Abstract
Natural killer (NK) cells can be divided into phenotypic subsets based on expression of receptors that bind self-MHC-I molecules, a concept termed licensing or education. Here we show NK cell subsets with different migratory, effector, and immunoregulatory functions in dendritic cell and antigen (ag)-specific CD8+ T cell responses during influenza and murine cytomegalovirus infections. Shortly after infection, unlicensed NK cells localized in draining lymph nodes and produced GM-CSF, which correlated with the expansion and activation of dendritic cells, and resulted in greater and sustained ag-specific T cell responses. In contrast, licensed NK cells preferentially migrated to infected tissues and produced IFN-γ. Importantly, human NK cell subsets exhibited similar phenotypic characteristics. Collectively, our studies demonstrate a critical demarcation between the functions of licensed and unlicensed NK cell subsets, with the former functioning as the classical effector subset and the latter as the stimulator of adaptive immunity helping to prime immune responses.
NK cells can be divided into subsets with different effector roles during viral infection, including both anti-viral and immunoregulatory roles.
Introduction
Natural killer (NK) cells are large granular lymphocytes that provide innate defense against certain virally infected and transformed cells (1). NK cells rapidly respond against target cells in the absence of prior sensitization, owing to the expression of germline-encoded inhibitory and activating receptors (2–4). In both humans and mice, NK cells survey various tissue environments and integrate signals through their extensive repertoire of receptors, which can bind both host- and pathogen-encoded ligands to ultimately determine their activity and function (5). Two families of these receptors, Ly49 in mice and killer-cell immunoglobulin-like receptors (KIRs) in humans, bind major histocompatibility complex (MHC) class I molecules and play a critical role in regulating NK cell activation and self-tolerance (6, 7). Alterations in the expression of MHC class I molecules, as is often seen in virally infected and neoplastic cells, renders cells susceptible to NK cell cytotoxicity (2, 8). Activated NK cells can either directly kill target cells or produce inflammatory cytokines, such as IFN-γ, providing an early innate immune response that helps clear and prevent viral spread. In addition to the classical role of NK cells, a series of recent studies suggest NK cells can also function as immune regulators by affecting adaptive immune responses during the course of viral infection (9–12). Whether NK cells possess a universal mechanism to regulate the adaptive immune response remains unclear. Several studies have demonstrated that NK cells can directly lyse ag-specific T cells (9, 10), resulting in suppressed adaptive immunity. Others, however, showed that NK cells indirectly heighten (11, 13) adaptive immune responses by directing dendritic cell (DC) expansion and migration (14, 15). Furthermore, it is unclear whether the various immune regulatory capabilities of NK cells result from differences in their location, the type of pathogenic challenge, the kinetics of the immune response, or the existence of distinct subpopulations of NK cells with different effector functions.
NK cells can be divided into subpopulations based on their expression of inhibitory receptors that have varying binding affinities to self-MHC-I molecules. Those expressing inhibitory receptors capable of binding to self-MHC class I molecules are deemed licensed (16), educated (17), or functional NK cells. In contrast, the unlicensed population of NK cells fail to express inhibitory receptors or express inhibitory receptors that have low binding affinities to host MHC class I molecules and are thus hyporesponsive to targets in terms of cytotoxicity and cytokine production (16, 18, 19). Recent reports have further characterized functional and intrinsic differences between the licensed and unlicensed NK cells in terms of target binding and recognition (20), antiviral responses in both mice (21–23) and humans (18, 24), and retention of DCs (12). There have been conflicting reports on the role of licensing in antiviral responses. A recent study suggested that unlicensed NK cells are the predominant anti–murine cytomegalovirus (anti-MCMV) NK subset (21). Conversely, others suggested that the licensed NK cell subset exhibits greater anti-MCMV activity following hematopoietic stem cell transplantation (HSCT) or after CD25 depletion (22, 23). Although it is well established that NK cells play a critical role in direct cytotoxic antiviral immune responses, only recently have studies begun to explore and expand on the immune regulatory role of NK cells. Furthermore, it is still unclear what the exact contributions of the licensed and unlicensed NK cell subsets are during viral infection, and whether these differences can be attributed to direct or indirect antiviral responses.
To address these questions, we assessed licensed and unlicensed NK cell subsets in multiple strains of mice (with disparate MHC haplotypes) and in the context of 2 pathogens: MCMV and influenza virus. We hypothesized that licensing determines the specific role of NK cell subpopulations during the course of viral infection and that this may explain the conflicting results regarding differential mechanisms of immune regulation and antiviral capabilities. In this study, we report that self-recognition and inhibitory receptor expression regulates NK cell functionality throughout the course of infection and that the differences in NK cell subset function can, at least partially, be attributed to differences in tissue localization and inherent differences in effector functions between these subsets. Shortly after infection, licensed NK cells served as early innate effectors by producing IFN-γ in inflamed parenchymal tissues and mediated direct antiviral responses. Conversely, the unlicensed NK cell subset localized in the draining lymph nodes (DLNs) and helped promote DC activation and expansion, resulting in sustained ag-specific CD8+ T cell responses. Collectively, these results clearly demonstrated that licensing delineates NK cell subsets into effector and helper subpopulations based on their unique phenotypic and functional characteristics.
Results
Differential localization of unlicensed and licensed NK cells following viral infection.
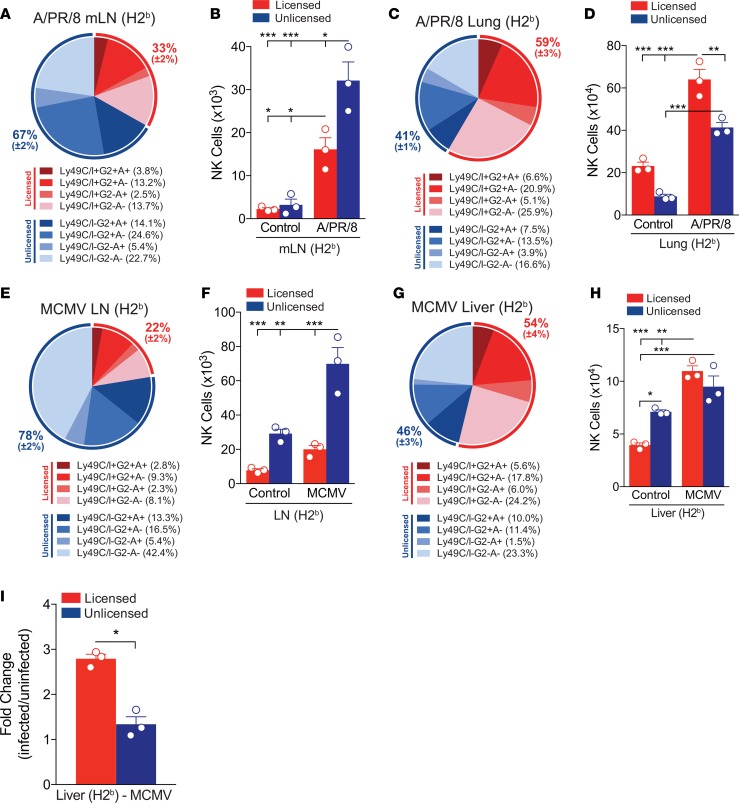
Under steady-state conditions, NK cells are distributed among both hematopoietic and nonhematopoietic tissues, with the highest frequencies being found in the spleen and nonlymphoid organs such as the liver and lung (25), and very low frequencies (0.2%–0.5%) being found in LNs (26). However, following immunization or infection, NK cells can enter DLNs (26–29). To assess the distribution and kinetics of the NK cell response to different viral infections, we challenged C57BL/6 (H-2b) and B10.D2 (H-2d) mice with either influenza strain A/Puerto Rico/8/34 (referred to as APR8) or MCMV and monitored changes in NK cell number in both the target organs (lung and liver for APR8 and MCMV, respectively) and LNs throughout the course of infection (Supplemental Figure 1, A–E; supplemental material available online with this article; https://doi.org/10.1172/jci.insight.87032DS1). As early as 1 day post-infection (d.p.i.), and peaking between 5 and 7 d.p.i., there was a preferential recruitment of NK cells to the DLNs (mediastinal LNs [mLNs] for APR8 and see Methods section for MCMV) (Supplemental Figure 1, A, B, and D). By 5 d.p.i., there was a statistically significant 10-fold increase in the number of NK cells within the mLNs, with no changes observed in control nondraining LNs (ndLNs) after APR8 challenge (Supplemental Figure 1, A and B). Interestingly, while the NK cells in the LNs of noninfected mice predominantly had an immature (CD27+CD11b–) phenotype, similar to what has been reported by others (30, 31), those in the DLNs at 5 d.p.i. had a more mature (CD27+CD11b+) phenotype (Supplemental Figure 2A). Additionally, there was a 3- to 4-fold increase in total NK cell numbers in the lung and liver of APR8- or MCMV-challenged mice, respectively (Supplemental Figure 1, C and E).
In both mice and humans, subsets of NK cells exhibit distinct tissue localization, effector functions, and receptor expression associated with LN trafficking (32–34). NK cells expressing Ly49C and/or Ly49I (referred to here as Ly49C/I) bind most strongly to H2Kb and are licensed in C57BL/6 (H2b) mice (16), whereas they are unlicensed in B10.D2 (H2d) mice. Conversely, NK cells that express Ly49G2 or Ly49A bind most strongly to H2Dd, are unlicensed in C57BL/6 (H2b) mice (16, 35), and licensed in B10.D2 (H2d) mice. In contrast to the well-characterized differences in cytotoxicity and IFN-γ production, it is unclear whether these NK cell subsets also exhibit differences in trafficking and tissue distribution, as well as other cytokine and immunoregulatory responses after infection. To investigate whether licensed and unlicensed NK cell subsets, based on the Ly49 receptor expression on distinct NK cells (Supplemental Figure 3), exhibit distinct migratory capacities, we utilized mice differing in MHC-haplotype expression [C57BL/6 (H2b) and B10.D2 (H2d) mice] and quantified the frequency and total number of each subset at the peak of the NK cell response to both APR8 and MCMV in various tissues. In both C57BL/6 and B10.D2 mice, we observed that the unlicensed NK cell subset accounted for 60%–78% of the total DLN NK cell population (Figure 1, A and E, and Supplemental Figure 4A) and the absolute number of unlicensed NK cells significantly increased compared with both uninfected controls and the licensed NK subset (Figure 1, B and F, and Supplemental Figure 4B) at the peak of NK cell migration into the DLNs (5 d.p.i.). Many of the unlicensed NK cells in the DLNs at 5 d.p.i. had an immature phenotype; however, a large portion of the unlicensed NK cells consisted of the more mature (CD27+CD11b+ and CD27–CD11b+) phenotypes (Supplemental Figure 2B). Given the increases in both immature and mature NK cells in the LNs, we reasoned the increase was due to a preferential localization of unlicensed NK cells into the DLNs; however, owing to the larger majority of immature NK cells in the DLNs prior to infection (Supplemental Figure 2A), we cannot rule out in situ maturation of NK cells in the DLNs.
Figure 1. Differential distribution of unlicensed and licensed NK cells following viral infection.
(A) Frequency or (B) absolute number of licensed (CD3–NK1.1+Ly49C/I+G2+A+, Ly49C/I+G2+A–, Ly49C/I+G2-A+, or Ly49C/I+G2–A–; shades of red) and unlicensed (CD3–NK1.1+Ly49C/I–G2+A+, Ly49C/I–G2+A–, Ly49C/I–G2–A+, or Ly49C/I–G2–A–; shades of blue) NK cell subsets in the mediastinal lymph nodes (mLNs) of C57BL/6 (H2b) mice on day 5 after APR8 infection. (C) Frequency or (D) absolute number of licensed (shades of red) and unlicensed (shades of blue) NK cell subsets in the lung of C57BL/6 (H2b) mice on day 5 after APR8 infection. (E) Frequency or (F) absolute number of licensed (shades of red) and unlicensed (shades of blue) NK cell subsets in the draining lymph nodes (DLNs) of C57BL/6 (H2b) mice on day 5 after MCMV infection. (G) Frequency or (H) absolute number of licensed (shades of red) and unlicensed (shades of blue) NK cell subsets in the liver of C57BL/6 (H2b) mice on day 5 after MCMV infection. (I) Fold change of the absolute numbers of licensed or unlicensed NK cells subsets from MCMV-infected livers on day 5 over the absolute number of licensed or unlicensed NK cells subsets from uninfected livers. n = 3 mice per group, representative of 2 to 3 experiments. (B, D, F, and H) Two-way ANOVA with Tukey post-test used to compare groups. (I) Unpaired 2-tailed Student’s t test used to compare groups. *P < 0.05, **P < 0.01, ***P < 0.001.
In contrast, NK cells isolated from the lungs of APR8-challenged mice contained increased frequencies (Figure 1C and Supplemental Figure 4C) and significantly increased absolute numbers (Figure 1D and Supplemental Figure 4D) of licensed NK cells compared with both the uninfected controls and unlicensed subset. Although the frequency and absolute number of licensed and unlicensed NK cells was comparable in the liver after MCMV challenge (Figure 1, G and H), the relative increase in the number of licensed NK cells after MCMV challenge was greater compared with the unlicensed subset (Figure 1I). Thus, licensed and unlicensed NK cell subsets are differentially represented at the target and priming sites after viral infection.
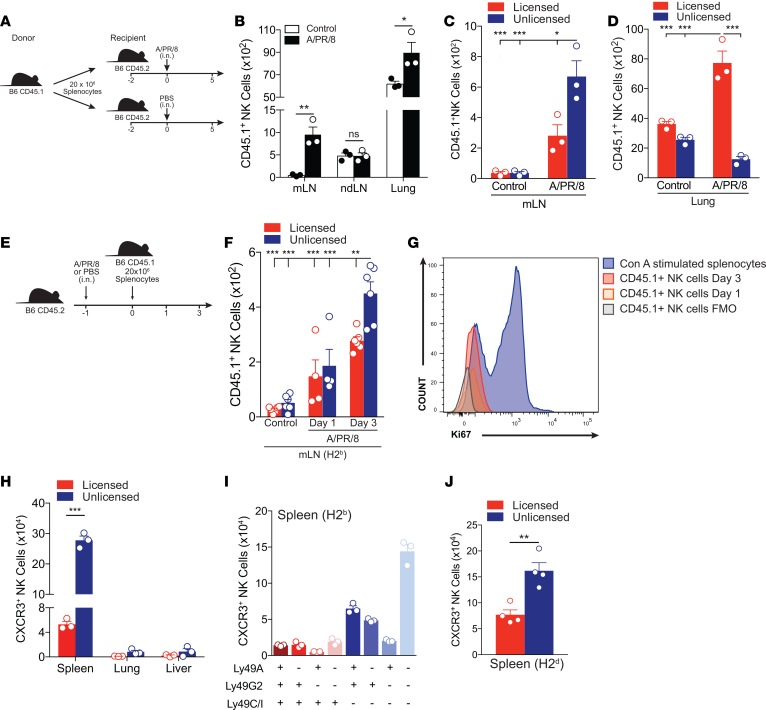
Adoptively transferred unlicensed NK cells preferentially migrate to the DLNs.
Given the low frequency of NK cells in the LNs of naive mice and the rapid kinetics of NK cell accumulation in the DLNs following infection, we hypothesized that the unlicensed NK cells were actively being recruited to the DLNs rather than gradually proliferating from resident populations in situ. To test this hypothesis, we isolated splenocytes from naive CD45.1 congenic mice and adoptively transferred 20 × 106 splenocytes to C57BL/6 (CD45.2) recipients. Two days following the adoptive transfer, we challenged the recipient mice with APR8 and on day 5 after challenge we determined the absolute numbers of donor (CD45.1) NK cells in the mLNs, ndLNs, and lungs (Figure 2, A and B). After APR8 challenge, donor NK cells preferentially trafficked to the mLNs and lungs of recipient mice (Figure 2B). We observed a 10-fold increase in the absolute number of donor NK cells in the mLNs of APR8-challenged mice compared with control, whereas no difference was observed in the ndLNs of these same groups. Moreover, the absolute number of total donor unlicensed NK cells was significantly increased compared with both the uninfected controls and licensed subset in the mLNs at 5 d.p.i. (Figure 2C). In contrast to the trafficking of the unlicensed subset to the mLNs, the licensed NK cell subset preferentially migrated to the lung following APR8 challenge compared with the unlicensed NK cells and increased 2-fold compared with control (Figure 2D). To further determine whether the increased number of donor unlicensed NK cells in the mLNs was due to trafficking and not local proliferation, we challenged recipient (CD45.2) C57BL/6 mice with APR8 and the following day adoptively transferred 20 × 106 splenocytes from naive CD45.1 congenic mice (Figure 2E). On days 1 and 3 after challenge, we determined the absolute numbers of donor (CD45.1) NK cells in the mLNs. While not significantly increased at day 1 after transfer, the absolute number of total donor unlicensed NK cells was significantly increased compared with both the uninfected controls and licensed subset in the mLNs at 3 d.p.i. (Figure 2F). Furthermore, the increase in unlicensed NK cells was due to increased trafficking and not proliferation of local NK cells, as the adoptively transferred NK cells did not express Ki-67 at either day 1 or day 3 after transfer (Figure 2G).
Figure 2. Adoptively transferred unlicensed NK cells preferentially traffic to draining lymph nodes following viral infection.
(A) Schema of adoptive transfer study where 20 × 106 splenocytes from naive CD45.1 congenic mice were i.v. adoptively transferred to naive C57BL/6 (CD45.2) recipients. Two days after adoptive transfer, recipient mice were challenged with either 100 PFU of APR8 in 40 μl of PBS or 40 μl of PBS (control) intranasally (i.n.). Five days later, the absolute number of donor (CD45.1) NK cells in the mediastinal lymph nodes (mLNs), nondraining lymph nodes (ndLNs), and lung were determined. (B) Absolute number of transferred (CD45.1+) NK cells (CD3-NK1.1+) in the mLNs, ndLNs, or lung of C57BL/6 (CD45.2) mice at 5 days postinfection (d.p.i.). (C) Absolute number of transferred (CD45.1+) licensed or unlicensed NK cell subsets in the mLNs of C57BL/6 (CD45.2) mice at 5 d.p.i. (D) Absolute number of transferred (CD45.1+) licensed or unlicensed NK cell subsets in the lung of C57BL/6 (CD45.2) mice at day 5 d.p.i. (E) Schema of adoptive transfer study where C57BL/6 (CD45.2) mice were challenged with 100 PFU of APR8 in 40 μl of PBS or 40 μl of PBS (control) i.n. One day later, 20 × 106 splenocytes from naive CD45.1 congenic mice were i.v. adoptively transferred to challenged C57BL/6 (CD45.2) recipients. One and 3 days after adoptive transfer, the absolute number of donor (CD45.1) NK cells in the mLNs was determined. (F) Absolute number of transferred (CD45.1+) licensed or unlicensed NK cell subsets in the mLN of C57BL/6 (CD45.2) mice at 1 and 3 d.p.i. (G) Histograms showing the level of Ki-67 of transferred (CD45.1+) NK cells in the mLNs of C57BL/6 (CD45.2) mice at 1 and 3 d.p.i. Three-day concanavalin A (Con A)–stimulated splenocytes were included as a positive control. (H) Absolute number of licensed or unlicensed NK cell subsets that express the chemokine receptor CXCR3 in the spleen, lung, or liver of naive C57BL/6 (H2b) mice. (I) Absolute number of NK cell subsets (CD3–NK1.1+Ly49C/I+G2+A+, Ly49C/I+G2+A–, Ly49C/I+G2–A+, Ly49C/I+G2–A–, Ly49C/I–G2+A+, Ly49C/I–G2+A–, Ly49C/I–G2–A+, or Ly49C/I–G2–A–) that express CXCR3 in the spleen of naive C57BL/6 mice. (J) Absolute number of licensed or unlicensed NK cell subsets that express CXCR3 in the spleen of naive B10.D2 (H2d) mice. n = 3–6 mice per group, representative of 2 to 3 experiments. (C, D, and F) Two-way ANOVA with Tukey post-test used to compare groups. (B, H, and J) Unpaired 2-tailed Student’s t test used to compare groups. *P < 0.05, **P < 0.01, ***P < 0.001.
Several recent studies have implicated the chemokine receptor CXCR3 as a key mediator of NK cell trafficking to the DLNs (27, 28, 36). To gain further insight into the mechanism(s) underlying the preferential trafficking of the unlicensed NK subset to the DLNs, we analyzed the expression of CXCR3 on both the licensed and unlicensed NK subsets in various tissues of uninfected C57BL/6 (Figure 2, H and I) and B10.D2 mice (Figure 2J). We observed a significantly greater number of CXCR3-expressing unlicensed NK cells compared with licensed NK cells in the spleens of naive mice, which primarily consisted of unlicensed NK cells with a Ly49G2+A+C/I– or Ly49G2–A–C/I– phenotype (Figure 2I). However, further congenic adoptive transfer studies using CXCR3-knockout mice showed that this was not the dominant mechanism mediating unlicensed NK cell trafficking to LNs (data not shown). Despite CXCR3 being found predominantly on unlicensed NK cells compared with licensed, there is still a greater frequency of unlicensed NK cells that do not express CXCR3, leading us to believe that additional chemokine receptors and adhesion molecules likely play a role in unlicensed LN homing in our viral models. Collectively, these findings indicate that NK cell subsets exhibit differential trafficking patterns shortly after viral infection, and can be delineated based on licensing patterns.
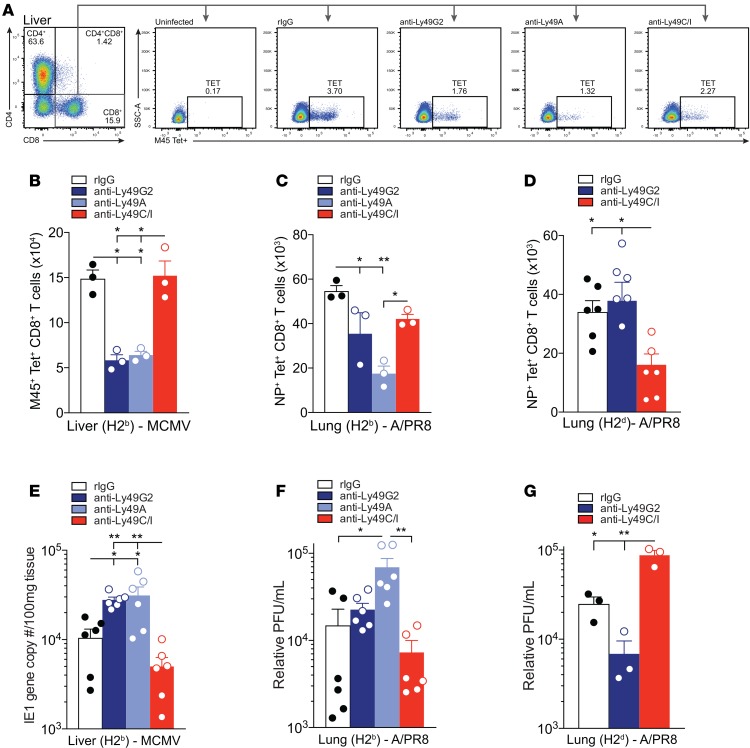
Depletion of unlicensed NK cells results in diminished ag-specific CD8+ T cell numbers and viral control.
Several recent studies suggest that in addition to playing a major role in direct antiviral defense, NK cells may actually play a more critical role in orchestrating an effective immune response during viral infection (11, 37, 38). Whether NK cells play a predominant role in enhancing (11) or suppressing (39) the adaptive immune response during viral infection remains controversial, as studies utilizing different MCMV-resistant or -susceptible strains of mice have provided conflicting results. However, the majority of these studies focus on the role of NK cell–activating receptors on viral control, DC-NK interactions, and the kinetics of the early CD8+ T cell response. To assess whether these discrepancies may partially be explained by differences in the ability of the licensed or unlicensed NK cell subsets to either promote or restrict early ag-specific CD8+ T cell responses, we used monoclonal antibodies to deplete C57BL/6 (Supplemental Figure 5) or B10.D2 (Supplemental Figure 6) mice of their licensed (Ly49C/I in C57BL/6 and Ly49G2 in B10.D2) or unlicensed (Ly49G2 and Ly49A in C57BL/6 or Ly49C/I in B10.D2) NK cell subsets 2 days before viral challenge and evaluated T cell responses by flow cytometry (Figure 3A). Interestingly, 7 days after challenging with MCMV, we saw a significant decrease in the ag-specific CD8+ T cell response in C57BL/6 mice depleted of their unlicensed NK cell subsets compared with control (rIgG) (Figure 3B). In contrast, we did not observe any difference in ag-specific CD8+ T cell numbers in mice whose licensed NK cells were depleted (Figure 3B). Importantly, the ag-specific CD8+ T cell response 7 days after APR8 challenge was significantly reduced in both C57BL/6 (Figure 3C) and B10.D2 (Figure 3D) mice depleted of their unlicensed NK cell subsets, indicating a general antiviral effect of the subsets.
Figure 3. Depletion of unlicensed NK cells results in diminished antigen-specific CD8+ T cell numbers and viral control.
(A) Representative MHC-I tetramer (Tet) gating strategy and staining on CD3+CD8+Tet+ T cells for each group. Antibody depletions were performed 2 days prior to infection. (B) Absolute number of M45+CD8+ Tet+ T cells in the liver 7 days after MCMV infection in C57BL/6 (H2b) mice. (C and D) Absolute number of NP+CD8+Tet+ T cells in the lung after 7 days of APR8 infection in C57BL/6 (C) or B10.D2 (D) mice. (E) Viral loads in livers of C57BL/6 mice at day 7 after MCMV infection determined by qPCR of the IE1 gene copy number per 100 mg of tissue. (F and G) Relative PFU/ml of APR8 determined by qPCR at day 7 after infection in the lungs of C57BL/6 (F) or B10.D2 (G) mice. n = 3 or 6 mice per group, representative of 2 to 3 experiments. One-way ANOVA with Tukey post-test used to compare groups. *P < 0.05, **P < 0.01.
Because of the observed decrease in ag-specific CD8+ T cells in mice depleted of their unlicensed NK subsets and the role of CD8+ T cells in providing antiviral immunity (38, 40), we next examined whether these mice were less effective at eliminating virus in target tissues. Depletion of the unlicensed NK cell subsets (anti-Ly49G2 and anti-Ly49A in C57BL/6) resulted in liver viral titers ~2.5-fold higher than in undepleted controls (Figure 3E), consistent with results from previous studies (21). Similarly, depletion of the unlicensed NK cell subset (anti-Ly49A in C57BL/6 and anti-Ly49C/I in B10.D2) resulted in lung viral titers ~4.5-fold higher in C57BL/6 (Figure 3F) and ~3.5-fold higher in B10.D2 (Figure 3G) compared with undepleted controls. This increase in titer was similar to that in T cell depletion studies (data not shown). These findings suggest that unlicensed NK cells function to promote sustained ag-specific CD8+ T cell responses that impact viral resistance.
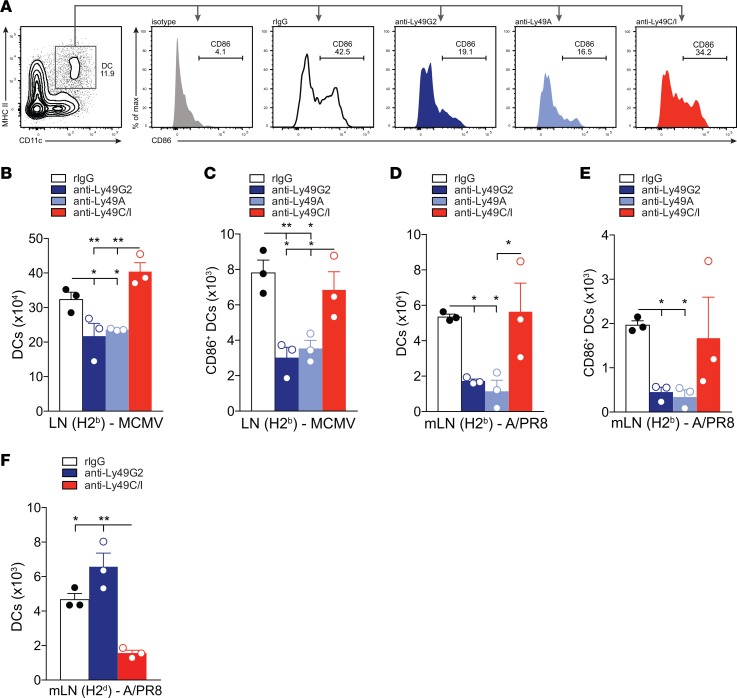
Unlicensed NK cell presence is correlated with increased DC responses in DLNs.
Because of the important role of DCs in priming T cells in the DLNs (28, 41) and our findings showing that depletion of the unlicensed NK cell subset reduces the number of ag-specific CD8+ T cells, we next investigated whether depletion of unlicensed or licensed NK subsets had a negative impact on DCs at the site of T cell priming. Depletion of the unlicensed NK cell subset resulted in significantly diminished total numbers of DLN DCs in C57BL/6 mice challenged with MCMV compared with undepleted controls (Figure 4B). Further, this phenomenon was maintained with mLN DCs when both C57BL/6 and B10.D2 mice were challenged with APR8 (Figure 4, D and F). Additionally, the number of activated DLN DCs was significantly reduced in mice depleted of their unlicensed NK cell subsets and challenged with either MCMV (Figure 4C) or APR8 (Figure 4, A and E). Collectively, these results suggest that the unlicensed NK cell subsets enhance the adaptive immune response against viral infection by augmenting DC activation and numbers, and this is correlated with expansion of antigen-specific CD8+ T cells in the DLNs.
Figure 4. Depletion of unlicensed NK cells results in diminished DC number and maturation/activation.
(A) Representative DC gating strategy and staining on CD45+CD19–CD11c+MHCII+ DCs and CD45+CD19–CD11c+MHCII+86+ activated DCs in the draining lymph nodes (DLNs) for each group. Antibody depletions were performed 2 days prior to infection. (B–F) Absolute number of CD45+CD19–CD11c+MHCII+ DCs (B, D, and F) or CD45+CD19–CD11c+MHCI–I+CD86+ activated DCs (C and E) in the DLNs of C57BL/6 mice at day 7 after MCMV infection (B and C), in the mediastinal lymph nodes (mLNs) of C57BL/6 at day 5 after APR8 infection (D and E), or in the mLNs of B10.D2 at day 7 after APR8 (F). n = 3 mice per group, representative of 2 to 3 experiments. One-way ANOVA with Tukey post-test used to compare groups. *P < 0.05, **P < 0.01.
Licensed and unlicensed NK cell subset cytokine profiles correlate with their effector functions and tissue distribution.
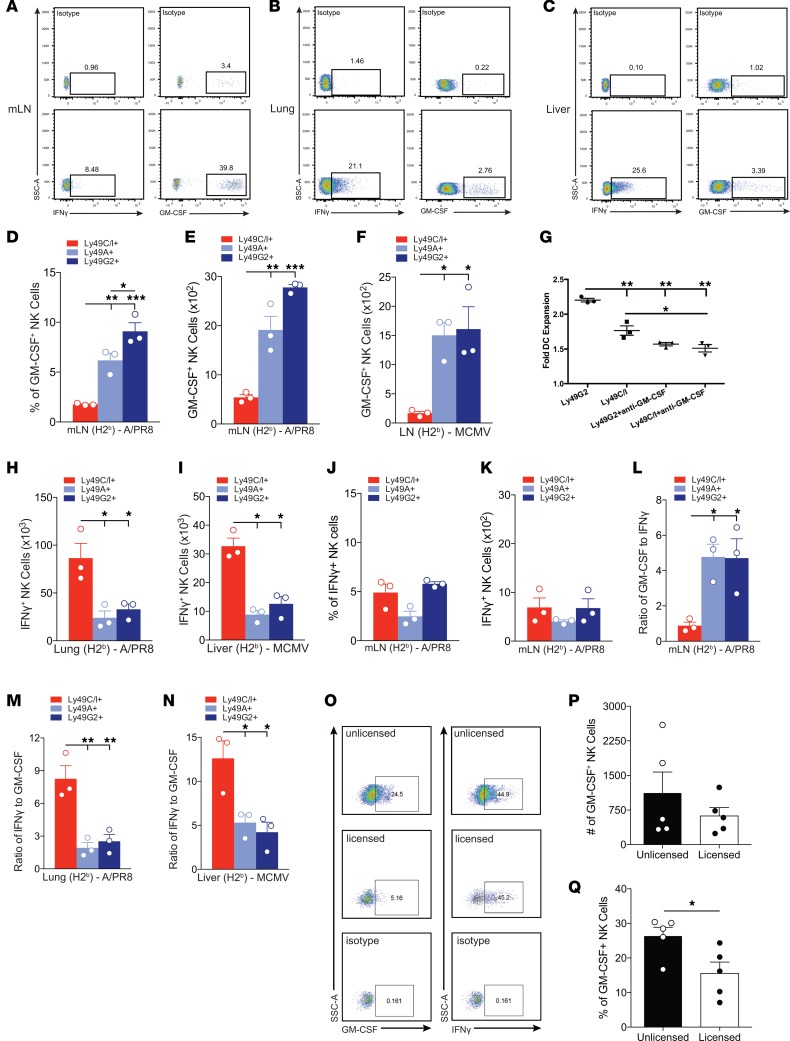
To assess the effector mechanisms driving the differential function of licensed and unlicensed NK cells, cytokine production by these NK cell subsets was examined following MCMV and APR8 challenge in both target (liver and lung, respectively) and priming tissues (DLNs) (Figure 5, A–C). At 5 d.p.i., a time point corresponding with significantly increased numbers of NK cells in the DLNs, lungs, and liver (Supplemental Figure 1), we observed significantly greater total numbers of unlicensed NK cells producing GM-CSF, compared with licensed NK cells in the DLNs of C57BL/6 mice challenged with either APR8 (Figure 5, D and E) or MCMV (Figure 5F). Similar results were obtained using the B10.D2 strain of mouse (Supplemental Figure 7A).
Figure 5. Cytokine profiles of licensed and unlicensed NK cell subsets.
(A–C) Representative staining of NK cells that are GM-CSF+ or IFN-γ+ from mediastinal lymph nodes (mLNs) (A) or lung (B) after APR8 infection or liver (C) after MCMV infection for 5 days. (D) Frequency and (E) absolute number of either CD3–NK1.1+Ly49G2+, Ly49A+, or Ly49C/I+ among GM-CSF+ NK cells at day 5 after APR8 infection in mLNs of C57BL/6, or draining lymph nodes (DLNs) 5 days after MCMV infection (F). (G) DC expansion following in vitro NK-DC coculture with neutralizing antibody against GM-CSF. Numbers of DCs were determined through flow cytometry (CD19–CD11c+MHCII+) and compared with cultures lacking NK cells to determine the fold expansion that occurred. (H and I) Absolute number of NK cells that are IFN-γ+ and either CD3–NK1.1+Ly49G2+, Ly49A+, or Ly49C/I+ at day 5 after APR8 infection (H) or MCMV infection (I) in lung (H) or liver (I) of C57BL/6 mice. (J) Frequency and (K) absolute number of NK cells that are IFN-γ+ and either CD3–NK1.1+Ly49G2+, Ly49A+, or Ly49C/I+ at day 5 after APR8 infection in mLNs of C57BL/6. (L) Ratio of GM-CSF to IFN-γ production by NK subsets in mLNs of day 5 APR8-infected C57BL/6 mice. (M and N) Ratio of IFN-γ to GM-CSF production by NK subsets in lung of day 5 APR8-infected C57BL/6 mice (M) or liver of day 5 MCMV-infected C57BL/6 mice (N). (O) Representative staining after single-cell exclusion gating for the evaluation of intracellular GM-CSF and IFN-γ production by human NK cell subsets. NK cells were identified as CD3–CD56+ cells. Intracellular IFN-γ, and GM-CSF production by NK cells exclusively expressing a single KIR was detected by excluding cells expressing other KIRs. HLA class I genotyping and KIR ligand assignment were performed for each sample to distinguish licensed versus unlicensed single-positive KIR (spKIR) NK cells. (P) Absolute number of unlicensed and licensed NK cells expressing GM-CSF from 5 distinct individuals lacking the HLA-C2 ligand for KIR2DL1 (HLA-C1/C1, Bw4/Bw6). Unlicensed NK cells were operationally defined as NKG2A–spKIR2DL1+ or NKG2A–KIR–; licensed NK cells expressed spKIR2DL3 and/or spKIR3DL1. (Q) Aggregate percentage of NK cell populations that are GM-CSF+. n = 3 mice per group, representative of 2 to 3 experiments. One-way ANOVA with Tukey post-test used to compare groups. Human data: Experiments were performed in duplicate; 1–3 aggregated single-positive NK cell subsets per unlicensed or licensed group. Five distinct individuals were used to obtain NK cells from peripheral blood mononuclear cells. *P < 0.05, **P < 0.01, ***P < 0.001.
To determine whether unlicensed NK cell–derived GM-CSF plays a role in DC expansion, we utilized an in vitro system involving coculturing bone marrow–derived DCs with purified NK cells enriched for either the licensed (96% ± 0.6% purity) or unlicensed (95% ± 1% purity) subset. The unlicensed subset of NK cells demonstrated significantly greater ability to expand DCs after 72 hours of coculture compared with the licensed subset, which was abrogated by GM-CSF neutralization (Figure 5G). Taken together, these data suggest that unlicensed NK cells act as helper cells by aiding in DC expansion, in part through GM-CSF production.
Furthermore, we observed significantly greater total numbers of licensed NK cells producing IFN-γ, compared with unlicensed NK cells, in the lung (Figure 5H and Supplemental Figure 7B) and liver (Figure 5I) of mice challenged with either APR8 or MCMV, respectively. Recent studies have suggested that NK cell recruitment to the DLNs may enhance TH1 polarization, owing to the ability of NK cells to produce high levels of IFN-γ (28, 42). To assess whether the licensed or unlicensed NK cell subsets in the DLNs also produced IFN-γ in our viral models, we quantified IFN-γ production by DLN NK cells. Although we were able to detect IFN-γ production by ~8% of our total NK cell population in the mLNs at this time point (Figure 5A), a far greater frequency and number of NK cells produced GM-CSF (~40%) (Figure 5A). The total number and frequency of NK subsets producing IFN-γ in the mLNs (Figure 5, J and K) was also lower than subsets producing GM-CSF (Figure 5, D and E). Additionally, when we compared the ratio of GM-CSF to IFN-γ production by NK cell subsets, the unlicensed NK cells produced significantly greater GM-CSF than IFN-γ in the DLNs following viral challenge (Figure 5L). The opposite trend was observed in parenchymal tissues following challenge with APR8 (Figure 5M) or MCMV (Figure 5N), with preferential production of IFN-γ over GM-CSF by licensed NK cell subsets. Thus, our results suggest that in addition to the licensed and unlicensed NK cell subsets differentially trafficking to target and priming sites, there is also a difference in the cytokine production by these subsets following viral infection.
Human NK cell subsets exhibit cytokine profiles similar to those of murine NK cell subsets.
In order to determine whether human NK cell subsets can also be delineated based on licensing, cytokine production between licensed (single KIR+ NK cells in subject with cognate HLA class I ligand present) and unlicensed (single KIR+ NK cells in subject lacking cognate HLA class I ligand or KIR–NKG2A– NK cells) human NK cells from the peripheral blood were examined. Licensing was determined through MHC-I haplotype genotyping and flow cytometry to identify NK cell populations with specific KIR/KIR-ligand binding capability. Unlicensed human NK cell subsets demonstrated increased production of GM-CSF compared with the licensed human NK cell subset (Figure 5, O–Q, and Supplemental Figure 7, C and D). Furthermore, the ratio of GM-CSF to IFN-γ production between the 2 subsets indicated that the unlicensed subset predominantly produced GM-CSF, while licensed human NK cells produce greater amounts of IFN-γ (Supplemental Figure 7C). Additional phenotypic characterization between the unlicensed and licensed human NK cell subsets to include NKG2A expression did not alter the differences in frequency between the unlicensed and licensed NK cell subsets producing GM-CSF (Supplemental Figure 7D). Thus, similar to the results from our mouse models, human NK cells also show differential cytokine production expression based on NK cell subset licensing.
Naive unlicensed and licensed NK cell subsets have unique transcriptional signatures.
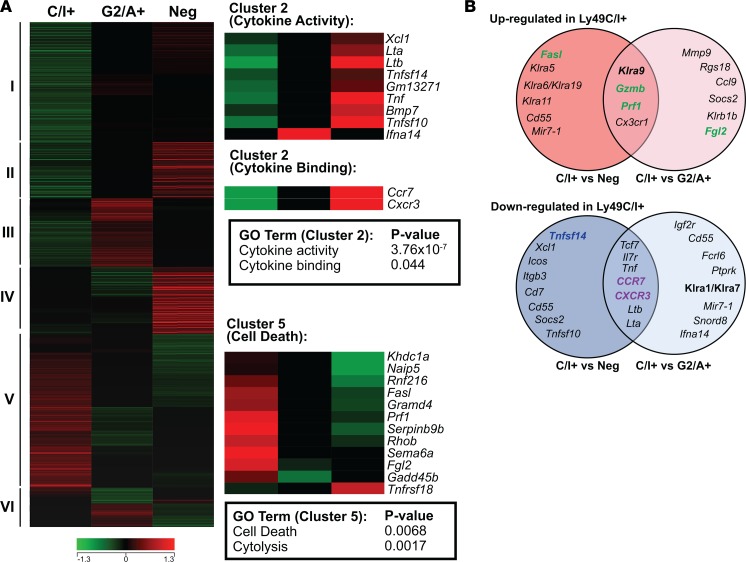
To confirm the relevance of the unique phenotypic and functional characteristics between licensed and unlicensed NK cells, we compared the gene expression profiles using whole-transcript microarrays of 2 FACS-sorted unlicensed splenic NK populations from C57BL/6 mice: CD3–CD122+Ly49G2+Ly49A+Ly49C/I– (Ly49G2/A+) and CD122+Ly49G2–A–Ly49C/I– (Ly49-negative), and a licensed population: CD3–CD122+Ly49G2–A–Ly49C/I+ (Ly49C/I+). As an initial and exploratory measure of NK subset identity, we performed hierarchical clustering of differentially expressed genes having a 1.25-fold or greater change between the different NK cell subsets, which resulted in 6 major clusters of coexpressed genes (Figure 6A). For each cluster, we performed functional gene-enrichment analysis using DAVID tools to reveal enrichment for gene ontology and pathway annotations based on gene-gene functional relationships. Within cluster V, “cell death” and “cytolysis” were by far the most statistically significant terms associated with the licensed (Ly49C/I+) NK subset signature, with the majority of the genes having known effector functions associated with cytotoxicity (Figure 6B). Conversely, within cluster II, “cytokine activity, “cytokine binding,” and “tumor necrosis factor receptor binding” were the most statistically significant terms associated with the unlicensed (Ly49-negative and Ly49G2/A+) NK cell subset signature (Figure 6B). Collectively, and in agreement with our in vivo murine models, these results support the notion that licensing can be used to delineate NK cell subsets into effector and helper subpopulations based on their unique phenotypic and functional characteristics.
Figure 6. Microarray gene expression profiling of NK cell subsets.
Total RNA was isolated from pools of licensed (CD3–CD122+Ly49G2–A–Ly49C/I+) and unlicensed (CD3–CD122+Ly49G2+Ly49A+Ly49C/I– or CD3–CD122+Ly49G2–Ly49A–Ly49C/I–) NK cell populations isolated by FACS of splenocytes from 30 naive C57BL/6 mice. Gene expression profiling was performed with Affymetrix mouse Gene ST microarrays as described in the Methods. Genes differentially expressed between the 3 NK cell subsets (Ly49-negative, Ly49-C/I+, and Ly49-G2/A+) were determined and subsequently utilized in downstream hierarchical clustering and gene functional classification. (A) Differentially expressed genes between the NK cell subsets were identified and clustered according to similarities in their expression patterns. The different clusters are labeled I to VI (left of heatmap). Heatmaps also depict the relative expression of genes comprising significantly enriched functional genes (i.e., gene ontology terms) within the licensed (cluster V) and unlicensed (cluster II) NK subset expression signatures (right-hand side of panel). (B) Overlap of genes that are commonly upregulated (top Venn diagram) in the Ly49C/I+ NK cell subset compared with either the Ly49-negative (left circle) or Ly49G2/A+ (right circle), or that are downregulated (bottom Venn diagram) in the Ly49C/I+ NK cell subset compared with either the Ly49-negative (left circle) or Ly49G2/A+ (right circle). Color indicates function of molecule encoded: green, effector molecule; purple, trafficking molecule; blue, secreted molecule.
Discussion
We have shown here that NK cell subsets exist based on the concept of self-recognition and licensing, which allows for differences in Ly49 receptor expression to be used as a broad means to phenotypically distinguish between licensed and unlicensed NK cell subsets. Moreover, this phenotypic classification can be used to further delineate the unlicensed and licensed NK cell subsets based on their unique functional characteristics, where the licensed NK cell subset functions as the effector subpopulation and the unlicensed NK cell subset functions as the helper/immunoregulatory subpopulation during viral infection. Utilizing mice that differ in their MHC-haplotype expression (i.e., H2b-expressing C57BL/6 and H2d-expressing B10.D2 mice), we have shown that the licensed and unlicensed subsets have unique trafficking, tissue distribution, and effector and immunoregulatory functions during viral infection that are conserved across mouse strains.
We found that unlicensed NK cells preferentially localize in the DLNs early after infection, whereas licensed NK cells localize in viral target tissues. This finding was further supported by adoptive transfer experiments showing a preferential trafficking of unlicensed NK cells to DLNs, rather than in situ proliferation of NK cells already present in the LNs. Similar to published studies supporting the role of CXCR3 in mediating NK cell trafficking to the DLNs (27, 28, 36), we found that naive mice have high frequencies and total numbers of CXCR3-expressing unlicensed NK cells in their spleens. Although this was not found to play a dominant role in the preferential trafficking observed, it remains a useful biomarker to differentiate NK subsets. Additionally, we found that the unlicensed NK cells found in the DLNs following infection had both mature and immature phenotypes. However, owing to a larger proportion of immature NK cells being present in the DLN prior to infection, we cannot rule out the potential of in situ NK maturation, or the role that maturation status of NK cells has in driving this LN localization. The licensed population was also found to be increased in DLNs following infection, although to a much lower extent that the unlicensed NK cell. Nonetheless, future studies will have to examine what role this subset plays in the LNs and whether it can compensate for a lack of unlicensed NK cells.
Although previous studies have suggested that the primary role of NK cell trafficking to the DLNs is to provide increased levels of IFN-γ (28, 42) and enhance TH1 polarization, these studies used different infectious disease models — either parasitic infections (using Leishmania major) (42) or LPS-stimulated mature DCs to induce NK cell trafficking — and did not exclude potential contributions from the other cytokines known to be produced by NK cells. Although we were also able to detect slightly elevated levels of IFN-γ production in the NK cells that trafficked to the DLNs in our models, GM-CSF was elevated to a much greater extent. In support of our findings that GM-CSF production may enhance DC maturation and activation, which subsequently helps prime ag-specific CD8+ T cell functions, a recent study suggests that in addition to the well-established role of GM-CSF in promoting hematopoietic stem cell development into immature DCs, GM-CSF can also function to support DC differentiation, survival, proliferation, and preferentially promotes MHC class I but not MHC class II expression, indicating the pleiotropic roles of this cytokine (43). Another interesting finding from our microarray studies was the increased enrichment of “tumor necrosis factor receptor binding” genes associated with the unlicensed (Ly49-negative and Ly49G2/A+) NK cell subset signature. A recent study looking to determine the mechanisms governing NK-DC interactions in humans found that the gene expression profile for licensed (defined by their increased expression of CD107 following coculture with K562 tumor cells) human NK cells was enriched for genes associated with “cytokine, chemokines, and TNFSF” (44). Many of the enriched genes in their licensed NK subset (most importantly, the TNFSF14 gene) overlapped with those expressed by our unlicensed NK cell subsets. In their study, the authors conclude that NK cells aid in DC maturation via TNSFSF14. Interestingly, we observed significant enrichment of the TNFSF14 gene in our unlicensed NK cell subset. Although this study and our study conflict with regard to which subset of NK cells aid in DC maturation, one possible explanation is the use of different cell-surface markers to define licensed and unlicensed NK cells, as there is less consensus on which markers should be used to phenotypically characterize human NK cell subsets.
Although we clearly demonstrated that the unlicensed NK cell subsets in the DLNs produced increased GM-CSF, while the licensed NK cells in the parenchymal tissues (liver and lung) produced elevated levels of IFN-γ, these are a matter of degree and we cannot rule out the possibility that the differences in effector function and cytokine production are the result of tissue distribution rather than inherent differences between licensed and unlicensed NK cell subsets. Based on the unique transcriptional signatures identified by our microarray studies, especially our findings that the unlicensed NK cell subset signature was enriched for genes involved with cytokine production and chemokine receptor expression and the licensed NK cell subset signature was characterized by genes enriched for effector cytotoxic function, this suggests that even within the same tissue the unlicensed and licensed NK cells have unique effector functions.
Several recent studies have begun to shed light on the involvement of NK cells in the interplay between the innate and adaptive immune responses, and suggest that NK cells may play a primary role in orchestrating adaptive immune responses during viral infection (11, 39). Although it remains to be determined whether NK cells play a predominant role in enhancing or suppressing the adaptive immune response, our studies support the notion that different subpopulations of NK cells may play opposing roles throughout the course of viral infection. The majority of the studies looking to decipher the mechanism behind NK modulation of the adaptive immune response have focused on the ability of NK cell–activating receptors to either directly or indirectly regulate T cell responses (9, 10, 39). Based on the findings from our studies, a more comprehensive assessment of both activating and inhibitory receptors expressed on NK cells may be warranted to better delineate the beneficial or detrimental effects of distinct NK cell populations on adaptive immune responses. Our findings suggest that, given the preferential trafficking of the unlicensed NK cells to the DLNs, they may function primarily to help prime the adaptive immune response. In support of this, we found that depletion of the unlicensed NK cell subset resulted in drastically reduced ag-specific CD8+ T cell numbers and increased viral loads in target tissues.
Similar to findings in our murine models, human NK cells showed similar patterns of cytokine production and chemokine receptor expression based on licensing and self-recognition. Additionally, others have shown that subpopulations of human NK cells exist that differentially express various chemokine receptors and exhibit distinct tissue localization (45), further supporting our findings that licensed and unlicensed NK cell subsets differentially distribute in mice. This is of significant clinical importance for the therapeutic utilization of NK cells, as specific populations can either be targeted or adoptively transferred to aid with particular disease states or immune responses.
Methods
Mice.
Female C57BL/6 (H2b), B10.D2 (H2d), and B6.SJL-Ptprca Pep3b/BoyJ (CD45.1) mice that were 6–12 weeks old were purchased from the Jackson Laboratory. For adoptive transfer experiments, total splenocytes (20 × 106 cells) from CD45.1 mice were i.v. injected into CD45.2 mice either 2 days prior to infection with 100 PFU APR8 influenza, or 1 day after infection. All mice were housed in AAALAC-approved specific pathogen–free facilities with free access to food and water. All animal studies and protocols were approved by the UC Davis IACUC.
Viruses.
MCMV Smith strain was obtained from American Type Culture Collection and maintained by repeated salivary gland passage. MCMV (3 × 103 to 5 × 103 PFU) was administered i.p. in 0.2 ml of RPMI (Gibco Laboratories. The A/PR/8 influenza strain was maintained as previously described (46, 47). Mice were lightly anesthetized by isoflurane inhalation and sublethally infected with 20 PFU or 100 PFU A/PR/8 influenza virus in 40 μl of PBS (control mice were mock infected 40 μl of PBS) by intranasal inoculation. Two days prior to infections, depletions were done with either 300 μg purified anti-Ly49G2 (4D11), anti-Ly49A (YE1/32), anti-Ly49C/I (5E6), or rat IgG (Jackson ImmunoResearch) in 0.2 ml of PBS i.p. Depleting antibodies were produced by purifying supernatant from hybridoma cultures.
Flow cytometry.
Conjugated antibodies purchased from commercial vendors were used to stain samples prior to analysis on an LSR Fortessa (BD Biosciences) flow cytometer. Data analysis was performed by using FlowJo software (Tree Star). Percentages and total cell numbers were determined for different phenotypic markers and cell types studied. See also supplemental experimental procedures for a more detailed flow cytometry reagent list.
Viral titer determination.
Quantification of MCMV (48, 49) and A/PR/8 influenza (50) virus using real-time PCR was performed as previously described. Briefly, for MCMV quantification DNA was extracted from livers using the DNeasy Tissue Kit (Qiagen) and the MCMV IE1 gene was amplified using the HotStarTaq Master Mix (Qiagen) and forward and reverse primers. Plasmid was purified, quantified, and serial 10-fold dilutions of the IE1-containing plasmid were amplified using the forward and reverse primers and probe. A standard curve was constructed by plotting the cycle threshold (CT) value against the log of the target template molecules obtained from the plasmid, followed by a sum of least square regression analysis. Target copy numbers in the tissue samples were then calculated using the equation obtained by least squares regression analysis. Results were expressed as IE1 gene copies/100 mg of tissue.
A/PR/8 influenza quantification was performed by extracting total RNA from lung homogenates using QIAamp Viral RNA Kit (Qiagen). Viral mRNA for nuclear protein was amplified and quantified by single-round quantitative PCR with an ABI 7900 real-time PCR system. Relative plaque-forming units were determined from lung samples by comparison to a standard curve established by amplification of serial dilutions of the purified virus.
In vitro DC and NK coculture.
Bone marrow was harvested from C57BL/6 mice and cultured with 20 ng/ml GM-CSF and 10 ng/ml IL-4 for 3 days. NK cells were negatively selected by using EasySep magnet (Stemcell Technologies) from splenocytes of mice that had been depleted of the opposite subsets to have an enriched subpopulation of NK cells. NK cells were cultured with bone marrow–derived DCs for 3 days with 1 μg/ml anti–GM-CSF or 1 μg/ml rIgG before flow cytometric analysis. Cells were plated at a 1:2 ratio of NK cells to DCs.
Microarray analysis.
NK cell subsets were FACS sorted for unlicensed (CD3–CD122+Ly49G2+Ly49A+Ly49C/I– and CD3–CD122+Ly49G2–A–Ly49C/I–) and licensed (CD3–CD122+Ly49G2–A–Ly49C/I+) NK cell subpopulations from the spleens of 30 naive C57BL/6 mice and subsequently pooled (into 1 of the 3 groups outlined above). Purity of sorted cell populations was greater than 98%. Total RNA was extracted using the RNeasy Kit (Qiagen). RNA quality was assessed on the Agilent 2100 Bioanalyzer (Agilent Technologies). Microarray gene expression profiling was performed by whole-transcript analysis on Affymetrix GeneChip Mouse Gene 2.0 Sense Target (ST) Arrays. All downstream microarray processing procedures were performed according to Affymetrix standard protocols. Microarray data analysis was performed with GeneSpring GX (Agilent Technologies). All original microarray data were deposited in the NCBI’s Gene Expression Omnibus (GEO GSE72632). See also supplemental experimental procedures.
Human NK cell analysis.
Peripheral blood from healthy volunteers was obtained from leukocyte reduction filters (Blood Centers of the Pacific). Peripheral blood mononuclear cells (PBMCs) were separated by density gradient centrifugation (Histopaque-1077, Sigma-Aldrich) and genomic DNA was extracted for KIR and HLA genotyping using QIAmp DNA Blood Mini Kit (Qiagen). PBMCs were stimulated for 16 hours with 50 ng of phorbol 12-myristate 13-acetate (PMA) and 1 μg ionomycin before intracellular staining for IFN-γ, and GM-CSF production. NK cell subsets exclusively expressing a single inhibitory KIR (spKIR2DL2/3, spKIR2DL1, spKIR3DL1) were detected after gating on the CD3–CD56+ NK cell population, excluding NKG2A. To confirm the presence of KIR and to distinguish licensed (single KIR+ NK cells in subjects with cognate HLA class I ligand present) from unlicensed (spKIR+NKG2A– NK cells in subjects lacking cognate HLA class I ligand or KIR–NKG2A–) NK cell subsets, we performed KIR and HLA class I genotyping using a combination of sequence-specific primers and PCR-specific oligonucleotide probes, as previously described (51, 52). HLA-C and -B alleles were segregated into KIR ligand groups: C1 (HLA-CAsn80), C2 (HLA-CLys80), and HLA-Bw4 or Bw6.
Statistics.
Data are shown as averages ± SEM. Statistical significance was determined by using 2-tailed Student’s t test and 2 or 1-way ANOVA with Bonferroni or Tukey post-test analysis when appropriate. P values were considered statistically significant when P was less than 0.05.
Study approval.
All animal studies and protocols were approved by the UC Davis IACUC (protocol numbers 17167 and 18940). Viral studies were approved by UC Davis biohazard use authorization (BUA) number R1592. Human sample experiments were approved by UCSF BUA number BU097781.
Author contributions
AEZ, EGA, and CMS all designed research studies, conducted experiments, acquired/analyzed data, and wrote the manuscript. LTK, CD, GRL, and JD conducted experiments and acquired/analyzed data. CP, BRB, DLL, JMV, and NB designed research studies and provided feedback on the manuscript. WJM designed research studies, analyzed data, and wrote the manuscript.
Supplementary Material
Acknowledgments
We would to thank Weihong Ma, Monja Metcalf, and Yajarayma Tang-Feldman for their technical help. We would also like to thank the other members in the Murphy lab for providing feedback and suggestions during preparation of the manuscript. We would also like to acknowledge the UC Davis Comprehensive Cancer Center’s Genomics Shared Resource core facility for their help with performing and analyzing the microarray data. The UC Davis Comprehensive Cancer Center Genomics Shared Resource is supported by Cancer Center Support Grant P30 CA093373 from the National Cancer Institute (NCI). We would also like to thank the NIH tetramer core facility at Emory University for providing the tetramers utilized in the studies. This work was funded by NIH grants R01 HL08990 and R01 CA72669. This project has been funded in whole or in part with federal funds from the NCI, NIH, under contract HHSN26120080001E. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government. This research was supported in part by the Intramural Research Program of the NIH, NCI, Center for Cancer Research.
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Reference information:JCI Insight. 2017;2(10):e87032. https://doi.org/10.1172/jci.insight.87032.
Contributor Information
Cordelia Dunai, Email: cdunai@ucdavis.edu.
G. Raymond Lochhead, Email: rayloch@hotmail.com.
Juan Du, Email: JDu@medicine.ucsf.edu.
Claire Pomeroy, Email: claire.pomeroy@ucdmc.ucdavis.edu.
Bruce R. Blazar, Email: blaza001@umn.edu.
Dan L. Longo, Email: danl_longo@dfci.harvard.edu.
Nicole Baumgarth, Email: nbaumgarth@ucdavis.edu.
William J. Murphy, Email: wmjmurphy@ucdavis.edu.
References
- 1.Miller JS. Biology of natural killer cells in cancer and infection. Cancer Invest. 2002;20(3):405–419. doi: 10.1081/CNV-120001185. [DOI] [PubMed] [Google Scholar]
- 2.Kärre K, Ljunggren HG, Piontek G, Kiessling R. Selective rejection of H-2-deficient lymphoma variants suggests alternative immune defence strategy. Nature. 1986;319(6055):675–678. doi: 10.1038/319675a0. [DOI] [PubMed] [Google Scholar]
- 3.Igarashi H, Gregory SC, Yokota T, Sakaguchi N, Kincade PW. Transcription from the RAG1 locus marks the earliest lymphocyte progenitors in bone marrow. Immunity. 2002;17(2):117–130. doi: 10.1016/S1074-7613(02)00366-7. [DOI] [PubMed] [Google Scholar]
- 4.Pegram HJ, Andrews DM, Smyth MJ, Darcy PK, Kershaw MH. Activating and inhibitory receptors of natural killer cells. Immunol Cell Biol. 2011;89(2):216–224. doi: 10.1038/icb.2010.78. [DOI] [PubMed] [Google Scholar]
- 5.Lanier LL. NK cell recognition. Annu Rev Immunol. 2005;23:225–274. doi: 10.1146/annurev.immunol.23.021704.115526. [DOI] [PubMed] [Google Scholar]
- 6.Parham P. MHC class I molecules and KIRs in human history, health and survival. Nat Rev Immunol. 2005;5(3):201–214. doi: 10.1038/nri1570. [DOI] [PubMed] [Google Scholar]
- 7.Raulet DH. Missing self recognition and self tolerance of natural killer (NK) cells. Semin Immunol. 2006;18(3):145–150. doi: 10.1016/j.smim.2006.03.003. [DOI] [PubMed] [Google Scholar]
- 8.Bix M, Liao NS, Zijlstra M, Loring J, Jaenisch R, Raulet D. Rejection of class I MHC-deficient haemopoietic cells by irradiated MHC-matched mice. Nature. 1991;349(6307):329–331. doi: 10.1038/349329a0. [DOI] [PubMed] [Google Scholar]
- 9.Waggoner SN, Cornberg M, Selin LK, Welsh RM. Natural killer cells act as rheostats modulating antiviral T cells. Nature. 2011;481(7381):394–398. doi: 10.1038/nature10624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lang PA, et al. Natural killer cell activation enhances immune pathology and promotes chronic infection by limiting CD8+ T-cell immunity. Proc Natl Acad Sci USA. 2012;109(4):1210–1215. doi: 10.1073/pnas.1118834109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Robbins SH, et al. Natural killer cells promote early CD8 T cell responses against cytomegalovirus. PLoS Pathog. 2007;3(8):e123. doi: 10.1371/journal.ppat.0030123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stadnisky MD, Xie X, Coats ER, Bullock TN, Brown MG. Self MHC class I-licensed NK cells enhance adaptive CD8 T-cell viral immunity. Blood. 2011;117(19):5133–5141. doi: 10.1182/blood-2010-12-324632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wong JL, Berk E, Edwards RP, Kalinski P. IL-18-primed helper NK cells collaborate with dendritic cells to promote recruitment of effector CD8+ T cells to the tumor microenvironment. Cancer Res. 2013;73(15):4653–4662. doi: 10.1158/0008-5472.CAN-12-4366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cooper MA, Fehniger TA, Fuchs A, Colonna M, Caligiuri MA. NK cell and DC interactions. Trends Immunol. 2004;25(1):47–52. doi: 10.1016/j.it.2003.10.012. [DOI] [PubMed] [Google Scholar]
- 15.Degli-Esposti MA, Smyth MJ. Close encounters of different kinds: dendritic cells and NK cells take centre stage. Nat Rev Immunol. 2005;5(2):112–124. doi: 10.1038/nri1549. [DOI] [PubMed] [Google Scholar]
- 16.Kim S, et al. Licensing of natural killer cells by host major histocompatibility complex class I molecules. Nature. 2005;436(7051):709–713. doi: 10.1038/nature03847. [DOI] [PubMed] [Google Scholar]
- 17.Fernandez NC, Treiner E, Vance RE, Jamieson AM, Lemieux S, Raulet DH. A subset of natural killer cells achieves self-tolerance without expressing inhibitory receptors specific for self-MHC molecules. Blood. 2005;105(11):4416–4423. doi: 10.1182/blood-2004-08-3156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Foley B, et al. Human cytomegalovirus (CMV)-induced memory-like NKG2C(+) NK cells are transplantable and expand in vivo in response to recipient CMV antigen. J Immunol. 2012;189(10):5082–5088. doi: 10.4049/jimmunol.1201964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim S, et al. HLA alleles determine differences in human natural killer cell responsiveness and potency. Proc Natl Acad Sci USA. 2008;105(8):3053–3058. doi: 10.1073/pnas.0712229105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Thomas LM, Peterson ME, Long EO. Cutting edge: NK cell licensing modulates adhesion to target cells. J Immunol. 2013;191(8):3981–3985. doi: 10.4049/jimmunol.1301159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Orr MT, Murphy WJ, Lanier LL. ‘Unlicensed’ natural killer cells dominate the response to cytomegalovirus infection. Nat Immunol. 2010;11(4):321–327. doi: 10.1038/ni.1849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sungur CM, et al. Murine natural killer cell licensing and regulation by T regulatory cells in viral responses. Proc Natl Acad Sci USA. 2013;110(18):7401–7406. doi: 10.1073/pnas.1218767110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sungur CM, Tang-Feldman YJ, Zamora AE, Alvarez M, Pomeroy C, Murphy WJ. Murine NK-cell licensing is reflective of donor MHC-I following allogeneic hematopoietic stem cell transplantation in murine cytomegalovirus responses. Blood. 2013;122(8):1518–1521. doi: 10.1182/blood-2013-02-483503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Béziat V, et al. NK cell responses to cytomegalovirus infection lead to stable imprints in the human KIR repertoire and involve activating KIRs. Blood. 2013;121(14):2678–2688. doi: 10.1182/blood-2012-10-459545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Grégoire C, et al. The trafficking of natural killer cells. Immunol Rev. 2007;220:169–182. doi: 10.1111/j.1600-065X.2007.00563.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bajénoff M, et al. Natural killer cell behavior in lymph nodes revealed by static and real-time imaging. J Exp Med. 2006;203(3):619–631. doi: 10.1084/jem.20051474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Watt SV, Andrews DM, Takeda K, Smyth MJ, Hayakawa Y. IFN-gamma-dependent recruitment of mature CD27(high) NK cells to lymph nodes primed by dendritic cells. J Immunol. 2008;181(8):5323–5330. doi: 10.4049/jimmunol.181.8.5323. [DOI] [PubMed] [Google Scholar]
- 28.Martín-Fontecha A, et al. Induced recruitment of NK cells to lymph nodes provides IFN-gamma for T(H)1 priming. Nat Immunol. 2004;5(12):1260–1265. doi: 10.1038/ni1138. [DOI] [PubMed] [Google Scholar]
- 29.Fehniger TA, et al. CD56bright natural killer cells are present in human lymph nodes and are activated by T cell-derived IL-2: a potential new link between adaptive and innate immunity. Blood. 2003;101(8):3052–3057. doi: 10.1182/blood-2002-09-2876. [DOI] [PubMed] [Google Scholar]
- 30.Desbois M, Rusakiewicz S, Locher C, Zitvogel L, Chaput N. Natural killer cells in non-hematopoietic malignancies. Front Immunol. 2012;3:395. doi: 10.3389/fimmu.2012.00395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Marquardt N, Wilk E, Pokoyski C, Schmidt RE, Jacobs R. Murine CXCR3+CD27bright NK cells resemble the human CD56bright NK-cell population. Eur J Immunol. 2010;40(5):1428–1439. doi: 10.1002/eji.200940056. [DOI] [PubMed] [Google Scholar]
- 32.Hayakawa Y, Smyth MJ. CD27 dissects mature NK cells into two subsets with distinct responsiveness and migratory capacity. J Immunol. 2006;176(3):1517–1524. doi: 10.4049/jimmunol.176.3.1517. [DOI] [PubMed] [Google Scholar]
- 33.Freud AG, et al. A human CD34(+) subset resides in lymph nodes and differentiates into CD56bright natural killer cells. Immunity. 2005;22(3):295–304. doi: 10.1016/j.immuni.2005.01.013. [DOI] [PubMed] [Google Scholar]
- 34.Hayakawa Y, Huntington ND, Nutt SL, Smyth MJ. Functional subsets of mouse natural killer cells. Immunol Rev. 2006;214:47–55. doi: 10.1111/j.1600-065X.2006.00454.x. [DOI] [PubMed] [Google Scholar]
- 35.Fahlén L, Lendahl U, Sentman CL. MHC class I-Ly49 interactions shape the Ly49 repertoire on murine NK cells. J Immunol. 2001;166(11):6585–6592. doi: 10.4049/jimmunol.166.11.6585. [DOI] [PubMed] [Google Scholar]
- 36.Pak-Wittel MA, Yang L, Sojka DK, Rivenbark JG, Yokoyama WM. Interferon-γ mediates chemokine-dependent recruitment of natural killer cells during viral infection. Proc Natl Acad Sci USA. 2013;110(1):E50–E59. doi: 10.1073/pnas.1220456110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vivier E, et al. Innate or adaptive immunity? The example of natural killer cells. Science. 2011;331(6013):44–49. doi: 10.1126/science.1198687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mitrović M, et al. The NK cell response to mouse cytomegalovirus infection affects the level and kinetics of the early CD8(+) T-cell response. J Virol. 2012;86(4):2165–2175. doi: 10.1128/JVI.06042-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Andrews DM, et al. Innate immunity defines the capacity of antiviral T cells to limit persistent infection. J Exp Med. 2010;207(6):1333–1343. doi: 10.1084/jem.20091193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Thomas PG, Keating R, Hulse-Post DJ, Doherty PC. Cell-mediated protection in influenza infection. Emerging Infect Dis. 2006;12(1):48–54. doi: 10.3201/eid1201.051237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ballesteros-Tato A, León B, Lund FE, Randall TD. Temporal changes in dendritic cell subsets, cross-priming and costimulation via CD70 control CD8(+) T cell responses to influenza. Nat Immunol. 2010;11(3):216–224. doi: 10.1038/ni.1838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Scharton TM, Scott P. Natural killer cells are a source of interferon gamma that drives differentiation of CD4+ T cell subsets and induces early resistance to Leishmania major in mice. J Exp Med. 1993;178(2):567–577. doi: 10.1084/jem.178.2.567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lutz MB. Buy one, get one free: additional functions of GM-CSF in DC maturation. Eur J Immunol. 2012;42(1):35–38. doi: 10.1002/eji.201142271. [DOI] [PubMed] [Google Scholar]
- 44.Holmes TD, et al. Licensed human natural killer cells aid dendritic cell maturation via TNFSF14/LIGHT. Proc Natl Acad Sci USA. 2014;111(52):E5688–E5696. doi: 10.1073/pnas.1411072112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Berahovich RD, Lai NL, Wei Z, Lanier LL, Schall TJ. Evidence for NK cell subsets based on chemokine receptor expression. J Immunol. 2006;177(11):7833–7840. doi: 10.4049/jimmunol.177.11.7833. [DOI] [PubMed] [Google Scholar]
- 46.Baumgarth N, Brown L, Jackson D, Kelso A. Novel features of the respiratory tract T-cell response to influenza virus infection: lung T cells increase expression of gamma interferon mRNA in vivo and maintain high levels of mRNA expression for interleukin-5 (IL-5) and IL-10. J Virol. 1994;68(11):7575–7581. doi: 10.1128/jvi.68.11.7575-7581.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Baumgarth N, Kelso A. In vivo blockade of gamma interferon affects the influenza virus-induced humoral and the local cellular immune response in lung tissue. J Virol. 1996;70(7):4411–4418. doi: 10.1128/jvi.70.7.4411-4418.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Collins T, Pomeroy C, Jordan MC. Detection of latent cytomegalovirus DNA in diverse organs of mice. J Infect Dis. 1993;168(3):725–729. doi: 10.1093/infdis/168.3.725. [DOI] [PubMed] [Google Scholar]
- 49.Tang-Feldman YJ, Wojtowicz A, Lochhead GR, Hale MA, Li Y, Pomeroy C. Use of quantitative real-time PCR (qRT-PCR) to measure cytokine transcription and viral load in murine cytomegalovirus infection. J Virol Methods. 2006;131(2):122–129. doi: 10.1016/j.jviromet.2005.07.013. [DOI] [PubMed] [Google Scholar]
- 50.Bourgeois C, Stockinger B. CD25+CD4+ regulatory T cells and memory T cells prevent lymphopenia-induced proliferation of naive T cells in transient states of lymphopenia. J Immunol. 2006;177(7):4558–4566. doi: 10.4049/jimmunol.177.7.4558. [DOI] [PubMed] [Google Scholar]
- 51.Hsu KC, Liu XR, Selvakumar A, Mickelson E, O’Reilly RJ, Dupont B. Killer Ig-like receptor haplotype analysis by gene content: evidence for genomic diversity with a minimum of six basic framework haplotypes, each with multiple subsets. J Immunol. 2002;169(9):5118–5129. doi: 10.4049/jimmunol.169.9.5118. [DOI] [PubMed] [Google Scholar]
- 52.Vilches C, Castaño J, Gómez-Lozano N, Estefanía E. Facilitation of KIR genotyping by a PCR-SSP method that amplifies short DNA fragments. Tissue Antigens. 2007;70(5):415–422. doi: 10.1111/j.1399-0039.2007.00923.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.