Abstract
Metabolic dysregulation promotes cancer growth through not only energy production, but also epigenetic reprogramming. Here, we report that a critical node in methyl donor metabolism, nicotinamide N-methyltransferase (NNMT), ranked among the most consistently overexpressed metabolism genes in glioblastoma relative to normal brain. NNMT was preferentially expressed by mesenchymal glioblastoma stem cells (GSCs). NNMT depletes S-adenosyl methionine (SAM), a methyl donor generated from methionine. GSCs contained lower levels of methionine, SAM, and nicotinamide, but they contained higher levels of oxidized nicotinamide adenine dinucleotide (NAD+) than differentiated tumor cells. In concordance with the poor prognosis associated with DNA hypomethylation in glioblastoma, depletion of methionine, a key upstream methyl group donor, shifted tumors toward a mesenchymal phenotype and accelerated tumor growth. Targeting NNMT expression reduced cellular proliferation, self-renewal, and in vivo tumor growth of mesenchymal GSCs. Supporting a mechanistic link between NNMT and DNA methylation, targeting NNMT reduced methyl donor availability, methionine levels, and unmethylated cytosine, with increased levels of DNA methyltransferases, DNMT1 and DNMT3A. Supporting the clinical significance of these findings, NNMT portended poor prognosis for glioblastoma patients. Collectively, our findings support NNMT as a GSC-specific therapeutic target in glioblastoma by disrupting oncogenic DNA hypomethylation.
N-methyl nicotinamide methyltransferase is overexpressed in glioblastoma, with preferential expression in mesenchymal GSCs residing in areas surrounding necrosis, regions of restricted nutrient availability.
Introduction
Glioblastoma (GBM) is one of the most lethal of all human cancers, with current therapies offering only palliation (1). Despite extensive genomic analysis that has informed the GBM genetic landscape, these findings have not been translated into clinical benefit. GBMs are extremely heterogeneous with striking differences between tumors (intertumoral heterogeneity) and within tumors (intratumoral and cellular heterogeneity) (2). Tumor genetics studies have identified a group of patients with mutations in isocitrate dehydrogenase 1 or 2 (IDH1/2) with a glioma-CpG island methylator phenotype (G-CIMP) that have a prolonged median survival (3). Other genetic lesions (including EGFR amplification and mutation, p53 mutation, and p16INK4A deletion) are less informative than IDH mutations, prompting the creation of gene expression signatures (proneural, neural, classical, and mesenchymal) that have been extensively investigated in laboratory studies but have yet to be informative in clinical settings (4–6). Within tumors, GBMs contain populations of stem-like tumor cells, called GBM stem cells (GSCs), which contribute to tumor malignancy through promotion of sustained tumor growth, angiogenesis, invasion, and therapeutic resistance (7, 8). While GSCs remain controversial due to unresolved questions of cell-of-origin and identification methods, GBM is one of the solid cancers for which cancer stem cells have been consistently described. Like normal stem cells, GSCs are functionally defined by stem cell markers, self-renewal, and recapitulation of parental tissues (in this case, a tumor). Recent studies have shown that proneural and mesenchymal GSCs differ significantly in gene expression profiles, regulatory mechanisms, and responses to therapy (9, 10). Thus, interrogation of molecular drivers of GSCs may inform therapeutic strategies.
IDH1 mutations have provided the most concrete link in brain tumors between metabolism and tumor initiation (11). IDH1 catalyzes a reversible oxidative decarboxylation of isocitrate to yield α-ketoglutarate (α-KG) in the tricarboxylic acid cycle. Mutations in IDH1 or IDH2 cause both a loss of normal enzymatic function and gain of neomorphic activity to generate the oncometabolite, 2-hydroxyglutarate (2-HG) (12). 2-HG inhibits the enzymatic function of several α-KG–dependent dioxygenases, including histone and DNA demethylases, such as the ten-eleven translocation (TET) enzymes involved in the oxidation of 5-methylcytosine (5mC) to 5-hydroxymethylcytosine (5hmC); thus, extensive changes in histone and DNA methylation landscape promote tumorigenesis. These findings have proven exciting but may not be relevant to the majority of GBMs, which do not harbor IDH mutations. GBM patients with WT IDH1, non–G-CIMP tumors display relative global DNA hypomethylation and worse clinical outcomes (3, 11). Further, patients with hypomethylation commonly have an unmethylated promoter of MGMT (O6-methylguanine-methyltransferase), permitting the expression of MGMT, a resistance mechanism to the oral methylator chemotherapy, temozolomide (TMZ). Based on this background, we hypothesized that IDH WT GBMs may harbor other metabolic alterations that may function, in part, through altering DNA methylation.
Embryonic stem cells share several metabolic features with cancer to maintain their rapid cell cycles and epigenetic landscape necessary for multilineage potency (13–15). As with stem cells, tumor cells require concerted dysregulation of metabolism and epigenetic cell states to simultaneously accommodate the bioenergetic demands of increased proliferation and aberrant tumor cell fate through epigenetic modification. Tumor cells co-opt and dysregulate metabolic pathways through oncogenic mutations (e.g., MYC amplification or PTEN mutation) and/or altered expression of metabolic regulators (16). Disorders of amino acid metabolism, including homocysteine and methionine, impair the developing nervous system through toxic effects on normal brain cells (17, 18). Based on this background, we employed a combined in silico and GSC-based discovery approach to determine potentially important metabolic regulators of GBM epigenetic maintenance.
Results
GBMs overexpress nicotinamide (NAM) N-methyltransferase (NNMT).
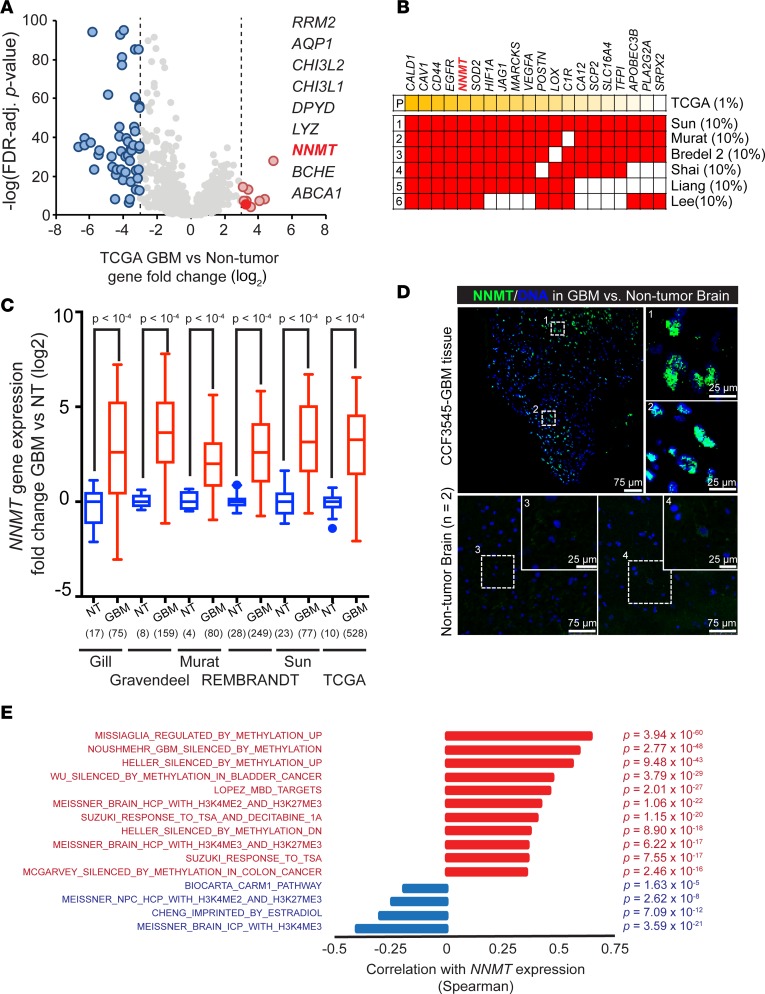
Metabolic reprogramming is not solely a passenger in cancer formation, but it also drives transformation and modifies tumor epigenetics (19, 20). To investigate derangement of metabolic programs in GBM, we interrogated the expression of 2,030 metabolism-related genes in The Cancer Genome Atlas (TCGA) GBM patient database compared with normal brain (Supplemental Table 1; supplemental material available online with this article; https://doi.org/10.1172/jci.insight.90019DS1) (21). Among these, 850 genes were upregulated, while 1,180 genes were downregulated, suggesting that metabolism was not globally activated. We found 9 genes whose mRNA expression increased at least 8-fold in GBMs compared with nonmalignant brain tissues, which we considered as potentially oncogenic in GBM-associated metabolism (Figure 1A). Several of these 9 genes have already been investigated in GBM (e.g., CHI3L1, also known as YKL-40, is a key marker of mesenchymal GBMs). Results from a single database can often be compromised by technical or patient-selection biases; therefore, to refine our target gene selection, we maximized the generalizability of our results by screening expression across several glioma datasets. We interrogated the top 1% of all expressed genes in the TCGA database across 6 major databases to identify genes that were consistently expressed at high levels in each set (Figure 1B). Targets identified included well-known oncogenic drivers involved in hypoxic responses and GSCs, including CD44, EGFR, HIF1A, VEGFA, and POSTN, supporting our strategy. Strikingly, NNMT fulfilled all the criteria: it was one of the 9 metabolic genes upregulated, a highly ranked gene among total upregulated genes in the TCGA GBM dataset, and consistently overexpressed in each dataset (odds ratio > 2, P < 0.001, via fold-change) (Figure 1B). Moreover, NNMT mRNA levels were consistently overexpressed relative to nontumor specimens in each glioma database (Figure 1C) (4, 22–26). To validate NNMT protein expression in GBM, we performed immunofluorescence for NNMT in GBM patient specimens and nonmalignant brain tissues, confirming higher expression of NNMT in GBM (Figure 1D). As NNMT is an essential cytosolic enzyme transferring the methyl group from S-adenosylmethionine (SAM) to nicotinamide (NAM), we tested whether the consistent enrichment of NNMT expression in GBMs affected epigenetic regulation in these tumors by performing gene ontology (GO) analysis of transcriptional signatures strongly correlated with NNMT mRNA levels in TCGA GBM data set. We found an exceptionally strong positive correlation with gene signatures whose expressions are repressed by DNA methylation in cancer and a strong anticorrelation with DNA methylation in normal stem or differentiated cancer cells (Figure 1E and Supplemental Table 2) (3, 27–35).
Figure 1. Overexpression of NNMT in glioblastoma.
(A) Fold-change (log2) of metabolic gene expression between glioblastoma and nontumor specimens in the TCGA GBM microarray dataset. (B) Top 10% of genes overexpressed in glioblastomas vs. nontumor samples in 6 glioblastoma expression databases cross-referenced with top 1% of genes overexpressed in glioblastomas vs. nontumor samples from the TCGA dataset by the unsupervised analysis tool in Oncomine. (C) Fold-change of NNMT mRNA expression between glioblastoma (GBM) and nontumor brain (NT) in glioblastoma expression datasets. Sample sizes as indicated on the figure. (D) Representative images of patient glioblastoma tissues with NNMT staining, out of 2 total experiments. Frozen glioblastoma sections were stained with anti-NNMT antibody and DAPI. Scale bars: 75 μm and 25 μm, respectively. (E) Waterfall plot of correlation between NNMT mRNA expression and cancer or neural precursor methylation signature scores in the TCGA GBM microarray dataset. Spearman correlation coefficients with FDR-adjusted P < 0.001 and R > 0.2 or R < –0.2 were considered significant.
Methyltransferases, like NNMT, are categorized based on structure, substrate, and target. Histone and DNA/RNA methyltransferases are ubiquitously expressed and frequently studied in stem cell and cancer biology. In contrast, natural product methyltransferases (NPMTs) comprise a diverse group of enzymes that frequently transfer methyl groups to small molecules from SAM as the methyl donor. Many NPMTs, including NNMT, are specifically expressed in nervous system tissues, and defects in function are associated with neurological disorders (summarized in Supplemental Figure 1A) (36–47). We interrogated the relative expression levels and prognostic significance of 11 NPMTs in the 2 largest GBM datasets, TCGA and REMBRANDT. Five enzymes (NNMT, HNMT, COMT, PEMT, and TRMT5) were upregulated in GBM compared with normal brain, while the others (GNMT, PNMT, GHMT, GAMT, ASMT, and CARNMT1) were downregulated in GBM (Supplemental Figure 1B). Further, only NNMT and ASMT were associated with poor prognosis (Supplemental Figure 1A). These findings further suggest that the high expression of NNMT in GBM is not a phenomenon general to all methyltransferases.
NNMT expression is enriched in mesenchymal GSCs.
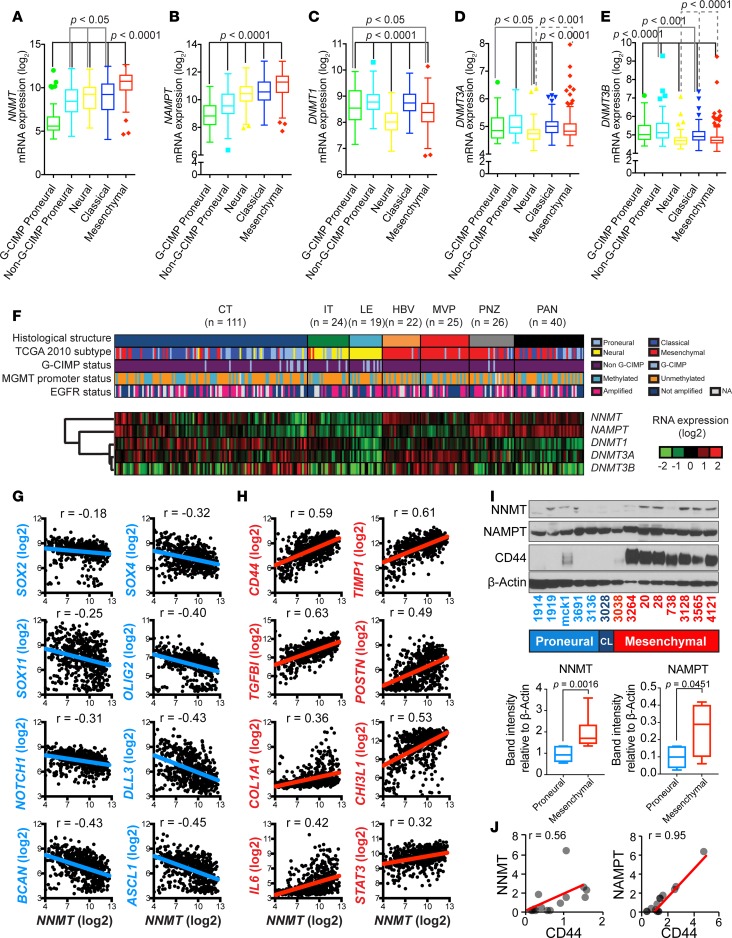
Because GBMs comprise heterogeneous tumors, we examined whether NNMT upregulation is enriched in specific patient cohorts. In GBM, transcriptional profiles of bulk tumors have yielded numerous profiles, with the greatest differences between proneural and mesenchymal tumors (4). During development of therapeutic resistance, some tumors will undergo a proneural-to-mesenchymal transition, mimicking epithelial-to-mesenchymal transition (EMT). Based on predictions that NNMT expression would be associated with a hypomethylated state, we investigated NNMT expression in the context of G-CIMP tumors. As expected, NNMT mRNA expression was strongly downregulated in G-CIMP tumors (Figure 2A). In contrast, NNMT mRNA expression was consistently upregulated in mesenchymal GBMs, which exhibit a high level of DNA hypomethylation (Figure 2A). Nicotinamide phosphoribosyltransferase (NAMPT), which is a key member of the NAD salvage pathway that catalyzes NAM modification to produce NAD+, was similarly upregulated in mesenchymal GBMs and downregulated in G-CIMP tumors (Figure 2B). DNA methyltransferases (DNMTs) catalyze transfer of a methyl group from SAM to cytosine to augment DNA methylation. Given the potentially opposing effects of NNMT and DNMTs on DNA methylation, we interrogated DNMT expression in GBMs. Concordant with antagonistic functions of DNMTs relative to NNMT, DNMTs were modestly elevated in proneural tumors relative to mesenchymal GBMs (Figure 2, C–E). GBMs display remarkable intratumoral heterogeneity, prompting us to examine NNMT mRNA expression in the Ivy GBM Atlas (Ivy GAP) dataset (http://glioblastoma.alleninstitute.org/), in which gene expression in GBM specimens is characterized by in situ hybridization and RNA sequencing (RNAseq). The highest expression levels of NNMT mRNA were found in pseudopallisading cells around necrosis, with more modest expression in areas of microvascular proliferation and cellular tumor, but with lowest expression in the leading edge and invasive tumor (Figure 2F and Supplemental Figure 2). NAMPT shared an expression pattern with NNMT, while the DNMTs were preferentially expressed in vascular tumor microenvironments (Figure 2F). Thus, mesenchymal GBMs and tumor microenvironments favorable to mesenchymal GSCs (9, 10, 48, 49) highly express NNMT and downregulate DNMTs.
Figure 2. NNMT and NAMPT enrichment in mesenchymal glioblastoma stem cells.
(A–E) NNMT, NAMPT, DNMT1, DNMT3A, and DNMT3B mRNA expression distribution by molecular subtypes (G-CIMP proneural, n = 41; non–G-CIMP proneural, n = 97; neural, n = 84; classical, n = 145; mesenchymal, n = 156) in TCGA GBM microarray dataset, respectively. (F) Supervised hierarchical clustering of NNMT, NAMPT, DNMT1, DNMT3A, and DNMT3B mRNA expression based on grouping by histological structure in the Ivy GAP RNAseq dataset. Sample size of each histological region as indicated. (G and H) Pairwise correlation between NNMT expression was performed with (G) proneural and (H) mesenchymal GSC markers in TCGA database as indicated. Numbers represent Pearson coefficient values. (I) Immunoblot analysis using indicated antibodies in indicated proneural, classical (CL), and mesenchymal GSCs. Quantification of NNMT and NAMPT protein levels was performed with ImageJ. Mann-Whitney U test was used to test differences in distribution. (J) Correlation between NNMT or NAMPT and CD44 protein levels shown in I. Pearson correlation coefficients (r values) were calculated with Microsoft Excel.
Having confirmed preferential expression of NNMT in mesenchymal tumors, which are considered more aggressive than proneural tumors, we hypothesized that NNMT is preferentially expressed within the most aggressive tumor cell population, GSCs. In TCGA data, NNMT mRNA levels were negatively correlated with proneural GSC markers (SOX2, SOX4, SOX11, OLIG2, NOTCH1, DLL3, BCAN, and ACSL1) (Figure 2G) but positively correlated with mesenchymal GSC markers (CD44, TIMP1, TGFBI, POSTN, COL1A1, CHI3L1, IL6, and STAT3) (Figure 2H). To directly confirm these findings at the protein level, we immunoblotted 14 patient-derived GSC models that we previously functionally validated as cancer stem cells and whose subtypes have been confirmed (Figure 2, I and J). NNMT and NAMPT were preferentially expressed in mesenchymal GSCs compared with proneural GSCs, significantly correlated with high expression of the mesenchymal marker CD44 (Pearson correlation coefficient: 0.56 and 0.95 for NNMT and NAMPT, respectively) (Figure 2J). These results support specific NNMT expression in mesenchymal GSCs.
Overexpression of specific genes in the NAM and nicotinate metabolic pathways promote depletion of methyl donor groups in GBM.
NAM is the amide derivative of nicotinic acid (vitamin B3, niacin) that functions as both a methyl sink and a substrate of NAD production. NNMT has a unique role in NAM metabolism, but the full metabolic regulation of NAM involves a large transcriptional program comprising dozens of genes. According to TCGA GBM dataset, the coding sequence of the gene NNMT was rarely altered, with amplification and mutation found in less than 1.6% of tumors in 6 glioma datasets (data not shown). Therefore, the action of the WT enzyme could act in concert with other NAM-related enzymes to selectively produce specific metabolites. Based on the KEGG database (http://www.genome.jp/kegg/pathway.html), 25 enzymes were involved in NAM metabolism. In our analysis of their expression levels in several brain tumor databases, 8 NAM pathway enzymes, including NNMT, NAMPT, NADK, PNP, BST1, NT5E, NADSYN1, and ENPP1, were significantly upregulated in GBM, whereas the expression of NAM pathway enzymes that act to augment methyl donor availability were decreased compared with normal brain (Supplemental Figure 3A). These multiple alterations can be mapped to a pathway analysis of NAM metabolism to potentially alter 3 distinct metabolic streams in GBM: i) Production of 1-methylnicotinamide (1-MNA), which may affect to fate of methyl donors; ii) accumulation of NAM-derived metabolites from nicotinate-related ones (nicotinic acid, NA); and iii) alteration of the methionine salvage pathway as a methyl group receiver (Supplemental Figure 3B). The reaction driven by NNMT is irreversible, and 1-MNA, its enzymatic product, is secreted into the extracellular environment, resulting in a futile cycle of SAM and NAM consumption. Our observations are concordant with a recent report that NNMT impairs macromolecule methylation in cancer cells by consuming methyl groups from its substrates NAM and SAM (50). Therefore, NNMT-expressing cancer cells displayed altered epigenetic states with hypomethylated histones and other proteins. Taken together, these findings suggest that collective changes of NNMT and other metabolic enzymes in NAM and nicotinate metabolism deplete available methyl donors.
Mesenchymal GSCs rapidly consume NAM to support NAD+ utilization and DNA hypomethylation.
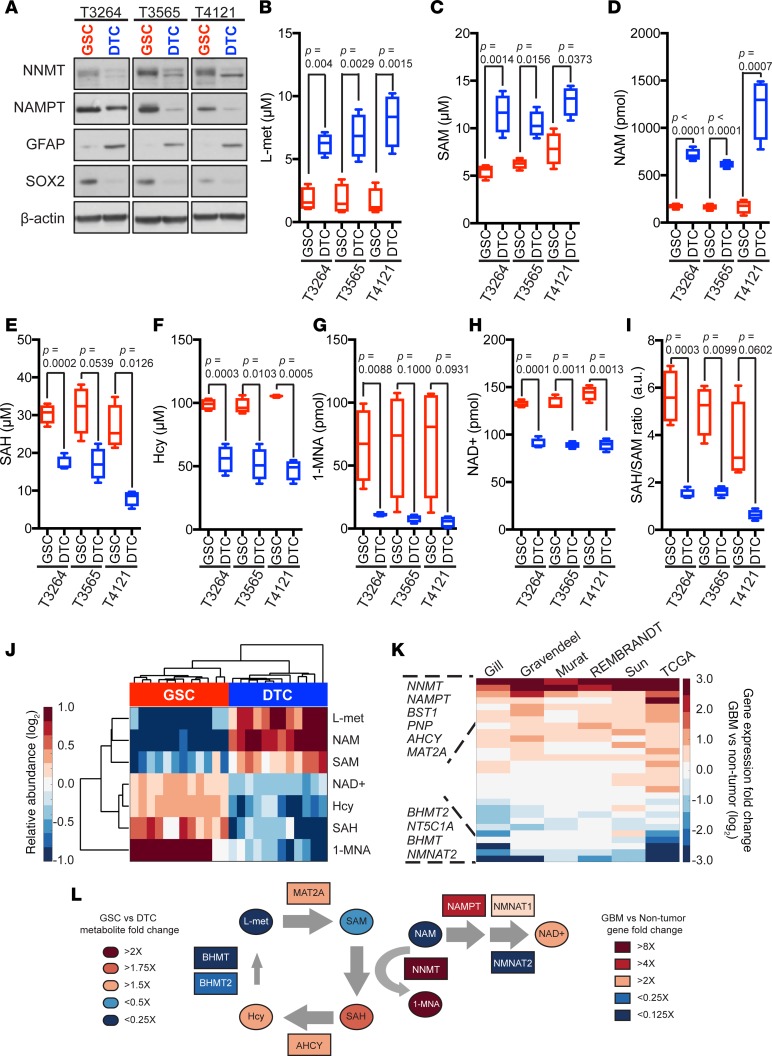
Based on the preferential expression of NNMT in mesenchymal GSCs, we hypothesized that NNMT promotes maintenance of stemness of these cells. To determine a potential role for NNMT in GSCs, we examined its expression within their cellular hierarchy in 3 mesenchymal GSC models (T3264, T3565, and T4121) and their differentiated progenies. Both NNMT and NAMPT expressions were higher in GSCs compared with their matched differentiated tumor cells (DTCs) (Figure 3A). Indeed, NNMT mRNA was also upregulated in other solid tumor cancer stem cells (30, 35), suggesting that NNMT upregulation may be a GSC-specific phenotype (Supplemental Figure 4). We then measured metabolites in the NAM and nicotinate metabolic pathways in matched mesenchymal GSCs and DTCs using conventional assay systems and liquid chromatography-mass spectrometry (LC-MS/MS) analysis. Aligned with our predicted model, levels of methionine, SAM, and NAM in 3 mesenchymal GSCs were lower than those in DTCs (Figure 3, B–D). In contrast, NAD+, homocysteine, S-adenosylhomocysteine (SAH), and MNAM levels were higher in the GSCs compared with the DTCs (Figure 3, E–H). SAH/SAM ratios were calculated to determine the potential methylation status in GSCs compared with DTCs. GSCs displayed higher SAH/SAM flux than DTCs, suggesting hypomethylation of NNMT-expressing GSCs (Figure 3I). Unsupervised hierarchical clustering of relative abundance of metabolites revealed a clear segregation of GSCs from DTCs (Figure 3J). Based on the biochemical activities of NNMT to convert SAM to SAH and NAM to 1-MNA, coupled with gene expression changes between GBMs and normal brain (Figure 3K), we created a predictive model of the metabolic status of NAD+, methionine, and homocysteine in GSCs (Figure 3L). NNMT-expressing tumor cells reduce methylation potential by consuming SAM as a methyl donor (50). Methionine and homocysteine metabolism regulate pluripotent stem cell maintenance and differentiation in their roles as methyl donors and metabolic regulators (51). Therefore, our findings support DNA hypomethylation and NAD+ utilization in mesenchymal GSCs. Moreover, mesenchymal GSC may indirectly drive the metabolism of precursors for methyl donors, including methionine and homocysteine.
Figure 3. Reduced availability of methyl donors in mesenchymal glioblastoma stem cells.
(A) Immunoblot analysis using indicated antibodies in mesenchymal GSCs versus DTCs. (B–I) Metabolite analysis by specific assay kits and liquid chromatography-mass spectrometry (LC-MS/MS) (E and I) in GSCs vs. DTCs. LC-MS/MS was performed with 1 × 107 cells for NAM and 1-MNA measurements. Data represented as Tukey’s box and whisker plot of 4 technical replicates. (J) Unsupervised hierarchical clustering of metabolite relative abundance in GSCs and DTCs from T3264, T3565, and T4121 tumors. Relative abundance for each metabolite was determined as fold-change compared with median of each metabolite across all samples in log2 values. (K) Rank-ordered list of genes (n = 29) from the KEGG nicotinate and nicotinamide metabolism gene set and 4 genes from the methionine salvage pathway (AHCY, BHMT, BHMT2, and MAT2A) based on fold-change between glioblastoma and nontumor specimens in indicated datasets. (L) Integrated pathways of methyl donors and NAD metabolism affected by NNMT and NAMPT gene expression in GSCs. L-met, L-methionine; SAM, S-adenosylmethionine; NAM, nicotinamide; 1-MNA, 1-methylnicotinamide; SAH, S-adenosylhomocysteine; Hcy, homocysteine.
Restricting methionine inhibits tumor growth with reduced expression of DNMT1 and DNMT3A.
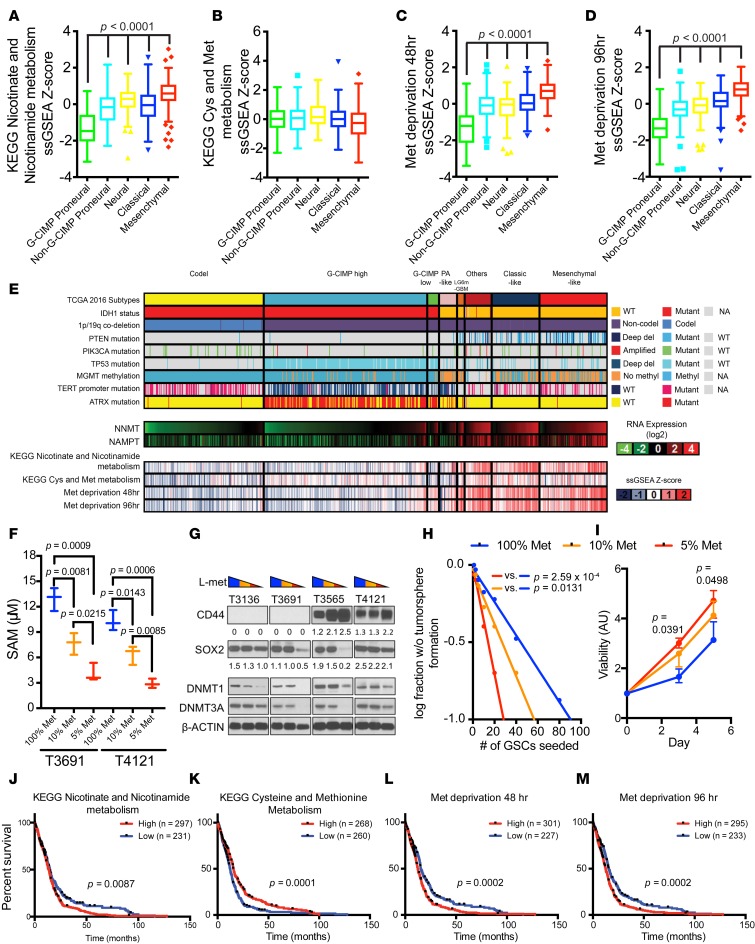
Tumors require carbon sources to build the structural components of cells during proliferation, yet areas of restricted nutrient availability are associated with increased malignancy. We recently demonstrated that restriction of glucose, the dominant carbon source for the brain, increases GSC frequency due to both preferential survival of GSCs under stress and reprogramming of DTCs to become GSCs (52). We hypothesized that restriction of the common source of methyl donor groups, methionine, could deplete methyl sources to augment malignancy. A connection between methionine and stemness is supported by findings that methionine deprivation rapidly reduces intracellular SAM levels in pluripotent stem cells to alter expression of methionine-related metabolic enzymes, including DNMT3B (51). Concordantly, mesenchymal GBMs were positively correlated with the KEGG nicotinate and NAM metabolism gene set signature (Figure 4A and Supplemental Figure 5). Though the KEGG methionine and cysteine metabolism gene set signature scores were lower in mesenchymal tumors than in proneural tumors (Figure 4B), the nonsignificance of this trend may be explained by the common co-occurrence of MTAP, an important methionine salvage pathway gene, and CDKN2A, a commonly deleted tumor suppressor in GBMs (Supplemental Figure 6). In contrast, signatures derived from cells cultured in methionine-depleted media (53) were positively correlated with mesenchymal tumors and inversely correlated with G-CIMP tumors (Figure 4, C and D).
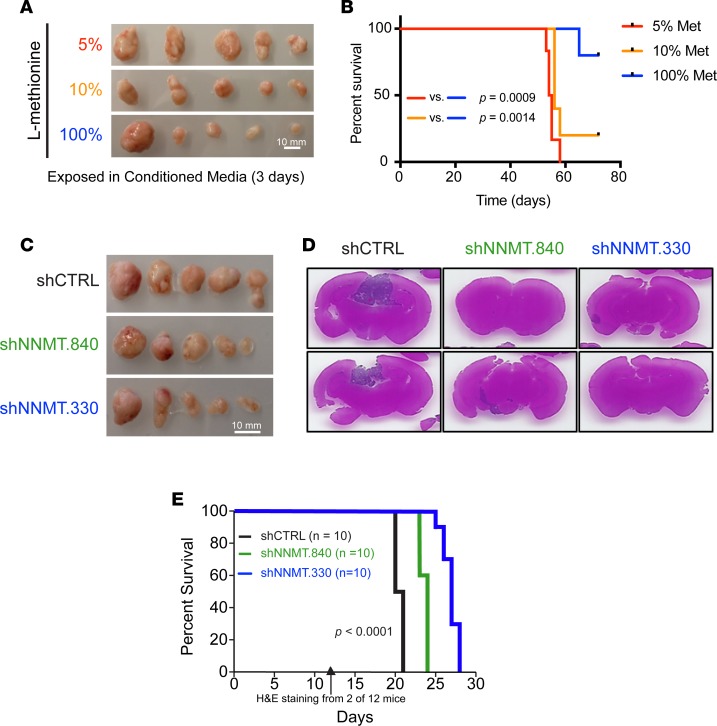
Figure 4. Reduced expression of DNMT1 and DNMT3A by methionine depletion.
(A–D) KEGG nicotinate and nicotinamide metabolism, KEGG cysteine (Cys) and methionine (Met) metabolism, Met deprivation 48 hr, and Met deprivation 96 hr gene set signature score distribution by molecular subtypes (G-CIMP proneural, n = 41; non–G-CIMP proneural, n = 97; neural, n = 84; classical, n = 145; mesenchymal, n = 156) in TCGA GBM microarray dataset. (E) NNMT and NAMPT mRNA expression and KEGG nicotinate and nicotinamide metabolism, KEGG cysteine and Met metabolism, Met deprivation 48 hr, and Met deprivation 96 hr gene set signature score distribution in TCGA pan-glioma (GBMLGG) RNAseq dataset (codel, n = 167; G-CIMP high, n = 236; G-CIMP low, n = 17; PA-like, n = 11; others, n = 39; classical-like, n = 69; mesenchymal-like, n = 98) (6). (F) SAM measurements 3 days after culturing in media of different Met concentrations (100% Met, 115.5 μM; 10% Met, 11.5 μM; 5% Met, 5.75 μM). (G) Immunoblot performed using indicated antibodies under the 3 media conditions. T3136 and T3691 are proneural GSCs; T3565 and T4121 are mesenchymal GSCs. CD44 and SOX2 band intensities were normalized to respective β-actin band intensities. (H) In vitro limiting dilution assay and (I) cell viability assay of T4121 GSCs cultured in different Met-restricted conditions. (J–M) Patient survival data in the TCGA GBM microarray dataset. A given gene signature was evaluated as high (ssGSEA Z score > 0) or low (ssGSEA Z score < 0) (Supplemental Figure 5).
GSCs are found in 2 distinct niches, hypoxic (perinecrotic) regions and the perivascular niche (54–58). Vascular regions would be expected to have high levels of methionine, while perinecrotic regions would be expected to have low levels of methionine due to unreliable blood supply. Therefore, methionine availability may determine methylation status and DNMT expression levels. We interrogated the Ivy GAP RNAseq data, which revealed enrichment of methionine restriction signatures in tumor microenvironments with high NNMT expression (Figure 4E and Supplemental Figure 2). To examine whether low methionine levels in GSCs could functionally regulate DNMTs, we cultured GSCs under reduced methionine concentrations. Methionine deprivation diminished intracellular SAM levels in GSCs (Figure 4F) and expression of DNMT1 and DNMT3A in a concentration-dependent manner (Figure 4G). Further, methionine restriction decreased SOX2 expression and induced CD44, suggesting methionine restriction may promote proneural-to-mesenchymal transition (Figure 4G). The reduction in methionine translated into functional changes with increased in vitro cell growth, although complete removal of methionine decreased cell viability (Figure 4H and data not shown). Collectively, these findings suggest that restricted availability of methionine can induce GSC transition to a mesenchymal state associated with loss of DNMT1 and DNMT3A expression and increased tumor growth. To determine the clinical significance of these findings, we interrogated the TCGA dataset with the NAM metabolism, methionine metabolism, and methionine depletion signatures, finding that each informed patient prognosis, supporting the potential that methionine restriction augments tumor growth in patients (Figure 4, I–M).
Targeting NNMT reduces mesenchymal GSC growth and self-renewal in vitro.
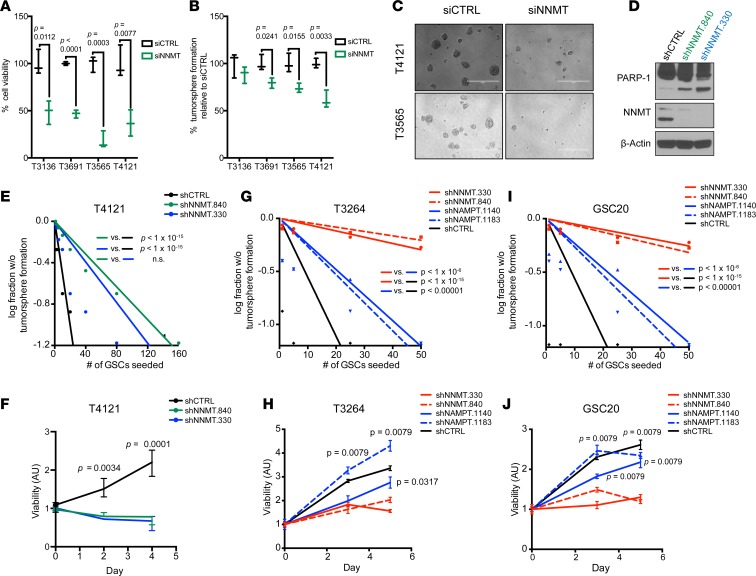
To evaluate the contribution of NNMT to tumor growth, we inhibited NNMT expression using siRNAs and then assayed cell viability and tumorsphere formation in a panel of 2 proneural (T1919 and T3691) and 2 mesenchymal (T3565 and T4121) GSCs. Transient transfection with NNMT siRNA pool decreased cell growth and tumorsphere formation, suggesting reduced self-renewal capacity (Figure 5, A–C). While the impact of the siRNA was similar across models, there was modestly greater effect against mesenchymal models. To discount potential off-target effects of siRNAs, we depleted NNMT using 2 nonoverlapping shRNAs (shNNMT.840 and shNNMT.330). Lentiviral transduction of these shNNMTs enhanced cleavage of PARP-1, indicating induction of apoptosis and reduction of GSC proliferation and sphere formation (Figure 5, D–F). To distinguish the contributions of NNMT and NAMPT in mesenchymal GSCs, we targeted either NNMT or NAMPT expression through 2 nonoverlapping lentiviral shRNAs for each target in mesenchymal and proneural GSCs, revealing a greater effect of targeting NNMT than NAMPT in the inhibition of tumorsphere formation, indicating reduced self-renewal capacity, and cell viability (Figure 5, G–J). Befitting the preferential expression of both NNMT and NAMPT in mesenchymal tumors, targeting their expression in proneural GSCs revealed modest efficacy (Supplemental Figure 7). These phenotypes in NNMT-depleted GSCs demonstrate that targeting NNMT reduces in vitro measures of mesenchymal tumor growth and GSC stemness.
Figure 5. Decreased in vitro mesenchymal GSC growth and self-renewal capacity upon targeting NNMT.
(A and B) Measurement of cell viability and sphere number formation after transient transfection with either siCTRL or siNNMT in proneural and mesenchymal GSCs. Data represented as box and whisker plots of at least 3 independent experiments. (C) Display of sphere size with siNNMT in T3565 and T4121 GSCs. Scale bars: 300 μm. (D) Apoptosis analysis using by PARP-1 cleavage in T4121 GSCs transduced with shCTRL, shNNMT.840, or shNNMT.330 lentivirus. (E) In vitro limiting dilution assay and (F) cell viability assay of T4121 GSCs transduced with shCTRL, shNNMT.840, or shNNMT.330 lentivirus. (G–J) In vitro limiting dilution assay and cell viability assay of (G and H) T3264 or (I and J) GSC20 GSCs transduced with shCTRL, shNNMT.840, shNNMT.330, shNAMPT.1140, or shNAMPT.1183 lentivirus. χ2 test was used for pair-wise differences in stem population frequency. Nonparametric Mann-Whitney U test was used to determine significance in differences between median of shNNMT and shNAMPT treated samples.
NNMT promotes DNA hypomethylation and reduces expression of DNMT1 and DNMT3A in a methionine-dependent manner.
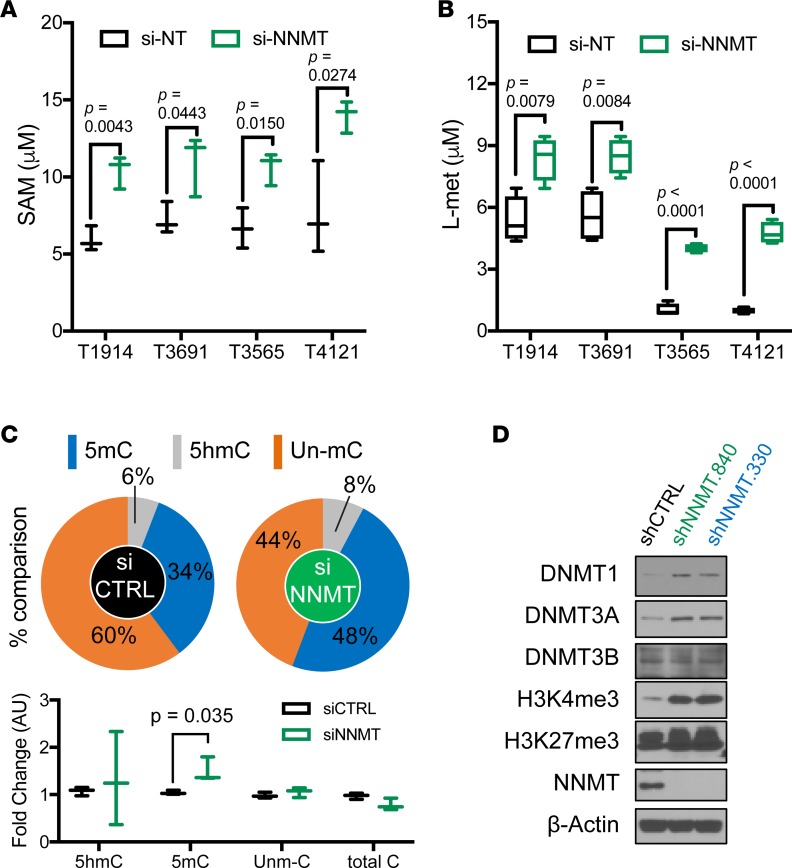
To determine the potential mechanism induced by loss of NNMT, we interrogated DNA methylation status after NNMT inhibition. First, we compared levels of methyl donors, SAM and L-methionine, in 2 proneural GSCs (T1914 and T3691) and 2 mesenchymal GSCs (T3565 and T4121) with and without NNMT depletion, which increased SAM and L-methionine levels (Figure 6, A and B). Notably, even with NNMT RNA interference (RNAi), mesenchymal GSCs exhibited lower levels of L-methionine compared with their proneural counterparts. NNMT depletion increased 5mC levels from 34%–48%, representing a shift toward increased DNA methylation (Figure 6C). Building on the reciprocal expression pattern between NNMT and DNMTs in GBM samples, we confirmed an inverse relationship between transcript expression of NNMT and the 3 DNMTs in TCGA GBM specimens (Supplemental Figure 5 and 9). We, therefore, sought direct evidence of inverse regulation between NNMT and DNMTs. Stable lentiviral transduction of shNNMTs (shNNMT.840 and shNNMT.330) increased expression of DNMT1 and DNMT3A, but not DNMT3B (Figure 6D). In a xenograft derived from mesenchymal GSCs, silencing of NNMT was associated with DNMT upregulation (Figure 6D). The reciprocal regulation of NNMT and DNMTs suggests that NNMT may have a dual mechanism to decrease DNA methylation through reduction of methyl donor availability and downregulation of DNMT1 and DNMT3A.
Figure 6. Reduced DNA methylation by depleting NNMT.
(A and B) SAM and L-methionine levels under transient transfection of siNNMT for 3 days in indicated GSCs. Data represent as box and whisker plot of 3 independent experiments. Differences of mean tested with 2-tailed Student’s t test. (C) 5-Methylcytosine measurement under transient transfection of siNNMT for 3 days in T4121 GSCs. 5hmC, 5-Hydroxymethylcytosine; un-mC, un-methylcytosine. Pie and Tukey’s box and whisker graphs were developed from same data for visualization. Data were represented as box and whisker plot of 3 technical replicates. Differences of mean tested with 2-tailed Student’s t test. (D) Immunoblot analysis of indicated antibodies in T4121 GSCs transduced with shCTRL, shNNMT.840, or shNNMT.330 lentivirus.
NNMT upregulation and DNMT1 downregulation in GBMs promote DNA hypomethylation of mesenchymal subtype genes.
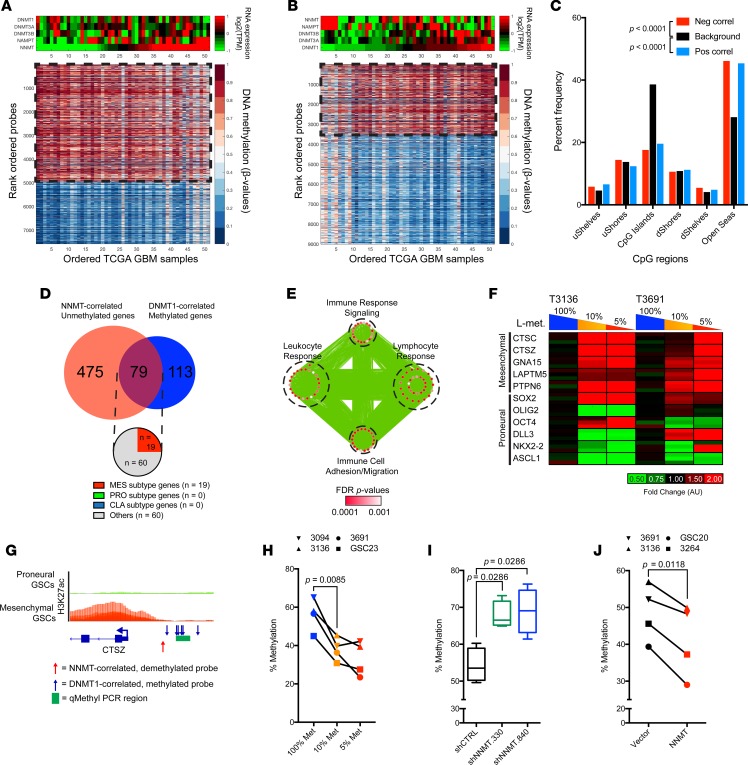
The changes we observed in DNMT expression in response to methionine restriction and increased DNA methylation upon NNMT knockdown suggested an epigenetic adaptation to metabolic stress to promote a proneural-to-mesenchymal transition. To interrogate this hypothesis, we first examined TCGA GBM RNAseq and DNA methylation array datasets to determine specific loci in GBM genomes that may be affected by reciprocal NNMT and DNMT1 expression levels (Figure 7, A and B). CpG probes whose methylation levels change significantly with coincident increased NNMT expression or DNMT1 downregulation were mostly present in the open sea region of the CpG landscape, while CpG islands remained relatively stable (Figure 7C). Among 192 genes that significantly correlated positively with DNMT1 expression and 554 genes that significantly anticorrelated with NNMT expression, 79 gene were common (Figure 7D). GO terms associated with these potential common targets of NNMT- and DNMT1-mediated epigenetic regulation were enriched in inflammatory responses and immune cell migration pathways (Figure 7E). Interestingly, ~25% of these genes were members of the mesenchymal GBM molecular subtype signature. Furthermore, DNA methylation at the CpG probes annotated for these genes were significantly anticorrelated with their respective genes, suggesting repression of these mesenchymal subtype signature genes through DNA methylation (Supplemental Figure 8). Thus, with reciprocal regulation of NNMT and DNMT1 expression, the epigenetic landscape of GBMs may promote the mesenchymal transcriptional program.
Figure 7. NNMT and DNMT1 affect DNA methylation of inflammatory genes.
(A) CpG probes significantly anticorrelated with NNMT mRNA expression in TCGA DNA methylation 450K array and RNAseq datasets (n = 51). (B) CpG probes with significant positive correlation with DNMT1 mRNA expression in TCGA DNA methylation 450K array and RNAseq datasets (n = 51). (C) Distribution of CpG region in probes with significant positive or negative correlation with NNMT expression (pos correl; neg correl) compared with nonsignificantly correlated CpG probes. Differences in distribution tested with 2-way ANOVA. (D) Annotated genes with significant negative correlation with NNMT mRNA expression and significant positive correlation with DNMT1 mRNA expression. (E) Gene ontology analysis of the common genes among those with significant negative correlation with NNMT mRNA expression and significant positive correlation with DNMT1 mRNA expression. Size of each node indicates the size of each gene set; thickness of each edge corresponds with the number of genes shared between connected nodes. (F) RT-PCR analysis of mesenchymal subtype genes in proneural T3136 and T3691 cultured in different methionine restriction conditions (100% Met, 115.5 μM; 10% Met, 11.5 μM; 5% Met, 5.75 μM). (G) Schema for methylation-sensitive (qMethyl) RT-PCR. Blue arrows indicate CpG probes significantly correlated with DNMT1 expression. Red arrow indicates CpG probe significantly correlated with NNMT expression. H3K27ac ChIP-seq data of proneural and mesenchymal GSCs (Stephen C. Mack, unpublished observations) visualized on IGV 2.3.80. (H) Quantification of DNA methylation levels of CpG loci in the CTSZ promoter region in 4 proneural GSC models (T3094, T3136, T3691, GSC23) cultured in different methionine restriction conditions (100% Met, 115.5 μM; 10% Met, 11.5 μM; 5% Met, 5.75 μM). Two-tailed Student’s t test used to determine changes in mean between conditions. (I and J) Quantification of DNA methylation levels of CpG loci in the CTSZ promoter region in 2 proneural GSC models (T3136 and T3691) and 2 mesenchymal GSC models (T3264 and GSC20) (I) transduced with nontargeting control or shRNA clones targeting NNMT or (J) transduced with vector control or NNMT overexpression constructs.
To validate this hypothesis, we cultured proneural GSCs in different methionine restriction conditions for 3 days and determined their mesenchymal and proneural subtype gene expression (Figure 7F). Proneural GSCs upon restricted methionine increased expression of predicted mesenchymal subtype genes (CTSC, CTSZ, GNA15, LAPTM5, and PTPN6) while simultaneously reducing core proneural subtype genes such as OLIG2 and ASCL1. To determine whether such increase in expression of mesenchymal subtype genes upon methionine restriction is due to changes in DNA methylation of their regulatory regions, we assayed the methylation level of the CTSZ promoter region predicted to be affected by NNMT and DNMT1 expression levels via methylation-sensitive real-time PCR (RT-PCR) (Figure 7G). We could confirm that methionine restriction reduced the methylation level of CpG loci in the CTSZ promoter region 4 proneural GSC models (T3094, T3136, T3691, and GSC23) (Figure 7H). As methionine-restricted GSCs upregulate NNMT expression, we examined whether NNMT level modulates DNA methylation of regulatory regions of mesenchymal subtype genes by knocking down or overexpressing NNMT in proneural and mesenchymal GSCs. The methylation level of the CTSZ promoter region was increased upon NNMT knockdown and reduced with NNMT overexpression (Figure 7, I and J). Thus, methionine restriction promotes a mesenchymal transcriptional program in proneural GSCs through DNA demethylation of regulatory regions of mesenchymal subtype genes through modulation of NNMT-associated methyl sink.
The core mesenchymal transcriptional factor C/EBPβ regulates NAM metabolism genes.
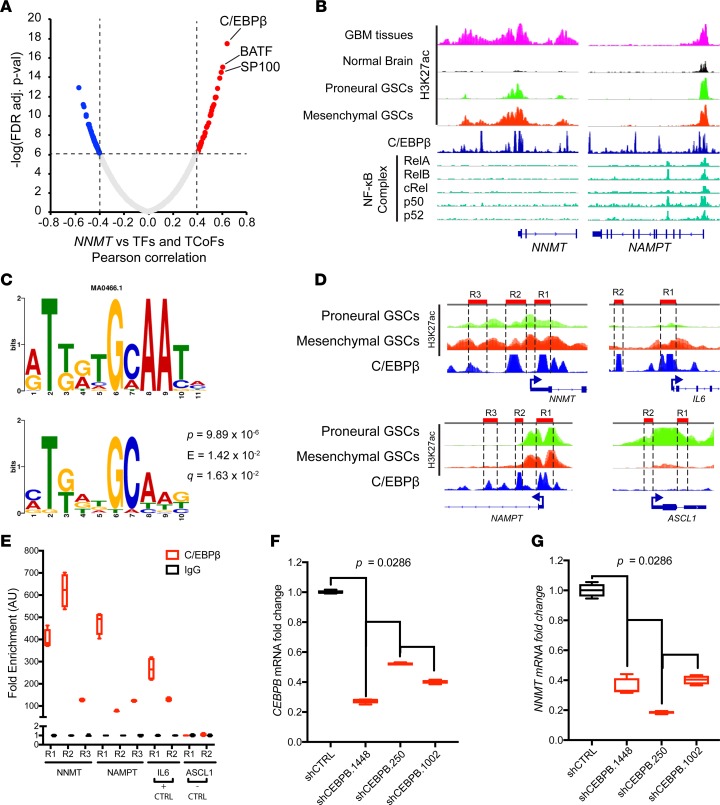
To determine how NNMT and other NAM metabolism genes are upregulated in mesenchymal GBMs, we interrogated TCGA GBM RNAseq database to find potential transcription factors and transcription coactivators that are significantly correlated with NNMT mRNA expression level (Figure 8A). We found that C/EBPβ, a core transcription factor associated with mesenchymal subtype program (59, 60), was the factor most significantly correlated with NNMT. Moreover, other transcription factors such as BATF and SP100 that are also mesenchymal subtype genes were very strongly correlated with NNMT expression. To further ascertain that mesenchymal transcription factors promote expression of other NAM metabolism genes, we examined the promoter regions of genes upregulated in the NAM metabolic pathway genes such as NNMT and NAMPT. In analyzing ChIP-seq data in GBMs and other human tissues, we found that both NNMT and NAMPT regulatory regions harbored C/EBPβ peaks, but NF-κB complex peaks were only present in the NAMPT promoter region (Figure 8B). We further examined for commonly enriched motives in the enhancer regions of upregulated genes of the NAM metabolism pathway (NNMT, NAMPT, MAT2A, AHCY, BST1, and BST2) and found that the C/EBPβ motif (MA0466.1 CEBPB) is the most significantly enriched motif across all the promoter regions (Figure 8C). We performed ChIP-PCR for C/EBPβ in mesenchymal GSCs to validate C/EBPβ interaction with NAM metabolism genes (Figure 8D). Indeed, C/EBPβ interaction was enriched in the regulatory regions of NNMT and NAMPT, as well as the classical target of C/EBPβ, the IL6 gene. On the other hand, C/EBPβ did not interact with the promoter region of the core proneural subtype transcription factor ASCL1 (Figure 8E). Knocking down CEBPB through shRNAs in GSCs resulted in decreased NNMT expression, as confirmed by RT-PCR (Figure 8, F and G). Thus, the core mesenchymal transcription factor C/EBPβ upregulates critical genes of the NAM metabolic pathway that promote DNA hypomethylation.
Figure 8. C/EBPβ promotes upregulation of nicotinamide metabolism genes.
(A) Correlation between NNMT and transcription factors and transcription coactivators in TCGA GBM RNAseq dataset. Pearson correlation test was used to evaluate relationship. TF, transcription factor; TCoF, transcription cofactor. (B) ChIP-seq analysis of glioblastoma samples and published datasets visualized on IGV 2.3.80: H3K27ac ChIP-seq data from glioblastoma tissues (n = 5), nonmalignant brain tissue (n = 5), proneural (n = 8 ), and mesenchymal GSCs (n = 8) marking enhancer regions (Stephen C. Mack, unpublished observations); C/EBPβ ChIP-seq from ENCODE database (91, 92); NF-κB complex ChIP-seq data from previously published studies (93, 94). (C) Motif analysis of enhancer regions of upregulated genes of the nicotinamide and nicotinate metabolism pathway (NNMT, NAMPT, MAT2A, AHCY, BST1, and BST2). P value represents the motif offset probability that the match occurred by random chance according to the null model. E value represents the expected number of times that the given query sequence would be expected to match a target motif as well or better than the observed match in a randomized target database of the given size. Q value is the match false discovery rate (84–86). (D) Schema for ChIP-PCR primer design in C/EBPβ ChIP peak-enriched regions in NNMT, NAMPT, IL6, and ASCL1 regulatory regions. (E) ChIP-PCR analysis of NNMT, NAMPT, IL6, and ASCL1 loci in mesenchymal T4121 GSCs. (F and G) RT-PCR of CEBPB and NNMT mRNA in T4121 GSCs transduced with shRNAs targeting CEBPB and NNMT and nontargeting controls (shCTRL).
Targeting NNMT extends survival of mice bearing mesenchymal GSCs.
In vivo tumor growth remains the gold standard of GSC functional assays. To determine if the roles of methionine restriction and NNMT extended into in vivo tumor growth, we performed a series of tumor growth studies. First, we tested methionine depletion on in vivo tumor growth; mesenchymal GSCs were grown under low methionine conditions for 3 days and then implanted into immunodeficient mice. Concordant with the in vitro findings, low methionine conditions augmented in vivo tumor growth (Figure 9A). Moreover, methionine restriction enhanced malignancy of proneural T3136, resulting in reduced survival of orthotopically xenografted mice (Figure 9B). We then targeted NNMT expression using 2 nonoverlapping shRNAs (shNNMT.840 or shNNMT.330) or a nontargeting controls (shCTRL) in both a mesenchymal and proneural GSC models. To directly monitor in vivo tumor growth, we initially implanted cells in the flanks of immunocompromised mice and monitored tumor growth. Concordant with the effects of NNMT silencing in vitro, in vivo tumor growth was substantially impaired with the loss of NNMT expression (Figure 9C). As NNMT may interact with the tumor microenvironment, we tested the impact of NNMT inhibition on orthotopic tumor growth. To determine the impact on tumor size, we sacrificed a cohort of tumor-bearing mice from each group and performed a histologic evaluation. As expected, tumors expressing shCTRL showed substantial in vivo tumor growth, whereas tumors transduced with shNNMT had substantially reduced volumes (Figure 9D). In parallel, we determined the impact of NNMT depletion on survival of tumor-bearing mice and noted a significant extension of survival (Figure 9E), supporting a role of NMMT in tumor growth.
Figure 9. In vivo mesenchymal GSC growth and mice survival rate by methionine restriction or targeting NNMT.
(A) Tumor mass from s.c. implantation of T4121 GSCs cultured under different methionine restriction conditions (100%, 115.5 μM; 10%, 11.5 μM; 5%, 5.75 μM) for 3 days before injection. (B) Kaplan-Meier curve of mice bearing intracranial xenograft of T3136 GSCs cultured for 3 days under different methionine restriction conditions (100%, 115.5 μM; 10%, 11.5 μM; 5%, 5.75 μM) before injection. (C) Tumor mass from s.c. implantation of T4121 GSCs transduced with shCTRL, shNNMT.840, or shNNMT.330 lentivirus. Scale bars: 10 mm. (D and E) H&E staining of tumor-bearing brains and survival of tumor-bearing mice bearing intracranially xenografted T4121 GSCs transduced with shCTRL, shNNMT.840, or shNNMT.330 lentivirus. Cohorts of 12 mice were implanted with tumors from 2 mice in each experimental arm that were randomly selected for sacrifice for H&E staining 2 weeks after implantation, and 10 were used to observe mice survival rate. The P value represents the Mantel-Cox log-rank test of survival analysis.
NNMT and NAMPT expression levels correlate with poor prognosis for GBM patients.
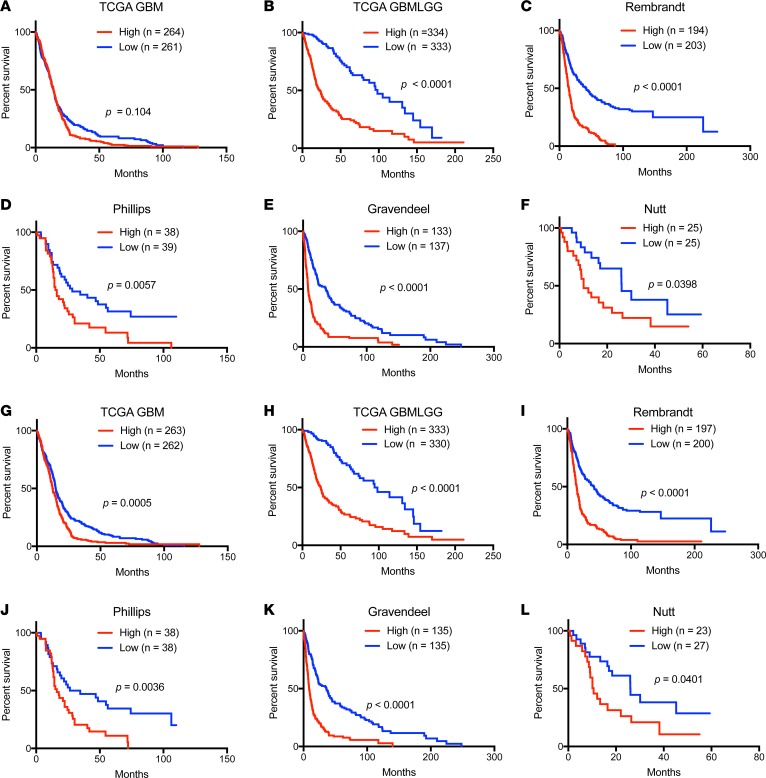
Based on the biologic effects of NNMT, we hypothesized that NNMT may inform the prognosis of patients afflicted with GBM. Therefore, we interrogated several available glioma datasets to correlate each target with patient outcome. We generated Kaplan-Meier estimates of survival for each gene based on 2 strategies: i) stratification based on median expression, and ii) stratification by the top and bottom quartiles, which could reveal the impact of expression of outliers. In each of 6 databases (TCGA GBM, REMBRANDT, TCGA GBMLGG, Phillips, Gravendeel, and Nutt) (4, 6, 22, 23, 61, 62) revealed that high expression of NNMT is correlated with poor patient survival both in the median (Figure 10, A–F) and the quartile cut-off (Supplemental Figure 9, A–F). The significance for patient survival was present even with the exclusion of G-CIMP patients, who have significantly better prognosis (Supplemental Figure 11). Likewise, NAMPT expression was associated with poor patient survival, using either the median (Figure 10, G–L) and the quartile cut-off (Supplemental Figure 10, G–L). In contrast, the DNMTs were not informative for patient survival (data not shown). Taken together, core regulators in NAM metabolism, NNMT and NAMPT, are negative prognostic factors for human glioma patients.
Figure 10. Kaplan-Meier survival curves of NNMT and NAMPT mRNA levels in glioblastoma patients.
Patient survival data based on expression of NNMT (A–G) and NAMPT (H–M) were evaluated by high vs. low expression levels, with the median value used as cutoff in indicated databases.
Discussion
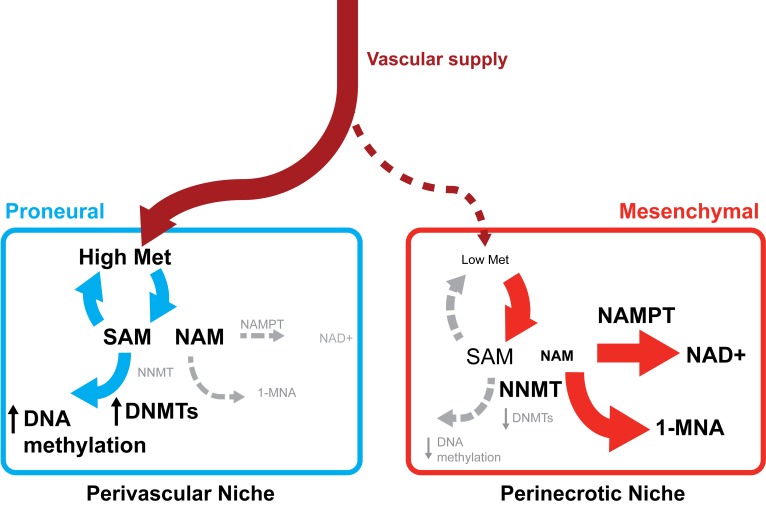
Metabolic dependencies in cancer have commonly centered on provision of cellular energy and building blocks for replication, but increasing evidence implicates epigenetic regulation based on selected metabolites functioning as substrates for chromatin modifying enzymes. Here, we show that mesenchymal GSCs preferentially activate 2 distinctive features ─ DNA hypomethylation and NAD+ utilization ─ due to a switch in NAM metabolism regulated by high levels of NNMT and NAMPT (Supplemental Figure 2B). To date, functional contributions of NNMT to diet-induced obesity, insulin resistance, glucose, lipid, and cholesterol metabolism have been delineated (63, 64). However, the oncogenic mechanisms of NNMT are poorly understood, despite high expression levels in many cancers, including GBM. GBMs display striking regional and cellular heterogeneity in metabolism and gene expression. Regions of tumors may be rich in methyl donors, while other areas that are often the most resistant to therapy may have low pools of methyl donors, supporting a heterogeneous DNA methylation profile, including MGMT methylation. Further, ROS and NAD+/NADH levels will likely vary within the tumor, creating differential redox potentials that could manifest in variable therapeutic responses. The NNMT-based observations suggest a new therapeutic model for malignant GBM treatment due to multiple factors: i) mesenchymal GBMs have smaller pools of methyl donors due to elevated NNMT expression; ii) within mesenchymal tumors, GSCs display elevated levels of NNMT and NAD+, with reduced DNMT expression and DNA hypomethylation relative to DTCs; and iii) within GBMs, regions of necrosis have elevated NNMT levels, reduced methionine availability, and lower DNMT expression, leading to DNA hypomethylation (Figure 11). This suggested mechanism is supported by the recent report showing that NNMT expression promotes histone hypomethylation by consuming SAM and methionine (50). In addition, intracellular SAM levels are maintained by methionine, a precursor of SAM, in a stem cell environment (51). Though NNMT upregulation in GBMs may not be exclusive to GSC populations, our results collectively suggest that GBMs harbor regions with differential metabolic and epigenetic regulation with key enzymes that could inform novel therapeutic targeting.
Figure 11. Proposed model of NNMT dependence in glioblastoma stem cells.
Mesenchymal GSCs are enriched in the perinecrotic niche where compromised vasculature delivers unreliable supplies of methionine and NAM. Reduced nutrient availability promotes upregulation of NNMT and NAMPT that converts the available methyl donor SAM and the small molecule NAM to SAH and 1-MNA or NAD+, respectively. The paucity of methyl donor pool and reduced expression of DNMTs reduce DNA methylation, promoting the mesenchymal transcriptional program. In the perivascular niche, relatively richer supply of nutrients including methionine and NAM promotes DNA methylation and increased expression of DNMTs, leading to enriched proneural GSCs in this microenvironment. Met, methionine; NAM, nicotinamide; 1-MNA, 1-methylnicotinamide; SAM, S-adenosyl methionine.
Necrotic regions within GBMs independently portend poor prognosis and have limited response to conventional therapy. Traditionally, the explanation for these areas has been that tumors outgrow their blood supplies. However, one could envision that the frequent intravascular blood clots and inflammatory responses in tumor regions could provide a significant advantage to long-term tumor growth. We previously showed that nutrient restriction, which would be expected to occur in hypovascular regions, promotes GSC maintenance (52). We now find that methionine restriction can increase tumor growth and is associated with poor patient outcome. These results could suggest that a tumor evolves to create regions of stress that favor the maintenance of GSCs through metabolic and epigenetic reprogramming. A recent report using mostly established cell lines suggested that methionine deprivation compromises proliferation, but the authors used complete removal of methionine, rather than a gradient as we examined (Nota bene we found that complete removal of methionine decreased proliferation) (65). Restriction of methionine and other epigenetic cofactors may promote reduction of available methyl donors and inhibit DNMT expression to shift the DNA methylation toward a more aggressive phenotype.
Although N-methylation of histones is closely associated with functional changes of chromatin and, thus, is involved in cell biology (63, 66), N-methylation of metabolites may serve as a point of fragility for tumor cells due to simultaneous effects on both metabolism and epigenetic modifications, as demonstrated by efficacy of NAMPT inhibitors in IDH1 mutant or MYC-amplified gliomas (67, 68). Further, NAMPT may regulate GBM sphere-forming cells through regulation of the inhibitor of differentiation (ID) pathway (69). Although NNMT and NAMPT are both involved in NAM metabolism, they are nonredundant with different enzymatic activities and functions. In contrast to NAMPT, NNMT is not part of the NAM salvage pathway and more directly depletes the methyl donor, SAM, which is reflected in its role in induction of DNA hypomethylation. NNMT methylates NAM to produce 1-MNA, thereby consuming SAM as a critical methyl source and increasing SAH (50, 51). In human studies and animal models of oxidative stress, 1-MNA has been suggested to increase oxidative stress to promote antiinflammatory M2 polarization of macrophages, reduced proliferative exhaustion, and lifespan extension (70–73). NNMT is required for low SAM levels and reduced histone methylation (histone 3 lysine 27 tri-methylation, H3K27me3) status in the epigenetic landscape of human embryonic stem cells (51). Targeting NNMT has proven beneficial for protection against diet-induced obesity through regulation of the SIRT1/NAMPT/NAD+ pathway (64, 74). Given the potential utility of NAMPT inhibitors in some gliomas, future studies may demonstrate the benefit of dual targeting of NNMT and NAMPT for glioma therapy (67, 68).
Translating our findings into clinically actionable efforts can be leveraged by recent efforts to apply NAM and its derivatives to drug discovery and patient treatment in human diseases. NAM, as an anticancer drug, has clinical efficacy in a phase III randomized trial of chemoprevention of nonmelanoma skin cancers (75). Based on the KEGG pathway and the functional analyses of NAM metabolism, enzymatic regulators, and their metabolites biochemically and functionally connect a methionine cycling pathway as a precursor of SAM. SAM is a major methyl donor, and its de novo synthesis pathways are recycled by methionine and homocysteine pathways. These metabolites have been implicated in various biological significances, including human diseases (65, 76, 77). SAM, popularly called SAMe, has been used in complementary and holistic medicine as a potential therapy for numerous conditions, including cancer, suggesting that it is safe. SAM supplementation may have limited impact due to delivery into poorly vascularized tumor regions but could serve as a complement to other therapies. In addition, NAD+, as a product of NAM metabolism, is regulated by an enzymatic reaction by NAMPT. Inhibitors of the NAMPT, such as FK866 and other small molecules, are currently under clinical trial to evaluate an effect on metabolic perturbations in various cancers (78, 79).
Clinical development of NNMT (or NAMPT) inhibitors should be informed by several opportunities and challenges. First, hypomethylation of the MGMT promoter is both prognostic and predictive of TMZ sensitivity. A suicide inhibitor of MGMT, O6-benzylguanine, has been used in clinical trials, with activity limited by severe toxicity (80). Dose-dense TMZ may reduce MGMT expression, but clinical trials have failed (81). Future studies may permit the use of NNMT or NAMPT inhibitors to increase MGMT promoter methylation, potentially sensitizing tumor cells to TMZ. Another potential targeting approach could leverage the differential regulation of NNMT and the DNMTs. Targeting NNMT expression is associated with increased DNMT expression. Therefore, NNMT inhibition may be combined or sequenced with DNMT inhibitors, including azacytidine and decitabine, to augment tumor control. In conclusion, the ability to target aberrant metabolic regulation could have several beneficial effects on tumor control and offer new therapies that could be combined with conventional therapies or other targeted therapies. Our bioinformatics findings support a precision medicine model to apply these efforts to patients with IDH WT, mesenchymal tumors with necrotic regions.
Methods
Culture of human GBM specimens and cells.
As previously described (8), patient-derived GSCs were cultured in Neurobasal medium supplemented with B27 (without Vitamin A, Invitrogen), basic fibroblast growth factor (bFGF, 20 ng/ml, R&D Systems), and epidermal growth factor (EGF, 20 ng/ml, R&D Systems). All specimens were validated as a unique cell by short tandem repeat (STR) analysis. All cell lines are routinely tested for mycoplasma contamination and were negative.
Intracranial and s.c. xenograft implantation.
Cells were intracranially or s.c. implanted into female and male NOD SCID γ (NSG, JAX, Charles River Laboratories) mice (4–6 weeks old). Development of neurological signs was considered end-point in all intracranial xenograft experiments. In parallel survival experiments, mice were monitored until they had neurological signs. For H&E staining, 2 mice in each group were sacrificed 2 weeks after the intracranial injection, and the staining was performed with Cleveland Clinic imaging core service.
Transfection of siRNA and transduction of shRNA or NNMT overexpression lentiviral particles.
Cells were transfected and Lipofectamine RNAiMAX (for siRNA, Invitrogen) according to the manufacturer’s reverse transfection protocol. siRNAs against the following genes were purchased from Santa Cruz Biotechnology Inc.: NNMT and a nontargeting control. shRNA lentiviral clones against NNMT (shNNMT.840, TRCN0000294436; shNNMT.330, TRCN0000035226), CEBPB (shCEBPB.1448, TRCN0000007440; shCEBPB.250, TRCN0000007441; shCEBPB.1002, TRCN0000007443), NAMPT (shNAMPT.1140, TRCN0000424413; shNAMPT.1183, TRCN0000454907), and a nontargeting control (shCTRL, SHC002) were purchased from Sigma-Aldrich. NNMT overexpression construct (HsCD00442343) and control vectors were purchased from DNASU plasmid repository (Arizona State University, Tempe, Arizona, USA). Cells were transduced per recommendations from the manufacturer. Lentiviral particles were generated in 293FT cells with cotransfection with the packaging vectors pCMV-dR8.2 dvpr (Addgene) and pCI-VSVG (Addgene) by Lipofectamine 2000 (Invitrogen). Stable clones were selected with puromycin dihydrochloride (Sigma-Aldrich) for 3 days and processed for downstream experiments.
5mC measurement.
Detection of 5mC and 5hmC was performed using EpiMark 5-hmC and 5-mC Analysis Kit (New England Biolabs) (82). Genomic DNA was treated with T4-BGT, generating glucosylated 5hmC (5ghmC) from 5hmC. Restriction endonuclease digestion was performed by MspI or HpaII on genomic DNA in accordance with the manufacturer’s instructions. Unmodified DNA was analyzed without T4-BGT by the same process. Amplification of the glucosylated DNA and mock target DNA with primers flanking CCGG sites were performed. Real-time PCR was performed for the amplification.
Methylation-sensitive RT-PCR.
DNA methylation levels in promoter regions of interest were measured using the OneStep qMethyl kit (Zymo Research). Primers for CTSZ region of interest were designed per manufacturer recommendations (Supplemental Table 3). Genomic DNA from GSCs was isolated using Quick-DNA Miniprep Plus Kit (Zymo Research). RT-PCR of restriction enzyme–treated samples and reference samples was performed with Radiant SYBR Green qPCR kits (Alkali Scientific Inc.).
ChIP-PCR.
C/EBPβ ChIP was performed as previously described (83). Briefly, 1 × 107 cells were fixed with 1% formaldehyde (Sigma-Aldrich) for 7 minutes and quenched with 125 mM glycine (Sigma-Aldrich). Mouse anti-C/EBPβ (5 μg, Santa Cruz Biotechnology Inc., sc-7962X) or mouse IgG control (Santa Cruz Biotechnology Inc., sc-2025) antibodies were used to bind sonicated nucleosomes and incubated overnight in 4°C. After washes with 0.25 M LiCl solution (Sigma-Aldrich), DNA was decrosslinked from nucleosomes in 0.1 M sodium bicarbonate 1% SDS solution (Sigma-Aldrich) in 65°C for 12 hours. Genomic DNA was isolated using the QIAquick PCR Purification kit (Qiagen, catalog 28104). DNA (50 ng) was used per PCR reaction with Radiant SYBR Green qPCR kits (Alkali Scientific Inc.), with primers designed for each C/EBPβ-enriched regions and negative control regions (Supplemental Table 3).
Motif analysis.
Promoter regions of genes upregulated in the NAM metabolism pathway were analyzed with the MEME-DREME suite v4.11.2 to identify enriched DNA binding motifs as previously described (84–86). DNA sequences from hg19 assembly were scanned for gapped motifs using GLAM2. The best motif as enriched by GLAM2 was cross-referenced with a known database of eukaryotic motifs using TOMTOM.
Metabolite measurement.
For measurement of L-methionine, homocysteine, SAM, SAH, and NAD+ conventional assay kits were used for their detection as follows: Bridge-It L-methionine fluorescence assay kit (Mediomics); Homosysteinie ELISA, SAM and SAH ELISA combo kit (Cell Biolabs); and NAD/NADH quantitation colorimetric kit (Biovision). All assays were performed as detailed in the manufacturers’ protocols under the indicated conditions. SAH/SAM ratios were mathematically calculated using SAH and SAM levels. For detection of NAM and methyl-NAM, LC-MS/MS was performed by the Cleveland Clinic core service. As recommended, cell pellets were suspended in 80% methanol and centrifuged to isolate supernatants. The supernatant was evaporated and concentrated by nitrogen. Nicotinamide-d4 (CND Isotope) and N-Methylnicotinamide-2,4,5,6-d4 (CND Isotope) were used for internal standard in LC-MS/MS.
Western blotting.
Proteins were extracted by CelLytic M (Sigma-Aldrich) cell lysis buffer supplemented with protease inhibitor cocktail and phosphatase inhibitors. Protein levels were determined by Western blotting using conventional protocols. Proteins were detected using specific primary antibodies from DNMT1 (Cell Signaling Technology, 5119), DNMT3A (Cell Signaling Technology, 3598), and DNMT3B (Cell Signaling Technology, 67259); PARP-1 (Cell Signaling Technology, 9542); NNMT (Abcam, ab58743 for immunoblot; Santa Cruz Biotechnology Inc., sc-376048 for IF), NAMPT (Abcam, ab45890), H3K4-me3 (Abcam, ab8895), and H3K27-me3 (Abcam, ab6002); β-actin (Sigma-Aldrich, A5316); GFAP (Covance, SMI-21R); SOX2 (R&D Systems, MAB2018) and CD44 (R&D Systems, MAB6127); and subsequently with the appropriate horseradish peroxidase–conjugated (HRP-conjugated) secondary antibodies (mouse-HRP: Cell Signaling Technology, 7076; Rb-HRP: Cell Signaling Technology, 7074).
Immunofluorescent staining.
Immunofluorescence assays of paraffin embedded and frozen patient GBM specimens, nonneoplastic epileptic brain frozen sections, and xenografted tumor tissues were performed with conventional protocols. After deparaffinization or fixation, slides were incubated with indicated primary antibodies and subsequently with the appropriate Alexa Fluor 488– (Thermo Fisher, catalog A-11001, 1:1,000) or -568–conjugated (Thermo Fisher, catalog A-11011, 1:1,000) secondary antibodies. DAPI (1 μg/ml) was stained for nuclei. Samples were photographed with a Leica confocal microscopy.
Cell viability assay.
GSC viability was assayed using CellTiter-Glo Reagent (Promega) per manufacturer recommendations. Briefly, after equilibration to room temperature for 30 minutes, cells were lysed with CellTiter-Glo reagent at 1:1 volume-wise ratio with cell culture media. After stabilizing the luminescent signal at room temperature for 10 minutes, the cell plate was read on the multiplate reader (Perkin Elmer Victor3), used for signal detection.
In silico analysis.
All gene expression, DNA methylation array, and clinical datasets were downloaded from GlioVis (http://gliovis.bioinfo.cnio.es/) and TCGA2STAT package on R (http://www.liuzlab.org/TCGA2STAT). Mutational status of target genes in TCGA GBMLGG dataset was analyzed and integrated with clinical information on cBioPortal (87, 88). Gene signatures analyzed were obtained from the Molecular Signatures Database (http://software.broadinstitute.org/gsea/msigdb) (89). Gene signature scores were calculated by deriving Z-scores across all the tumor specimens from single sample GSEA results for each gene set (89, 90). For survival analysis based on gene signature scores, high enrichment of a gene signature was determined as Z-score > 0 and low enrichment as Z-score < 0. Hierarchical clustering of gene expression was performed with median-centered gene expression values on Matlab using Euclidean distances and optimal leaf ordering. For gene rank analysis by Oncomine (http://www.oncomine.org), the top 10% of genes overexpressed in glioblastomas vs. nontumor samples in 6 glioblastoma expression data sets were crossreferenced with the top 1% of genes overexpressed in glioblastomas compared with nontumor samples in the TCGA dataset through the unbiased analysis tool from Oncomine.
Statistics.
All data presented as Tukey’s box and whisker plots (the box marked by the median, bounded by the 75th and 25th percentiles) were analyzed with Mann-Whitney U test when possible (n > 3) and 2-tailed unpaired Student’s t test to analyze differences in mean between groups (n = 3). To determine the correlation between mRNA expression and DNA methylation levels, Spearman correlation test was performed and FDR adjusted using the Benjamini-Hochberg procedure. Chi-square test was performed to examine differences in CpG probe distribution among CpG landscape. The statistical analyses were performed with Matlab 2016b, GraphPad Prism, and Microsoft Excel software. Kaplan-Meier curves were generated by using GraphPad Prism and NCSS softwares. Mantel-Cox log-rank test was used to test significance in survival outcomes, with p < 0.05 considered statistically significant.
Study approval.
GBM and nonmalignant brain tissues were obtained from excess surgical materials with full informed consent from patients at the Cleveland Clinic as approved by the IRB of the Cleveland Clinic Foundation (IRB protocol 2955). All animal experiments were performed as approved by the IACUC of the Lerner Research Institute in the Cleveland Clinic Foundation (IACUC protocol 2016-1566).
Author contributions
JJ, LJYK, and JNR designed the overall experiments, analyzed data, and wrote the manuscript. JJ, LJYK, XW, QW, TS, and LCW performed in vitro or animal experiments. JJ, LJYK, CGH, BCP, XJ, and SCM performed bioinformatics analysis of published expression datasets. CGH, XJ, and SCM provided intellectual input. All authors provided scientific input and edited and approved the final manuscript.
Supplementary Material
Acknowledgments
The authors thank Angela H. Ting for kind discussions regarding measurement of DNA methylation. We would like to thank the Cleveland Clinic Lerner Research Institute imaging core and proteomics core service teams. We also thank members of the Rich lab for input about the manuscript. Finally, we would like to thank our funding sources: the NIH grants CA203101 (LK), CA189647 (CGH), CA197718, CA154130, CA169117, CA171652, NS087913, and NS089272 (JNR); General Program of the National Natural Science Foundation of China (no. 81572891) (XJ); and Canadian Institutes of Health Research Banting Fellowship (SCM).
Footnotes
JJ’s present address is: Neuro-Oncology Branch, National Cancer Institute, NIH, Bethesda, Maryland, USA.
JNR’s current address: Division of Regenerative Medicine, Department of Medicine, University of California, San Diego, San Diego, California, USA.
Conflict of interest: The authors have declared that no conflict of interest exists.
Reference information:JCI Insight. 2017;2(10):e90019. https://doi.org/10.1172/jci.insight.90019.
Contributor Information
Jinkyu Jung, Email: skycross00@gmail.com.
Xiuxing Wang, Email: xiuxing81@gmail.com.
Qiulian Wu, Email: wuq2@ccf.org.
Tanwarat Sanvoranart, Email: tanwaratm@gmail.com.
Lisa C. Wallace, Email: WALLACL3@ccf.org.
Xun Jin, Email: jinx2354@gmail.com.
References
- 1. doi: 10.1002/cncr.24950. Linz U. Commentary on Effects of radiotherapy with concomitant and adjuvant temozolomide versus radiotherapy alone on survival in glioblastoma in a randomised phase III study: 5-year analysis of the EORTC-NCIC trial (Lancet Oncol. 2009;10:459-466). Cancer. 2010;116(8):1844–1846. [DOI] [PubMed] [Google Scholar]
- 2.Kim Y, et al. Platelet-derived growth factor receptors differentially inform intertumoral and intratumoral heterogeneity. Genes Dev. 2012;26(11):1247–1262. doi: 10.1101/gad.193565.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Noushmehr H, et al. Identification of a CpG island methylator phenotype that defines a distinct subgroup of glioma. Cancer Cell. 2010;17(5):510–522. doi: 10.1016/j.ccr.2010.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Verhaak RG, et al. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell. 2010;17(1):98–110. doi: 10.1016/j.ccr.2009.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sturm D, et al. Hotspot mutations in H3F3A and IDH1 define distinct epigenetic and biological subgroups of glioblastoma. Cancer Cell. 2012;22(4):425–437. doi: 10.1016/j.ccr.2012.08.024. [DOI] [PubMed] [Google Scholar]
- 6.Ceccarelli M, et al. Molecular Profiling Reveals Biologically Discrete Subsets and Pathways of Progression in Diffuse Glioma. Cell. 2016;164(3):550–563. doi: 10.1016/j.cell.2015.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Visvader JE, Lindeman GJ. Cancer stem cells in solid tumours: accumulating evidence and unresolved questions. Nat Rev Cancer. 2008;8(10):755–768. doi: 10.1038/nrc2499. [DOI] [PubMed] [Google Scholar]
- 8.Bao S, et al. Glioma stem cells promote radioresistance by preferential activation of the DNA damage response. Nature. 2006;444(7120):756–760. doi: 10.1038/nature05236. [DOI] [PubMed] [Google Scholar]
- 9.Mao P, et al. Mesenchymal glioma stem cells are maintained by activated glycolytic metabolism involving aldehyde dehydrogenase 1A3. Proc Natl Acad Sci USA. 2013;110(21):8644–8649. doi: 10.1073/pnas.1221478110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Piao Y, Liang J, Holmes L, Henry V, Sulman E, de Groot JF. Acquired resistance to anti-VEGF therapy in glioblastoma is associated with a mesenchymal transition. Clin Cancer Res. 2013;19(16):4392–4403. doi: 10.1158/1078-0432.CCR-12-1557. [DOI] [PubMed] [Google Scholar]
- 11.Turcan S, et al. IDH1 mutation is sufficient to establish the glioma hypermethylator phenotype. Nature. 2012;483(7390):479–483. doi: 10.1038/nature10866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dang L, et al. Cancer-associated IDH1 mutations produce 2-hydroxyglutarate. Nature. 2009;462(7274):739–744. doi: 10.1038/nature08617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen CL, et al. NANOG Metabolically Reprograms Tumor-Initiating Stem-like Cells through Tumorigenic Changes in Oxidative Phosphorylation and Fatty Acid Metabolism. Cell Metab. 2016;23(1):206–219. doi: 10.1016/j.cmet.2015.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee E, et al. Metabolic stress induces a Wnt-dependent cancer stem cell-like state transition. Cell Death Dis. 2015;6:e1805. doi: 10.1038/cddis.2015.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sperber H, et al. The metabolome regulates the epigenetic landscape during naive-to-primed human embryonic stem cell transition. Nat Cell Biol. 2015;17(12):1523–1535. doi: 10.1038/ncb3264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jones RG, Thompson CB. Tumor suppressors and cell metabolism: a recipe for cancer growth. Genes Dev. 2009;23(5):537–548. doi: 10.1101/gad.1756509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim D, et al. SHMT2 drives glioma cell survival in ischaemia but imposes a dependence on glycine clearance. Nature. 2015;520(7547):363–367. doi: 10.1038/nature14363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mattson MP, Shea TB. Folate and homocysteine metabolism in neural plasticity and neurodegenerative disorders. Trends Neurosci. 2003;26(3):137–146. doi: 10.1016/S0166-2236(03)00032-8. [DOI] [PubMed] [Google Scholar]
- 19.Cairns RA, Harris IS, Mak TW. Regulation of cancer cell metabolism. Nat Rev Cancer. 2011;11(2):85–95. doi: 10.1038/nrc2981. [DOI] [PubMed] [Google Scholar]
- 20.Lu C, Thompson CB. Metabolic regulation of epigenetics. Cell Metab. 2012;16(1):9–17. doi: 10.1016/j.cmet.2012.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Possemato R, et al. Functional genomics reveal that the serine synthesis pathway is essential in breast cancer. Nature. 2011;476(7360):346–350. doi: 10.1038/nature10350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gravendeel LA, et al. Intrinsic gene expression profiles of gliomas are a better predictor of survival than histology. Cancer Res. 2009;69(23):9065–9072. doi: 10.1158/0008-5472.CAN-09-2307. [DOI] [PubMed] [Google Scholar]
- 23.Madhavan S, Zenklusen JC, Kotliarov Y, Sahni H, Fine HA, Buetow K. Rembrandt: helping personalized medicine become a reality through integrative translational research. Mol Cancer Res. 2009;7(2):157–167. doi: 10.1158/1541-7786.MCR-08-0435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sun L, et al. Neuronal and glioma-derived stem cell factor induces angiogenesis within the brain. Cancer Cell. 2006;9(4):287–300. doi: 10.1016/j.ccr.2006.03.003. [DOI] [PubMed] [Google Scholar]
- 25.Gill BJ, et al. MRI-localized biopsies reveal subtype-specific differences in molecular and cellular composition at the margins of glioblastoma. Proc Natl Acad Sci USA. 2014;111(34):12550–12555. doi: 10.1073/pnas.1405839111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Murat A, et al. Stem cell-related “self-renewal” signature and high epidermal growth factor receptor expression associated with resistance to concomitant chemoradiotherapy in glioblastoma. J Clin Oncol. 2008;26(18):3015–3024. doi: 10.1200/JCO.2007.15.7164. [DOI] [PubMed] [Google Scholar]
- 27.Meissner A, et al. Genome-scale DNA methylation maps of pluripotent and differentiated cells. Nature. 2008;454(7205):766–770. doi: 10.1038/nature07107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Heller G, et al. Genome-wide transcriptional response to 5-aza-2’-deoxycytidine and trichostatin a in multiple myeloma cells. Cancer Res. 2008;68(1):44–54. doi: 10.1158/0008-5472.CAN-07-2531. [DOI] [PubMed] [Google Scholar]
- 29.Missiaglia E, Donadelli M, Palmieri M, Crnogorac-Jurcevic T, Scarpa A, Lemoine NR. Growth delay of human pancreatic cancer cells by methylase inhibitor 5-aza-2’-deoxycytidine treatment is associated with activation of the interferon signalling pathway. Oncogene. 2005;24(1):199–211. doi: 10.1038/sj.onc.1208018. [DOI] [PubMed] [Google Scholar]
- 30.Lopez-Serra L, et al. Unmasking of epigenetically silenced candidate tumor suppressor genes by removal of methyl-CpG-binding domain proteins. Oncogene. 2008;27(25):3556–3566. doi: 10.1038/sj.onc.1211022. [DOI] [PubMed] [Google Scholar]
- 31.Wu G, et al. LOXL1 and LOXL4 are epigenetically silenced and can inhibit ras/extracellular signal-regulated kinase signaling pathway in human bladder cancer. Cancer Res. 2007;67(9):4123–4129. doi: 10.1158/0008-5472.CAN-07-0012. [DOI] [PubMed] [Google Scholar]
- 32.Suzuki H, et al. A genomic screen for genes upregulated by demethylation and histone deacetylase inhibition in human colorectal cancer. Nat Genet. 2002;31(2):141–149. doi: 10.1038/ng892. [DOI] [PubMed] [Google Scholar]
- 33.Weber M, et al. Chromosome-wide and promoter-specific analyses identify sites of differential DNA methylation in normal and transformed human cells. Nat Genet. 2005;37(8):853–862. doi: 10.1038/ng1598. [DOI] [PubMed] [Google Scholar]
- 34.Schlesinger Y, et al. Polycomb-mediated methylation on Lys27 of histone H3 pre-marks genes for de novo methylation in cancer. Nat Genet. 2007;39(2):232–236. doi: 10.1038/ng1950. [DOI] [PubMed] [Google Scholar]
- 35.Cheng AS, et al. Epithelial progeny of estrogen-exposed breast progenitor cells display a cancer-like methylome. Cancer Res. 2008;68(6):1786–1796. doi: 10.1158/0008-5472.CAN-07-5547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Aoyama K, et al. Nicotinamide-N-methyltransferase is higher in the lumbar cerebrospinal fluid of patients with Parkinson’s disease. Neurosci Lett. 2001;298(1):78–80. doi: 10.1016/S0304-3940(00)01723-7. [DOI] [PubMed] [Google Scholar]
- 37.Thomas MG, Saldanha M, Mistry RJ, Dexter DT, Ramsden DB, Parsons RB. Nicotinamide N-methyltransferase expression in SH-SY5Y neuroblastoma and N27 mesencephalic neurones induces changes in cell morphology via ephrin-B2 and Akt signalling. Cell Death Dis. 2013;4:e669. doi: 10.1038/cddis.2013.200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Palada V, et al. Histamine N-methyltransferase Thr105Ile polymorphism is associated with Parkinson’s disease. Neurobiol Aging. 2012;33(4):836.e1–836.e3. doi: 10.1016/j.neurobiolaging.2011.06.015. [DOI] [PubMed] [Google Scholar]
- 39.Müller T. Catechol-O-methyltransferase inhibitors in Parkinson’s disease. Drugs. 2015;75(2):157–174. doi: 10.1007/s40265-014-0343-0. [DOI] [PubMed] [Google Scholar]
- 40.Jacobs RL, et al. Impaired de novo choline synthesis explains why phosphatidylethanolamine N-methyltransferase-deficient mice are protected from diet-induced obesity. J Biol Chem. 2010;285(29):22403–22413. doi: 10.1074/jbc.M110.108514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Selley ML. A metabolic link between S-adenosylhomocysteine and polyunsaturated fatty acid metabolism in Alzheimer’s disease. Neurobiol Aging. 2007;28(12):1834–1839. doi: 10.1016/j.neurobiolaging.2006.08.003. [DOI] [PubMed] [Google Scholar]
- 42.Powell CA, et al. TRMT5 Mutations Cause a Defect in Post-transcriptional Modification of Mitochondrial tRNA Associated with Multiple Respiratory-Chain Deficiencies. Am J Hum Genet. 2015;97(2):319–328. doi: 10.1016/j.ajhg.2015.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yang CP, et al. Characterization of the neuropsychological phenotype of glycine N-methyltransferase-/- mice and evaluation of its responses to clozapine and sarcosine treatments. Eur Neuropsychopharmacol. 2012;22(8):596–606. doi: 10.1016/j.euroneuro.2011.12.007. [DOI] [PubMed] [Google Scholar]
- 44.Mann MB, Wu S, Rostamkhani M, Tourtellotte W, MacMurray J, Comings DE. Phenylethanolamine N-methyltransferase (PNMT) gene and early-onset Alzheimer disease. Am J Med Genet. 2001;105(4):312–316. doi: 10.1002/ajmg.1363. [DOI] [PubMed] [Google Scholar]
- 45.Stabler SP, Marcell PD, Podell ER, Allen RH, Savage DG, Lindenbaum J. Elevation of total homocysteine in the serum of patients with cobalamin or folate deficiency detected by capillary gas chromatography-mass spectrometry. J Clin Invest. 1988;81(2):466–474. doi: 10.1172/JCI113343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schmidt A, et al. Severely altered guanidino compound levels, disturbed body weight homeostasis and impaired fertility in a mouse model of guanidinoacetate N-methyltransferase (GAMT) deficiency. Hum Mol Genet. 2004;13(9):905–921. doi: 10.1093/hmg/ddh112. [DOI] [PubMed] [Google Scholar]
- 47.Etain B, et al. Genetic and functional abnormalities of the melatonin biosynthesis pathway in patients with bipolar disorder. Hum Mol Genet. 2012;21(18):4030–4037. doi: 10.1093/hmg/dds227. [DOI] [PubMed] [Google Scholar]
- 48.Heddleston JM, Li Z, McLendon RE, Hjelmeland AB, Rich JN. The hypoxic microenvironment maintains glioblastoma stem cells and promotes reprogramming towards a cancer stem cell phenotype. Cell Cycle. 2009;8(20):3274–3284. doi: 10.4161/cc.8.20.9701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ricci-Vitiani L, et al. Mesenchymal differentiation of glioblastoma stem cells. Cell Death Differ. 2008;15(9):1491–1498. doi: 10.1038/cdd.2008.72. [DOI] [PubMed] [Google Scholar]
- 50.Ulanovskaya OA, Zuhl AM, Cravatt BF. NNMT promotes epigenetic remodeling in cancer by creating a metabolic methylation sink. Nat Chem Biol. 2013;9(5):300–306. doi: 10.1038/nchembio.1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shiraki N, et al. Methionine metabolism regulates maintenance and differentiation of human pluripotent stem cells. Cell Metab. 2014;19(5):780–794. doi: 10.1016/j.cmet.2014.03.017. [DOI] [PubMed] [Google Scholar]
- 52.Flavahan WA, et al. Brain tumor initiating cells adapt to restricted nutrition through preferential glucose uptake. Nat Neurosci. 2013;16(10):1373–1382. doi: 10.1038/nn.3510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kokkinakis DM, et al. Mitotic arrest, apoptosis, and sensitization to chemotherapy of melanomas by methionine deprivation stress. Mol Cancer Res. 2006;4(8):575–589. doi: 10.1158/1541-7786.MCR-05-0240. [DOI] [PubMed] [Google Scholar]
- 54.Gilbertson RJ, Rich JN. Making a tumour’s bed: glioblastoma stem cells and the vascular niche. Nat Rev Cancer. 2007;7(10):733–736. doi: 10.1038/nrc2246. [DOI] [PubMed] [Google Scholar]
- 55.Hu B, et al. Epigenetic Activation of WNT5A Drives Glioblastoma Stem Cell Differentiation and Invasive Growth. Cell. 2016;167(5):1281–1295.e18. doi: 10.1016/j.cell.2016.10.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cheng L, et al. Glioblastoma stem cells generate vascular pericytes to support vessel function and tumor growth. Cell. 2013;153(1):139–152. doi: 10.1016/j.cell.2013.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Li Z, et al. Hypoxia-inducible factors regulate tumorigenic capacity of glioma stem cells. Cancer Cell. 2009;15(6):501–513. doi: 10.1016/j.ccr.2009.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Seidel S, et al. A hypoxic niche regulates glioblastoma stem cells through hypoxia inducible factor 2 alpha. Brain. 2010;133(Pt 4):983–995. doi: 10.1093/brain/awq042. [DOI] [PubMed] [Google Scholar]
- 59.Carro MS, et al. The transcriptional network for mesenchymal transformation of brain tumours. Nature. 2010;463(7279):318–325. doi: 10.1038/nature08712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bhat KP, et al. Mesenchymal differentiation mediated by NF-κB promotes radiation resistance in glioblastoma. Cancer Cell. 2013;24(3):331–346. doi: 10.1016/j.ccr.2013.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Phillips HS, et al. Molecular subclasses of high-grade glioma predict prognosis, delineate a pattern of disease progression, and resemble stages in neurogenesis. Cancer Cell. 2006;9(3):157–173. doi: 10.1016/j.ccr.2006.02.019. [DOI] [PubMed] [Google Scholar]
- 62.Nutt CL, et al. Gene expression-based classification of malignant gliomas correlates better with survival than histological classification. Cancer Res. 2003;63(7):1602–1607. [PubMed] [Google Scholar]
- 63.Singhal NK, et al. Changes in Methionine Metabolism and Histone H3 Trimethylation Are Linked to Mitochondrial Defects in Multiple Sclerosis. J Neurosci. 2015;35(45):15170–15186. doi: 10.1523/JNEUROSCI.4349-14.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kraus D, et al. Nicotinamide N-methyltransferase knockdown protects against diet-induced obesity. Nature. 2014;508(7495):258–262. doi: 10.1038/nature13198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Palanichamy K, et al. Methionine and Kynurenine Activate Oncogenic Kinases in Glioblastoma, and Methionine Deprivation Compromises Proliferation. Clin Cancer Res. 2016;22(14):3513–3523. doi: 10.1158/1078-0432.CCR-15-2308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kouzarides T. Chromatin modifications and their function. Cell. 2007;128(4):693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 67.Tateishi K, et al. Extreme Vulnerability of IDH1 Mutant Cancers to NAD+ Depletion. Cancer Cell. 2015;28(6):773–784. doi: 10.1016/j.ccell.2015.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Tateishi K, et al. Myc-Driven Glycolysis Is a Therapeutic Target in Glioblastoma. Clin Cancer Res. 2016;22(17):4452–4465. doi: 10.1158/1078-0432.CCR-15-2274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Gujar AD, et al. An NAD+-dependent transcriptional program governs self-renewal and radiation resistance in glioblastoma. Proc Natl Acad Sci USA. 2016;113(51):E8247–E8256. doi: 10.1073/pnas.1610921114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Imai S, Guarente L. NAD+ and sirtuins in aging and disease. Trends Cell Biol. 2014;24(8):464–471. doi: 10.1016/j.tcb.2014.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Montserrat-de la Paz S, Naranjo MC, Lopez S, Abia R, Muriana FJ, Bermudez B. Niacin and its metabolites as master regulators of macrophage activation. J Nutr Biochem. 2017;39:40–47. doi: 10.1016/j.jnutbio.2016.09.008. [DOI] [PubMed] [Google Scholar]
- 72.James EL, Lane JA, Michalek RD, Karoly ED, Parkinson EK. Replicatively senescent human fibroblasts reveal a distinct intracellular metabolic profile with alterations in NAD+ and nicotinamide metabolism. Sci Rep. 2016;6:38489. doi: 10.1038/srep38489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Schmeisser K, et al. Role of sirtuins in lifespan regulation is linked to methylation of nicotinamide. Nat Chem Biol. 2013;9(11):693–700. doi: 10.1038/nchembio.1352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hong S, et al. Nicotinamide N-methyltransferase regulates hepatic nutrient metabolism through Sirt1 protein stabilization. Nat Med. 2015;21(8):887–894. doi: 10.1038/nm.3882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Chen AC, et al. A Phase 3 Randomized Trial of Nicotinamide for Skin-Cancer Chemoprevention. N Engl J Med. 2015;373(17):1618–1626. doi: 10.1056/NEJMoa1506197. [DOI] [PubMed] [Google Scholar]
- 76.Dudman NP, Wilcken DE, Wang J, Lynch JF, Macey D, Lundberg P. Disordered methionine/homocysteine metabolism in premature vascular disease. Its occurrence, cofactor therapy, and enzymology. Arterioscler Thromb. 1993;13(9):1253–1260. doi: 10.1161/01.ATV.13.9.1253. [DOI] [PubMed] [Google Scholar]
- 77.Ko DC, et al. Functional genetic screen of human diversity reveals that a methionine salvage enzyme regulates inflammatory cell death. Proc Natl Acad Sci USA. 2012;109(35):E2343–E2352. doi: 10.1073/pnas.1206701109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Tolstikov V, Nikolayev A, Dong S, Zhao G, Kuo MS. Metabolomics analysis of metabolic effects of nicotinamide phosphoribosyltransferase (NAMPT) inhibition on human cancer cells. PLoS ONE. 2014;9(12):e114019. doi: 10.1371/journal.pone.0114019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Sampath D, Zabka TS, Misner DL, O’Brien T, Dragovich PS. Inhibition of nicotinamide phosphoribosyltransferase (NAMPT) as a therapeutic strategy in cancer. Pharmacol Ther. 2015;151:16–31. doi: 10.1016/j.pharmthera.2015.02.004. [DOI] [PubMed] [Google Scholar]
- 80.Quinn JA, et al. Phase II trial of temozolomide plus o6-benzylguanine in adults with recurrent, temozolomide-resistant malignant glioma. J Clin Oncol. 2009;27(8):1262–1267. doi: 10.1200/JCO.2008.18.8417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Gilbert MR, et al. Dose-dense temozolomide for newly diagnosed glioblastoma: a randomized phase III clinical trial. J Clin Oncol. 2013;31(32):4085–4091. doi: 10.1200/JCO.2013.49.6968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Tahiliani M, et al. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science. 2009;324(5929):930–935. doi: 10.1126/science.1170116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Lara-Astiaso D, et al. Immunogenetics. Chromatin state dynamics during blood formation. Science. 2014;345(6199):943–949. doi: 10.1126/science.1256271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Grant CE, Bailey TL, Noble WS. FIMO: scanning for occurrences of a given motif. Bioinformatics. 2011;27(7):1017–1018. doi: 10.1093/bioinformatics/btr064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Frith MC, Saunders NF, Kobe B, Bailey TL. Discovering sequence motifs with arbitrary insertions and deletions. PLoS Comput Biol. 2008;4(4):e1000071. doi: 10.1371/journal.pcbi.1000071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Gupta S, Stamatoyannopoulos JA, Bailey TL, Noble WS. Quantifying similarity between motifs. Genome Biol. 2007;8(2):R24. doi: 10.1186/gb-2007-8-2-r24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Cerami E, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2(5):401–404. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Gao J, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6(269):pl1. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Subramanian A, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102(43):15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Barbie DA, et al. Systematic RNA interference reveals that oncogenic KRAS-driven cancers require TBK1. Nature. 2009;462(7269):108–112. doi: 10.1038/nature08460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.ENCODE Project Consortium. A user’s guide to the encyclopedia of DNA elements (ENCODE) PLoS Biol. 2011;9(4):e1001046. doi: 10.1371/journal.pbio.1001046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.ENCODE Project Consortium. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489(7414):57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Iannetti A, et al. Regulation of p53 and Rb links the alternative NF-κB pathway to EZH2 expression and cell senescence. PLoS Genet. 2014;10(9):e1004642. doi: 10.1371/journal.pgen.1004642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Zhao B, et al. The NF-κB genomic landscape in lymphoblastoid B cells. Cell Rep. 2014;8(5):1595–1606. doi: 10.1016/j.celrep.2014.07.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.