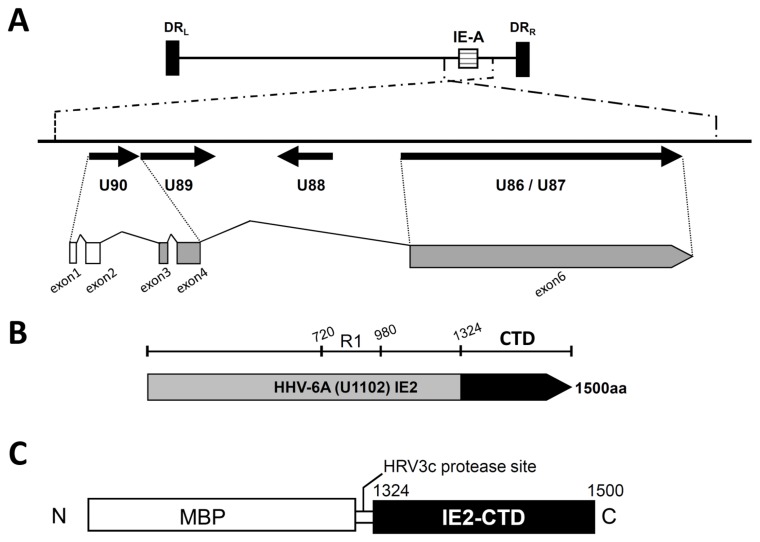
Immediate early protein 2 (IE2) of HHV-6A is an essential transactivator that promotes viral gene expression. IE2 is one of the first proteins expressed at the beginning of the lytic infection, and it promotes the expression of viral proteins involved in genome replication. IE2 is encoded at the IE-A locus of the viral genome, and the IE2 transcript consists of a 5′ region from the putative ORF U90 and a 3′ region from the putative ORF U86/87 (Figure 1A) (12). The transcript encodes a protein of 1500 amino acids that has SSRA/D repeats in the central R1 region (Figure 1B). Biochemical analysis of HHV-6A IE2 revealed that the N-terminal part preceding the repeat region, the repeat region itself and the following C-terminal section are all required for the full transactivation potency of IE2, although the contribution of the repeat region appears to be less crucial (13). The C-terminal region of IE2 is highly conserved in HCMV IE2 and HHV-6A IE2 (14), while the other regions are highly variable. Analysis of HCMV IE2 revealed that the C-terminal region includes a DNA binding site (14), a dimerization site (14) and an interaction site for transcription factors, namely, transcription factor II B (TFIIB) and TATA binding protein (TBP) (15, 16). Thus, the C-terminal region of IE2 is considered as the essential functional module required for transactivation activity.
Figure 1.
HHV-6A U1102 IE2 and the C-terminal structural domain IE2-CTD. (A) A schematic diagram of the immediate early A locus of HHV-6A strain U1102. The upper panel shows the entire HHV-6A strain U1102 genome including direct repeat regions left (DRL) and right (DRR) and the major immediate early A locus (IE-A). The lower panel is an enlarged view of the ORFs at the IE-A locus (represented by arrows). Exons 1 and 2 (white) are non-coding, and exons 3, 4 and 6 (grey) generate the IE2 protein. Exons 3 and 4 are derived from U90, and exon 6 is from U86/87. (B) The full length HHV-6A IE2. The black region is the C-terminal domain of IE2 (IE2-CTD), the focus of this study. R1 (720–980) indicates the serine/arginine repeat region. (C) The HHV-6A IE2-CTD construct used for expression in Escherichia coli. The 1324–1500 region of HHV-6A (IE2-CTD) is fused to maltose binding protein (MBP) with a human rhinovirus (HRV) 3c protease site at the domain boundary.
To address the issue of how the homologous C-terminal region of IE2 facilitates the multifunctional transactivation activity, structural information is required, since this is not currently available for homologues of IE2. In the present study, we expressed and purified a recombinant structural domain of the IE2 C-terminal domain (IE2-CTD). The purified protein was crystallized, X-ray diffraction data were collected and structure determination was initiated.
MATERIALS AND METHODS
Construction of the HHV-6A IE2-CTD expression plasmid
The secondary structure of the C-terminal region of HHV-6A IE2 strain U1102 was predicted using the Jpred4 webserver (17; http://www.compbio.dundee.ac.uk/jpred/index.html). The results of the prediction indicated that the 1324–1500 region formed a distinct IE2 C-terminal domain (IE2-CTD). A codon-optimized nucleotide sequence encoding this region was amplified by PCR with primers 5′-acaGGATCCctggaggtgctgttccagggccccaagcagatccccaagaag-3′ and 5′-actGTCGACtcagcacttggacagggtg-3′ that included BamHI and SalI restriction enzyme sites (represented by capital letters) and the HRV3c protease recognition site (underlined). The resulting fragment was subcloned into the E. coli expression vector pMAL-c2 (NEW ENGLAND BioLabs) between the BamHI and SalI sites for expression of a fusion protein with an N-terminal MBP. The construct was verified by sequencing on a 3130 Genetic Analyser (Applied Biosystems).
Expression and purification of IE2-CTD
The IE2-CTD expression plasmid described above was used to transform E. coli strain BL21. Colonies were used to inoculate a 150 ml lysogeny broth (LB) starter culture containing 100 μg ml−1 carbenicillin, and cultivation was performed at 37°C overnight. Cultures were diluted in 1 l of LB medium containing 100 μg ml−1 carbenicillin and cultivated until the optical density at 600 nm (OD600) reached ~0.5–0.6, at which point the temperature was lowered to 30°C and protein expression was induced by adding isopropyl-β-D-thiogalactopyranoside (IPTG) to a final concentration of 0.3 mM. After cultivation for a further 16 h, cells were harvested by centrifugation at 8000× g for 10 min and stored at −80°C until needed. For preparation of the selenomethionine (SeMet) derivative of IE2-CTD, E. coli strain B834 was used in replacement of BL21. The starter culture in LB medium containing 100 μg ml−1 carbenicillin was grown in two steps, firstly in 10 ml, which was then increased to 150 ml. Cells were collected by centrifugation for 10 min at 6000 × g, resuspended in M9 medium supplemented with 100 μg ml−1 carbenicillin and 100 μg ml−1 SeMet, and cultured, induced and harvested as described above for unlabelled protein.
Purification of IE2-CTD
All purification steps were performed at 4°C or on ice unless otherwise stated. Purification of unlabelled IE2-CTD and the SeMet derivative was carried out identically. Cells were thawed on ice and resuspended in lysis buffer (20 mM Tris-HCl, pH 8.0; 1 M NaCl; 20 mM MgCl2; 0.2 mg ml−1 Lysozyme; 1 mM EDTA; and 1 mM DTT) and incubated for 30 min. Triton X-100 was added to a final concentration of 0.5% (v/v) and rocked gently for 20 min. Cells were disrupted by ultrasonication and clarified by centrifugation at 19,000 × g for 30 min. The supernatant was incubated with amylose resin (NEW ENGLAND BioLabs) overnight and poured into a column and washed with column buffer (20 mM Tris-HCl, pH 8.0; 1 mM EDTA; 1 mM DTT; 20 mM MgCl2) supplemented with 1 M NaCl and 0.5% Triton X-100. MBP-tagged IE2-CTD was eluted with column buffer containing 10 mM maltose and 1 M NaCl, and eluted protein was cleaved with HRV3c protease (Accelagen) for 36–48 h. Protein was concentrated using an Amicon centrifugal concentrator with a 30 KDa molecular weight cut-off (Millipore), diluted 5-fold with column buffer to reduce the NaCl concentration and applied to a sulphopropyl (SP) sepharose column (GE healthcare) equilibrated with the column buffer supplemented with 0.2 M NaCl. After washing with 10 column volumes of column buffer containing 0.2 M NaCl, IE2-CTD was eluted by stepwise increments in NaCl concentration. The purity was assessed by sodium dodecyl sulphate polyacrylamide gel electrophoresis (SDS-PAGE). The concentration of IE2-CTD was determined by measuring the absorbance at 280 nm and using a value of 0.775 for 1 mg ml−1 protein as determined from the amino acid composition by ProtParam (18). Purified protein was concentrated to 1.2–1.5 mg ml−1 using an Amicon centrifugal concentrator with a 30 KDa molecular weight cut-off (Millipore) and stored at 4°C until needed.
Crystallization
Suitable crystallization conditions for unlabelled IE2-CTD were searched using the Index HT screening kit (Hampton Research) and the JBScreen Wizard 1 and 2 kits (Jena Biosciences). Crystallization was performed using the sitting-drop vapour diffusion technique with 96-well plates at 4°C. Drops containing 0.5 μl of 0.9 mg ml−1 IE2-CTD and 0.5 μl of reservoir solution were mixed and equilibrated over 70 μl of reservoir solution. Initial crystals were obtained in condition 90 of the Index HT screen. The Additive Screen HT (Hampton Research) was then used to improve the crystallization condition according to the manufacturer’s protocol. The best crystals were obtained in an optimized solution containing 24% polyethylene glycol 3,350, 0.2 M sodium formate, 20 mM Tris-HCl (pH 8.8), 20 mM CaCl2 and 1 mM DTT. Larger crystals of the SeMet derivative were obtained with better reproducibility using the same conditions; hence only derivative crystals were used for X-ray diffraction experiments. Crystals appeared within 2 or 3 days, and continued to grow for 1 or 2 weeks.
X-ray diffraction and structure determination
X-ray diffraction data were collected on beamline BL38B1 at SPring-8, Harima, Japan. Crystals were transferred to cryoprotectant solution prepared by adding ethylene glycol to reservoir solution to a final concentration of 20%, and crystals were flash frozen in liquid nitrogen. Diffraction data were collected under a N2 stream at 100 K using a Rayonix MX225HE CCD detector at the Se K-edge peak wavelength. Details of data collection are shown in Table I. Diffraction data were processed using XDS (19). In the integration step, the 3.70–3.64 Å resolution shell was excluded to avoid an ice ring. Experimental phasing was performed using the single anomalous diffraction method implemented in Phenix.autoSol (20, 21), and electron density was visualized using Coot (22).
Table I.
Statistics for X-ray diffraction data collection and processing.
| Parameter | |
|---|---|
| Data Collection | |
| Wavelength (Å) | 0.97905 |
| Crystal-to-camera distance (mm) | 200 |
| Exposure time (sec) | 20 |
| Oscillation angle (°) | 1.0 |
| Total angle (°) | 360 |
| Data Processing | |
| Space group | P21 |
| Unit cell parameters | |
| a, b, c (Å) | 102.84, 39.71, 103.73 |
| α, β, γ (°) | 90, 110.8, 90 |
| Resolution (Å)a | 42.51–2.50 (2.59–2.50) |
| Total reflectionsa | 169806 (15626) |
| Unique reflectionsa | 27197 (2672) |
| Rmergea | 0.138 (0.343) |
| Rmeasa | 0.150 |
| Mean I/sigma (I)a | 9.47 (5.56) |
| Completeness (%)a | 97.88 (99.96) |
| Multiplicitya | 6.3 (5.8) |
| CC1/2a | 0.984 (0.910) |
| Experimental Phasing | |
| Se sites found | 21 |
| Average figure of merit | 0.336 |
Values in parentheses are for the outermost resolution shell.
RESULTS
Design, expression and purification of HHV-6A IE2-CTD
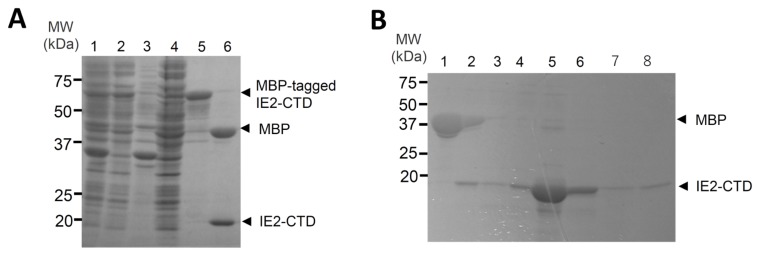
A structural domain in the IE2 C-terminal region was identified with the help of the secondary structure prediction program Jpred4 (17). The predicted C-terminal domain spanning residues 1324–1500 is rich in secondary structural elements (Figure 2), and includes most of the homologous region shared with HHV-6A IE2, HHV-6B IE2, HHV-7 IE2 and HCMV IE2 (Figure 2). This region was expressed as an N-terminally tagged MBP fusion protein in E. coli without noticeable degradation (Figure 3A), and was therefore named the C-terminal domain of IE2 (IE2-CTD). Purification using amylose resin and strong cation exchange afforded a protein with a theoretical isoelectric point (pI) of 9.29 (as calculated by ProtParam) (18), consistent with its DNA binding function. Indeed, a high salt concentration (1 M NaCl and 20 mM MgCl2) during lysis and purification on amylose resin was required to prevent nonspecific interaction with nucleic acids (data not shown). The fusion protein was readily cleaved with HRV3c protease (Figure 3A, lane 6), and tag-free protein was purified to homogeneity by cation exchange chromatography (Figure 3B) to give a yield of 0.5 mg per liter of culture for unlabelled protein, and 0.3 mg per liter of culture for SeMet-labelled protein.
Figure 2.
Sequence alignment of IE2 homologue C-terminal regions. Amino acid sequences of HHV-6A IE2 and homologues from beta-herpesviruses were aligned by Clustal Omega (23). Fully conserved or highly similar positions are shaded black and grey, respectively. Predicted α helix (“H”) and β strand (“E”) are shown above the alignment. The region corresponding to HHV-6A IE2-CTD is indicated by a bar at the top of the alignment. Abbreviations and accession numbers are as follows; HHV-6A, HHV-6A U1102, Q77Z83.2; HHV-6B, HHV-6B HST, gi|4996073; HHV-7, HHV-7 UCL1, gi|541905786; HCMV, HCMV AD169, P19893.2.
Figure 3.
Purification of HHV-6A IE2-CTD. (A) Expression and purification of MBP-tagged IE2-CTD. Lanes are as follows: 1, cell lysate; 2, supernatant; 3, pellet; 4, fraction not bound to amylose beads; 5, eluate from amylose beads; 6, fraction cleaved by HRV-3c protease. (B) Purification by cation exchange column chromatography. Lanes are as follows: 1, fraction not bound to SP sepharose beads; 2, 0.2 M NaCl elution; 3, 0.3 M NaCl elution; 4, 0.4 M NaCl elution; 5, 0.5 M NaCl elution; 6, 0.6 M NaCl elution; 7, 0.8 M NaCl elution; 8, 1 M NaCl elution. Molecular weight (MW) markers are shown at the left, and the positions of MBP-tagged IE2-CTD, MBP and IE2-CTD are indicated on the right.
Crystallization of HHV-6A IE2
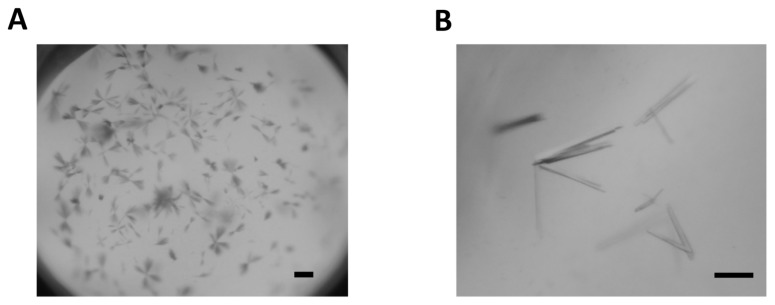
Initial crystallization produced needle clusters of unlabelled IE2-CTD (Figure 4A), and, following additive screening, CaCl2 significantly improved crystallization. Additionally, the SeMet derivative of IE2-CTD produced larger crystals than the unlabelled protein, and was subsequently used for further optimization and final crystallization. In the optimized conditions, rod-shaped crystals suitable for the X-ray diffraction studies were obtained (Figure 4B). A single crystal separated from the cluster was used for the X-ray experiment. The dimensions of the best crystals were approximately 200 × 20 × 10 μm.
Figure 4.
Crystals of HHV-6A IE2-CTD. (A) Needle clusters of unlabelled IE2-CTD obtained in the initial crystallization screens. Scale bar = 50 μm. (B) Rod clusters of IE2-CTD obtained after optimization of the crystallization condition. Scale bar = 50 μm.
X-ray diffraction
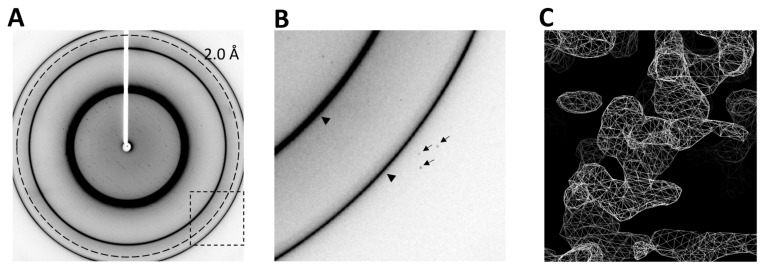
X-ray diffraction data were collected using synchrotron radiation at the SPring-8 BL38B1 beamline. The selenium peak wavelength was searched for using X-ray absorption fine structure (XAFS) analysis and determined to be 0.97905 Å. Details of data collection are summarized in Table I. Although the diffraction images indicated the presence of ice rings (Figure 5A, B), sufficient diffraction data for structure analysis were obtained by excluding the main ice ring from the data processing step (Table I). The space group was P21, and the maximum resolution was 2.5 Å (Table I).
Figure 5.
X-ray diffraction of IE2-CTD crystals. (A) Representative diffraction image collected from an IE2-CTD crystal. The dashed circle represents the resolution at 2.0 Å. The dashed rectangle delineates the area shown in (B). The two sharp rings correspond to ice crystals. (B) Close-up view of (A). High-resolution diffraction spots are indicated by arrows, and ice rings are indicated by arrowheads. (C) Preliminary electron density map (2Fo-Fc) after SAD phasing. An α-helix is clearly visible, and the backbone structure can be easily traced. The map is contoured at 1.5 σ.
Experimental phasing was performed using the single wavelength anomalous dispersion (SAD) method, and a valid solution was obtained (Table I). The electron density map generated during preliminary structure refinement was readily decipherable, and the IE2-CTD backbone structure could be clearly traced (Figure 5C). Full structure determination and analysis is now underway and will be reported in the near future.
DISCUSSION
In this study, a structural domain from the C-terminal region of HHV-6A IE2 was purified and crystallized. The 1324–1500 region, defined as IE2-CTD, was purified to homogeneity by amylose affinity and cation exchange chromatography. The predicted amino acid sequence had a calculated pI of 9.29, consistent with the predicted DNA binding function that has been confirmed for the homologous C-terminal region of HCMV IE2 (14). Although the homologous region shared with HCMV IE2 and HHV-6A IE2 extends beyond this fragment towards the N-terminus, corresponding to 1290–1500 in IE2-CTD (Figure 2), the fact that we could successfully purify and crystallize IE2-CTD strongly indicates that residues 1324–1500 form a stable domain. In a transactivation analysis performed by Tomoiu et al. (2006), deletion of the C-terminal 70 residues of HHV-6A IE2 resulted in a complete loss of function (13). Given that IE2-CTD includes these 70 amino acids, the loss of function may be attributed to structural disruption in this region. HCMV IE2 possesses transactivation factor TBP and TFIIB binding sites at 290–504 and 290–542, respectively (15, 16), and HHV-6A IE2-CTD includes the region corresponding to 380–555 of HCMV IE2. The 290–390 region of HCMV IE2 is insufficient for binding of both TBP and TFIIB (15, 16); thus the corresponding region in IE2-CTD is likely to be at least partially responsible for this interaction. Furthermore, the 505–542 region of HCMV IE2 is essential for interaction with TFIIB, since the truncated 290–504 protein displayed severely impaired interaction ability (15, 16). Thus, the region corresponding to IE2-CTD may also contribute to interactions with transcription factors.
The electron density for HHV-6A IE2-CTD clearly allows tracing of the protein backbone (Figure 5C). Further refinement and model building are in progress, and the resulting atomic structure of IE2-CTD will facilitate the characterization of this essential domain of HHV-6A IE2.
ACKNOWLEDGEMENTS
We thank Prof. Huamin Tang (Nanjing Medical University) and Mr. Bochao Wang (Kobe University Graduate School of Medicine) for the technical assistance. Synchrotron radiation experiments were performed at BL38B1 in SPring-8 with the approval of RIKEN (Proposal No. 2016A2556).
REFERENCES
- 1.Yamanishi K, Okuno T, Shiraki K, et al. Identification of human herpesvirus-6 as a causal agent for exanthem subitum. Lancet. 1988;1:1065–1067. doi: 10.1016/s0140-6736(88)91893-4. [DOI] [PubMed] [Google Scholar]
- 2.Aubin JT, Collandre H, Candotti D, et al. Several groups among human herpesvirus 6 strains can be distinguished by Southern blotting and polymerase chain reaction. Journal of clinical microbiology. 1991;29:367–372. doi: 10.1128/jcm.29.2.367-372.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Campadelli-Fiume G, Guerrini S, Liu X, Foa-Tomasi L. Monoclonal antibodies to glycoprotein B differentiate human herpesvirus 6 into two clusters, variants A and B. The Journal of general virology. 1993;74:2257–2262. doi: 10.1099/0022-1317-74-10-2257. [DOI] [PubMed] [Google Scholar]
- 4.Wyatt LS, Balachandran N, Frenkel N. Variations in the replication and antigenic properties of human herpesvirus 6 strains. The Journal of infectious diseases. 1990;162:852–857. doi: 10.1093/infdis/162.4.852. [DOI] [PubMed] [Google Scholar]
- 5.Mori Y. Recent topics related to human herpesvirus 6 cell tropism. Cell Microbiol. 2009;11:1001–1006. doi: 10.1111/j.1462-5822.2009.01312.x. [DOI] [PubMed] [Google Scholar]
- 6.Adams MJ, Carstens EB. Ratification vote on taxonomic proposals to the International Committee on Taxonomy of Viruses. Arch Virol. 2012;157:1411–1422. doi: 10.1007/s00705-012-1299-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tang H, Serada S, Kawabata A, et al. CD134 is a cellular receptor specific for human herpesvirus-6B entry. Proc Natl Acad Sci U S A. 2013;110:9096–9099. doi: 10.1073/pnas.1305187110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jasirwan C, Furusawa Y, Tang H, et al. Human herpesvirus-6A gQ1 and gQ2 are critical for human CD46 usage. Microbiol Immunol. 2014;58:22–30. doi: 10.1111/1348-0421.12110. [DOI] [PubMed] [Google Scholar]
- 9.Ablashi D, Agut H, Alvarez-Lafuente R, et al. Classification of HHV-6A and HHV-6B as distinct viruses. Archives of virology. 2014;159:863–870. doi: 10.1007/s00705-013-1902-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Behzad-Behbahani A, Mikaeili MH, Entezam M, et al. Human herpesvirus-6 viral load and antibody titer in serum samples of patients with multiple sclerosis. J Microbiol Immunol Infect. 2011;44:247–251. doi: 10.1016/j.jmii.2010.08.002. [DOI] [PubMed] [Google Scholar]
- 11.Voumvourakis KI, Kitsos DK, Tsiodras S, et al. Human herpesvirus 6 infection as a trigger of multiple sclerosis. Mayo Clin Proc. 2010;85:1023–1030. doi: 10.4065/mcp.2010.0350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Papanikolaou E, Kouvatsis V, Dimitriadis G, et al. Identification and characterization of the gene products of open reading frame U86/87 of human herpesvirus 6. Virus Res. 2002;89:89–101. doi: 10.1016/s0168-1702(02)00126-0. [DOI] [PubMed] [Google Scholar]
- 13.Tomoiu A, Gravel A, Flamand L. Mapping of human herpesvirus 6 immediate-early 2 protein transactivation domains. Virology. 2006;354:91–102. doi: 10.1016/j.virol.2006.06.030. [DOI] [PubMed] [Google Scholar]
- 14.Chiou CJ, Zong J, Waheed I, et al. Identification and mapping of dimerization and DNA-binding domains in the C terminus of the IE2 regulatory protein of human cytomegalovirus. J Virol. 1993;67:6201–6214. doi: 10.1128/jvi.67.10.6201-6214.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Caswell R, Hagemeier C, Chiou CJ, et al. The human cytomegalovirus 86K immediate early (IE) 2 protein requires the basic region of the TATA-box binding protein (TBP) for binding, and interacts with TBP and transcription factor TFIIB via regions of IE2 required for transcriptional regulation. J Gen Virol. 1993;74:2691–2698. doi: 10.1099/0022-1317-74-12-2691. [DOI] [PubMed] [Google Scholar]
- 16.Hagemeier C, Caswell R, Hayhurst G, et al. Functional interaction between the HCMV IE2 transactivator and the retinoblastoma protein. EMBO J. 1994;13:2897–2903. doi: 10.1002/j.1460-2075.1994.tb06584.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Drozdetskiy A, Cole C, Procter J, et al. JPred4: a protein secondary structure prediction server. Nucleic Acids Res. 2015;43:W389–394. doi: 10.1093/nar/gkv332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gasteiger E, Hoogland C, Gattiker A, et al. The Proteomics Protocols Handbook. Humana Press; 2006. Protein Identification and Analysis Tools on the ExPASy Server; pp. 571–607. [Google Scholar]
- 19.Kabsch W. Xds. Acta crystallographica Section D, Biological crystallography. 2010;66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Adams PD, Afonine PV, Bunkoczi G, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta crystallographica Section D, Biological crystallography. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Terwilliger TC, Adams PD, Read RJ, et al. Decision-making in structure solution using Bayesian estimates of map quality: the PHENIX AutoSol wizard. Acta crystallographica Section D, Biological crystallography. 2009;65:582–601. doi: 10.1107/S0907444909012098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Emsley P, Lohkamp B, Scott WG, et al. Features and development of Coot. Acta crystallographica Section D, Biological crystallography. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sievers F, Wilm A, Dineen D, et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol. 2011;7:539. doi: 10.1038/msb.2011.75. [DOI] [PMC free article] [PubMed] [Google Scholar]