Abstract
Oral squamous cell carcinoma (OSCC) accounting for about 90% of malignant oral lesions is the 6th most common malignancy worldwide. Diagnostic delay may contribute to dismal survival rate therefore, there is a need for developing specific and sensitive biomarkers to improve early detection. Hungarian population occupies the top places of statistics regarding OSCC incidence and mortality figures therefore, we aimed at finding potential salivary protein biomarkers suitable for the Hungarian population. In this study we investigated 14 proteins which were previously reported as significantly elevated in saliva of patients with OSCC. In case of IL-1α, IL-1β, IL-6, IL-8, TNF-α and VEGF a Luminex-based multiplex kit was utilized and the salivary concentrations were determined. In case of catalase, profilin-1, S100A9, CD59, galectin-3-bindig protein, CD44, thioredoxin and keratin-19, SRM-based targeted proteomic method was developed and the relative amount of the proteins was determined in the saliva of patients with OSCC and controls. After several rounds of optimization and using stable isotope-containing peptides, we developed an SRM-based method for rapid salivary protein detection. The validation of the selected potential biomarkers by ELISA revealed salivary protein S100A9 and IL-6 as useful protein biomarkers for OSCC detection improving the diagnostic accuracy for OSCC in the Hungarian population.A noninvasive diagnostic method to detect biomarkers useful for the early diagnosis of OSCC was developed. This can be an attractive strategy in screening saliva samples collected in a nation-wide multi-centric study in order to decrease morbidity, mortality, to enhance survival rate and to improve quality of life. The heterogeneity of protein biomarkers found in different ethnic groups presented in the literature highlights the importance of identification of population-tailored protein biomarkers.
Introduction
With 350,000–400,000 new cases annually, oral cavity squamous cell carcinomas (OSCC) represent a major health-care problem worldwide. The increasing incidence of OSCC among women and young and middle-aged males is particularly challenging [1–4]. Despite advances in surgical and radiation treatments and chemotherapy, the average 5-year overall survival rate of advanced OSCC is not much higher than 50% [5]. Hungarian women and men occupy the top places of statistics regarding global OSCC incidence and mortality figures with an embarrassing fourfold elevation of overall OSCC mortality rate in Hungary by the new millennium since the 1970-ies [6]. In contrast to other European countries where mortality rates from OSCC started to decline after 2000, the untoward trend of rising OSCC mortality figures in Hungary did not stop [7].
Standard diagnostic methods, i.e. expert clinical examination and histological evaluation of suspicious areas fail to detect the majority of patients with early stage OSCC so improvement of the diagnostic methods is required [5,8]. Salivary protein biomarkers may represent a promising tool for improving clinical outcome of patients with OSCC.
Saliva is a very dilute body fluid which contains electrolytes, nitrogenous products and proteins. The protein content of saliva has been analyzed by several workgroups, and more than 2000 salivary proteins have been identified [9]. The most abundant proteins in saliva are α-amylase, cystatins, proline rich peptides, serum albumin and mucins [9]. Besides the highly abundant proteins, the low-abundance proteins of saliva have diagnostic values in oral diseases like bisphosphonate-related osteonecrosis of the jaw [10] and also in systemic diseases such as breast cancer [11]. In the past decade, several studies identified a series of proteins, mostly NF-κB-dependent cytokines, with elevated concentration in the saliva of patients with OSCC using antibody-based techniques, such as enzyme-linked immunosorbent assay (ELISA) as the gold standard for protein biomarker detection [12,13]. Unfortunately, this methodology does not allow the identification of protein biomarkers in great numbers, it is time consuming and expensive.
Selected reaction monitoring (SRM)-based targeted proteomics allows the identification and quantification of multiple proteins with high specificity and great detection dynamics [14]. It can be applied practically for any kind of samples but it has high importance for the study of different body fluids such as of patients with cancer [15].
Application of SRM-based targeted proteomics platform was already used for salivary protein biomarker detection [16] and our aim was to test if some of the potential salivary protein biomarkers already described in scientific literature can be used in the Hungarian population for OSCC detections.
In this prospective study we report on the detection of salivary biomarkers in accordance to the Clinical Proteomic Technologies for Cancer (CPTC) initiative guidelines established by the National Cancer Institute [17]. The guidelines suggest the application of SRM-based targeted proteomics for verification of potential biomarkers identified in the discovery phase, and after verification, the biomarkers can be subjected to the validation step. For validation, only few proteins should be chosen and tested using ELISA or other antibody-based method [17]. Candidate biomarkers reported in the literature were selected and SRM-based targeted proteomics platform along with Luminex-based multiplex assay was used to monitor the level of these biomarkers on a test set of samples followed by the verification of the potential biomarkers whose level was significantly altered in the OSCC group compared to the controls by ELISA. ELISA tests specific for the potential biomarkers were carried out on a reference set of samples and the IL-6 and S100A9 was shown to be specific for OSCC in the studied Hungarian patient cohort.
Materials and methods
All the reagents used in this work were of analytical or LC-MS grade and were purchased from Sigma-Aldrich unless stated otherwise.
Patients, sample collection
Unstimulated saliva samples were collected from 108 donors between 9 a.m. and 11 a.m. at the Faculty of Dentistry, University of Debrecen. Patient enrollment, sample collection and processing were carried out respecting the Declaration of Helsinki. Ethical approval was obtained from the University of Debrecen Ethics Committee (No. 3385–2011) and the subjects gave written informed consent. In parallel to sample collection a questionnaire containing questions on smoking habits, alcohol consumption was filled out by the patients. Patients were asked to avoid eating, drinking, smoking, or using oral hygiene products for at least 1 hour before sample collection.
A two-step sample collection was applied: 1) the test set (collection between 2011.06.30.- 2012.04.13.) contained 29 OSCC, 25 age-matched and 8 young controls for method development and biomarker identifications and 2) a reference set (collection between 2013.05.09–2016.02.29) containing 26 OSCC, 12 age-matched and 7 young controls for biomarker verification. Saliva samples were kept on ice throughout the collection and processing—no more than 60 minutes elapsed from sample collection to freezing. Samples of the test set were centrifuged at 4100 x g for 15 min at 4°C at the Genomic Medicine and Bioinformatics Core Facility (University of Debrecen). The supernatants were transferred to fresh tubes and the aliquots were stored at -70°C until further processing. The samples of the reference set were filtered using a PVDF membrane-containing filter unit (5 μm pore size, Millipore SLSV025LS) and the filtered saliva was aliquoted and stored at -70°C until further processing. All donors were patients of the University of Debrecen, Faculty of Dentistry. Consecutive patients with diagnosed OSCC were recruited into the study. Age-matched controls were consecutive patients admitted to the Faculty of Dentistry for regular dental checkup. The young controls were students of the University of Debrecen admitted to the Faculty of Dentistry for regular dental checkup. OSCC was diagnosed by histopathological evaluation. Treatment was started based on positive histology result and was not influenced by saliva sample collection and evaluation. Periodontal condition was evaluated by a periodontist from Department of Periodontology; none of the patients and healthy volunteers had diabetes mellitus, human papilloma virus infection or any autoimmune diseases. The study population was a consecutive series of patients and volunteers according to the above presented criteria.
Study design
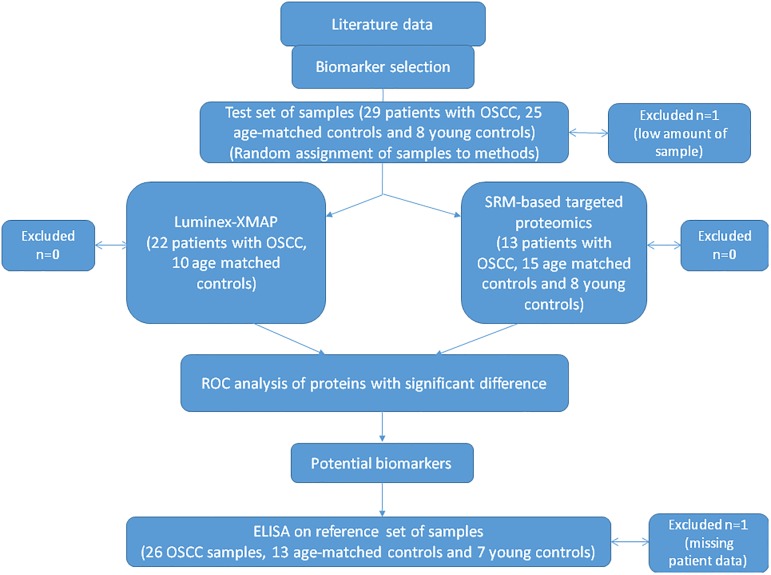
In this prospective study we did not compare two laboratory methods rather we wanted to apply the methodology of proteomics to determine proteins with high sensitivity and specificity in saliva samples from patients with OSCC. Data collection was planned before sampling and performing the examinations. Three types of examinations were applied according to the Fig 1. During study design the CPTAC guidelines were followed: first; SRM-based biomarker verification was carried out followed by the ELISA analysis of the three selected potential biomarkers on larger patient cohort.
Fig 1. Study design.
The samples for the Luminex assay and SRM-based assay were randomly selected from the test set of samples, and the samples for validation by ELISA were also randomly selected from the reference set of samples (S1 Table).
All the clinical evaluations of patients and controls were done by expert health care professionals (IT and AS). The laboratory examinations were done by well-trained, graduated molecular biologists (GK, PL, BM, and EC). This was a non-interventional study and the results of the performed methods did not influence in any means the treatment of patients. The sampling procedure was non-invasive and completely harmless to the study subjects. Therefore, no adverse events were related to performing the laboratory examinations.
Cytokine assay
The multiplex immunobead Luminex x-MAP-based cytokine assay was carried out on a Custom 6plex Milliplex kit (Merck-Millipore) containing antibodies against IL-1α, IL-1β, IL-6, IL-8, TNF-α and VEGF. 25 μl of the saliva samples of patients with OSCC and age-matched controls were analyzed in duplicates. The assay was carried out based on the protocol provided by the manufacturer and the data acquisition was done on BioPlex 2.0 Workstation (Bio-Rad) operated by the Bioplex Manager 4.0 software. The level of IL-1α, IL-1β, IL-6, IL-8, TNF-α and VEGF was calculated by the Bioplex Manager software based on the recorded 7-point calibration curve. For curve fitting a logistic regression model was used.
Design of SRM-based targeted proteomics method
The amino acid sequences of the examined proteins were utilized from the UniProt database (www.uniprot.org) and were subjected to in silico trypsin digestion. In order to determine the unique protein-specific tryptic sequences BLASTp analysis (http://blast.ncbi.nlm.nih.gov) was carried out searching the NCBI non-redundant protein sequence database. SRM transition design was performed by the Skyline software [18] (www.brendanx-uw1.gs.washington.edu) on the protein-specific tryptic peptide sequences. All possible transitions of singly charged “y” ions were tested on digested saliva samples from patients suffering from OSCC. Peptides which gave reproducible SRM spectra with good peak shape were chosen for further analyses and their stable isotope-labeled synthetic forms were obtained from the JPT Peptide Technologies GmbH, Germany. The quality of the synthetic peptides was assessed in our laboratory by mass spectrometry analyzes. The SRM spectra of all fragment ions were recorded and the two best transitions were selected for further analyzes.
Sample preparation for mass spectrometry
200 μl filtered saliva was dried in speedvac and redissolved in 50 mM ammonium bicarbonate buffer. Protein concentration of the samples was determined using the Bradford method [19]. Sample blocking was carried out before trypsin digestion; one randomly selected OSCC sample was grouped with one randomly selected age-matched and one young control sample and the groups were processed together on the same day. The proteins were denatured with 6 M urea and then reduced with 10 mM dithiothreitol. The samples were alkylated with 20 mM iodo-acetamide and diluted with 25 mM ammonium bicarbonate in order to decrease the urea concentration to 1 M. Trypsin digestion was performed at 37°C overnight using MS grade modified trypsin (ABSciex) in 1:25 enzyme to protein ratio. The digested samples were dried in speedvac and redissolved in 1% formic acid. The samples were desalted using Pierce C18 Tips (Thermo Scientific) and the eluates were dried and dissolved in 1% formic acid.
Mass spectrometry analysis
SRM-based analysis of saliva samples were carried out on a 4000 QTRAP (ABSciex) mass spectrometer using a NanoSpray II MicroIon Source and controlled by the Analyst 1.4.2 software (ABSciex). The spray voltage was 2800V, the ion source gas was 50 psi, the curtain gas was 20 psi and the source temperature was 70°C. The dwell time was 20 msec and the cycle time was 1.7 sec allowing the collection of approximately 15 data points/chromatographic peak. The chromatographic separation was done on an EasynLC II system (Bruker) and the peptide mixture was first loaded and desalted onto an in-line trap column (5 x 0.3mm, 5μm particle size, 300 Å pore size Zorbax 300SB-C18,) followed by separation on a Zorbax 300SB-C18 analytical column (150 mm x 75μm 3.5μm particle size, 300 Å pore size) using a 90 min acetonitrile/water gradient with a slow increase in acetonitrile concentration from 0% to 100% during 60 min. Solvent A was 0.1% formic acid in LC water, solvent B was LC acetonitrile containing 0.1% formic acid.
For SRM analysis 20 μg digested protein spiked with the stable isotope-labeled reference peptides was introduced to the mass spectrometer. All SRM analyses were carried out in duplicates.
ELISA
Each saliva sample from patients with OSCC, matched control and young control subjects were analyzed in duplicate by ELISA using Human ELISA Kit. The concentration of IL-6 and thioredoxin (EK0410 and EK1254, respectively, Boster Biological Technology Co., Pleasanton, USA) and S100A9 (E-EL-H1290, Elabscience Biotechnology Co., Wuhan, China) in saliva were determined by sandwich enzyme-linked immune-sorbent assay technique according to manufacturer’s protocol. Absorbance was measured at 450 nm and concentrations were calculated based on the recorded 6-point (thioredoxin and S100A9) and 7-point (IL-6) calibration curves, respectively.
Data analysis
The SRM data were analyzed using the Skyline software [18]. All data where the AUC value was less than 10 were excluded from further analyses. The Skyline data are publicly available through the Panorama [20] web site:
(https://panoramaweb.org/labkey/project/University%20of%20Debrecen/OSCC%20saliva/begin.view?)
The primary data were transformed into appropriate format of MSstats R-package [21–23] by an in-house developed software. Before the statistical analysis peptides with insufficient amount of data across the set of samples were removed and for further analyses only those samples were used where both technical replicates were recorded. After normalization based on heavy standards and log2 transformation of measured abundances the group differences were investigated by mixed-effect variance analysis [22,24] for all proteins. The groups were used as the fixed effect and the subject level variance were modeled as random effects. The raw p-values of group differences were adjusted by the Benjamini and Hochberg type false discovery rate (FDR) method for multiple testing issues [25]. Besides the adjusted p-values the log2 fold changes, standard error and T values were examined. For evaluation of test performances multivariate receiver operating characteristic (ROC) curve analyses [26] were constructed by the Epi R-package [27], the accuracy and the 95% confidence intervals were calculated.
Results and discussion
Detection of salivary biomarkers which can help in the early diagnosis of OSCC is of high importance in improving survival rates of patients with OSCC. More than 35 proteins were identified by different groups as potential salivary biomarkers but a high variability in the results was observed [28]. Protein biomarkers identified in one population may not necessarily be suitable in another population. For example, IL-8 was identified as a biomarker in the USA [12,29–32], India [13] and Serbia [33] but not in Japan and Iran [34,35]. Similarly, S100A9 proved to be a suitable OSCC biomarker in the USA but not in China [36,37]. In USA catalase was shown to be biomarker by Hu et. al. [36] but not by de Jong et. al. [38]. The difference of the useful biomarkers studied in different populations (Table 1) raises the question if biomarkers validated in one population have predictive value in another population [12,29–37,39–50,38,51–60]. The prevalence of OSCC in Hungary is the highest in Europe so our aim was to examine if some of the previously described OSCC biomarkers can be used in the Hungarian population.
Table 1. List of examined salivary protein biomarkers for OSCC in different countries.
The proteins included in the studies carried out in different countries are indicated. The proteins in bold were found to be potential biomarkers for OSCC. The abbreviations of protein names are used according to gene name.
| Country | Examined proteins | Reference |
|---|---|---|
| Brazil | C1R, C3, C4B, CFB, FAM49B, LCN2, LRG1, SERPINA1, SLPI, TAGLN2 | [16] |
| China | CEACAM5, S100A7, S100A8, S100A9 | [37,59] |
| Croatia | FGF2, IL6 | [51] |
| Hungary | CAT, CD44, CD59, IL1A, IL1B, IL6, IL8, KRT19, LGALS3BP, PROF1, S100A9, TNF, TXN, VEGF | present study |
| India | LDH**, MUC16, IL-8 | [13,58,64] |
| Iran | IL1A, IL6, IL8, TNF | [35] |
| Israel | ALB, CEACAM5, CCNDBP1, KRT19, EGFR, IGG, IGF1, LDH, MKI67, MMP2, MMP9, MUC16, OGG1, SRC, SERPINB5, sIgA**, SERPINB3, TPA*** | [54–56] |
| Italy | STATH, BIRC5 | [52,53] |
| Japan | DEFA1, IL1B, IL6, IL8, SPP1 | [34,39,40] |
| Serbia | IL1B, IL8, M2BP | [33] |
| Taiwan | ALB, AMY**, ANXA2, APOA1, APOA2, APOA4, C4BPA, C6, C9, CCNDBP1, CD109, CHIT1, EGFR, EPHX1, FSCN1, FABP, GSTM1, HEP2, HEXA, HIST1H2AA, HPRT, HSD17B4, ICAM3, IGHA2, IGHV1-2, IGKV1D-12, IGKV4-1, IGLV1-47, ITIH2, ITIH4, NKIRAS1, KNG1, PRCP, RETN, RPL7, S100A8, SERPINA6, SLC4A1, TF, TMEM132A, TUBA1C, VTN, ZN497, ZN501 | [42–45] |
| United Kingdom | P53ab | [41] |
| USA | ACT**, FGF2, CAT*, CD44, CD59, EDN1, H1F0, IL1A, IL1B, IL6, IL8, INL, KRT19, LGALS3BP, MSN, MYL**, PROF1, RAB7A, S100A12, S100A7, S100A9, S100P, TNF, VEGFA | [29–32,36,46–50,38,65] |
*In case of catalase protein there are controversial data; in the study carried out by Hu et. al. [36] it was reported as potential biomarker but the study carried out by de Jong et. al. [38] could not confirm this finding.
** Specific isoforms are undistinguishable.
*** Mixture of fragments of cytokeratins 8, 18 and 19.
ACT: actin, ALB: serum albumin, AMY: α-amylase, ANXA2: annexin A2, APOA1: apolipoprotein A1, APOA2: apolipoprotein A2, APOA4: apolipoprotein A–IV, BIRC5: Baculoviral IAP repeat-containing protein 5, C1R: complement C1r subcomponent, C4B: complement C4-B, C4BPA: C4b-binding protein α chain, C3: complement C3, C6: complement C6, C9: complement C9, CAT: catalase, CCNDBP1: cyclin D1-binding protein 1, CD44: CD44 antigen, CD59: CD59 antigen, CD109: CD109 antigen, CEACAM5: carcinoembryonic antigen-related cell adhesion molecule 5, CFB: complement factor B, CHIT1: chitotriosidase-1, DEFA1: human neutrophil defensin 1, EGFR: epidermal growth factor receptor, EDN1: endothelin-1, EPHX1: epoxide hydrolase 1, FABP4: fatty acid-binding protein adipocyte, FAM49: protein FAM49B, FGF2: basic fibroblast growth factor 2, FSCN1: fascin, GSTM1: glutathione S-transferase Mu1, H1F0: histone H1.0, HEP2: heparin cofactor 2, HEXA: β-hexosaminidase subunit α, HIST1H2AA: histone H2A type 1-A, HPRT: haptoglobin-related protein, HSD17B4: peroxisomal multifunctional enzyme type 2, ICAM3: intercellular adhesion molecule 3, IGF1: insulin-like growth factor I, IGG: immunoglobulin G, IGHA2: immunoglobulin alpha-2 chain C region, IGHV1-2: immunoglobulin heavy variable 1–2, IGKV1D-12: immunoglobulin kappa variable 1D-12, IGKV4-1: immunoglobulin kappa variable 4–1, IGLV1-47: immunoglobulin lambda variable 1–47, IL1A: interleukin-1α, IL1B: interleukin-1β, IL6: interleukin-6, IL8: interleukin-8, INL: involucrin, ITIH2: inter-α-trypsin inhibitor heavy chain H2, ITIH4: inter-α-trypsin inhibitor heavy chain H4, KNG1: kininogen-1, KRT19: cytokeratin-19, LCN2: neutrophil gelatinase-associated lipocalin, LDH: lactate dehydrogenase, LGALS3BP: galectin-3-binding protein, LRG1: leucine-rich alpha-2-glycoprotein, MKI67: proliferation marker protein Ki-67, MMP2: matrix metalloproteinase 2, MMP9: matrix metalloproteinase 9, MSN: moesin, MUC16: cancer antigen 125, MYL: myosin regulatory light polypeptide, NKIRAS1: NF-kappa-B inhibitor-interacting Ras-like protein 1, OGG1: N-glycosylase/DNA lyase, SPP1: osteopontin, SRC: Proto-oncogene tyrosine-protein kinase Src, P53ab: p53 autoantibody, PRCP: lysosomal Pro-X carboxypeptidase, PROF1: profiling 1, RAB7A: Ras-related protein Rab-7a, RETN: resistin, RPL7: 60S ribosomal protein L7, S100A7: protein S100-A7, S100A8: protein S100-A8, S100A9: protein S100-A9, S100P: protein S100-P, SERPINA1: alpha-1-antitrypsin, SERPINA6: corticosteroid binding globulin, SERPINB3: serpin B3, SERPINB5: serpin B5, sIgA: secreted immunoglobulin A, SLC4A1: band 3 anion transporter, SLPI: antileukoproteinase, STATH: statherin, TAGLN2: transgelin-2, TF: serotransferrin, TMEM132A: transmembrane protein 132A, TNF: tumor necrosis factor, TUBA1C: tubulin-α-1C chain, TPA: tissue polypeptide antigen, TXN: thioredoxin, VEGFA: vascular endothelial growth factor A, VTN: vitronectin, ZN497: zinc finger protein 497, ZN501: zinc finger protein 501.
Based on literature data, 14 proteins were chosen and their levels were examined in the saliva of patients with OSCC and controls originating from the North-Eastern part of Hungary. Oral hygiene of age-matched controls was quite poor, characterized by the presence of various oral inflammatory conditions, independently from the presence of OSCC, as expected from earlier stomatoepidemiologic studies investigating this population [61,62]. Therefore, recruitment of healthy young controls with good oral hygiene, absent from oral inflammatory lesions was considered important.
Multiplex analyses were carried out; the level of IL-1α, IL-1β, IL-6, IL-8, TNF-α and VEGF was examined using a custom Luminex assay taking the benefit of high sensitivity of the antibody-based methods. We have chosen these cytokines as far as their levels were observed to be elevated in saliva from patients with OSCC compared to the levels observed in controls [12,29–31,33–35,40,46]. However, in some other studies no significant changes could be observed in the levels of IL-6 and IL-8 [29,34,35]. In order to examine the level of catalase, profilin-1, S100A9, CD59, galectin-3-bindig protein, CD44 and keratin-19 identified as biomarkers by different groups [36,50,54,63], targeted proteomics method was developed and optimized.
After careful examination of the recorded metadata originating from each patient, a considerable difference regarding the alcohol consumptions and smoking habits between the groups was observed. Smoking was more abundant in patients with OSCC; they consumed more alcohol and more of them consumed alcohol on a daily base. Accordingly, the cariological and periodontal status of patients was poor; the DMFT, DMFS, gingival index and plaque index values were higher in the OSCC group compared to controls (Table 2).
Table 2. Patient data.
| Test set | Reference set | |||||||
|---|---|---|---|---|---|---|---|---|
| OSCC patients (BioPlex) | Age-matched control patients (Bioplex) | OSCC patients (mass spectrometry) | Age-matched control patients (MC—mass spectrometry) | Young healthy control patients (HC—mass spectrometry) |
OSCC patients (ELISA) | Age-matched control patients (MC-ELISA) | Young healthy control patients (YC-ELISA) |
|
| Total number | 22 | 10 | 13 | 15 | 8 | 26 | 12 | 7 |
| Age | 61.9±7.8 (49–77) | 63.5±6.7 (53–73) | 60.8±7.5 (52–77) | 64.9±10.4 (52–87) | 24.3±1.9 (22–28) | 58.2±9.7 (44–77) | 59.3± 5.6 (50–68) | 24.4± 1.3 (22–26) |
| Sex M/F | 18/4 | 9/1 | 11/2 | 11/4 | 3/5 | 20/6 | 3/9 | 4/3 |
| Socioeconomical status | ||||||||
| · low/middle/ high | 6/14/0 | 2/6/2 | 4/7/0 | 0/10/5 | 0/0/8 | 7/17/1 | 2/8/0 | 0/0/5 |
| Smoking habit | ||||||||
| · Never smoker | 5% | 40% | 7.7% | 79% | 62.5% | 4% | 41% | 43% |
| · Ex-smoker | 22% | 10% | 23% | 21% | 12.5% | 23% | 25% | 14% |
| · Current smoker | 68% | 50% | 61.6% | - | 12.5% | 69% | 17% | 14% |
| · No response | 5% | - | 7.7% | - | 12.5% | 4% | 17% | 29% |
| Alcohol consumption | ||||||||
| · Never/rarely | 2/3 | 2/1 | 1/0 | 4/3 | 0/3 | 4/6 | 4/2 | 0/3 |
| · Monthly/ weekly | 4/4 | 4/3 | 4/1 | 3/3 | 3/1 | 3/8 | 0/4 | 2/0 |
| · Daily | 8 | - | 6 | 0 | 0 | 4 | 0 | 0 |
| · Intake (mL/day) | beer 285 mL/day/person |
beer 171 mL/day/person |
beer 426 mL/day/person |
beer 30 mL/day/person |
beer 33 mL/day/person |
beer 162 mL/day/person |
beer 171 mL/day/person |
beer 0 mL/day/person |
| wine / champagne 94 mL/day/person |
wine / champagne 34 mL/day/person |
wine / champagne 130 mL/day/person | wine / champagne 11 mL/day/person |
wine / champagne 11 mL/day/person | wine / champagne 84 mL/day/person |
wine / champagne 31 mL/day/person |
wine / champagne 23 mL/day/person |
|
| spirits 25 mL/day/person |
spirits 0 mL/day/person |
spirits 28 mL/day/person |
spirits 9 mL/day/person |
spirits 10 mL/day/person |
spirits 24 mL/day/person |
spirits 0 mL/day/person |
spirits 0 mL/day/person |
|
| Periodontal assessment | ||||||||
| · Gingival index | 0.7 | 0.3 | 0.9 | 0.5 | 0.5 | 0.7 | 0.2 | 0.3 |
| · Plaque index | 1.4 | 0.4 | 1.6 | 0.3 | 0.2 | 1.3 | 0.6 | 0.3 |
| Cariological assessment | ||||||||
| · DMFT | 23.1 | 19.7 | 23 | 18.9 | 7.5 | 26.9 | 26 | 8.3 |
| · DMFS | 99.8 | 81.6 | 67.5 | 79.9 | 17.5 | 121.6 | 117.8 | 18.0 |
| Comorbidities | ||||||||
| ·Hypertonia+Cardiovascular disease | 1 | 2 | 1 | 1 | - | 6 | 2 | - |
| · Hypertonia | 4 | 2 | 4 | 4 | - | 8 | 3 | - |
| · Cardiovascular disease | 6 | 0 | 4 | 0 | - | 2 | - | |
Antibody-based analysis of potential OSCC salivary biomarkers
Investigating salivary appearance of proinflammatory cytokines in OSCC patients is relevant and clinically meaningful. Cancer and chronic inflammation are interrelated with a link represented by cytokines and chemokines [66]. Both tumor cells and tumor-infiltrating immune cells produce these regulatory compounds offering thereby multiple therapeutic targets for immunomodulatory treatment of patients with oral and head and neck squamous cell carcinoma [67–69]. However, the source of the investigated cytokines in the saliva of patients with chronic oral inflammation, independent from OSCC, can be from immune cells and stimulated gingival and oral mucosal tissue [70].
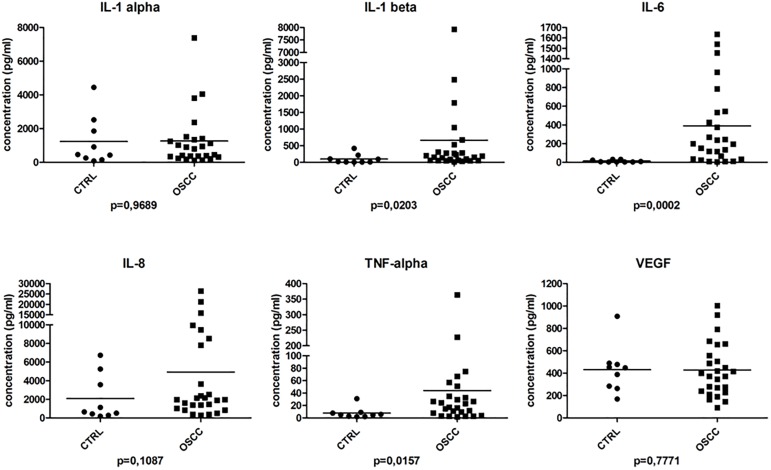
The saliva samples of randomly selected 26 patients and 9 age-matched controls were analyzed in duplicates and the levels of IL-1α, IL-1β, IL-6, IL-8, TNF-α and VEGF was determined (Fig 2). In our study the levels of IL-1β, IL-6 and TNF-α were significantly elevated in the OSCC group compared to the age-matched controls being in accordance with the previous results reported by different groups [28,33]. Examining the distribution of data points, it was observed that the results for IL-1β in the lower concentration range overlapped between the OSCC and control groups and despite the significant difference they could not discriminate between the two groups. Based on our results, it seems that among the studied cytokines only IL-6 and TNF-α can be used as potential biomarkers in the Hungarian population.
Fig 2. Salivary cytokine levels determined with Luminex-based multiplex assay.
The concentration (pg/ml) of the studied cytokines is indicated in case of control (CTRL) and OSCC group. Below each graph the p value is indicated.
IL-6 is expressed by OSCC tumor cells and stromal cells and it has been shown to play a crucial role in OSCC carcinogenesis, progression and recurrence involving the IL-6, IL-6R, STAT3 pathway [71–73]. Using a different signal transduction pathway resulting in NF-κB activation, TNF-α has also been related to oral carcinogenesis [74]. Factors, known to be associated with poor oral hygiene and oral inflammation, such as advanced age and smoking have been shown to correlate with elevated salivary IL-6 levels [75]. Therefore, not every cytokine may serve as a suitable diagnostic salivary biomarker of OSCC in different populations.
The age-matched controls did not show any signs of precancerous lesions in their oral cavity but as a result of oral inflammatory conditions we could not see any significant differences in the levels of other cytokines between the OSCC and control groups. These results might be explained by the fact that oral hygiene in the age matched group was compromised resulting in inflammation without any signs or symptoms of OSCC. Cheng and coworkers have demonstrated that the levels of IL-6 and IL-8 were significantly higher in the saliva of patients with OSCC compared to those who have chronic periodontitis [46]. Our results confirmed these findings. IL-6 level proved to be significantly higher in patients with OSCC than in controls exhibiting a compromised oral health condition and the same trend was true in case of IL-8 (Fig 2). OSCC patients formed two subgroups with respect to salivary IL-8 concentration, 7 patients had above-average and 19 patients had below average IL-8 levels. Similar to IL-1β, IL-8 level in the lower concentration range overlapped between the OSCC and control groups. Although salivary levels of IL-8 tended to be higher in OSCC patients than in age-matched controls, the difference in this cohort was not significant. A similar dual distribution of serum IL-8 concentration and IL-8 expression by the tumor cells in patients with OSCC was observed recently by Fujita et al. High serum IL-8 concentrations, and intensive IL-8 expression by tumor cells were significantly correlated with poor disease outcome measures [76].
Design of targeted proteomics method
An SRM-based targeted mass spectrometry method was designed for the examination of catalase, profilin-1, thioredoxin, protein S100A9, CD59 glycoprotein, CD44 antigen, galectin-3-bindig protein and keratin-19. The sequences were retrieved from the Uniprot database (P04040, P07737, Q99757, P06702, P13987, P16070, Q08380, and P08727, respectively) and in silico digested with trypsin. The tryptic peptides were searched against the NCBInr database using the Blastp software in order to identify the peptides which are specific for the studied proteins. Using the protein-specific sequences SRM transitions have been designed with the help of the Skyline software. The stabile isotope-labeled (SIL) synthetic counterparts of the specific peptides were ordered and spiked into the samples in known amounts. For collision energy (CE) and declustering potential (DP) optimization the synthetic peptides were used. All the possible transitions of singly charged “y” ions were optimized with the help of the Skyline software and the best two transitions per peptide were selected. The optimized parameters (Table 3) were used for the SRM method development and further used for the examination of saliva samples.
Table 3. Optimized SRM parameters used for the mass spectrometric examination of salivary proteins.
The Q1 and Q3 m/z values, the type of “y” ion used, the dwell time, declustering potential (DP) and collision energy (CE) is indicated in case of each studied peptide.
| Protein | Peptide | Q1 (m/z) | Q3 (m/z) | Ion | Dwell time (msec) | DP | CE |
|---|---|---|---|---|---|---|---|
| Catalase | LNVITVGPR | 484.798 | 642.393 | y6 | 20 | 63.0 | 23.4 |
| 484.798 | 529.309 | y5 | 20 | 63.0 | 23.4 | ||
| 484.798 | 428.261 | y4 | 20 | 63.0 | 23.4 | ||
| LNVITVGPR* | 489.802 | 652.401 | y6 | 20 | 63.0 | 23.4 | |
| 489.802 | 539.317 | y5 | 20 | 84.8 | 23.4 | ||
| 489.802 | 438.269 | y4 | 20 | 84.8 | 23.4 | ||
| CD44 antigen | NDVTGGR | 359.677 | 489.277 | y5 | 20 | 84.8 | 16.2 |
| 359.677 | 390.209 | y4 | 20 | 84.8 | 16.2 | ||
| NDVTGGR* | 364.681 | 499.286 | y5 | 20 | 84.8 | 16.2 | |
| 364.681 | 400.217 | y4 | 20 | 84.8 | 16.2 | ||
| TEAADLCK | 425.702 | 620.307 | y6 | 20 | 63.1 | 20.0 | |
| 425.702 | 549.270 | y5 | 20 | 63.1 | 20.0 | ||
| 425.702 | 478.232 | y4 | 20 | 63.1 | 20.0 | ||
| TEAADLCK* | 429.709 | 628.321 | y6 | 20 | 63.1 | 20.0 | |
| 429.709 | 557.284 | y5 | 20 | 66.6 | 20.0 | ||
| 429.709 | 486.247 | y4 | 20 | 66.6 | 20.0 | ||
| CD59 glycoprotein | AGLQVYNK | 446.747 | 651.346 | y5 | 20 | 66.6 | 21.2 |
| 446.747 | 523.287 | y4 | 20 | 66.6 | 21.2 | ||
| AGLQVYNK* | 450.755 | 659.360 | y5 | 20 | 66.6 | 21.2 | |
| 450.755 | 531.301 | y4 | 20 | 66.6 | 21.2 | ||
| Keratin-19 | APSIHGGSGGR | 498.254 | 627.295 | y7 | 20 | 79.9 | 24.1 |
| 498.254 | 490.236 | y6 | 20 | 79.9 | 24.1 | ||
| 498.254 | 433.215 | y5 | 20 | 79.9 | 24.1 | ||
| 498.254 | 376.193 | y4 | 20 | 79.9 | 24.1 | ||
| APSIHGGSGGR* | 503.258 | 637.304 | y7 | 20 | 64.3 | 24.1 | |
| 503.258 | 500.245 | y6 | 20 | 64.3 | 24.1 | ||
| 503.258 | 443.223 | y5 | 20 | 64.3 | 24.1 | ||
| 503.258 | 386.202 | y4 | 20 | 64.3 | 24.1 | ||
| Profilin-1 | TLVLLMGK | 437.775 | 660.411 | y6 | 20 | 60.6 | 20.7 |
| 437.775 | 561.342 | y5 | 20 | 60.6 | 20.7 | ||
| 437.775 | 448.258 | y4 | 20 | 60.6 | 20.7 | ||
| TLVLLMGK* | 441.782 | 668.425 | y6 | 20 | 60.6 | 20.7 | |
| 441.782 | 569.357 | y5 | 20 | 63.0 | 20.7 | ||
| 441.782 | 456.273 | y4 | 20 | 63.0 | 20.7 | ||
| SSFYVNGLTLGGQK | 735.882 | 887.494 | y9 | 20 | 63.0 | 37.7 | |
| 735.882 | 773.451 | y8 | 20 | 63.0 | 37.7 | ||
| 735.882 | 603.346 | y6 | 20 | 84.8 | 37.7 | ||
| SSFYVNGLTLGGQK* | 739.890 | 895.508 | y9 | 20 | 84.8 | 37.7 | |
| 739.890 | 781.465 | y8 | 20 | 84.8 | 37.7 | ||
| 739.890 | 611.360 | y6 | 20 | 84.8 | 37.7 | ||
| Protein S100A9 | DLQNFLK | 439.242 | 649.366 | y5 | 20 | 84.8 | 20.8 |
| 439.242 | 521.308 | y4 | 20 | 84.8 | 20.8 | ||
| DLQNFLK* | 443.249 | 657.380 | y5 | 20 | 63.1 | 20.8 | |
| 443.249 | 529.322 | y4 | 20 | 63.1 | 20.8 | ||
| LTWASHEK | 486.250 | 757.362 | y6 | 20 | 63.1 | 23.5 | |
| 486.250 | 571.283 | y5 | 20 | 63.1 | 23.5 | ||
| 486.250 | 500.246 | y4 | 20 | 66.6 | 23.5 | ||
| LTWASHEK* | 490.257 | 765.376 | y6 | 20 | 66.6 | 23.5 | |
| 490.257 | 579.297 | y5 | 20 | 66.6 | 23.5 | ||
| 490.257 | 508.260 | y4 | 20 | 66.6 | 23.5 | ||
| Thioredoxin | TAFQEALDAAGDK | 668.822 | 760.383 | y8 | 20 | 66.6 | 33.9 |
| 668.822 | 689.346 | y7 | 20 | 66.6 | 33.9 | ||
| TAFQEALDAAGDK* | 672.829 | 768.397 | y8 | 20 | 79.9 | 33.9 | |
| 672.829 | 697.360 | y7 | 20 | 79.9 | 33.9 | ||
| VGEFSGANK | 454.727 | 623.314 | y6 | 20 | 79.9 | 21.7 | |
| 454.727 | 476.246 | y5 | 20 | 79.9 | 21.7 | ||
| VGEFSGANK* | 458.734 | 631.328 | y6 | 20 | 64.3 | 21.7 | |
| 458.734 | 484.260 | y5 | 20 | 64.3 | 21.7 | ||
| Galectin-3-binding protein | LASAYGAR | 404.719 | 624.310 | y6 | 20 | 64.3 | 18.8 |
| 404.719 | 466.240 | y4 | 20 | 64.3 | 18.8 | ||
| LASAYGAR* | 409.723 | 634.318 | y6 | 20 | 60.6 | 18.8 | |
| 409.723 | 476.249 | y4 | 20 | 60.6 | 18.8 |
* indicates the stable isotope-labeled, heavy amino acid.
In order to gain information regarding the concentration range where the peptide amount introduced into the mass spectrometer is linear with the intensity of the signal recorded, a dilution series of approximately 60 fmol-1 pmol of SIL peptides spiked into saliva samples were analyzed. For keratin-19 and CD44 antigen peptides no linearity could be observed. For galectin 3 binding protein LASAYGAR, protein S100A9 LTWASHEK, thioredoxin VGEFSGANK and profilin-1 TLVLLMGK peptides the curve was linear between approximately 120 fmol-0.5 pmol range, while in case of catalase LNVITVGPR, CD59 glycoprotein AGLWVYNK peptides the linear range was slightly broader; between approximately 120 fmol-1 pmol. The thioredoxin TAFQEALDAAGDK peptide shows linearity between approximately 60 fmol-0.5 pmol concentrations, while the whole studied range was covered only by protein S100A9 DLQNFLK peptide (approximately 60 fmol-1 pmol concentrations). The broad concentration range observed permits the study of high-amplitude changes often present in biological systems.
Targeted proteomic analysis of the selected potential OSCC biomarkers
The SRM transitions for endogenous peptides and SIL counterparts were recorded and using the amount of the SIL peptide as a reference the relative levels of endogenous peptides were calculated. In order to assess which peptides and which transitions can be successfully utilized in an eventual screening, saliva samples of randomly selected 15 patients with OSCC, 15 age-matched controls (MC) and 10 healthy young controls (HC) were analyzed using the developed targeted mass spectrometry method. In all cases where the area under the curve values for the transitions were less than 10, the results were omitted and not used for further calculations.
Some of the transitions did not give consistent results in the analyzed samples so they were omitted from the calculations. This means that for CD44 antigen, CD59 glycoprotein, galectin-3-bindig protein and keratin-19 the best transitions used during the optimization did not provide good quality data in most of the analyzed saliva samples so they were not used for the statistical analysis. The catalase LNVITVGPR peptide, the SSFYVNGLTLGGYK and TLVLLMGK peptides of profilin-1, the DLYNFLK peptide from protein S100A9 and TAFQEALDAAGDK peptide from thioredoxin gave data of acceptable quality in most of the samples and were further used for statistical analysis.
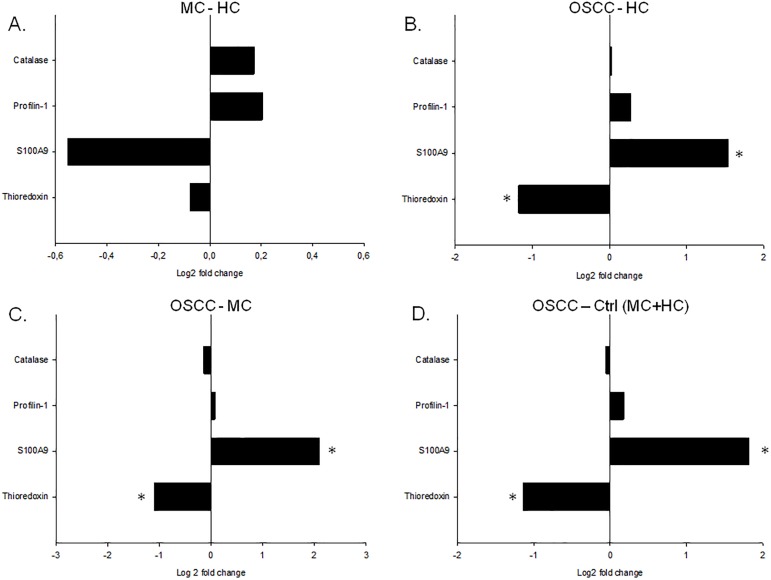
The statistical analyses indicated no significant differences between age-matched (MC) and healthy young (HC) controls (Fig 3A) while significant differences were observed in case of S100A9 and thioredoxin between patients with OSCC and controls (Fig 3B–3D, Table 4). These data suggest that these changes were specific for OSCC and were not age-dependent. In the saliva of patients with OSCC the level of S100A9 was significantly elevated while the level of thioredoxin was significantly reduced compared to both healthy young and age matched controls. S100A9 is a general marker of cancerous lesions, it was identified in different tumors including tumors of tongue, prostate, breast, lung, in colorectal cancer, gastric cancer, squamous cervical cancer, etc. and it is considered as a primary target for drug development [77–84]. Thioredoxin is part of the antioxidant system and it was shown to have increased expression in tumor samples [85–87]. We do not have information regarding the thioredoxin levels in the salivary gland or tumor tissue itself, but it cannot be excluded that the reduction of thioredoxin levels in saliva may arise from the fact that this protein accumulates in the tissues resulting in a reduced secretion.
Fig 3. Quantitative analysis of salivary proteins by SRM.
The log2 fold change in case of thioredoxin, S100A9, profilin and catalase is shown. The comparison between the (A) age matched controls (MC) and healthy controls (HC), (B) OSCC group and healthy control (HC) group, (C) OSCC group and age matched (MC) group and (D) OSCC group and both healthy and matched control groups is indicated. * indicates significant change, p<0.05.
Table 4. Results of mixed-effect variance analysis of OSCC and MC group differences.
The log2 fold change (logFC) standard error (SE), T values (Tvalue) and the FDR corrected p-values are shown.
| Protein | Log2FC | SE | Tvalue | Adjusted p-value |
|---|---|---|---|---|
| Catalase | -0.14 | 0.25 | -0.56 | 0.66 |
| Profilin-1 | 0.07 | 0.17 | 0.44 | 0.66 |
| S100A9 | 2.09 | 0.41 | 5.14 | <0.0001 |
| Thioredoxin | -1.10 | 0.39 | -2.79 | 0.01 |
ROC analysis
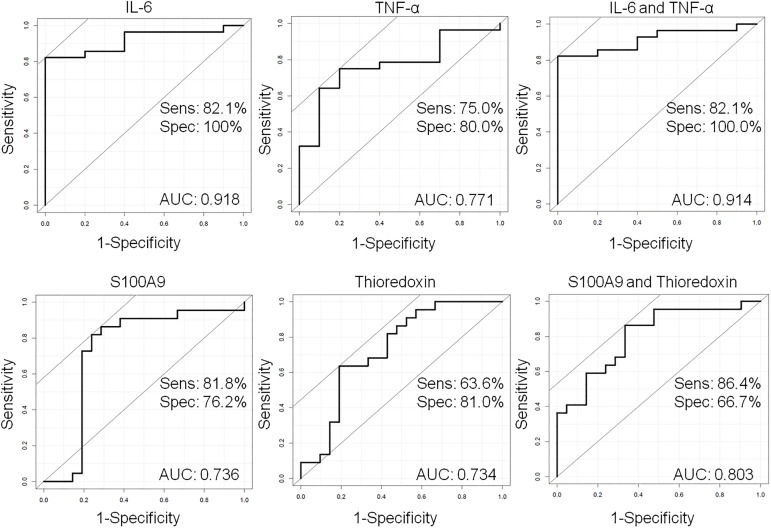
The results of either the mass spectrometry-based or Luminex-based experiments provided information about the changes of the studied proteins in patients with OSCC compared to those in the controls. To test which proteins with a significant difference between the studied groups can be used as possible biomarkers, a ROC curve analysis was performed and the area under the curve (AUC) was calculated. Values of AUC close to 1.0 suggest a perfectly performing biomarker; while values close to 0.5 indicate that the biomarker performs not better than random.
In our experiments the AUC value for S100A9 was 0.74 (accuracy 0.75, 95% confidence interval: 0.55–0.96) and for thioredoxin 0.73 (accuracy 0.79, 95% confidence interval: 0.61–0.96) suggesting equally performing potential biomarkers (Fig 4). In order to examine the sensitivity and specificity of the combination of S100A9 and thioredoxin multivariate ROC analysis was carried out. The S100A9 and thioredoxin together performed better than alone, the AUC value was 0.80 (accuracy 0.88, 95% confidence interval: 0.77–1.0) suggesting an additive effect of the two potential biomarkers on the discrimination of OSCC samples from the controls (Fig 4).
Fig 4. Estimation of predictive power of possible biomarkers using ROC curve analysis.
The sensitivity (y axis) was plotted versus 1−specificity (x axis) in case of each potential biomarker alone or in combinations. The area under the curve is indicated on each pane. The sensitivity and specificity was calculated for each biomarkers and biomarker combination.
In the next step we wanted to examine the sensitivity and specificity of the IL-6 and TNF-α alone or in combination. The AUC for IL-6 was 0.92 (accuracy 0.92, 95% confidence interval: 0.83–1.0) and for TNF-α was 0.77 (accuracy 0.77, 95% confidence interval: 0.6–0.93), while the AUC for the combination of IL-6 and TNF-α was 0.91 (accuracy 0.91, 95% confidence interval: 0.83–1.0) (Fig 4). These results indicate that the IL-6 alone seems to be the best potential biomarker able to distinguish between the OSCC samples and controls as it was shown in most of the studies presented in the literature (Table 1).
Verification of the potential biomarkers using ELISA
The level of IL-6, S100A9 and thioredoxin was examined using quantitative ELISA according to the protocol provided by the manufacturers in the saliva samples of patients with OSCC, age-matched controls and young controls. The 46 samples of the reference set were analyzed in duplicates (Fig 5) but for the statistical analysis data for only 45 samples were used (one age-matched control sample was excluded because of missing patient data).
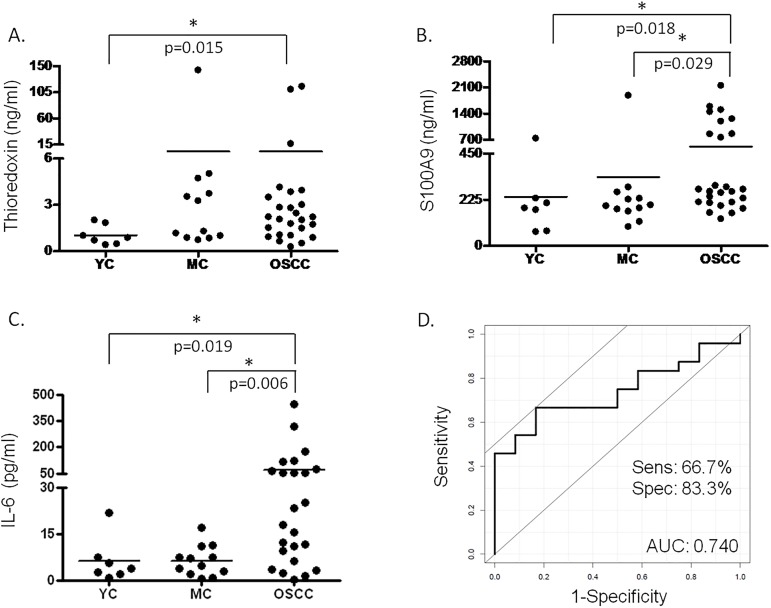
Fig 5. Salivary protein levels determined with ELISA.
The salivary concentration of (A) thioredoxin, (B) S100A9 protein and (C) IL-6 is indicated in case of young healthy controls (YC), age matched controls (MC) and OSCC group. In cases where significant difference was observed the p value is indicated. (D) ROC analysis of the combined effect of IL-6 and S100A9 protein.
According to the validation experiments in case of IL-6 and S100A9 there was significant difference observed between the OSCC and young controls and OSCC and age-matched controls, while in case of thioredoxin the difference was significant only between OSCC and young controls. ROC analysis was performed for proteins having significantly different levels between the OSCC and age-matched control groups. The AUC for IL-6 was 0.708 with 50% sensitivity and 100% specificity and for S100A9 the AUC was 0.463, the sensitivity was 96% and specificity was 16%. This means that despite the previous good sensitivity and specificity results obtained on the test set, the analyses carried out on the reference set, on a larger sample cohort indicate that either IL-6 or S100A9 alone are not well-enough performing biomarkers. However, their combination with AUC of 0.74, 67% sensitivity and 83% specificity show a well performing biomarker combination. In order to improve the sensitivity and specificity of OSCC detection further research should be done and extension of the analyses on larger populations would be beneficial.
Conclusion
In this study we developed and optimized an SRM-based targeted proteomic approach for monitoring the level of salivary biomarker candidate proteins and we could identify and verify potential salivary protein biomarkers suitable for the Hungarian population.
Four out of 14 potential salivary protein biomarkers included in our study were shown to have significant differences between the control and OSCC groups and three of them proved worth using as potential biomarkers for the Hungarian population. The validation of IL-6, S100A9 and thioredoxin revealed the potential utility of combination of IL-6 and S100A9.
Our results provide further evidence to literature data demonstrating that general protein biomarkers for OSCC which can be applied world-wide are very hard to find. It should be noted that on protein level there are discrepancies between the different studied groups which highlights the importance of population-tailored proteomics studies to find OSCC-specific protein biomarkers applicable for cost-effective screening purposes. In our opinion our findings are of remarkable importance since the incidence of OSCC is high and shows an increasing tendency not only in the old but in the young generation as well.
Supporting information
Samples in bold were used for both Bioplex and ELISA experiments.
(XLSX)
Acknowledgments
This work was supported in part by the János Bolyai Research Scholarship of the Hungarian Academy of Sciences, TÁMOP-4.2.2.D-15/1/KONV-2015-0016 project implemented through the New Széchenyi Plan co-financed by the European Social Fund, and Hungarian Scientific Research Fund OTKA K105034. Gergő Kalló is receiver of Astellas Pharma Ltd. Fellowship in form of support for chemicals and consumables which does not alter our adherence to PLOS ONE policies on sharing data and materials. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. The work of Dr. Mária Fera for sample collection is greatly acknowledged.
Data Availability
Data are publicly available through the Panorama website (https://panoramaweb.org/labkey/project/University%20of%20Debrecen/OSCC%20saliva/begin.view?).
Funding Statement
This work was supported in part by the János Bolyai Research Scholarship of the Hungarian Academy of Sciences, TÁMOP-4.2.2.D-15/1/KONV-2015-0016 project implemented through the New Széchenyi Plan co-financed by the European Social Fund, and Hungarian Scientific Research Fund OTKA K105034. Gergő Kalló is receiver of Astellas Pharma Ltd. Fellowship in form of support for chemicals and consumables, which does not alter our adherence to PLOS ONE policies on sharing data and materials. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.SEER Cancer Statistics Review 1975–2007—Previous Version—SEER Cancer Statistics [Internet]. Available: http://seer.cancer.gov/archive/csr/1975_2007/
- 2.Oral Cancer Facts—The Oral Cancer Foundation [Internet]. Available: http://www.oralcancer.org/facts/
- 3.La Vecchia C, Lucchini F, Negri E, Levi F. Trends in oral cancer mortality in Europe. Oral Oncol. 2004;40: 433–439. doi: 10.1016/j.oraloncology.2003.09.013 [DOI] [PubMed] [Google Scholar]
- 4.Garavello W, Bertuccio P, Levi F, Lucchini F, Bosetti C, Malvezzi M, et al. The oral cancer epidemic in central and eastern Europe. Int J Cancer. 2010;127: 160–71. doi: 10.1002/ijc.25019 [DOI] [PubMed] [Google Scholar]
- 5.Zhang H, Dziegielewski PT, Biron VL, Szudek J, Al-Qahatani KH, O’Connell DA, et al. Survival outcomes of patients with advanced oral cavity squamous cell carcinoma treated with multimodal therapy: a multi-institutional analysis. J Otolaryngol Head Neck Surg. 2013;42: 30 doi: 10.1186/1916-0216-42-30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Suba Z, Mihályi S, Takács D, Gyulai-Gaál S. [Oral cancer: morbus Hungaricus in the 21st century]. Fogorv Sz. 2009;102: 63–8. [PubMed] [Google Scholar]
- 7.Bonifazi M, Malvezzi M, Bertuccio P, Edefonti V, Garavello W, Levi F, et al. Age-period-cohort analysis of oral cancer mortality in Europe: the end of an epidemic? Oral Oncol. 2011;47: 400–7. doi: 10.1016/j.oraloncology.2010.06.010 [DOI] [PubMed] [Google Scholar]
- 8.Walsh T, Liu JLY, Brocklehurst P, Glenny A-M, Lingen M, Kerr AR, et al. Cochrane Database of Systematic Reviews The Cochrane database of systematic reviews. Chichester, UK: John Wiley & Sons, Ltd; 1996. [Google Scholar]
- 9.Csősz É, Kalló G, Márkus B, Deák E, Csutak A, Tőzsér J. Quantitative body fluid proteomics in medicine—A focus on minimal invasiveness. J Proteomics. 2016;1874–3919: 30364–5. [DOI] [PubMed] [Google Scholar]
- 10.Thumbigere-Math V, Michalowicz BS, de Jong EP, Griffin TJ, Basi DL, Hughes PJ, et al. Salivary proteomics in bisphosphonate-related osteonecrosis of the jaw. Oral Dis. 2015;21: 46–56. doi: 10.1111/odi.12204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Streckfus CF, Mayorga-Wark O, Arreola D, Edwards C, Bigler L, Dubinsky WP. Breast cancer related proteins are present in saliva and are modulated secondary to ductal carcinoma in situ of the breast. Cancer Invest. 2008;26: 159–67. doi: 10.1080/07357900701783883 [DOI] [PubMed] [Google Scholar]
- 12.Rhodus NL, Cheng B, Myers S, Miller L, Ho V, Ondrey F. The feasibility of monitoring NF-κB associated cytokines: TNF-α, IL-1α, IL-6, and IL-8 in whole saliva for the malignant transformation of oral lichen planus. Mol Carcinog. 2005;44: 77–82. doi: 10.1002/mc.20113 [DOI] [PubMed] [Google Scholar]
- 13.Punyani SR, Sathawane RS. Salivary level of interleukin-8 in oral precancer and oral squamous cell carcinoma. Clin Oral Investig. 2012;17: 517–524. doi: 10.1007/s00784-012-0723-3 [DOI] [PubMed] [Google Scholar]
- 14.Lange V, Picotti P, Domon B, Aebersold R. Selected reaction monitoring for quantitative proteomics: a tutorial. Mol Syst Biol. 2008;4: 222 doi: 10.1038/msb.2008.61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pan S, Chen R, Brand RE, Hawley S, Tamura Y, Gafken PR, et al. Multiplex Targeted Proteomic Assay for Biomarker Detection in Plasma: A Pancreatic Cancer Biomarker Case Study. J Proteome Res. 2012;11: 1937–1948. doi: 10.1021/pr201117w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kawahara R, Bollinger JG, Rivera C, Ribeiro ACP, Brandão TB, Leme AFP, et al. A targeted proteomic strategy for the measurement of oral cancer candidate biomarkers in human saliva. Proteomics. 2016;16: 159–173. doi: 10.1002/pmic.201500224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rodriguez H, Rivers R, Kinsinger C, Mesri M, Hiltke T, Rahbar A, et al. Reconstructing the pipeline by introducing multiplexed multiple reaction monitoring mass spectrometry for cancer biomarker verification: an NCI-CPTC initiative perspective. Proteomics Clin Appl. 2010;4: 904–14. doi: 10.1002/prca.201000057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.MacLean B, Tomazela DM, Shulman N, Chambers M, Finney GL, Frewen B, et al. Skyline: an open source document editor for creating and analyzing targeted proteomics experiments. Bioinformatics. 2010;26: 966–968. doi: 10.1093/bioinformatics/btq054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72: 248–54. [DOI] [PubMed] [Google Scholar]
- 20.Sharma V, Eckels J, Taylor GK, Shulman NJ, Stergachis AB, Joyner SA, et al. Panorama: a targeted proteomics knowledge base. J Proteome Res. 2014;13: 4205–10. doi: 10.1021/pr5006636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chang C-Y, Picotti P, Huttenhain R, Heinzelmann-Schwarz V, Jovanovic M, Aebersold R, et al. Protein Significance Analysis in Selected Reaction Monitoring (SRM) Measurements. Mol Cell Proteomics. 2011;11: M111.014662–M111.014662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Clough T, Thaminy S, Ragg S, Aebersold R, Vitek O. Statistical protein quantification and significance analysis in label-free LC-MS experiments with complex designs. BMC Bioinformatics. 2012;13 Suppl 1: S6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Choi M, Chang C-Y, Clough T, Broudy D, Killeen T, MacLean B, et al. MSstats: an R package for statistical analysis of quantitative mass spectrometry-based proteomic experiments. Bioinformatics. 2014;30: 2524–6. doi: 10.1093/bioinformatics/btu305 [DOI] [PubMed] [Google Scholar]
- 24.Bates D, Mächler M, Bolker B, Walker S. Fitting Linear Mixed-Effects Models Using lme4. J Stat Softw. 2015;67. [Google Scholar]
- 25.Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing on JSTOR [Internet]. Available: http://www.jstor.org/stable/2346101?seq=1#page_scan_tab_contents
- 26.Shultz EK. Multivariate receiver-operating characteristic curve analysis: prostate cancer screening as an example. Clin Chem. 1995;41: 1248–55. [PubMed] [Google Scholar]
- 27.Carstensen B, Plummer M. A Package for Statistical Analysis in Epidemiology. 2015; Available: https://cran.r-project.org/web/packages/Epi/index.html
- 28.Cheng Y-SL, Rees T, Wright J. A review of research on salivary biomarkers for oral cancer detection. Clin Transl Med. 2014;3: 3 doi: 10.1186/2001-1326-3-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.St John MAR, Li Y, Zhou X, Denny P, Ho C-M, Montemagno C, et al. Interleukin 6 and interleukin 8 as potential biomarkers for oral cavity and oropharyngeal squamous cell carcinoma. Arch Otolaryngol Head Neck Surg. 2004;130: 929–35. doi: 10.1001/archotol.130.8.929 [DOI] [PubMed] [Google Scholar]
- 30.Arellano-Garcia M, Hu S, Wang J, Henson B, Zhou H, Chia D, et al. Multiplexed immunobead-based assay for detection of oral cancer protein biomarkers in saliva. Oral Dis. 2008;14: 705–712. doi: 10.1111/j.1601-0825.2008.01488.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Elashoff D, Zhou H, Reiss J, Wang J, Xiao H, Henson B, et al. Prevalidation of salivary biomarkers for oral cancer detection. Cancer Epidemiol Biomarkers Prev. 2012;21: 664–72. doi: 10.1158/1055-9965.EPI-11-1093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Korostoff A, Reder L, Masood R, Sinha UK. The role of salivary cytokine biomarkers in tongue cancer invasion and mortality. Oral Oncol. 2011;47: 282–287. doi: 10.1016/j.oraloncology.2011.02.006 [DOI] [PubMed] [Google Scholar]
- 33.Brinkmann O, Kastratovic DA, Dimitrijevic M V, Konstantinovic VS, Jelovac DB, Antic J, et al. Oral squamous cell carcinoma detection by salivary biomarkers in a Serbian population. Oral Oncol. 2011;47: 51–5. doi: 10.1016/j.oraloncology.2010.10.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Katakura A, Kamiyama I, Takano N, Shibahara T, Muramatsu T, Ishihara K, et al. Comparison of salivary cytokine levels in oral cancer patients and healthy subjects. Bull Tokyo Dent Coll. 2007;48: 199–203. [DOI] [PubMed] [Google Scholar]
- 35.SahebJamee M, Eslami M, AtarbashiMoghadam F, Sarafnejad A. Salivary concentration of TNFalpha, IL1 alpha, IL6, and IL8 in oral squamous cell carcinoma. Med oral, Patol oral y cirugía bucal. 2008;13: E292–5. [PubMed] [Google Scholar]
- 36.Hu S, Arellano M, Boontheung P, Wang J, Zhou H, Jiang J, et al. Salivary proteomics for oral cancer biomarker discovery. Clin Cancer Res. 2008;14: 6246–52. doi: 10.1158/1078-0432.CCR-07-5037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jou Y-J, Hua C-H, Lin C-D, Lai C-H, Huang S-H, Tsai M-H, et al. S100A8 as potential salivary biomarker of oral squamous cell carcinoma using nanoLC–MS/MS. Clin Chim Acta. 2014;436: 121–129. doi: 10.1016/j.cca.2014.05.009 [DOI] [PubMed] [Google Scholar]
- 38.de Jong EP, Xie H, Onsongo G, Stone MD, Chen X-B, Kooren JA, et al. Quantitative proteomics reveals myosin and actin as promising saliva biomarkers for distinguishing pre-malignant and malignant oral lesions. PLoS One. 2010;5: e11148 doi: 10.1371/journal.pone.0011148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mizukawa N, Sugiyama K, Fukunaga J, Ueno T, Mishima K, Takagi S, et al. Defensin-1, a peptide detected in the saliva of oral squamous cell carcinoma patients. Anticancer Res. 18: 4645–9. [PubMed] [Google Scholar]
- 40.Sato J, Goto J, Murata T, Kitamori S, Yamazaki Y, Satoh A, et al. Changes in saliva interleukin-6 levels in patients with oral squamous cell carcinoma. Oral Surg Oral Med Oral Pathol Oral Radiol Endod. 2010;110: 330–6. doi: 10.1016/j.tripleo.2010.03.040 [DOI] [PubMed] [Google Scholar]
- 41.Warnakulasuriya S, Soussi T, Maher R, Johnson N, Tavassoli M. Expression of p53 in oral squamous cell carcinoma is associated with the presence of IgG and IgA p53 autoantibodies in sera and saliva of the patients. J Pathol. 2000;192: 52–57. doi: 10.1002/1096-9896(2000)9999:9999<::AID-PATH669>3.0.CO;2-C [DOI] [PubMed] [Google Scholar]
- 42.Chen Y-C, Li T-Y, Tsai M-F. Analysis of the saliva from patients with oral cancer by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. Rapid Commun Mass Spectrom. 2002;16: 364–9. doi: 10.1002/rcm.588 [DOI] [PubMed] [Google Scholar]
- 43.Jou Y-J, Lin C-D, Lai C-H, Chen C-H, Kao J-Y, Chen S-Y, et al. Proteomic identification of salivary transferrin as a biomarker for early detection of oral cancer. Anal Chim Acta. 2010;681: 41–8. doi: 10.1016/j.aca.2010.09.030 [DOI] [PubMed] [Google Scholar]
- 44.Jou Y-J, Lin C-D, Lai C-H, Tang C-H, Huang S-H, Tsai M-H, et al. Salivary zinc finger protein 510 peptide as a novel biomarker for detection of oral squamous cell carcinoma in early stages. Clin Chim Acta. 2011;412: 1357–65. doi: 10.1016/j.cca.2011.04.004 [DOI] [PubMed] [Google Scholar]
- 45.Wu C-C, Chu H-W, Hsu C-W, Chang K-P, Liu H-P. Saliva proteome profiling reveals potential salivary biomarkers for detection of oral cavity squamous cell carcinoma. Proteomics. 2015;15: 3394–404. doi: 10.1002/pmic.201500157 [DOI] [PubMed] [Google Scholar]
- 46.Lisa Cheng Y-S, Jordan L, Gorugantula LM, Schneiderman E, Chen H-S, Rees T. Salivary interleukin-6 and -8 in patients with oral cancer and patients with chronic oral inflammatory diseases. J Periodontol. 2014;85: 956–65. doi: 10.1902/jop.2013.130320 [DOI] [PubMed] [Google Scholar]
- 47.Gorugantula LM, Rees T, Plemons J, Chen H-S, Cheng Y-SL. Salivary basic fibroblast growth factor in patients with oral squamous cell carcinoma or oral lichen planus. Oral Surg Oral Med Oral Pathol Oral Radiol. 2012;114: 215–22. doi: 10.1016/j.oooo.2012.03.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pickering V, Jordan RCK, Schmidt BL. Elevated salivary endothelin levels in oral cancer patients—a pilot study. Oral Oncol. 2007;43: 37–41. doi: 10.1016/j.oraloncology.2005.12.027 [DOI] [PubMed] [Google Scholar]
- 49.Cheng Y-SL, Rees T, Jordan L, Oxford L, O’Brien J, Chen H-S, et al. Salivary endothelin-1 potential for detecting oral cancer in patients with oral lichen planus or oral cancer in remission. Oral Oncol. 2011;47: 1122–6. doi: 10.1016/j.oraloncology.2011.07.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Franzmann EJ, Reategui EP, Pedroso F, Pernas FG, Karakullukcu BM, Carraway KL, et al. Soluble CD44 is a potential marker for the early detection of head and neck cancer. Cancer Epidemiol Biomarkers Prev. 2007;16: 1348–55. doi: 10.1158/1055-9965.EPI-06-0011 [DOI] [PubMed] [Google Scholar]
- 51.Vucicević Boras V, Cikes N, Lukać J, Virag M, Cekić-Arambasin A. Salivary and serum interleukin 6 and basic fibroblast growth factor levels in patients with oral squamous cell carcinoma. Minerva Stomatol. 2005;54: 569–73. [PubMed] [Google Scholar]
- 52.Contucci A, Inzitari R, Agostino S, Vitali A, Fiorita A, Cabras T, et al. Statherin levels in saliva of patients with precancerous and cancerous lesions of the oral cavity: a preliminary report. Oral Dis. 2005;11: 95–99. doi: 10.1111/j.1601-0825.2004.01057.x [DOI] [PubMed] [Google Scholar]
- 53.Mascitti M SA. Detection Level of Salivary Survivin in Patients with OSCC. J Carcinog Mutagen. OMICS International; 2013;2013. [Google Scholar]
- 54.Nagler R, Bahar G, Shpitzer T, Feinmesser R. Concomitant analysis of salivary tumor markers—a new diagnostic tool for oral cancer. Clin Cancer Res. 2006;12: 3979–84. doi: 10.1158/1078-0432.CCR-05-2412 [DOI] [PubMed] [Google Scholar]
- 55.Shpitzer T, Hamzany Y, Bahar G, Feinmesser R, Savulescu D, Borovoi I, et al. Salivary analysis of oral cancer biomarkers. Br J Cancer. 2009;101: 1194–1198. doi: 10.1038/sj.bjc.6605290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shpitzer T, Bahar G, Feinmesser R, Nagler RM. A comprehensive salivary analysis for oral cancer diagnosis. J Cancer Res Clin Oncol. 2007;133: 613–617. doi: 10.1007/s00432-007-0207-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Balan JJ, Rao RS, Premalatha BR, Patil S. Analysis of tumor marker CA 125 in saliva of normal and oral squamous cell carcinoma patients: a comparative study. J Contemp Dent Pract. 13: 671–5. [DOI] [PubMed] [Google Scholar]
- 58.Shetty SR, Chadha R, Babu S, Kumari S, Bhat S, Achalli S. Salivary lactate dehydrogenase levels in oral leukoplakia and oral squamous cell carcinoma: a biochemical and clinicopathological study. J Cancer Res Ther. 2012;8 Suppl 1: S123–5. [DOI] [PubMed] [Google Scholar]
- 59.He H, Chen G, Zhou L, Liu Y. A joint detection of CEA and CA-50 levels in saliva and serum of patients with tumors in oral region and salivary gland. J Cancer Res Clin Oncol. 2009;135: 1315–21. doi: 10.1007/s00432-009-0572-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jessie K, Jayapalan JJ, Ong K-C, Abdul Rahim ZH, Zain RM, Wong K-T, et al. Aberrant proteins in the saliva of patients with oral squamous cell carcinoma. Electrophoresis. 2013;34: 2495–502. doi: 10.1002/elps.201300107 [DOI] [PubMed] [Google Scholar]
- 61.Hermann P, Gera I, Borbély J, Fejérdy P, Madléna M. Periodontal health of an adult population in Hungary: findings of a national survey. J Clin Periodontol. 2009;36: 449–57. doi: 10.1111/j.1600-051X.2009.01395.x [DOI] [PubMed] [Google Scholar]
- 62.Madléna M, Hermann P, Jáhn M, Fejérdy P. Caries prevalence and tooth loss in Hungarian adult population: results of a national survey. BMC Public Health. 2008;8: 364 doi: 10.1186/1471-2458-8-364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Thiel UJE, Feltens R, Adryan B, Gieringer R, Brochhausen C, Schuon R, et al. Analysis of differentially expressed proteins in oral squamous cell carcinoma by MALDI-TOF MS. J Oral Pathol Med. 2011;40: 369–79. doi: 10.1111/j.1600-0714.2010.00982.x [DOI] [PubMed] [Google Scholar]
- 64.Van Raemdonck G, Zegels G, Coen E, Vuylsteke B, Jennes W, Van Ostade X. Increased Serpin A5 levels in the cervicovaginal fluid of HIV-1 exposed seronegatives suggest that a subtle balance between serine proteases and their inhibitors may determine susceptibility to HIV-1 infection. Virology. 2014;458–459: 11–21. doi: 10.1016/j.virol.2014.04.015 [DOI] [PubMed] [Google Scholar]
- 65.Rhodus NL, Ho V, Miller CS, Myers S, Ondrey F. NF-κB dependent cytokine levels in saliva of patients with oral preneoplastic lesions and oral squamous cell carcinoma. Cancer Detect Prev. 2005;29: 42–45. doi: 10.1016/j.cdp.2004.10.003 [DOI] [PubMed] [Google Scholar]
- 66.Mantovani A, Allavena P, Sica A, Balkwill F. Cancer-related inflammation. Nature. 2008;454: 436–444. doi: 10.1038/nature07205 [DOI] [PubMed] [Google Scholar]
- 67.Eiró N, Vizoso FJ. Inflammation and cancer. World J Gastrointest Surg. Baishideng Publishing Group Inc; 2012;4: 62–72. doi: 10.4240/wjgs.v4.i3.62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Komohara Y, Fujiwara Y, Ohnishi K, Takeya M. Tumor-associated macrophages: Potential therapeutic targets for anti-cancer therapy. Adv Drug Deliv Rev. 2016;99: 180–185. doi: 10.1016/j.addr.2015.11.009 [DOI] [PubMed] [Google Scholar]
- 69.Bell RB, Leidner R, Feng Z, Crittenden MR, Gough MJ, Fox BA. Developing an Immunotherapy Strategy for the Effective Treatment of Oral, Head and Neck Squamous Cell Carcinoma. J Oral Maxillofac Surg. 2015;73: S107–S115. doi: 10.1016/j.joms.2015.05.026 [DOI] [PubMed] [Google Scholar]
- 70.Márton IJ, Kiss C. Overlapping Protective and Destructive Regulatory Pathways in Apical Periodontitis. J Endod. 2014;40: 155–163. doi: 10.1016/j.joen.2013.10.036 [DOI] [PubMed] [Google Scholar]
- 71.Jinno T, Kawano S, Maruse Y, Matsubara R, Goto Y, Sakamoto T, et al. Increased expression of interleukin-6 predicts poor response to chemoradiotherapy and unfavorable prognosis in oral squamous cell carcinoma. Oncol Rep. 2015;33: 2161–8. doi: 10.3892/or.2015.3838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Huang J-S, Yao C-J, Chuang S-E, Yeh C-T, Lee L-M, Chen R-M, et al. Honokiol inhibits sphere formation and xenograft growth of oral cancer side population cells accompanied with JAK/STAT signaling pathway suppression and apoptosis induction. BMC Cancer. 2016;16: 245 doi: 10.1186/s12885-016-2265-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Skrinjar I, Brailo V, Vidovic-Juras D, Vucicevic-Boras V, Milenovic A. Evaluation of pretreatment serum interleukin-6 and tumour necrosis factor alpha as a potential biomarker for recurrence in patients with oral squamous cell carcinoma. Med Oral Patol Oral Cir Bucal. 2015;20: e402–7. doi: 10.4317/medoral.20373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Piva MR, DE Souza LB, Martins-Filho PRS, Nonaka CFW, DE Santana Santos T, DE Souza Andrade ES, et al. Role of inflammation in oral carcinogenesis (Part II): CD8, FOXP3, TNF-α, TGF-β and NF-κB expression. Oncol Lett. Spandidos Publications; 2013;5: 1909–1914. doi: 10.3892/ol.2013.1302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Arduino PG, Menegatti E, Cappello N, Martina E, Gardino N, Tanteri C, et al. Possible role for interleukins as biomarkers for mortality and recurrence in oral cancer. Int J Biol Markers. 2015;30: 262–6. [DOI] [PubMed] [Google Scholar]
- 76.Fujita Y, Okamoto M, Goda H, Tano T, Nakashiro K, Sugita A, et al. Prognostic significance of interleukin-8 and CD163-positive cell-infiltration in tumor tissues in patients with oral squamous cell carcinoma. PLoS One. Public Library of Science; 2014;9: e110378 doi: 10.1371/journal.pone.0110378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.He Q-Y, Chen J, Kung H-F, Yuen AP-W, Chiu J-F. Identification of tumor-associated proteins in oral tongue squamous cell carcinoma by proteomics. Proteomics. 2004;4: 271–8. doi: 10.1002/pmic.200300550 [DOI] [PubMed] [Google Scholar]
- 78.Leanderson T, Ivars F. S100A9 and tumor growth. Oncoimmunology. 2012;1: 1404–1405. doi: 10.4161/onci.21027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Cross SS, Hamdy FC, Deloulme JC, Rehman I. Expression of S100 proteins in normal human tissues and common cancers using tissue microarrays: S100A6, S100A8, S100A9 and S100A11 are all overexpressed in common cancers. Histopathology. 2005;46: 256–69. doi: 10.1111/j.1365-2559.2005.02097.x [DOI] [PubMed] [Google Scholar]
- 80.Kawai H, Minamiya Y, Takahashi N. Prognostic impact of S100A9 overexpression in non-small cell lung cancer. Tumour Biol. 2011;32: 641–6. doi: 10.1007/s13277-011-0163-8 [DOI] [PubMed] [Google Scholar]
- 81.Duan L, Wu R, Ye L, Wang H, Yang X, Zhang Y, et al. S100A8 and S100A9 are associated with colorectal carcinoma progression and contribute to colorectal carcinoma cell survival and migration via Wnt/β-catenin pathway. PLoS One. 2013;8: e62092 doi: 10.1371/journal.pone.0062092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wu W, Juan WC, Liang CRMY, Yeoh KG, So J, Chung MCM. S100A9, GIF and AAT as potential combinatorial biomarkers in gastric cancer diagnosis and prognosis. Proteomics Clin Appl. 2012;6: 152–62. doi: 10.1002/prca.201100050 [DOI] [PubMed] [Google Scholar]
- 83.Chao A, Wang T-H, Lee Y-S, Hsueh S, Chao A-S, Chang T-C, et al. Molecular characterization of adenocarcinoma and squamous carcinoma of the uterine cervix using microarray analysis of gene expression. Int J Cancer. 2006;119: 91–8. doi: 10.1002/ijc.21813 [DOI] [PubMed] [Google Scholar]
- 84.Markowitz J, Carson WE. Review of S100A9 biology and its role in cancer. Biochim Biophys Acta. 2013;1835: 100–9. doi: 10.1016/j.bbcan.2012.10.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Raffel J, Bhattacharyya AK, Gallegos A, Cui H, Einspahr JG, Alberts DS, et al. Increased expression of thioredoxin-1 in human colorectal cancer is associated with decreased patient survival. J Lab Clin Med. 2003;142: 46–51. doi: 10.1016/S0022-2143(03)00068-4 [DOI] [PubMed] [Google Scholar]
- 86.Noike T, Miwa S, Soeda J, Kobayashi A, Miyagawa S. Increased expression of thioredoxin-1, vascular endothelial growth factor, and redox factor-1 is associated with poor prognosis in patients with liver metastasis from colorectal cancer. Hum Pathol. 2008;39: 201–8. doi: 10.1016/j.humpath.2007.04.024 [DOI] [PubMed] [Google Scholar]
- 87.Grogan TM, Fenoglio-Prieser C, Zeheb R, Bellamy W, Frutiger Y, Vela E, et al. Thioredoxin, a putative oncogene product, is overexpressed in gastric carcinoma and associated with increased proliferation and increased cell survival. Hum Pathol. 2000;31: 475–81. doi: 10.1053/hp.2000.6546 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Samples in bold were used for both Bioplex and ELISA experiments.
(XLSX)
Data Availability Statement
Data are publicly available through the Panorama website (https://panoramaweb.org/labkey/project/University%20of%20Debrecen/OSCC%20saliva/begin.view?).