Abstract
A family of 11 lytic transglycosylases in Pseudomonas aeruginosa, an opportunistic human pathogen, turn over the polymeric bacterial cell wall in the course of its recycling, repair and maturation. The functions of these enzymes are not fully understood. We disclose herein that SltB3 of P. aeruginosa is an exolytic lytic transglycosylase. We characterize its reaction and its products by the use of peptidoglycan-based molecules. The enzyme recognizes a minimum of four sugars in its substrate, but can process a substrate comprised of a peptidoglycan of 20 sugars. The ultimate product of the reaction is N-acetylglucosamine-1,6-anhydro-N-acetylmuramic acid. The X-ray structure of this enzyme is reported for the first time. The enzyme is comprised of four domains, arranged within an annular conformation. The polymeric linear peptidoglycan substrate threads through the opening of the annulus, as it experiences turnover.
Graphical Abstract
Cell wall, a crosslinked polymer that encases the entire bacterium, is critical for survival. The peptidoglycan, the major constituent of cell wall, is comprised of a linear polymer of repeating N-acetylglucosamine (NAG)-N-acetylmuramic acid (NAM) disaccharide, with a peptide stem appended to the NAM unit (Figure 1). Crosslinking of the neighboring peptidoglycan strands takes place at the peptide stems. The mature cell wall is a single molecule of significant complexity, which is assembled and recycled in a dynamic manner.1 Recycling of cell wall takes place in response to damage (due to antibiotics, for example), to construction of complex macromolecular assemblies such as the flagellum, or to processing during replication that requires changes to the cell wall.2, 3
Figure 1.
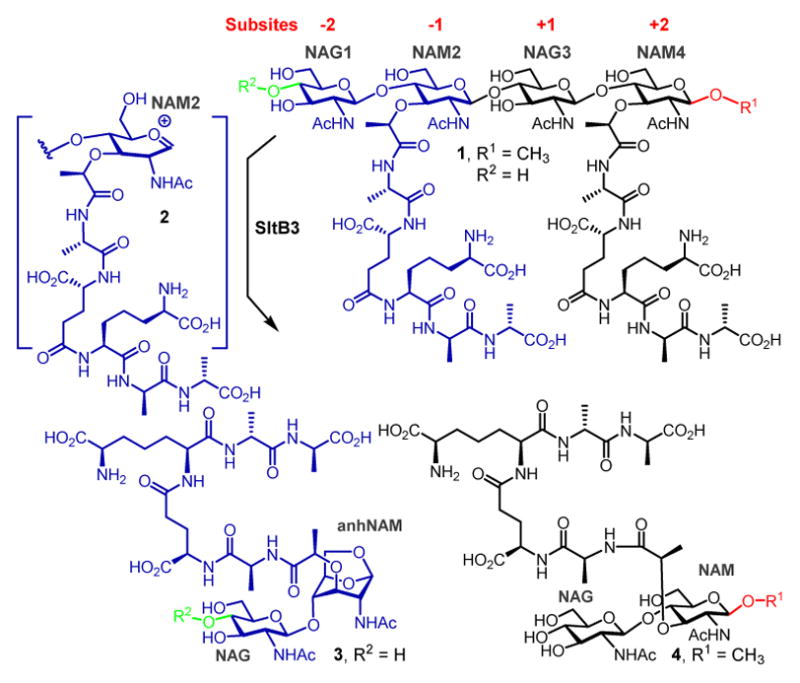
Reaction of a LT of Gram-negative bacteria. R1 and R2 continue with (NAG-NAM)n for the typical peptidoglycan. R1 = CH3 and R2 = H for compounds 1, 3 and 4.
The breakdown of the peptidoglycan for the recycling events is performed by lytic transglycosylases (LTs), enzymes that exist within the periplasmic space of Gram-negative bacteria in either soluble or membrane-bound forms. These enzymes perform important biological functions, which are the subjects of intense study. The importance of their functions is underscored by their multiplicity—Escherichia coli has eight3–5 and Pseudomonas aeruginosa has eleven6—and by redundancies of the reactions that they perform. For example, six of the enzymes of E. coli could be ablated without any apparent consequence, but the organism cannot survive the loss of seven.4 The mechanism of reaction of LT has been studied previously.3, 7, 8 These enzymes bind to the peptidoglycan and degrade it either by removal of a disaccharide from the ends of the linear polymer (the exolytic reaction) or by cleavage of the polymer in the middle (the endolytic reaction). Whereas the catalytic sites of LTs bear resemblance to the lysozyme structure, these enzymes are not hydrolytic. The peptidoglycan binds the active site by straddling the seat of the reaction, which is located between the so-called −1 and +1 subsites. The sugars in the peptidoglycan line up in both directions in what are referred to as the −i to +j subsites, the number of which could vary from enzyme to enzyme. The active-site glutamate (or aspartate) of LTs would protonate the oxygen of the scissile glycosidic bond, resulting in transient formation of an oxocarbenium species (2). This species entraps the C6-hydroxyl of the NAM unit, resulting in the 1,6-anhydromuramyl product (3). We describe here the properties of SltB3 (also referred to as SltH; the gene for which is annotated as PA3992), a soluble LT of P. aeruginosa. Our analysis of the reaction of this enzyme reveals it to be an exolytic enzyme. We also disclose three high-resolution X-ray structures for this enzyme in the apo form and in complexes with the product (3) of the reaction and with a synthetic peptidoglycan-based substrate mimetic (4). These structures reveal for the first time recognition of the peptidoglycan within the opening of the unusual annular structure of the enzyme, which leads to catalysis.
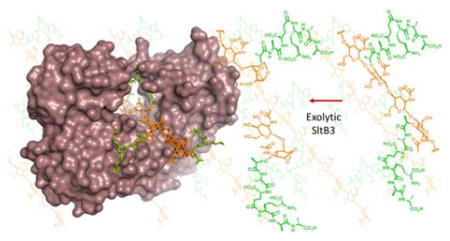
The reaction of SltB3 of P. aeruginosa had not been studied and it is presumed to be a LT based on sequence analysis. The gene was cloned and the protein was purified to homogeneity (Figure S1). We investigated the reaction of this enzyme with a synthetic fragment of the peptidoglycan from our laboratory. Compound 1 (Figures 1 and S2) is a minimal structural motif for the substrate with the backbone of NAG-NAM-NAG-NAM, and with the pentapeptide incorporated at each NAM. The purified recombinant SltB3 turned over 1 (Figure 2B), with the following kinetics parameters: kcat = 19.9 ± 0.8 s−1, Km = 200 ± 20 μM and kcat/Km = 99,500 ± 10,700 M−1s−1 (Figure S3). We prepared compounds 3 and 4 as synthetic standards for characterization of this reaction, which validated the reaction outcome. Hence, the minimal substrate for this enzyme is a tetrasaccharide. Compounds 1, 3 and 4 were prepared in 41–63 convergent synthetic steps, according to methods that we have published earlier.9 We have also reported the preparation of a glycan mixture from the cell wall,10 which was purified as the mixture 5 for this study. This mixture is a ladder of compounds (devoid of peptides), which are different from one another by two sugars (Figure 2C), spanning a minimum size of six sugars and a maximum of 20. When this mixture of the peptidoglycans was incubated with SltB3, the ladder of compounds collapsed to a single product, compound 6 (Figure 2D). The ladder of substrates lacks peptide at the NAM units, hence the presence of the peptide is not critical for this reaction. The outcome is compared to two enzymes from E. coli, one is MltA, a known exolytic enzyme, the other is MltE, a known endolytic enzyme. The reaction outcome with SltB3 was identical to that of MltA of E. coli and not to MltE (Figure 2D and 2E). The reaction outcomes were confirmed by comparison of reaction mixtures with synthetic standards (LC retention times, high-resolution mass-spectrometric analyses, and LC/MS/MS experiments; Supporting Information).
Figure 2.
Reactions of SltB3 with substrates 1 and 5. UPLC chromatograms of substrate 1 (A) before and (B) after the addition of SltB3. (C) LC/MS extracted-ion chromatograms of substrates 5, and the reaction with (D) SltB3, with (E) MltA and with (F) MltE. The numbers (1009.4, 1025.5, 993.4, 497.2, 957.3, 718.3) are m/z ratios of substrates and products.
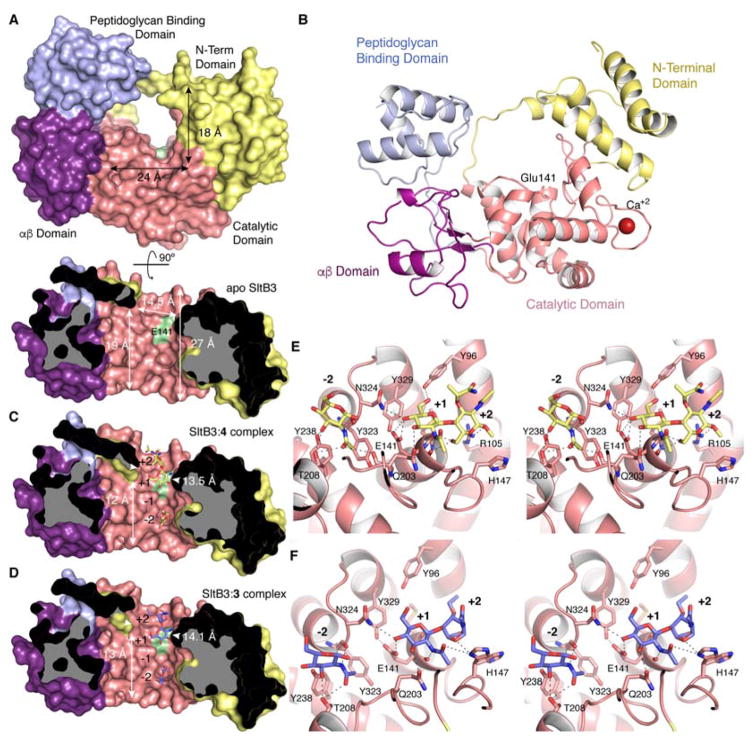
SltB3 was crystallized by the hanging drop method with 25% PEG 3350, 0.2 M NaCl, 0.1 M tris pH = 8.5 and the X-ray data were collected at 1.61 Å resolution. The structure of apo SltB3 reveals it to be an annular protein of four domains. The opening in the middle of the annulus (with dimensions of 24 Å × 18 Å), as will be elaborated in this report, is the site of binding to the strand of the peptidoglycan, as depicted in Figure 3. SltB3 monomer presents a unique arrangement of four well-defined domains, the N-terminal domain, the catalytic domain, the αβ-domain and the peptidoglycan-binding domain (Figures 3 and S4). The peptidoglycan-binding domain in SltB3 was named so based on PFAM annotation for this domain (PG_binding_1 (PF01471)). This domain has been found in the crystal structure of peptidoglycan amidases AmpDh211 and AmpDh3,12 both from P. aeruginosa, among others. Despite its name, no three-dimensional structure up to now has been determined in complex with peptidoglycan analogues. The catalytic domain is folded as in SltB1 from P. aeruginosa13 and Slt35 from E. coli 8, 14 (Figure S4). However, the unique modular arrangement found in SltB3 provides a 27 Å-long active site that narrows (to 14.5 Å) at its exit near the proposed catalytic glutamate (Figure 3).
Figure 3.
(A) The three-dimensional structure of apo SltB3 for the frontal view (top) and the cross-section view from 12 o’clock downward (bottom) drawn as Connolly surface. The N-terminal domain is in yellow, the catalytic domain is in pink, the αβ-domain is in magenta and the peptidoglycan-binding domain is in light blue. The surface position of the proposed catalytic Glu141 is indicated in green. (B) Ribbon representation of the crystal structure of apo SltB3. Each domain is colored as in panel A. (C) Same view of the active site for the SltB3:4 complex. Compound 4 is depicted as yellow sticks for carbons. (D) Active site of the SltB3:3 complex with compound 3 depicted as blue sticks for carbons. Panels B and C show how the peptidoglycan and its products of turnover would span the opening of the annulus, the site of catalysis. The protein dimensions within the cavity for the two complexes are different (indicated) compared to the apo structure. (E) Stereo view showing the interactions stabilizing compound 4 in the active site of SltB3. Substrate is depicted as capped sticks and colored in yellow for C atoms, red for O atoms and dark blue for N atoms. (F) Stereo view showing the interactions stabilizing compound 3 in the active site of SltB3. Ligand 3 is depicted as capped sticks and colored in light blue for C atoms, red for O atoms and dark blue for N atoms. The two conformational states for His147 are shown. Protein residues (in panels E and F) involved in stabilization are depicted as capped sticks and labeled. Interactions are represented with dashed lines.
We also have two complexes of SltB3 with synthetic cell-wall ligands. One is the complex with compound 4 at 2.23 Å resolution. Compound 4 is a β-methyl glycoside with the NAG-NAM backbone intact, as would be found in the regular peptidoglycan. Two molecules of 4 are bound at the catalytic groove (occupying positions −2, −1, and +1, +2, respectively). While full occupancy is observed for one molecule at subsites +1 and +2 (Figures 3B and S5A), only density for the NAG moiety at −2 is observed for the second molecule. As explained earlier, catalytic cleavage of the peptidoglycan backbone takes place between subsites −1 and +1. No electron density was observed for the peptide-stem in either of these ligands (Figure S5) indicating a high mobility of this moiety in the active site, and consistent with the lack of requirement of the peptide in the substrate for the reactions that we have characterized. However the NAG ring at position +1 presents an excellent electron density. This structure reveals that the proposed catalytic Glu141 establishes a strong hydrogen bond with the glycosidic oxygen (2.5 Å) (Figure 3E). This oxygen is further stabilized by interaction with the amide group of Asn324. The position of the side chain of the catalytic glutamate is stabilized through a strong hydrogen bond of the other oxygen atom of the carboxylate with the hydroxyl of Tyr329. The orientation of the NAG ring at position +1 during catalysis is secured by three additional polar interactions with the active site. The active site further narrows by 1 Å in this complex (14.5 Å in apo structure versus 13.5 Å in the SltB:4 complex) (Figure 3A and C). This defines how the length of the peptidoglycan backbone would sequester within the active site to give the reaction (Figure 4A) as it threads through the annulus (Figure 4B–4H). We considered whether an “open” and a “closed” conformation to this protein could exist. This would require that the first three helices of the N-terminal domain, together with the “bridge” region (that links these helices to the catalytic domain) would separate from the rest of the protein, unfolding the N-terminal domain, to allow entrance of the peptidoglycan onto the surface. However, this possibility is very unlikely in our opinion as these three N-terminal helices are strongly held to the other two helices of the N-terminal domain by a large core of hydrophobic residues and some polar interactions (Figure S6).
Figure 4.
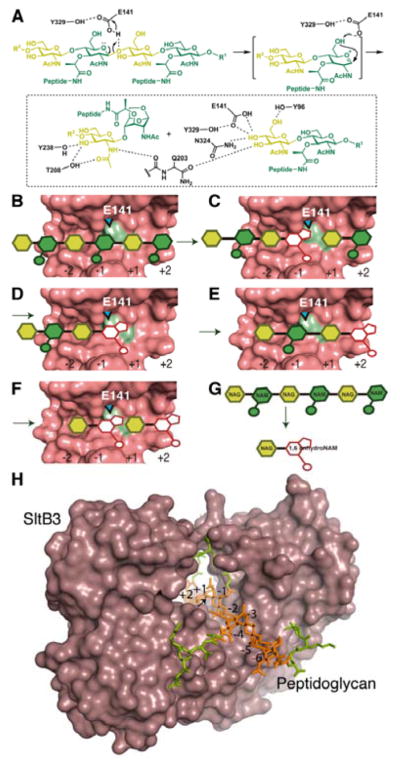
(A) The proposed catalytic scheme for SltB3. (B–F) A schematic depicting the exolytic reaction of SltB3, with the active site drawn as Connolly surface. (B) A hexasaccharide peptidoglycan binds to the active site (NAG in yellow and NAM in green). (C) The catalytic reaction produces the 1,6-anhydromuramyl moiety at the −1 subsite (in white) and a disaccharide product, which (D) departs from the active site. (E) The resulting tetrasaccharide translocates to occupy subsites −2 to +2, straddling the seat of the catalytic reaction. (F) The exolytic reaction produces two products with the 1,6-anhydromuramyl moiety. (G) The overall conversion of a hexasaccharide to the disaccharide is depicted. (H) A model of the molecular surface of SltB3 with the NMR structure of an octasaccharide peptidoglycan15 bound to the active site. The glycan chain is colored in orange and peptide stems in green. The glycosidic bond to be cleaved is indicated by an arrow.
Whereas the complex with 4 teaches us as to how the linear peptidoglycan substrate binds the surface of the catalytic domain (within the annulus), the complex with compound 3 discloses the unique set of interactions with the two product molecules of the enzymatic reaction for the exolytic function of SltB3 (Figure 3F). Figure 4A is the proposed mechanism based on the interactions observed in complexes with 3 and 4. This scheme is adapted from that for Slt35 proposed by van Asselt et al.8, 14 Main residues involved in catalysis in Slt35 are conserved in SltB3 (Figure S7).
As the scissile glycosidic bond is compromised by the aforementioned protonation event mediated by Glu141(Figures 3 and 4B), peptidoglycan generates the transient oxocarbenium species that entraps the C-6 hydroxyl in generating the 1,6-anhydromuramyl moiety of the product (Figure 4C). The disaccharide product 3 leaves the active site (Figure 4D), allowing for translocation of the peptidoglycan strand forward to reoccupy subsites −2 to +2 to initiate another cycle of catalysis (Figure 4E) in the exolytic reaction of the enzyme. Thus, translocation of peptidoglycan within the active site by two subsites predisposes the substrate for the next catalytic event by full occupancy of the active site from subsite −2 to +2. The terminal saccharide occupying the +2 subsite at this stage will be 1,6-anhydromuramyl moiety (Figure 4E). Catalysis generates the disaccharide 3 again, as peptidoglycan is degraded by two sugars at a time in the exolytic reaction, leading to the formation of the disaccharide product, as documented by the experiment of Figure 2D. The complex of 3 with SltB3 is that of the species that is depicted in Figure 4F. At this final stage of the reaction, less interactions are observed between compound 3 and SltB3 (Figure 3F) and the active-site narrowing widens to values close to the apo structure (Figure 3D).
Analysis of the reaction and X-ray crystal structures of SltB3 presented in this report documents it as an exolytic enzyme, for which the reaction takes place by the threading of the linear peptidoglycan through the annulus of the enzyme. The functional importance of catalysis by SltB3 in homeostasis of the polymeric cell wall remains to be elucidated, but the disclosure of its reaction and the structural aspects of interactions with the peptidoglycan is a first step toward this goal.
Methods
Cloning and Protein Purification
The sltB3 gene (PA3992) was cloned according to the methodology reported earlier.16 The construct was cloned in pET28a(+) (Novagen) using NdeI and XhoI restriction enzymes to generate an N-terminal His-tagged protein lacking the first 32 amino acids. The protein sequence of the His-tagged protein is given in Figure S1. The His-Tagged protein was purified also by HiTrap Chelating column.16 The yield of protein was 60 mg from 1 L of culture. SDS-PAGE analysis of His-tagged protein after affinity column showed that the protein was pure (>95%) as shown in Figure S1B, lane 1. The His-tag of the protein was removed by thrombin (Sigma) as per manufacture’s instruction for 18 h at 4 °C. The protein without His-tag was purified by HiTrap Chelating column and flow through was collected. SDS-PAGE analysis showed that the purity of SltB3 (after removal of His-tag) exceeded 95% (Figure S1B, lane 2). So it was used for crystallization without further purification. The fresh stock of protein was stored at 18 °C and was good for a week. The unused stock of protein can be frozen at −80 °C.
Preparations of Compounds 1, 3, 4, and 9
Substrate (compound 1), synthetic standard of two reaction products (3 and 4), the internal standard for kinetics (9) were synthesized in our laboratory using known methods that we described previously.9
Preparations of Compounds 5
The conversion of sacculus to mixture of glycan (devoide of peptides) by MltC of E. coli, followed by amidase AmpDh3 of P. aeruginosa was described previously.10 The glycan mixtures were purified by HPLC by using a Dionex Acclaim™ PolarAdvantage II C18 column (3 μm, 120 Å, 4.6 mm i.d. × 150 mm). The mobile phase (A = 0.1% formic acid in water; B = 0.1% formic acid in acetonitrile) gradient consisted of elution at 1 mL/min with 98% A/2% B for 5 min, followed by a 40-min linear gradient to 80% A/20% B. The fractions contain oligosaccharide, (NAG-NAM)n-NAG-anhNAM, n = 2–9 were collected, concentrated in vacuo, and reconstituted with water. The purified oligosaccharide (NAG-NAM)n-NAG-anhNAM, (n = 2–9)) is designated as compounds 5 and is free of peptides and dissacharide 6, confirmed by LC/MS.
Turnover of Compound 1 by SltB3
Turnover of compound 1 by SltB3 was carried out in 20 mM HEPES, 0.1 M NaCl, 0.1% Triton X, pH 7.0 at 25 °C. The reactions were initiated by the addition enzyme to the buffer solution of substrate and were stopped by the addition of 2% trifluoroacetic acid after 30 min incubation and the resulting reaction mixture was analyzed by LC/MS. Peaks of two products from chromatograms were compared to those of synthetic standards (compounds 3 and 4). As shown in Figure S2, product 1 corresponds to compound 3, and product 2 corresponds to compound 4 from analysis of LC/MS/MS. The LC-MS-MS instrument conditions are reported previously.16
For kinetic study, the reaction condition is as described above. The concentration of substrate (1) was varied between 25 and 600 μM (7 different concentrations), while the concentration of enzyme was kept at 8 nM in a total reaction volume of 20 μL. The reaction products were quantified with an internal standard (9, 1,6-anhydro-N-acetylmuramyl L-Ala-γ-D-Glu-m-DAP-D-Ala-D-Ala, see Supporting Information for chemical structure). The data were analyzed by nonlinear regression (Figure S3) using GraphPad Prism to get kinetic parameters, kcat (19.9 ± 0.8 s−1) and Km (200 ± 20 μM).
Turnover of Compounds 5 by SltB3
Turnover of compounds 5 by SltB3 was carried out in 20 mM HEPES, 0.1 M NaCl, 0.1% Triton X, pH 7.0 at 37 °C for 2 h. The reaction mixture was analyzed by LC/MS, as shown in Figure 2C, 2D, and 2E.
Crystallization
Apo Structure of SltB3
Crystals of SltB3 were obtained by the hanging drop method for 25% PEG 3350, 0.2 M NaCl, 0.1 M tris pH = 8.5 at a concentration of 7 mg/mL. Drops containing 1 μL of protein with 1 μL of reservoir solution were equilibrated against 150 μL of reservoir solution. The optimized crystallization conditions were obtained using the microseeding technique. X-ray data sets were collected from flash-cooled crystals at 100 K. Before cooling, the crystals were washed for a few seconds in a cryoprotectant solution that was obtained from the crystal growth solution by adding PEG 400 to obtain a 20% (v/v) concentration.
SltB3 complexes
Soaking experiments with apo SltB3 crystals were performed using N-acetylglucosamine-1,6-anhydro-N-acetylmuramyl L-Ala-γ-D-Glu-m-DAP-D-Ala-D-Ala; DAP for diaminopimelate (SltB3:3 complex), β-methoxy N-acetylglucosamine-N-acetylmuramyl L-Ala-γ-D-Glu-m-DAP-D-Ala-D-Ala for diaminopimelate (SltB3:4 complex). Crystals were soaked for 48 h in a solution with 15 mM of the ligand. The X-ray data sets were collected from flash-cooled crystals at 100 K. Before flash cooling, the crystals were washed for a few seconds in a cryoprotectant solution that was obtained by adding PEG 400 to the crystal growth solution.
Crystallographic data collection and processing
X-ray diffraction data set for apo SltB3 was collected on beamline PX at the Swiss Light Source (SLS) Villigen, Switzerland with 0.5° oscillation between images and up to 1.61 Å resolution. The data set was processed using iMOSFLM17 and scaled using AIMLESS from CCP418 program suite. SltB3 crystallized in space group P21212, with unit cell parameters a = 112.28, b = 61.84, c = 50.63 Å, α = β = γ = 90° and one monomer in the asymmetric unit.
Diffraction data sets for SltB3 complexes were collected at the beamline XALOC of the ALBA synchrotron (CELLS – ALBA, Spain), using a Pilatus 6M detector and fixed wavelength of 1.05739 Å and 0.97626 Å for SltB3:3 complex and SltB3:4 complex respectively. Collected images were processed using XDS19 and scaled using AIMLESS from CCP4.18 Crystals of SltB3:3 complex belonged to the orthorhombic space group P21212 with cell dimensions of a = 111.30, b = 61.53, c = 49.94 Å, α = β = γ = 90°, and one monomer in the asymmetric unit. Crystals of SltB3:4 complex belonged to the orthorhombic space group P21212 with cell dimensions of a =111.10, b = 61.44, c = 49.87 Å, α = β = β = 90°, and one monomer in the asymmetric unit.
Structure solution and refinement
The SltB3 structure was solved by the molecular replacement method using BALBES.20 The best initial model, as found by BALBES, was the crystal structure of lytic transglycosylase SltB1 from P. aeruginosa (PDB 4ANR). The model was completed manually (for part of the αβ domain and for the entire peptidoglycan-binding domain) using Coot21 and was refined with PHENIX.22 The Rwork converged to 0.18 and the Rfree to 0.20 in the final model. A summary of the refinement statistics is given in Table S2 (Supporting Information). The SltB3:3 complex was solved at 1.77 Å resolution and the SltB3:4 complex at 2.23 Å resolution. Both complexes were solved by the molecular replacement method with the apo SltB3 structure using MOLREP.23 Models were refined using Phenix22 and modeled using Coot.21 Data refinement results are summarized in Table S2.
Supplementary Material
Acknowledgments
This work was supported by a grant from the NIH (GM61629) and by grants BFU2014-59389-P (the Spanish Ministry of Economy and Competitiveness) and S2010/BMD-2457 (the Government of Community of Madrid).
Footnotes
Supporting Information. The Supporting Information is available free of charge on the ACS Publications website. Tables S1–S2, Figures S1–S7 (PDF). The crystallographic coordinates are deposited in the Protein Data Bank (PDB codes 5anz, 5ao7 and 5ao8).
References
- 1.Fisher JF, Mobashery S. Bacterial Cell Wall: Morphology and Biochemistry. In: Goldman E, Green LH, editors. Practical Handbook of Microbiology. CRC Press; Young, K. D: 2015. pp. 221–264. 2011. [Google Scholar]; eLS. John Wiley & Sons, Ltd; Chichester: Peptidoglycan. [Google Scholar]
- 2.Park JT, Uehara T. How bacteria consume their own exoskeletons (Turnover and recycling of cell wall peptidoglycan) Microbiol Mol Biol Rev. 2008;72:211–227. doi: 10.1128/MMBR.00027-07. [DOI] [PMC free article] [PubMed] [Google Scholar]; van Heijenoort J. Peptidoglycan hydrolases of Escherichia coli. Microbiol Mol Biol Rev. 2011;75:636–663. doi: 10.1128/MMBR.00022-11. [DOI] [PMC free article] [PubMed] [Google Scholar]; Vollmer W, Joris B, Charlier P, Foster S. Bacterial peptidoglycan (murein) hydrolases. FEMS Microbiol Rev. 2008;32:259–286. doi: 10.1111/j.1574-6976.2007.00099.x. [DOI] [PubMed] [Google Scholar]
- 3.Scheurwater E, Reid CW, Clarke AJ. Lytic transglycosylases: Bacterial space-making autolysins. Int J Biochem Cell Biol. 2008;40:586–591. doi: 10.1016/j.biocel.2007.03.018. [DOI] [PubMed] [Google Scholar]
- 4.Scheurwater EM, Clarke AJ. The C-terminal domain of Escherichia coli YfhD functions as a lytic transglycosylase. J Biol Chem. 2008;283:8363–8373. doi: 10.1074/jbc.M710135200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yunck R, Cho H, Bernhardt TG. Identification of MltG as a potential terminase for peptidoglycan polymerization in bacteria. Mol Microbiol. 2016;99:700–718. doi: 10.1111/mmi.13258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cavallari JF, Lamers RP, Scheurwater EM, Matos AL, Burrows LL. Changes to its peptidoglycan-remodeling enzyme repertoire modulate β-lactam resistance in Pseudomonas aeruginosa. Antimicrob Agents Chemother. 2013;57:3078–3084. doi: 10.1128/AAC.00268-13. [DOI] [PMC free article] [PubMed] [Google Scholar]; Jorgenson MA, Chen Y, Yahashiri A, Popham DL, Weiss DS. The bacterial septal ring protein RlpA is a lytic transglycosylase that contributes to rod shape and daughter cell separation in Pseudomonas aeruginosa. Mol Microbiol. 2014;93:113–128. doi: 10.1111/mmi.12643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Thunnissen AMWH, Rozeboom HJ, Kalk KH, Dijkstra BW. Structure of the 70-kDa soluble lytic transglycosylase complexed with bulgecin A. Implications for the enzymic mechanism. Biochemistry. 1995;34:12729–12737. doi: 10.1021/bi00039a032. [DOI] [PubMed] [Google Scholar]; van Straaten KE, Barends TRM, Dijkstra BW, Thunnissen A-MWH. Structure of Escherichia Coli lytic transglycosylase mlta with bound chitohexaose: implications for peptidoglycan binding and cleavage. J Biol Chem. 2007;282:21197–21205. doi: 10.1074/jbc.M701818200. [DOI] [PubMed] [Google Scholar]
- 8.van Asselt EJ, Kalk KH, Dijkstra BW. Crystallographic studies of the interactions of Escherichia coli lytic transglycosylase Slt35 with peptidoglycan. Biochemistry. 2000;39:1924–1934. doi: 10.1021/bi992161p. [DOI] [PubMed] [Google Scholar]
- 9.Hesek D, Lee M, Zhang WL, Noll BC, Mobashery S. Total synthesis of N-acetylglucosamine-1,6-anhydro-N-acetylmuramylpentapeptide and evaluation of Its turnover by AmpD from Citrabacter freundii. J Am Chem Soc. 2009;131:5187–5193. doi: 10.1021/ja808498m. [DOI] [PMC free article] [PubMed] [Google Scholar]; Lee M, Hesek D, Shah IM, Oliver AG, Dworkin J, Mobashery S. Synthetic peptidoglycan motifs for germination of bacterial spores. ChemBioChem. 2010;11:2525–2529. doi: 10.1002/cbic.201000626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Artola-Recolons C, Lee M, Bemardo-Garcia N, Blazquez B, Hesek D, Bartual SG, Mahasenan KV, Lastochkin E, Pi HL, Boggess B, Meindl K, Uson I, Fisher JF, Mobashery S, Hermoso JA. Structure and cell wall cleavage by modular lytic transglycosylase MltC of Escherichia coli. ACS Chem Biol. 2014;9:2058–2066. doi: 10.1021/cb500439c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Martinez-Caballero S, Lee M, Artola-Recolons C, Carrasco-Lopez C, Hesek D, Spink E, Lastochkin E, Zhang WL, Hellman LM, Boggess B, Mobashery S, Hermoso JA. Reaction products and the X-ray structure of AmpDh2, a virulence determinant of Pseudomonas aeruginosa. J Am Chem Soc. 2013;135:10318–10321. doi: 10.1021/ja405464b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lee M, Artola-Recolons C, Carrasco-Lopez C, Martinez-Caballero S, Hesek D, Spink E, Lastochkin E, Zhang W, Hellman LM, Boggess B, Hermoso JA, Mobashery S. Cell-wall remodeling by the zinc-protease AmpDh3 from Pseudomonas aeruginosa. J Am Chem Soc. 2013;135:12604–12607. doi: 10.1021/ja407445x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nikolaidis I, Izore T, Job V, Thielens N, Breukink E, Dessen A. Calcium-dependent complex formation between PBP2 and lytic transglycosylase SltB1. Microb Drug Resist. 2012;18:298–305. doi: 10.1089/mdr.2012.0006. [DOI] [PubMed] [Google Scholar]
- 14.van Asselt EJ, Dijkstra AJ, Kalk KH, Takacs B, Keck W, Dijkstra BW. Crystal structure of Escherichia coli lytic transglycosylase Slt35 reveals a lysozyme-like catalytic domain with an EF-hand. Structure. 1999;7:1167–1180. doi: 10.1016/s0969-2126(00)80051-9. [DOI] [PubMed] [Google Scholar]; van Asselt EJ, Dijkstra BW. Binding of calcium in the EF-hand of Escherichia coli lytic transglycosylase Slt35 is important for stability. FEBS Lett. 1999;458:429–435. doi: 10.1016/s0014-5793(99)01198-9. [DOI] [PubMed] [Google Scholar]; Van Asselt EJ, Perrakis A, Kalk KH, Lamzin VS, Dijkstra BW. Accelerated X-ray structure elucidation of a 36 kDa muramidase/transglycosylase using wARP. Acta Crystallogr D Biol Crystallogr. 1998;D54:58–73. doi: 10.1107/s0907444997010330. [DOI] [PubMed] [Google Scholar]
- 15.Meroueh SO, Bencze KZ, Hesek D, Lee M, Fisher JF, Stemmler TL, Mobashery S. Three-dimensional structure of the bacterial cell wall peptidoglycan. Proc Natl Acad Sci USA. 2006;103:4404–4409. doi: 10.1073/pnas.0510182103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee M, Hesek D, Llarrull LI, Lastochkin E, Pi H, Boggess B, Mobashery S. Reactions of all Escherichia coli lytic transglycosylases with bacterial cell wall. J Am Chem Soc. 2013;135:3311–3314. doi: 10.1021/ja309036q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Battye TGG, Kontogiannis L, Johnson O, Powell HR, Leslie AGW. iMOSFLM: a new graphical interface for diffraction-image processing with MOSFLM. Acta Crystallogr D Biol Crystallogr. 2011;D67:271–281. doi: 10.1107/S0907444910048675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Winn MD, Ballard CC, Cowtan KD, Dodson EJ, Emsley P, Evans PR, Keegan RM, Krissinel EB, Leslie AGW, McCoy A, McNicholas SJ, Murshudov GN, Pannu NS, Potterton EA, Powell HR, Read RJ, Vagin A, Wilson KS. Overview of the CCP4 suite and current developments. Acta Crystallogr D Biol Crystallogr. 2011;D67:235–242. doi: 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kabsch W. XDS. Acta Crystallogr D Biol Crystallogr. 2010;D66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Long F, Vagin AA, Young P, Murshudov GN. BALBES: a molecular-replacement pipeline. Acta Crystallogr D Biol Crystallogr. 2008;D64:125–132. doi: 10.1107/S0907444907050172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;D66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Afonine PV, Grosse-Kunstleve RW, Echols N, Headd JJ, Moriarty NW, Mustyakimov M, Terwilliger TC, Urzhumtsev A, Zwart PH, Adams PD. Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallogr D Biol Crystallogr. 2012;D68:352–367. doi: 10.1107/S0907444912001308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vagin A, Teplyakov A. Molecular replacement with MOLREP. Acta Crystallogr D Biol Crystallogr. 2010;D66:22–25. doi: 10.1107/S0907444909042589. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.