Abstract
Epithelial ovarian cancer (EOC) is a disease responsible for more deaths among women in the Western world than all other gynecologic malignancies. There is urgent need for new therapeutic targets and a better understanding of EOC initiation and progression. We have previously identified the polypeptide N-acetylgalactosaminyltransferase 3 (GALNT3) gene, a member of the GalNAc-transferases (GalNAc-Ts) gene family, as hypomethylated and overexpressed in high-grade serous EOC tumors, compared to low malignant potential EOC tumors and normal ovarian tissues. This data also suggested for a role of GALNT3 in aberrant EOC glycosylation, possibly implicated in disease progression. To evaluate differential glycosylation in EOC caused by modulations in GALNT3 expression, we used a metabolic labeling strategy for enrichment and mass spectrometry-based characterization of glycoproteins following GALNT3 gene knockdown (KD) in A2780s EOC cells. A total of 589 differentially expressed glycoproteins were identified upon GALNT3 KD. Most identified proteins were involved in mechanisms of cellular metabolic functions, post-translational modifications, and some have been reported to be implicated in EOC etiology.
The GALNT3-dependent glycoproteins identified by this metabolic labeling approach support the oncogenic role of GALNT3 in EOC dissemination and may be pursued as novel EOC biomarkers and/or therapeutic targets. Biological significance: Knowledge of the O-glycoproteome has been relatively elusive, and the functions of the individual polypeptide GalNAc-Ts have been poorly characterized. Alterations in GalNAc-Ts expression were shown to provide huge variability in the O-glycoproteome in various pathologies, including cancer. The application of a chemical biology approach for the metabolic labeling and subsequent characterization of O-glycoproteins in EOC using the Ac4GalNAz metabolite has provided a strategy allowing for proteomic discovery of GalNAc-Ts specific functions. Our study supports an essential role of one of the GalNAc-Ts — GALNT3, in EOC dissemination, including its implication in modulating PTMs and EOC metabolism. Our approach validates the use of the applied metabolic strategy to identify important functions of GalNAc-Ts in normal and pathological conditions.
Keywords: Epithelial ovarian cancer, GALNT3, Glycosylation, Metabolic labeling, Glycoproteomics, Pathway and network analysis
1. Introduction
Epithelial ovarian cancer (EOC) is a disease that is responsible for more cancer deaths among women in the Western world than all other gynecologic malignancies [1]. EOC lethality primarily stems from the inability to detect the disease at an early, organ-confined stage, and the lack of effective therapies for advanced-stage (metastatic) disease. Indeed, despite advances in cytotoxic therapies [2,3], only 30% of patients with advanced-stage EOC survive 5 years after initial diagnosis [1]. Thus, management of metastatic disease is a crucial problem for EOC treatment. One way to resolve this problem is to target metastasis-specific pathways with novel therapies. Hence, focused identification of novel pro-metastatic EOC pathways and molecules could improve the chances of discovering new and more effective EOC therapies.
Protein glycosylation is a highly heterogeneous and ubiquitous post-translational modification (PTM) representing glycan structures attached at asparagine (N-linked) or serine or threonine (O-linked) side chains [4]. Analysis of the SWISS-PROT database predicted that more than 50% of all proteins are glycosylated [5]. Glycoproteins are involved in many biological functions, such as intercellular communication [6], intracellular signaling [7] and protein stability [8]. Glycosylation changes characterized in cancer cells follow a variety of forms, as glycan alterations can be associated with loss or gain of expression, depending on the cell type and the specific glycan’s structure [9]. Aberrant glycosylation in cancer cells could affect certain ligand–receptor interactions and more importantly, could favor cancer cell proliferation, migration and invasion/metastasis [4,10]. Some of the most extensively examined mechanisms of aberrant glycosylation in cancer represent the alterations observed in the expression of glycotransferases, which are the enzymes responsible for the biosynthesis of glycoprotein and glycolipid sugar chains [11]. These enzymes are located in the endoplasmic reticulum and the Golgi apparatus [5]. Multiple studies have investigated the expression profiles of glycotransferases in different cancer types [11–14], and it was reported that oncogenic transformations of these enzymes are essentially regulated at their transcriptional level, thus emphasizing the importance of analyzing the glycotransferases expression patterns as inducers/markers for tumor progression and disease prognosis [12].
We have previously identified the polypeptide N-acetylgalactosaminyltransferase 3 (GALNT3) gene, a member of the GalNAc-transferases (GalNAc-Ts) gene family, as hypomethylated and overexpressed in high-grade serous EOC tumors, compared to low-malignant potential EOC tumors and normal ovarian tissues, as GALNT3 expression was functionally related to EOC progression [14]. Moreover, a short hairpin RNA- (shRNA)-mediated GALNT3 knockdown (KD) in the EOC cell line A2780s was associated with reduced mucin-1 (MUC1) protein expression, probably related to destabilization of the MUC1 protein due to the lack of GALNT3 glycosylation activity [14]. Taken together, our data are indicative of a strong oncogenic potential of the GALNT3 gene in advanced EOC, and suggest that GALNT3 overexpression may contribute to EOC dissemination through aberrant mucin O-glycosylation. Studying the role of individual GalNAc-T isoforms has been difficult in the past due to the small number of identified glycoproteins. O-Glycosylation directed by distinct GalNAc-Ts has been demonstrated to emerge as an important regulator of protein function [15], and thus a better understanding of the O-glycoproteome is vital for advancing our knowledge on the large GalNAc-T gene family and their role during cell differentiation and malignant progression.
In the present study, we performed a glycoproteomic analysis aimed to characterize GALNT3-dependent glycoprotein expression in EOC and its role in disease progression. Our methodological approach included the application of a bioorthogonal chemical reporter strategy for probing glycans [16,17]. The method is implemented by metabolical labeling of glycoproteins with a monosaccharide precursor attached to a functional azido group [18]. This approach allows the labeling of glycans bearing azido sugars followed by selective capture from cell lysates [18]. Following metabolic labeling of GALNT3-KD and control A2780s EOC cells with a peracetylated azido GalNAc analog (peraceetylated N-azidoacetylgalactosamine, Ac4GalNAz), glycoproteomics was performed by mass spectrometry (MS) analysis of enriched glycoproteins, to better understand the molecular mechanisms of GALNT3 in EOC dissemination.
We show herein that GALNT3 KD in A2780s cells was associated with the down-regulation of glycoproteins having significant functions in cancer progression including PTMs, cell adhesion and proliferation, regulation of protein transport, cholesterol biosynthesis, cellular metabolism, as well as with alterations of some cancer-related signaling pathways, including the TGFβ, MAPK and NF-kappa-B pathways. Our glycoproteomic analysis led to the identification of novel glycoproteins potentially relevant to EOC etiology and confirmed our previous data of the oncogenic role of GALNT3 in EOC dissemination.
2. Materials and methods
2.1. Cell culture
The GALNT3 KD and control A2780s clones used in this study were previously generated, as described [14]. Briefly, a shRNA targeting the GALNT3 sequence 5′-TACTGCTGAAGGAAATCAT-3′ was designed using the siRNA Ambion Target Finder software (http://www.ambion.com/techlib/misc/siRNA_finder.html), and subcloned into the pSilencer 4.1 puro vector (Ambion). A2780s cells were stably transfected with this construct and clones were isolated as previously shown [14]. In addition, A2780s cells were mock-transfected with the pSilencer 4.1 puro vector, and the stably-transfected clones were isolated and used as controls.
2.2. Metabolite labeling and protein enrichment
Four cell culture dishes (150 mm) were seeded with 100 μM of the Ac4GalNAz metabolite. Another four cell culture dishes (150 mm) were seeded with 100 μM Ac4GalNAc as a control. Cell suspensions of control and GALNT3 KD cells (2 × 105 cells/ml) were added to each tissue culture plate in Media 1 (RPMI 1640 media containing 10% FBS, 1% penicillin–streptomycin and 1 μg/ml puromycin). The cell culture dishes were incubated for 48 h at 37 °C in a humidified 5% CO2 incubator. Following incubation, the cell culture plates were washed with PBS; then Media 2 (RPMI 1640 media containing 1% penicillin–streptomycin and 1 μg/ml puromycin) was added to the aspirated plates. The cell culture dishes were incubated for an additional 48 h at 37 °C in a humidified 5% CO2 incubator. Media and cell lysates were harvested and concentrated as described previously [19,20]. Briefly, to obtain the “conditioned media fraction”, media from all conditions were harvested, cleared by centrifugation and spin concentrated (using Amicon, 15-ml 10-kDa spin filters) to a final volume of 1 ml. The concentrated residue was washed with PBS and transferred to an Eppendorf microcentrifuge tube. To obtain the “soluble” and “insoluble” fractions, adherent cells were washed with PBS and trypsinized for 5 min at 37 °C. Cells were harvested, centrifuged (300 g, 3 min), and washed with PBS. Cell pellets were resuspended in lysis buffer (2 ml: 10 mM HEPES, pH 7.9, 1.5 mM MgCl2, 10 mM KCl, 0.5% Triton X-100, 1× protease inhibitors, 1 μM thiamet G), swelled for 5 min on ice, and disrupted by Dounce homogenization using a tight glass hand pestle (20–30 strokes) as described previously [19,20]. Insoluble material was pelleted by centrifugation (3700 g, 10 min, at 4 °C). The supernatant was collected as the “soluble fraction” while the pellet was kept as the “insoluble fraction.” The insoluble fraction was resuspended in 1% RapiGest/PBS and briefly sonicated. The conditioned medium and soluble fractions were adjusted to 1% RapiGest/PBS as described before [19,20]. Protein concentration from the three fractions was measured by bicinchonic acid assay (Pierce). Western blots were performed as described in [13]. Affinity purification via copper-catalyzed azide-alkyne cycloaddition (CuAAC) with a biotin–alkyne probe was next performed as previously shown ([19,20]; see also [13]). Briefly, following CuAAC, protein aliquots were precipitated and solubilized. Streptavidin-agarose resin was added, and the resulting mixture was incubated for 12 h at 24 °C with rotation. Beads were pelleted by centrifugation, and the supernatant containing uncaptured proteins was separated as described in [13]. The beads were then washed, reduced and alkylated, followed by on-bead trypsin digestion as shown in [13].
2.3. Mass spectrometry
Peptides resulting from trypsin digestion were resuspended in 15 μl of acetonitrile 2%, trifluoroacetic acid 0.05%, and 1 μl of each sample was analyzed by nanoLC/MS/MS. Peptides were injected and separated by online reversed-phase (RP) nanoscale capillary liquid chromatography (nanoLC) and analyzed by electrospray mass spectrometry (ESI MS/MS). The experiments were performed with a Dionex UltiMate 3000 nanoRSLC chromatography system (Thermo Fisher Scientific/Dionex Softron GmbH, Germering, Germany) connected to an Orbitrap Fusion mass spectrometer (Thermo Fisher Scientific, San Jose, CA, USA) equipped with a nanoelectrospray ion source. Peptides were trapped at 20 μl/min in loading solvent (2% acetonitrile, 0.05% trifluoroacetic acid) on a 5 mm × 300 μm C18 pepmap cartridge pre-column (Thermo Fisher Scientific/Dionex Softron GmbH, Germering, Germany) during a 5 min period. Then, the pre-column was switched online with a self-made 50 cm × 75 μm internal diameter separation column packed with ReproSil-Pur C18-AQ 3-μm resin (Dr. Maisch HPLC GmbH, Ammerbuch-Entringen, Germany) and the peptides were eluted with a linear gradient from 5 to 40% solvent B (A: 0.1% formic acid, B: 80% acetonitrile, 0.1% formic acid) for 90 min, at 300 nl/min. Mass spectra were acquired using a data dependent acquisition mode using Thermo XCalibur software version 3.0.63. Full scan mass spectra (350 to 1800 m/z) were acquired in the Orbitrap using an Automatic Gain Control (AGC) target of 4e5, a maximum injection time of 50 ms and a resolution of 120,000. Internal calibration using lock mass on the m/z 445.12003 siloxane ion was used. Each MS scan was followed by acquisition of fragmentation MS spectra of the most intense ions for a total cycle time of 3 s (top speed mode). The selected ions were isolated using the quadrupole analyzer in a window of 1.6 m/z and fragmented by Higher-energy Collision-induced Dissociation (HCD) with 35% of collision energy. The resulting fragments were detected by the linear ion trap in rapid scan rate with an AGC target of 1e4 and a maximum injection time of 50 ms. Dynamic exclusion of previously fragmented peptides was set for a period of 20 s and a tolerance of 10 ppm.
2.4. Database searching and label free quantification
Spectra were searched against a human proteins database (Uniprot Complete Proteome – taxonomy Homo sapiens – 69165 sequences) using the Andromeda module of MaxQuant software v. 1.5.2.8 [11,21], as shown in [13]. For protein validation, a maximum false discovery rate of 1% at peptide and protein level was used based on a target/decoy search. MaxQuant was also applied for Label Free Quantification (LFQ), as described in [13].
2.5. Bioinformatic annotation & analysis
O- and N-glycoprotein prediction analysis using the NetOGlyc 4.0 and the NetNGlyc 1.0 servers was performed, as described in [13]. Gene ontology (GO) enrichment analysis was performed using AmiGO (http://amigo.geneontology.org), as shown in [13]. The GO term enrichment tool was used to determine the observed level of annotations for the set of proteins from our study and determine the significance in the context of all proteins annotated in the human proteome.
Functional, canonical pathway and network analyses were generated using the Ingenuity pathways analysis (IPA) software v7.1 (Ingenuity Systems, www.ingenuity.com). Proteins that met the expression ratio with a cut-off ≥2.0, and a p-value cut-off ≤0.05 for differential expression were considered for the analyses. Swiss-Prot accession numbers were inserted into the software along with corresponding comparison fold change ratios between our control and GALNT3 KD protein data sets. The corresponding Gene ID of each protein accession ID was then generated using the IPA software.
3. Results
3.1. Purification and LC/MS analysis of glycosylated proteins from control and GALNT3 KD A2780s cells
In order to analyze the differential expression of mucin-type glycoproteins upon GALNT3 KD in EOC cells, control and GALNT3 KD A2780s cells were separately labeled with Ac4GalNAz or tetraacetylated N-acetylgalactosamine (Ac4GalNAc, negative control) (See Fig. 1 in [13]). Ac4GalNAz is intracellularly deacetylated and used to incorporate N-azidoacetylgalactosamine (GalNAz) to tag mucin type O-linked glycans [5]. It has been demonstrated that GalNAz is transformed to UDP-GalNAz in vitro and in vivo, enabling the labeling for proteins with O-linked glycans [5]. The labeled control and GALNT3 KD A2780s cells were then subjected to subcellular fractionation to obtain the three fractions (conditioned media fraction, soluble fraction and insoluble fraction) followed by glycoprotein enrichment (see Fig. 1 in [13]). Incorporation of azido-sugar into glycoproteins from all three fractions was examined by Western blot analysis of the collected lysates. Anti-biotin signal was checked before affinity-capture (Load) and after affinity-capture on the fraction not bound to the beads (Supernatant), as well as on the fractions that included the bead after washing (Capture) from all there fractions collected from both the control and GALNT3 KD A2780s EOC cells (see Fig. 2 in [13]). Glycoproteins from the conditioned media fraction incubated with Ac4GalNAz showed a strong robust labeling, whereas labeling in the soluble and insoluble fractions were not as efficient, as evidenced in lane C in both control and GALNT3 KD blots (see Fig. 2 in [13]). Trypsin digestion was then performed and the released peptides were analyzed by LC-MS/MS mass spectrometry. The analysis of the three subcellular fractions from the control and GALNT3 KD A2780s cells cultured with Ac4GalNAz lead to the identification of a total of 5111 proteins by searching the data against the UniProt Complete Proteome Human database (see Supplementary Table 1 in [13]). Subtraction of proteins found in the Ac4GalNAc fraction, identified a total of 2120 proteins in the Ac4GalNAz treated samples (see Supplementary Table 1 in [13]). Prediction analysis of the 2120 proteins dataset using the NetOGlyc 4.0 server indicated that the majority of these proteins (1870 proteins, representing ~88%) could be O-glycosylated (Fig. 1A, also see Supplementary Table 2 in [13]). Moreover, we examined if our list of 2120 proteins also contain N-glycosylation prediction sites; for that we used the NetNGlyc 1.0 server, and interestingly 1576 (~74%) of the annotated proteins were predicted to be N-glycosylated (Fig. 1A, also see Supplementary Table 2 in [13]). Comparison of our list of annotated glycoproteins to previously published assigned glycopeptides from key references [19, 22–31], focused on isolating and/or sequencing a profound number of glycoproteins, indicated that 638 (~30%) of the 2120 proteins identified in our study have been formerly characterized as glycoproteins (Fig. 1B, also see Supplementary Table 3 in [13]), while the remaining proteins have not been previously known to be glycosylated, suggesting that many more glycoproteins, and particularly O-glycoproteins, remain to be discovered.
Fig. 1.
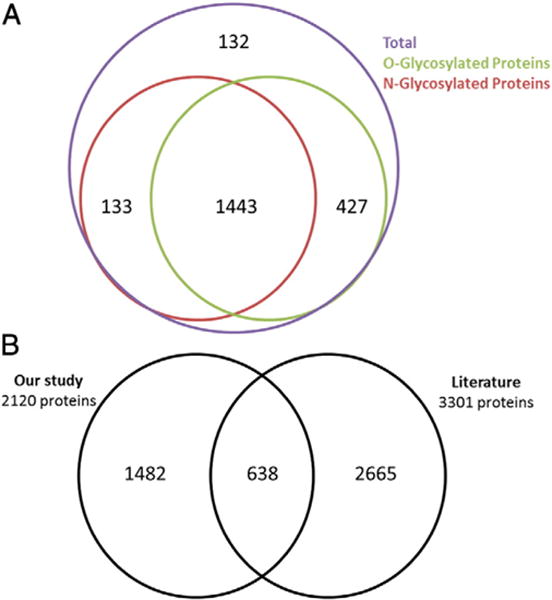
Glycoprotein predictions. A. Comparative analysis of O-glycoprotein and N-glycoprotein predictions using the NetOGlyc 4.0 and NetNGlyc 1.0 servers. B. Comparative analysis of the proteins identified in our study to glycoproteins previously identified in the literature.
3.2. Comparative proteomic analysis of differentially regulated glycoproteins identified between control and GALNT3 KD A2780s EOC cells
We further examined the differential regulation of glycoproteins upon GALNT3 KD in EOC cells, based on our identified set of 2120 proteins. Metabolic labeling and high mass accuracy MS were applied to retrieve a maximum level of proteomic data. We investigated the three subcellular fractions by quantitative proteomics using the MaxQuant software and Andromeda search engine (included in MaxQuant) [11,32]. Proteins were considered as differentially expressed between control and GALNT3 KD A2780s cells if they met the following criteria: Welch test p-value ≤0.05 and fold change in relative expression of ≥2. Based on these stringent selection criteria, we found a total of 275 glycoproteins to be up-regulated and 314 glycoproteins to be down-regulated following GALNT3 KD in A2780s cells (see Supplementary Table 4 in [13]). These criteria were similar to those described in the literature on the use of label-free quantification methods to define biological regulation evaluated at the proteome level [33–36]. Alternative statistical analysis applying z-score statistics (as described in [34]) gave almost identical results with 95% overlap of the data obtained with the previous analysis (see Supplementary Table 4 in [13]). Thus, data obtained with the initial selection criteria (Welch test p-value ≤0.05 and fold change ≥2), were used for all consecutive analyses (see [13] for a more detailed description of the statistical tests performed on the datasets). Following these analyses, a quantifiable difference between control A2780s and GALNT3 KD cells was found for 327, 118, and 144 proteins in the conditioned media, the soluble and the insoluble fractions, respectively (see Supplementary Table 4 in [13]). In the conditioned media fraction, 104 of the identified proteins were up-regulated and 223 were down-regulated (see Supplementary Table 4 in [13]). In the soluble fraction comparison, 63 of the identified proteins were up-regulated and 55 were down-regulated. In the insoluble fraction comparison, 108 of the identified proteins were up-regulated and 36 were down-regulated (see Supplementary Table 4 in [13]). Volcano plots −log10 (p-value) vs. log2 (fold change of GALNT3 KD/Control) were also constructed to graphically display the quantitative data from each of the three fractions (Fig. 2A–C).
Fig. 2.
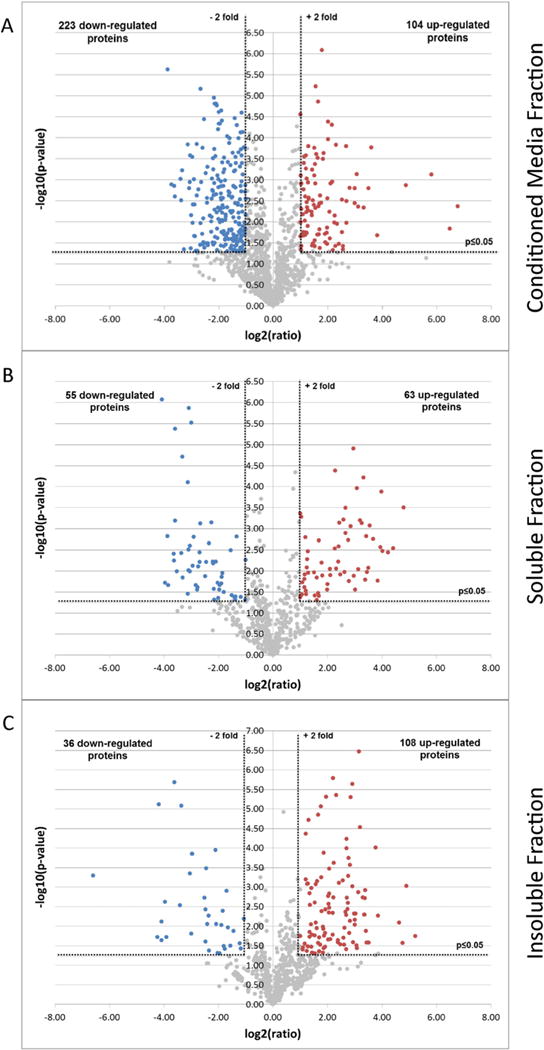
Volcano plots illustrate differentially regulated proteins. The quantified proteins in A. Conditioned media fraction, B. Soluble fraction and C. Insoluble fraction were plotted on Volcano graphs — The x-axis represents the log2 (ratio) where ‘ratio’ is the ratio of the averages of LFQ Intensities of GALNT3 KD over control. The y-axis represents the −log10 (p-value) where ‘p-value’ is the p-value associated to the statistical Welch test applied on the three technical replicates of each GALNT3 and control samples. The non-axial vertical dotted lines denote ±2-fold change while the non-axial horizontal line denotes p ≤ 0.05. The colored dots showed proteins considered as up- (red) or down- (blue) regulated, while the non-regulated proteins are display as gray dots.
3.3. Cellular classification of differentially regulated proteins identified between control and GALNT3 KD A2780s EOC cells
Cellular component GO analysis of the differentially regulated glycoproteins identified between the control and GALNT3 KD A2780s cells was performed on each of the identified fractions (conditioned media fraction, soluble fraction and insoluble fraction), and data were compared to the entire human proteome using the GO Consortium for enrichment analysis [12]. Analysis from the conditioned media fraction showed a clear overrepresentation in membrane type, extracellular, as well as organelle compartment proteins (Fig. 3; also see Fig. 3 and Supplementary Table 5 in [13]). Interestingly, cytoplasmic and ER proteins were also clearly represented in this fraction (Fig. 3; also see Fig. 3 and Supplementary Table 5 in [13]). O-glycosylation of ER type proteins has not been extensively examined, as our metabolic labeling approach has revealed a strong overrepresentation of glycoproteins in the ER organelle. In contrast, in addition to membrane-bound organelle proteins, proteins localized to the intracellular, organelle and cytoplasm compartments were significantly enriched in the soluble fraction (Fig. 3; also see Fig. 3 and Supplementary Table 5 in [13]). Finally, protein localization analysis from the insoluble fraction indicated enrichment in intracellular and organelle compartments, as expected (Fig. 3; also see Fig. 3 in (13)). These observations indicate that GALNT3 KD was predominantly and significantly associated with glycoproteins alterations in the membrane, cytoplasmic and secreted/extracellular space compartments (Fig. 3; also see Fig. 3 and Supplementary Table 5 in [13]). Western blot assays confirmed the enrichment of the glycoproteins in the conditioned media fraction (see Fig. 2 in [13]). Overall our data indicate that most of the labeled glycoproteins arising from the Ac4GalNAz treatment may be of the O-GalNAz-type, as judged by the relative enrichment of membrane glycoproteins in the conditioned media fraction (Fig. 3; also see Fig. 3 and Supplementary Table 5 in [13]).
Fig. 3.
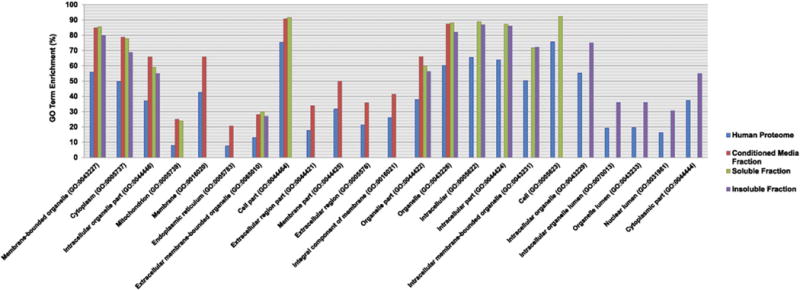
Cellular component Gene Ontology analysis. Bar graphs showing the cellular component GO terms that are significantly over represented from the differentially regulated proteins in our study, compared to the entire human proteome. Data were submitted to the Gene Ontology Consortium for enrichment analysis [12]. The analysis was performed on the differentially regulated proteins identified from each of the three fractions: Conditioned media fraction (red bars), Soluble fraction (green bars) and Insoluble fraction (purple bars) as compared to the entire human proteome (blue bars). All identified proteins annotated with GO cellular component terms are compared against the annotated human proteome. Categories were scored by a combination of percentage of enrichment and p-value (p ≤ 0.05). The enrichment p-values are corrected by Benjamini’s methods.
3.4. Pathways and network analyses following GALNT3 gene KD in EOC cells
The functional and canonical pathway and network analyses of differentially expressed glycoproteins identified upon GALNT3 KD in A2780s cells were explored using the IPA software. Here, the 589 identified up- and down-regulated glycoproteins (listed in Supplementary Table 4 in [13]) were uploaded in IPA and their matched gene symbols were used for consecutive analyses.
We illustrated the functional changes induced upon KD of the GALNT3 gene by separately comparing the functional categories that were up- and down-regulated in the three studied fractions (conditioned media, soluble and insoluble fractions). The top 15 most significantly perturbed up- and down-regulated molecular and cellular functions are presented in Fig. 4. As seen from Fig. 4A, the major up-regulated functional categories were related to essential cellular mechanisms including molecular transport, small molecule biochemistry, cell-to-cell signaling and interaction, cellular assembly and organization, cellular function and maintenance, cellular development, cellular movement, cell morphology, cellular compromise, protein trafficking, while notably, PTM-related pathways were found to be both induced and suppressed (Fig. 4A and B). Down-regulated functional pathways were mostly implicated in energy production and metabolism (including nucleic acid, lipid, amino acid and carbohydrate metabolism), and protein synthesis (Fig. 4B).
Fig. 4.
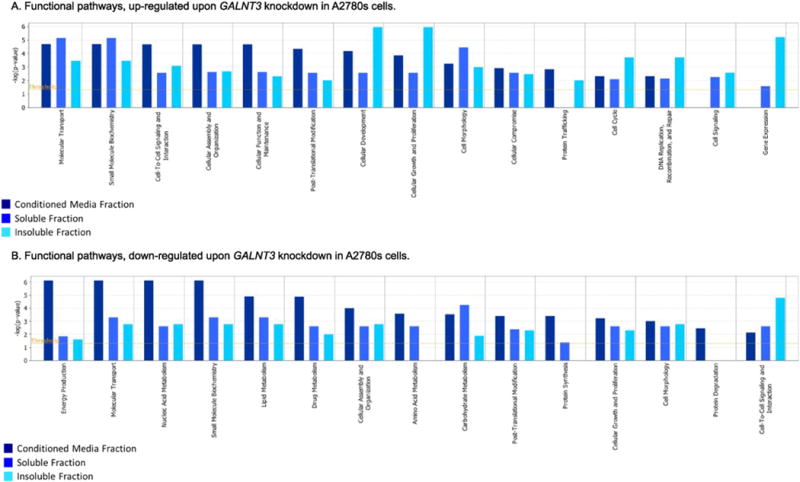
Functional pathway analysis. Functional pathway analysis for a dataset of differentially expressed glycoproteins (≥2-fold; as based on their matched gene symbols) following GALNT3 suppression in A2780s cells, as observed in the three fractions (conditioned media, soluble and insoluble fractions). A. Functional pathway analysis of up-regulated genes (275); B. Functional analysis of down-regulated genes (314). Dark blue indicates the conditioned media fraction functional network gene expression values, light blue indicates the soluble fraction functional network gene expression values; cyan blue indicates the insoluble fraction functional network gene expression values. The functional pathways included in this analysis are shown along the x-axis of the bar chart. The y-axis indicates the statistical significance on the left. Calculated using the right-tailed Fisher exact test, the p-value indicates which biologic annotations are significantly associated with the input molecules relative to all functionally characterized mammalian molecules. The yellow threshold line represents the default significance cutoff at p = 0.05.
IPA canonical pathway analysis supported these findings, as the top 18 up-regulated canonical pathways were mostly related to sphingosine 1-phosphate signaling, epithelial adherens junction signaling, ILK signaling, integrin signaling, ovarian cancer signaling, p38 MAPK signaling and AMPK signaling (Fig. 5A), while the top 18 down-regulated canonical pathways were associated with mitochondrial dysfunction, oxidative phosphorylation, cholesterol biosynthesis, leucine degradation, isoleucine degradation, valine degradation, autophagy, and UDP-N-acetyl-D-galactosamine biosynthesis I (Fig. 5B).
Fig. 5.
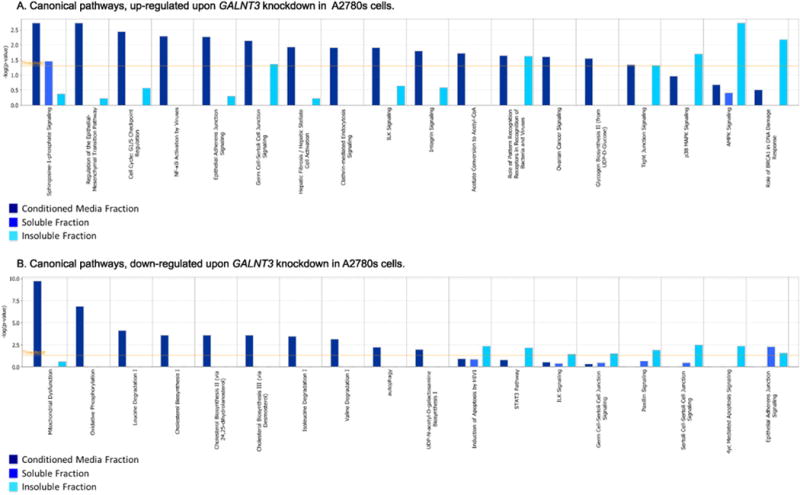
Canonical pathway analysis. Canonical pathway analysis for a dataset of differentially expressed genes (≥2-fold; as based on their matched gene symbols) following GALNT3 suppression in A2780s cells, as observed in the three fractions (conditioned media, soluble and insoluble fractions). A. Canonical pathway analysis of up-regulated genes (275); B. Canonical pathway analysis of down-regulated genes (314). Dark blue indicates the conditioned media fraction canonical network gene expression values, light blue indicates the soluble fraction canonical network gene expression values; cyan blue indicates the insoluble fraction canonical network gene expression values. The canonical pathways included in this analysis are shown along the x-axis of the bar chart. The y-axis indicates the statistical significance on the left. Calculated using the right-tailed Fisher exact test, the p-value indicates which biologic annotations are significantly associated with the input molecules relative to all functionally characterized mammalian molecules. The yellow threshold line represents the default significance cutoff at p = 0.05.
Network analyses confirmed the major functionally related gene groups that were found to be differentially expressed in the GALNT3 KD clone compared to the control A2780s cells from the three fractions examined. We identified 25 highly significant networks with score ≥13. The five top-scoring networks were associated with functions linked to PTM, cardiovascular disease, inflammatory disease, lipid metabolism, embryonic development, cell morphology, and cellular assembly and organization (Table 1). A common network obtained upon merging the five top-scoring networks (Fig. 6) recognized several important nodes linked with numerous interaction partners, including transforming growth factor beta (TGFβ1), mitogen-activated protein kinases (MAPK), phosphoinositide 3-kinase complex (PI3K), sphingomyelin phosphodiesterase 1 (SMPD1), tubulin beta 3 (TUBB3), magnesium superoxide dismutase (SOD2), mitochondrial folate-coupled dehydrogenase (MTHFD2), autophagy protein 5 (ATG5), microtubule-associated protein 1B (MAP1B), dolichyldiphosphooligosaccharide-protein glycosyltransferase (DDOST), dolichyl-diphosphooligosaccharide-protein glycosyltransferase subunit STT3B (STT3B), in addition to several prominent cancer-related genes stemming from the nuclear factor kappa-light-chain-enhancer of activated B complex (NF-Kappa B) such as metadherin (MTDH), annexin A4 (ANXA4), and ribonucleotide reductase M2 (RRMP2).
Table 1.
IPA networks identified via analysis of focus genes. Analysis of differentially expressed genes using IPA revealed a set of gene networks based on known interactions. Up-regulated genes are shown in (red) and down-regulated genes are shown in (green). Genes not altered in our signature are indicated in plain text. The table represents networks 1 to 5 as illustrated in Fig. 6.
| Molecules in network | Focus molecules | Top diseases and functions | p–Value |
|---|---|---|---|

|
31 |
Post–translational modification | (1.49E–09 – 4.73E–03) |
| Nervous system development and function | (1.16E–05 – 4.90E–02) | ||
| Neurological disease | (1.16E–05 – 4.70E–02) | ||

|
31 |
Cardiac Inflammation | (8.27E–06 – 5.24E–01) |
| Cardiovascular disease | (8.27E–06 – 5.32E–01) | ||
| Inflammatory disease | (8.27E–06 – 2.23E–01) | ||

|
28 |
Lipid metabolism | (4.27E–07 – 4.62E–02) |
| Molecular transport | (4.27E–07 – 4.62E–02) | ||
| Small molecule biochemistry | (4.60E–06 – 4.73E–02) | ||

|
27 |
Embryonic development | (4.00E–05 – 1.95E–01) |
| Nervous system development and Function | (4.00E–05 – 2.62E–02) | ||
| Organ development | (4.00E–05 – 4.01E–01) | ||

|
26 |
Cell morphology | (4.16E–04 – 4.26E–02) |
| Cellular assembly and organization, | (4.16E–04 – 4.72E–02) | ||
| Hereditary disorder | (5.61E–04 – 1.05E–02) |
Fig. 6.

Network analysis of dynamic gene expression in A2780s cells based on the 2-fold glycoprotein expression list obtained following GALNT3 KD (see Supplementary Table 4 in [13]). The five top-scoring networks of up- and down-regulated genes were merged and are displayed graphically as nodes (genes/gene products) and edges (the biological relationships between the nodes). Intensity of the node color indicates the degree of up- (red) or down-regulation (green). Nodes are displayed using various shapes that represent the functional class of the gene product (square, cytokine, vertical oval, transmembrane receptor, rectangle, nuclear receptor, diamond, enzyme, rhomboid, transporter, hexagon, translation factor, horizontal oval, transcription factor, circle, other). Edges are displayed with various labels that describe the nature of relationship between the nodes: __ binding only, → acts on. The length of an edge reflects the evidence supporting that node-to-node relationship, in that edges supported by article from literature are shorter. Dotted edges represent indirect interaction.
4. Discussion
Aberrant glycosylation has been previously described and investigated in EOC (reviewed in [37]). Thus, a number of studies suggest the involvement of N-glycosylation in EOC spreading and metastasis. By using a quantitative glycoproteomic analysis, Tian et al. have demonstrated that different N-linked glycoproteins are associated with/could represent biomarkers for the specific EOC subtypes, including high grade and low grade serous, mucinous, high grade and low grade endometrioid, clear-cell, transitional-cell, squamous-cell, mixed and undifferentiated subtypes, as some N-glycoproteins are exclusively expressed in normal ovaries [38]. It was also shown that the N-glycan-dependent mesothelin-MUC16 binding facilitates EOC peritoneal dissemination [39]. Two recent studies have described metastasis-related N-glycan alterations in EOC secretomes in vitro [40, 41], as definite N-glycan substructures and their complexes were found to be associated with specific epigenetic programming of synthetic enzymes in EOC [41]. Moreover, specific sialylated N-glycoproteins were exclusively identified in biological fluids from EOC patients [42]. Aberrant N-glycosylation has been also involved in EOC chemoresistance, as N-glycosylation inhibitors were shown to markedly reduce multidrug resistance [43].
Aberrant O-linked (mucin-type) glycosylation has been less extensively studied and is poorly defined in EOC. Several studies have demonstrated that MUC1 is overexpressed and aberrantly O-glycosylated in most ovarian adenocarcinomas [44], including primary and metastatic EOC tumors [45]. The major structural difference observed in tumor-associated MUC1, when compared to its normal counterpart, is that tumor-associated MUC1 contains shorter and less dense O-glycan chains, thus exposing more regions of the MUC1 protein core [46]. This feature of the tumor-associated MUC1 makes it a more accessible EOC therapeutic target, and thus delineates novel approaches for more effective EOC therapy. To date, 20 GalNAc-Ts have been identified that are responsible for the initiation of mucin type O-glycosylation. The differential glycosylation of mucins in cancer is caused by the dysregulation of the glycosylation machinery that include expression alterations of specific glycotransferases, including GalNAc-Ts, which consecutively leads to the formation of aberrant glycan structures implicated in cancer growth and metastasis.
Alterations of the expression of the large family of 20 GalNAc-Ts have been investigated using RNA interference approaches including small interfering RNA (siRNA), and shRNA in a number of cancers (reviewed in [37,47]), which reinforces the conviction that GalNAc-Ts may play essential roles in the mechanisms leading to tumor dissemination and disease progression. Indeed, a direct correlation has been revealed between elevated expression of GALNT6 in breast cancer cells and increased glycosylation and surface expression of MUC1 [48]. It has been also shown that GALNT14 contributes to the aberrant glycosylation and over-expression of MUC13 in EOC cells, suggesting a role of GALNT14 in EOC carcinogenesis [49]. We have previously demonstrated that GALNT3 KD is associated with increased adhesion and decreased MUC1 stability in EOC cells, suggesting for GALNT3 involvement in EOC progression via aberrant MUC1 glycosylation [14]. Overall, these data provide evidence that altered expression of different members of the GalNAc-Ts gene family can contribute to aberrant O-glycosylation associated with tumor progression and dissemination, also emphasizing the potential use of these enzymes as cancer biomarkers and/or therapeutic targets.
Global analysis of proteins and their modifications has been majorly dependent on mass spectrometry tools for sequencing N- and O-glycans from cells and tissues. Multiple factors have contributed to difficulties in specifically performing O-glycoprotein analysis and have thus complicated the advancement in determining and understanding the functions of O-glycosylation in normal and pathological conditions. More recently, advances in mass-spectrometry have overcome some of the challenges accompanied with the complexity of glycosylation, including enrichment protocols aimed at labeling O-linked glycosylation. Thus, extensive research has introduced some enrichment methods including lectin affinity chromatography, which has been used for specifically analyzing O-GlcNAc and O-GalNAc type glycoproteins [33,50], while other approaches have been developed towards a more targeted O-glycan selection including Vicia villosa agglutinin (VVA) enrichment for O-glycans combined with serial lectin chromatography methods [34].
In this study, we have characterized the role of the GALNT3 enzyme in aberrant O-glycosylation of mucin-like targets in EOC cells using a bioorthogonal chemical reporter strategy that involves metabolic labeling of glycans with a monosaccharide precursor (Ac4GalNAz) carrying a bio-orthogonal azido group, which allows a linker attachment to glycoproteins (including mucin-type O-linked glycoproteins) for consecutive labeling and isolation/purification ([36]; also see Fig. 1 in [13]). To the best of our knowledge, this is the first study that has used metabolic labeling to identify the impact of a GalNAc-T inhibition on altering the glycoproteins pattern in an EOC cell line. Moreover, the approach used allowed us to identify numerous novel glycoproteins differentially regulated in EOC cells.
To date, only limited numbers of glycoproteins have been characterized, which reflects the low percentage of glycoprotein entries annotated in SWISS-PROT [5]. This fact is mainly due to difficulties in performing experimental determination of glycosylation sites [5]. Here, we used the NetOGlyc 4.0 server, which is a developed glycosylation site prediction method for O-glycosylation [51]. Our prediction analysis estimated that more than 88% of the identified proteins may be O-glycosylated (Fig. 1A; also see Supplementary Table 2 in [13]). In addition to performing NetOGlyc prediction analysis, we also compared our list of annotated proteins to previously published studies [19, 22–31] (Fig. 1B), and results indicated that more than 30% of the identified proteins are O-glycosylated (Fig. 1B; also see Supplementary Table 3 in [13]). Together, our comparative analyses suggest that the O-glycoproteome is much larger than previously predicted, and a more extensive examination may reveal more interesting numbers of glycosylated proteins than those currently reported in SWISS-PROT. We believe that improved and more precise results can be obtained when a better integration of information pertaining to glycosite identification on the list of predicted glycoproteins is acquired, similar to the data, previously generated [19,20].
Additionally, using cellular fractionation, glycoprotein enrichment and high resolution LC/MS-based quantitative proteomics, we quantified over 2000 proteins. Among these proteins, we found several hundreds to be differentially expressed (≥2 fold, p-value ≤ 0.05) between the control and GALNT3 KD A2780s EOC cells (Fig. 2, also see Supplementary Table 4 in [13]). GO cellular component classifications revealed that these differentially regulated proteins were predominantly mapped to membrane, extracellular, cytoplasmic and intracellular organelle compartments (Fig. 3; also see Fig. 3 and Supplementary Table 5 in [13]). More interestingly, we observed a relative enrichment in proteins identified in the ER compartment, and recent evidence suggests that the GalNAc-Ts can actually relocate to the ER, indicating a possible role of the GalNAc-Ts in protein O-glycosylation of ER proteins [52].
It has been previously demonstrated that Ac4GalNAz treatment of human cells results in the metabolic labeling of mucin-type glycoproteins, in addition to the labeling of extracellular N-glycan and intracellular O-GlcNAcylated proteins [19,20,50,53]. The biosynthesis of both UDP-GalNAz and GlcNAz from Ac4GalNAz is conducted by the mammalian enzyme GALE, which interconverts UDP-N-acetylglucosamine (UDP-GlcNAc) and its C4 epimer, UDP-N-acetylgalactosamine (UDP-GalNAc) [18]. UDP-GalNAc is the common substrate of the GalNAc-Ts, which is derived primarily from UDP-GlcNAc via GALE and alternatively, UDP-GalNAc can be generated from GalNAc by the action of GalNAc 1-kinase and UDP-GalNAc pyrophosphorylase, representing enzymes of the salvage pathway [47]. Thus, metabolic labeling with Ac4GalNAz results in heterogeneous labeling of nuclear and cytoplasmic proteins, in addition to membrane-bound mucin-type glycoproteins; such a robust labeling by Ac4GalNAz can allow for a better comprehensive measurement of the GalNAc-Ts activities on diverse aspects of cellular functions, and especially in human diseases such as cancer. GO and Western blot analysis were indicative that in both control and GALNT3 KD cells, the majority of the Ac4GalNAz labeling occurred in the conditioned media fraction, as compared to lower labeling in the soluble and insoluble fractions (Fig. 3; also see Fig. 2, Fig. 3 and Supplementary Table 5 in [13]). This data indicates that EOC cells may have an inactive or defective GALE activity, as GALE deficiency has been reported to decrease the production of UDP-GalNAc from UDP-GlcNAc, which in turn affects glycolipid and glycoprotein biosynthesis [54]. Moreover, IPA pathway analyses illustrated that the GALE enzyme is significantly down-regulated in the GALNT3 KD cells, suggesting that GALE activity has been additionally perturbed by GALNT3 KD in A2780s EOC cells. Collectively, our data are indicative for GALNT3-mediated modulation of GALE expression in EOC, possibly associated with alterations in the GALE glycosylation pattern.
The IPA functional pathway analyses of our proteomics data have revealed that the GALNT3 gene suppression resulted in reduced expression of essential pathways related to lipid, carbohydrate and amino acid metabolism, also demonstrating that changes in glycogen metabolism can provoke crucial alterations in various EOC metabolic pathways (Fig. 4B). This was further confirmed by IPA canonical pathway analyses (Fig. 5), as the top five canonical pathways (PTM, cardiovascular and inflammatory disease, lipid metabolism, embryonic development and cell morphology, and cellular assembly) were found to be significantly perturbed upon GALNT3 KD (Table 1). Our data also support others’ findings for the implications of members of the GalNAc-Ts gene family in controlling cellular metabolism. Indeed, a role of GALNT2 in controlling lipid metabolism was postulated, as GALNT2 was functionally associated with the glycosylation of high-density lipoprotein cholesterol and/or triglycerides [55]. Interestingly, the canonical pathway associated with degradation of the branched-chain amino acids (BCAAs) leucine, isoleucine and valine was found to be among the significantly suppressed canonical pathways following GALNT3 KD (Fig. 5B). BCAAs degradation is regulated by the BCAT1 and BCAT2 enzymes [34]. We have recently shown that the suppression of the BCAT1 gene in EOC cells resulted in the down-regulation of numerous genes implicated in lipid production and protein synthesis, suggesting its important role in controlling EOC metabolism [56]. Our data suggest a similar effect imposed by the suppression of the GALNT3 gene.
Additionally, GALNT3 KD was associated with the down-regulation of the metabolism related gene nodes SOD2 and MTHFD2 (Fig. 6), previously shown to be overexpressed in different cancer types, including EOC [33,57]. SOD2 encodes the enzyme manganese superoxide dismutase (MnSOD), which is a mitochondrial enzyme responsible for the reduction of free radicals and thus acts by protecting cells from oxidative stress, and SOD2 over-expression was demonstrated to be involved in EOC development [33]. MTHFD2 represents an enzyme of the mitochondrial metabolic pathway shown to be implicated in cancer cell proliferation, as MTHFD2 has displayed strong overexpression in different tumors including EOC tumors [57]. Our findings suggest that metabolic pathways regulated via genes of the oxidative stress and/or mitochondrial enzymes might be controlled by PTMs such as glycosylation. GALNT3 KD also resulted in the upregulation of the SMPD1 gene node. SMPD1 represents an acid sphingomyelinase, responsible for the hydrolysis of sphingomyelin to phosphorylchole ceramide, and the increase in ceramide production has been reported to lead to cell death showing a link between ceramide expression and EOC [58]. SMPD1 expression correlates with enhanced cell death in GALNT3 KD cells compared to their corresponding controls, as reported from our previous study [14].
Moreover, two oligosaccharyltransferases (OST) were found to be down-regulated upon GALNT3 KD. The main function of OSTs is the transfer of lipid-linked oligosaccharides to selected asparagine residues within the consensus sequence Asn-Xaa-Ser/Thr on nascent polypeptides [59]. Several studies reported OST mediated glycosylation to be shared between N- and O-glycosylation pathways [60]. The OST subunits examined from our network analysis include DDOST and STT3B (Fig. 6), which were significantly down-regulated upon GALNT3 KD in EOC cells.
Network analysis also demonstrated that GALNT3 KD directs the down-regulation of several major gene nodes and corresponding pathways previously shown to be associated with EOC etiology, including TGF-β, MAPK, and NF-kappa-B (Fig. 6). Accordingly, some members of the NF-kappa-B pathway (MTDH, ANXA4 and RRM2; see Fig. 6) were found to be significantly suppressed in GALNT3 KD EOC cells. MTDH (also known as AEG1) was repeatedly shown to be implicated in EOC dissemination and chemoresistance [61–63] and has been recognized as an independent prognostic biomarker for metastatic EOC tumors [64]. ANXA4 was reported to be up-regulated in EOC, and is being used as molecular marker to distinguish the different EOC subtypes [65]. Similarly, RRM2 was shown to be over-expressed in EOC patients with advanced disease [66], and was recently identified as a potential target for pro-senescence EOC therapy [67]. Moreover, several gene nodes implicated in EOC chemoresistance (including TUBB3, MAP1B and ATG5; see Fig. 6), displayed a significant decrease upon GALNT3 KD. Indeed, augmented expression of TUBB3 and MAP1B has been associated with worse prognosis and paclitaxel resistance in EOC patients [68,69]. Also, knockdown of autophagy-related ATG5 gene was shown to increase cisplatin-induced growth inhibition by enhancing cisplatin-induced apoptosis in EOC [70].
Finally, the lipid kinases PI3K complex, known to be highly activated in human cancers [71], was up-regulated upon GALNT3 KD. The PI3K complex controls multiple cellular signaling pathways that play key regulatory roles in cell survival, differentiation, and proliferation [71, 72]. Interestingly, recent reports have shown that alterations in protein glycosylation can be implicated in the activation of multiple oncogenic signaling pathways including the PI3K pathway in different cancer types [71]. These glycosylation alterations are usually caused by a premature stop in proteins O-glycosylation due to sialylation, mediated by the cancer-associated sialyl-Tn antigen. Notably, it has been shown that in cancer, the enhanced expression of sialyltransferases is often associated with shorter O-GalNAc glycan structures [5]. Another gene node B4GALT1, previously reported as an oncogene, was also found to be up-regulated following GALNT3 KD. B4GALT1 synthesizes Galβ1-4GlcNAc (N-acetyllactosamine) by transferring galactose from UDP-Gal to an acceptor sugar molecule, and thus represents a key enzyme in glycobiology [73]. Several studies have reported B4GALT1 to play an essential role in the EOC dissemination [74,75]. The observed up-regulations of the PI3K complex and the B4GALT1 gene may be due to the frequently observed redundancy associated with overlapping functions of other GalNAc-Ts enzymes [76], partially compensating for the KD of GALNT3 in EOC cells.
5. Conclusions
In this study, we performed a quantitative proteomic analysis of the GalNAz-metabolically labeled proteins in both control and GALNT3 KD A2780s cells. The use of the azido analog GalNAz for metabolic labeling and enrichment of glycoproteins in control and GALNT3 KD A2780s cells, coupled with MS analysis, has allowed us to more quantitatively characterize the role of the GALNT3 enzyme in aberrant glycosylation of mucin-like targets in EOC cells and better understand the role of this transferase in EOC dissemination. This study has led to the identification of novel glycoproteins expressed in EOC and more importantly, sets of glycoproteins whose expression is altered by GALNT3 KD, indicative for a potential role of GALNT3 in modulating PTMs and metabolism pathways in EOC cells, which could significantly impact disease development. However, we do not exclude that certain compensatory changes may occur upon GALNT3 KD due to some level of redundancy reported among the family of 20 GalNAc-Ts. Further studies focused on these genes may provide valuable insights into the pathophysiological roles of the GalNAc-T enzymes in EOC. Consecutive analyses should also include validation of these data in EOC tumor sets displaying high or low GALNT3 expression.
Furthermore, changes in glycosylation patterns in cancer are associated not only with differential expression of glycoproteins [77], but also with alterations in their glycan structures [78], as a common feature of tumors is the overexpression of truncated O-glycans. We anticipate further expansion of our analyses by the application of targeted O-glycoproteomic strategies that do not reduce O-glycan structures, so that information about the site and glycan structures can be studied simultaneously [19,20]. The application of such an innovative approach could allow for a better comprehension of the role of the GALNT3 enzyme and other members of the GalNAc-T isoforms in aberrant EOC glycosylation, associated with disease dissemination.
Supplementary Material
Acknowledgments
This study was sustained by grant from the Cancer Research Society of Canada to D.B., as well as by grants from Jane Coffin Childs Fund to C.M.W., Burroughs Wellcome Fund CASI to C.M.W., National Institutes of Health (CA200423) to C.R.B. and Howard Hughes Medical Institute to C.R.B.
Abbreviations
- EOC
epithelial ovarian cancer
- PTM
post-translational modification
- GALNT3
N-acetylgalactosaminyltransferase 3
- GalNAc-Ts
GalNAc-transferases
- KD
knockdown
- shRNA
short hairpin RNA
- siRNA
small interfering RNA
- MUC1
mucin-1
- Ac4GalNAz
peraceetylated N-azidoacetylgalactosamine
- CuAAC
copper-catalyzed azide-alkyne cycloaddition
- nanoLC
nanoscale capillary liquid chromatography
- AGC
Automatic Gain Control
- LFQ
label free quantification
- IPA
ingenuity pathway analysis
- Ac4GalNAc
tetraacetylated N-acetylgalactosamine
- GalNAz
N-azidoacetylgalactosamine
- GALE
UDP-galactose 4′-epimerase
- GO
gene ontology
- ILK
integrin-linked kinase
- MAPK
mitogen-activated protein kinases
- AMPK
AMP-activated protein kinase
- TGFβ1
transforming growth factor beta
- PI3K
phosphoinositide 3-kinase complex
- SMPD1
sphingomyelin phosphodiesterase 1
- TUBB3
tubulin beta 3
- SOD2
magnesium superoxide dismutase
- MTHFD2
mitochondrial folate-coupled dehydrogenase
- ATG5
autophagy protein 5
- MAP1B
microtubule-associated protein 1B
- DDOST
dolichyl-diphosphooligosaccharide-protein glycosyltransferase
- STT3B
dolichyl-diphosphooligosaccharide–protein glycosyltransferase subunit
- NF-Kappa B
nuclear factor kappa-light-chain-enhancer of activated B cells
- MTDH
metadherin
- ANXA4
annexin A4
- RRMP2
ribonucleotide reductase M2
- VVA
Vicia villosa agglutinin
- UDP-GlcNAc
UDP-N-acetylglucosamine
- UDP-GalNAc
UDP-N-acetylgalactosamine
- BCAAs
branched-chain amino acids
- BCAT1
branched chain amino-acid transaminase 1
- BCAT2
branched chain amino-acid transaminase 2
- MnSOD
manganese superoxide dismutase
- ASM
acid sphingomyelinase
- OST
oligosaccharyltransferases
- B4GALT1
UDP-Gal: BetaGlcNAc Beta 1,4- Galactosyltransferase, Polypeptide 1
Footnotes
Conflict of interest
The authors declare no conflicts of interest.
References
- 1.Siegel R, Ward E, Brawley O, Jemal A. Cancer statistics, 2011: the impact of eliminating socioeconomic and racial disparities on premature cancer deaths. CA Cancer J Clin. 2011;61:212–236. doi: 10.3322/caac.20121. [DOI] [PubMed] [Google Scholar]
- 2.Fruscio R, Corso S, Ceppi L, Garavaglia D, Garbi A, Floriani I, et al. Conservative management of early-stage epithelial ovarian cancer: results of a large retrospective series. Ann Oncol. 2013;24:138–144. doi: 10.1093/annonc/mds241. [DOI] [PubMed] [Google Scholar]
- 3.Alouini S. Management of ovarian cancer has changed. Gynecol Oncol. 2012;126:313. doi: 10.1016/j.ygyno.2012.04.044. (author reply 4) [DOI] [PubMed] [Google Scholar]
- 4.Yarema KJ, Bertozzi CR. Characterizing glycosylation pathways. Genome Biol. 2001;2 doi: 10.1186/gb-2001-2-5-reviews0004. REVIEWS0004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Apweiler R, Hermjakob H, Sharon N. On the frequency of protein glycosylation, as deduced from analysis of the SWISS-PROT database. Biochim Biophys Acta. 1999;1473:4–8. doi: 10.1016/s0304-4165(99)00165-8. [DOI] [PubMed] [Google Scholar]
- 6.Paszek MJ, DuFort CC, Rossier O, Bainer R, Mouw JK, Godula K, et al. The cancer glycocalyx mechanically primes integrin-mediated growth and survival. Nature. 2014;511:319–325. doi: 10.1038/nature13535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zachara NE, Hart GW. Cell signaling, the essential role of O-GlcNAc! Biochim Biophys Acta. 1761;2006:599–617. doi: 10.1016/j.bbalip.2006.04.007. [DOI] [PubMed] [Google Scholar]
- 8.Parodi AJ. Protein glucosylation and its role in protein folding. Annu Rev Biochem. 2000;69:69–93. doi: 10.1146/annurev.biochem.69.1.69. [DOI] [PubMed] [Google Scholar]
- 9.Fuster MM, Esko JD. The sweet and sour of cancer: glycans as novel therapeutic targets. Nat Rev Cancer. 2005;5:526–542. doi: 10.1038/nrc1649. [DOI] [PubMed] [Google Scholar]
- 10.Pinho SS, Reis CA. Glycosylation in cancer: mechanisms and clinical implications. Nat Rev Cancer. 2015;15:540–555. doi: 10.1038/nrc3982. [DOI] [PubMed] [Google Scholar]
- 11.Cox J, Neuhauser N, Michalski A, Scheltema RA, Olsen JV, Mann M. Andromeda: a peptide search engine integrated into the MaxQuant environment. J Proteome Res. 2011;10:1794–1805. doi: 10.1021/pr101065j. [DOI] [PubMed] [Google Scholar]
- 12.Carbon S, Ireland A, Mungall CJ, Shu S, Marshall B, Lewis S. AmiGO: online access to ontology and annotation data. Bioinformatics. 2009;25:288–289. doi: 10.1093/bioinformatics/btn615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sheta R, Roux-Dalvai F, Woo CM, Fournier F, Bourassa S, Bertozzi CR, et al. Proteomic dataset for altered glycoprotein expression upon GALNT3 knockdown in ovarian cancer cells. Data Brief. 2016 doi: 10.1016/j.dib.2016.05.060. (submitted for publication) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang ZQ, Bachvarova M, Morin C, Plante M, Gregoire J, Renaud MC, et al. Role of the polypeptide N-acetylgalactosaminyltransferase 3 in ovarian cancer progression: possible implications in abnormal mucin O-glycosylation. Oncotarget. 2014;5:544–560. doi: 10.18632/oncotarget.1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bennett EP, Mandel U, Clausen H, Gerken TA, Fritz TA, Tabak LA. Control of mucin-type O-glycosylation: a classification of the polypeptide GalNAc-transferase gene family. Glycobiology. 2012;22:736–756. doi: 10.1093/glycob/cwr182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sletten EM, Bertozzi CR. Bioorthogonal chemistry: fishing for selectivity in a sea of functionality. Angew Chem Int Ed Eng. 2009;48:6974–6998. doi: 10.1002/anie.200900942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Laughlin ST, Bertozzi CR. Metabolic labeling of glycans with azido sugars and subsequent glycan-profiling and visualization via Staudinger ligation. Nat Protoc. 2007;2:2930–2944. doi: 10.1038/nprot.2007.422. [DOI] [PubMed] [Google Scholar]
- 18.Hang HC, Yu C, Kato DL, Bertozzi CR. A metabolic labeling approach toward proteomic analysis of mucin-type O-linked glycosylation. Proc Natl Acad Sci U S A. 2003;100:14846–14851. doi: 10.1073/pnas.2335201100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Woo CM, Iavarone AT, Spiciarich DR, Palaniappan KK, Bertozzi CR. Isotope-targeted glycoproteomics (IsoTaG): a mass-independent platform for intact N-and O-glycopeptide discovery and analysis. Nat Methods. 2015;12:561–567. doi: 10.1038/nmeth.3366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Woo CM, Bertozzi CR. Isotope targeted glycoproteomics (IsoTaG) to characterize intact, metabolically labeled glycopeptides from complex proteomes. Curr Protoc Chem Biol. 2016;8:59–82. doi: 10.1002/9780470559277.ch150185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cox J, Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat Biotechnol. 2008;26:1367–1372. doi: 10.1038/nbt.1511. [DOI] [PubMed] [Google Scholar]
- 22.Trinidad JC, Schoepfer R, Burlingame AL, Medzihradszky KF. N- and O-glycosylation in the murine synaptosome. Mol Cell Proteomics. 2013;12:3474–3488. doi: 10.1074/mcp.M113.030007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hornbeck PV, Kornhauser JM, Tkachev S, Zhang B, Skrzypek E, Murray B, et al. PhosphoSitePlus: a comprehensive resource for investigating the structure and function of experimentally determined post-translational modifications in man and mouse. Nucleic Acids Res. 2012;40:D261–D270. doi: 10.1093/nar/gkr1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Halim A, Ruetschi U, Larson G, Nilsson J. LC–MS/MS characterization of O-glycosylation sites and glycan structures of human cerebrospinal fluid glycoproteins. J Proteome Res. 2013;12:573–584. doi: 10.1021/pr300963h. [DOI] [PubMed] [Google Scholar]
- 25.Trinidad JC, Barkan DT, Gulledge BF, Thalhammer A, Sali A, Schoepfer R, et al. Global identification and characterization of both O-GlcNAcylation and phosphorylation at the murine synapse. Mol Cell Proteomics. 2012;11:215–229. doi: 10.1074/mcp.O112.018366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hahne H, Sobotzki N, Nyberg T, Helm D, Borodkin VS, van Aalten DM, et al. Proteome wide purification and identification of O-GlcNAc-modified proteins using click chemistry and mass spectrometry. J Proteome Res. 2013;12:927–936. doi: 10.1021/pr300967y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zaro BW, Yang YY, Hang HC, Pratt MR. Chemical reporters for fluorescent detection and identification of O-GlcNAc-modified proteins reveal glycosylation of the ubiquitin ligase NEDD4-1. Proc Natl Acad Sci U S A. 2011;108:8146–8151. doi: 10.1073/pnas.1102458108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chuh KN, Zaro BW, Piller F, Piller V, Pratt MR. Changes in metabolic chemical reporter structure yield a selective probe of O-GlcNAc modification. J Am Chem Soc. 2014;136:12283–12295. doi: 10.1021/ja504063c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Alfaro JF, Gong CX, Monroe ME, Aldrich JT, Clauss TR, Purvine SO, et al. Tandem mass spectrometry identifies many mouse brain O-GlcNAcylated proteins including EGF domain-specific O-GlcNAc transferase targets. Proc Natl Acad Sci U S A. 2012;109:7280–7285. doi: 10.1073/pnas.1200425109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hubbard SC, Boyce M, McVaugh CT, Peehl DM, Bertozzi CR. Cell surface glycoproteomic analysis of prostate cancer-derived PC-3 cells. Bioorg Med Chem Lett. 2011;21:4945–4950. doi: 10.1016/j.bmcl.2011.05.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Khidekel N, Ficarro SB, Peters EC, Hsieh-Wilson LC. Exploring the O-GlcNAc proteome: direct identification of O-GlcNAc-modified proteins from the brain. Proc Natl Acad Sci U S A. 2004;101:13132–13137. doi: 10.1073/pnas.0403471101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Steentoft C, Vakhrushev SY, Joshi HJ, Kong Y, Vester-Christensen MB, Schjoldager KT, et al. Precision mapping of the human O-GalNAc glycoproteome through SimpleCell technology. EMBO J. 2013;32:1478–1488. doi: 10.1038/emboj.2013.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sun N, Pan C, Nickell S, Mann M, Baumeister W, Nagy I. Quantitative proteome and transcriptome analysis of the archaeon Thermoplasma acidophilum cultured under aerobic and anaerobic conditions. J Proteome Res. 2010;9:4839–4850. doi: 10.1021/pr100567u. [DOI] [PubMed] [Google Scholar]
- 34.Ramus C, Hovasse A, Marcellin M, Hesse AM, Mouton-Barbosa E, Bouyssie D, et al. Spiked proteomic standard dataset for testing label-free quantitative software and statistical methods. Data Brief. 2016;6:286–294. doi: 10.1016/j.dib.2015.11.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang P, Guo Z, Zhang Y, Gao Z, Ji N, Wang D, et al. A preliminary quantitative proteomic analysis of glioblastoma pseudoprogression. Proteome Sci. 2015;13:12. doi: 10.1186/s12953-015-0066-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gautier V, Mouton-Barbosa E, Bouyssie D, Delcourt N, Beau M, Girard JP, et al. Label-free quantification and shotgun analysis of complex proteomes by one-dimensional SDS-PAGE/NanoLC-MS: evaluation for the large scale analysis of inflammatory human endothelial cells. Mol Cell Proteomics. 2012;11:527–539. doi: 10.1074/mcp.M111.015230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Abbott KL. Glycomic analysis of ovarian cancer: past, present, and future. Cancer Biomark. 2010;8:273–280. doi: 10.3233/CBM-2011-0218. [DOI] [PubMed] [Google Scholar]
- 38.Tian Y, Yao Z, Roden RB, Zhang H. Identification of glycoproteins associated with different histological subtypes of ovarian tumors using quantitative glycoproteomics. Proteomics. 2011;11:4677–4687. doi: 10.1002/pmic.201000811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gubbels JA, Belisle J, Onda M, Rancourt C, Migneault M, Ho M, et al. Mesothelin-MUC16 binding is a high affinity, N-glycan dependent interaction that facilitates peritoneal metastasis of ovarian tumors. Mol Cancer. 2006;5:50. doi: 10.1186/1476-4598-5-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang X, Wang Y, Qian Y, Wu X, Zhang Z, Liu X, et al. Discovery of specific metastasis-related N-glycan alterations in epithelial ovarian cancer based on quantitative glycomics. PLoS ONE. 2014;9:e87978. doi: 10.1371/journal.pone.0087978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Anugraham M, Jacob F, Nixdorf S, Everest-Dass AV, Heinzelmann-Schwarz V, Packer NH. Specific glycosylation of membrane proteins in epithelial ovarian cancer cell lines: glycan structures reflect gene expression and DNA methylation status. Mol Cell Proteomics. 2014;13:2213–2232. doi: 10.1074/mcp.M113.037085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kuzmanov U, Musrap N, Kosanam H, Smith CR, Batruch I, Dimitromanolakis A, et al. Glycoproteomic identification of potential glycoprotein biomarkers in ovarian cancer proximal fluids. Clin Chem Lab Med. 2013;51:1467–1476. doi: 10.1515/cclm-2012-0642. [DOI] [PubMed] [Google Scholar]
- 43.Wojtowicz K, Szaflarski W, Januchowski R, Zawierucha P, Nowicki M, Zabel M. Inhibitors of N-glycosylation as a potential tool for analysis of the mechanism of action and cellular localisation of glycoprotein P. Acta Biochim Pol. 2012;59:445–450. [PubMed] [Google Scholar]
- 44.Taylor-Papadimitriou J, Burchell J, Miles DW, Dalziel M. MUC1 and cancer. Biochim Biophys Acta. 1999;1455:301–313. doi: 10.1016/s0925-4439(99)00055-1. [DOI] [PubMed] [Google Scholar]
- 45.Wang L, Ma J, Liu F, Yu Q, Chu G, Perkins AC, et al. Expression of MUC1 in primary and metastatic human epithelial ovarian cancer and its therapeutic significance. Gynecol Oncol. 2007;105:695–702. doi: 10.1016/j.ygyno.2007.02.004. [DOI] [PubMed] [Google Scholar]
- 46.Price MR, Hudecz F, O’Sullivan C, Baldwin RW, Edwards PM, Tendler SJ. Immunological and structural features of the protein core of human polymorphic epithelial mucin. Mol Immunol. 1990;27:795–802. doi: 10.1016/0161-5890(90)90089-i. [DOI] [PubMed] [Google Scholar]
- 47.Sheta R, Bachvarov D. Role of aberrant glycosylation in ovarian cancer dissemination. Biochem Rev. 2014;25:83–92. [Google Scholar]
- 48.Park JH, Nishidate T, Kijima K, Ohashi T, Takegawa K, Fujikane T, et al. Critical roles of mucin 1 glycosylation by transactivated polypeptide N-acetylgalactosaminyltransferase 6 in mammary carcinogenesis. Cancer Res. 2010;70:2759–2769. doi: 10.1158/0008-5472.CAN-09-3911. [DOI] [PubMed] [Google Scholar]
- 49.Wang R, Yu C, Zhao D, Wu M, Yang Z. The mucin-type glycosylating enzyme polypeptide N-acetylgalactosaminyltransferase 14 promotes the migration of ovarian cancer by modifying mucin 13. Oncol Rep. 2013;30:667–676. doi: 10.3892/or.2013.2493. [DOI] [PubMed] [Google Scholar]
- 50.Boyce M, Carrico IS, Ganguli AS, Yu SH, Hangauer MJ, Hubbard SC, et al. Metabolic cross-talk allows labeling of O-linked beta-N-acetylglucosamine-modified proteins via the N-acetylgalactosamine salvage pathway. Proc Natl Acad Sci U S A. 2011;108:3141–3146. doi: 10.1073/pnas.1010045108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Julenius K, Molgaard A, Gupta R, Brunak S. Prediction, conservation analysis, and structural characterization of mammalian mucin-type O-glycosylation sites. Glycobiology. 2005;15:153–164. doi: 10.1093/glycob/cwh151. [DOI] [PubMed] [Google Scholar]
- 52.Gill DJ, Clausen H, Bard F. Location, location, location: new insights into O-GalNAc protein glycosylation. Trends Cell Biol. 2011;21:149–158. doi: 10.1016/j.tcb.2010.11.004. [DOI] [PubMed] [Google Scholar]
- 53.Vocadlo DJ, Hang HC, Kim EJ, Hanover JA, Bertozzi CR. A chemical approach for identifying O-GlcNAc-modified proteins in cells. Proc Natl Acad Sci U S A. 2003;100:9116–9121. doi: 10.1073/pnas.1632821100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lai K, Elsas LJ, Wierenga KJ. Galactose toxicity in animals. IUBMB Life. 2009;61:1063–1074. doi: 10.1002/iub.262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Schjoldager KT, Vakhrushev SY, Kong Y, Steentoft C, Nudelman AS, Pedersen NB, et al. Probing isoform-specific functions of polypeptide GalNAc-transferases using zinc finger nuclease glycoengineered SimpleCells. Proc Natl Acad Sci U S A. 2012;109:9893–9898. doi: 10.1073/pnas.1203563109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wang ZQ, Faddaoui A, Bachvarova M, Plante M, Gregoire J, Renaud MC, et al. BCAT1 expression associates with ovarian cancer progression: possible implications in altered disease metabolism, 6. Oncotarget. 2015:31522–31543. doi: 10.18632/oncotarget.5159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Nilsson R, Jain M, Madhusudhan N, Sheppard NG, Strittmatter L, Kampf C, et al. Metabolic enzyme expression highlights a key role for MTHFD2 and the mitochondrial folate pathway in cancer. Nat Commun. 2014;5:3128. doi: 10.1038/ncomms4128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Smith EL, Schuchman EH. Acid sphingomyelinase overexpression enhances the antineoplastic effects of irradiation in vitro and in vivo. Mol Ther. 2008;16:1565–1571. doi: 10.1038/mt.2008.145. [DOI] [PubMed] [Google Scholar]
- 59.Pan S, Chen R, Tamura Y, Crispin DA, Lai LA, May DH, et al. Quantitative glycoproteomics analysis reveals changes in N-glycosylation level associated with pancreatic ductal adenocarcinoma. J Proteome Res. 2014;13:1293–1306. doi: 10.1021/pr4010184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Dell A, Galadari A, Sastre F, Hitchen P. Similarities and differences in the glycosylation mechanisms in prokaryotes and eukaryotes. Int J Microbiol. 2010;2010:148178. doi: 10.1155/2010/148178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhou B, Yang J, Shu B, Liu K, Xue L, Su N, et al. Overexpression of astrocyte-elevated gene-1 is associated with ovarian cancer development and progression. Mol Med Rep. 2015;11:2981–2990. doi: 10.3892/mmr.2014.3056. [DOI] [PubMed] [Google Scholar]
- 62.Haug S, Schnerch D, Halbach S, Mastroianni J, Dumit VI, Follo M, et al. Metadherin exon 11 skipping variant enhances metastatic spread of ovarian cancer. Int J Cancer. 2015;136:2328–2340. doi: 10.1002/ijc.29289. [DOI] [PubMed] [Google Scholar]
- 63.Li C, Liu J, Lu R, Yu G, Wang X, Zhao Y, et al. AEG-1 overexpression: a novel indicator for peritoneal dissemination and lymph node metastasis in epithelial ovarian cancers. Int J Gynecol Cancer. 2011;21:602–608. doi: 10.1097/IGC.0b013e3182145561. [DOI] [PubMed] [Google Scholar]
- 64.Li C, Chen K, Cai J, Shi QT, Li Y, Li L, et al. Astrocyte elevated gene-1: a novel independent prognostic biomarker for metastatic ovarian tumors. Tumour Biol. 2014;35:3079–3085. doi: 10.1007/s13277-013-1400-0. [DOI] [PubMed] [Google Scholar]
- 65.Kim A, Enomoto T, Serada S, Ueda Y, Takahashi T, Ripley B, et al. Enhanced expression of Annexin A4 in clear cell carcinoma of the ovary and its association with chemoresistance to carboplatin. Int J Cancer. 2009;125:2316–2322. doi: 10.1002/ijc.24587. [DOI] [PubMed] [Google Scholar]
- 66.Wang LM, Lu FF, Zhang SY, Yao RY, Xing XM, Wei ZM. Overexpression of catalytic subunit M2 in patients with ovarian cancer. Chin Med J. 2012;125:2151–2156. [PubMed] [Google Scholar]
- 67.Aird KM, Li H, Xin F, Konstantinopoulos PA, Zhang R. Identification of ribonucleotide reductase M2 as a potential target for pro-senescence therapy in epithelial ovarian cancer. Cell Cycle. 2014;13:199–207. doi: 10.4161/cc.26953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ferrandina G, Zannoni GF, Martinelli E, Paglia A, Gallotta V, Mozzetti S, et al. Class III beta-tubulin overexpression is a marker of poor clinical outcome in advanced ovarian cancer patients. Clin Cancer Res. 2006;12:2774–2779. doi: 10.1158/1078-0432.CCR-05-2715. [DOI] [PubMed] [Google Scholar]
- 69.Yu Y, Gaillard S, Phillip JM, Huang TC, Pinto SM, Tessarollo NG, et al. Inhibition of spleen tyrosine kinase potentiates paclitaxel-induced cytotoxicity in ovarian cancer cells by stabilizing microtubules. Cancer Cell. 2015;28:82–96. doi: 10.1016/j.ccell.2015.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wang J, Wu GS. Role of autophagy in cisplatin resistance in ovarian cancer cells. J Biol Chem. 2014;289:17163–17173. doi: 10.1074/jbc.M114.558288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Liu P, Cheng H, Roberts TM, Zhao JJ. Targeting the phosphoinositide 3-kinase pathway in cancer. Nat Rev Drug Discov. 2009;8:627–644. doi: 10.1038/nrd2926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Engelman JA, Luo J, Cantley LC. The evolution of phosphatidylinositol 3-kinases as regulators of growth and metabolism. Nat Rev Genet. 2006;7:606–619. doi: 10.1038/nrg1879. [DOI] [PubMed] [Google Scholar]
- 73.Pennell DJ, Underwood SR, Ell PJ. Symptomatic bradycardia complicating the use of intravenous dipyridamole for thallium-201 myocardial perfusion imaging. Int J Cardiol. 1990;27:272–274. doi: 10.1016/0167-5273(90)90170-a. [DOI] [PubMed] [Google Scholar]
- 74.Yamashita H, Kubushiro K, Ma J, Fujii T, Tsukazaki K, Iwamori M, et al. Alteration in the metastatic potential of ovarian cancer cells by transfection of the antisense gene of beta-1,4-galactosyltransferase. Oncol Rep. 2003;10:1857–1862. [PubMed] [Google Scholar]
- 75.Ma J, Kubushiro K, Tashima Y, Tsukazaki K, Udagawa Y, Nozawa S, et al. Expression of human beta 1,4-galactosyltransferase in gynecological cancer cell lines. Int J Oncol. 1997;11:117–122. doi: 10.3892/ijo.11.1.117. [DOI] [PubMed] [Google Scholar]
- 76.Plantner JJ, Lentrichia BB, Kean EL. Biogenesis and content of rhodopsin in the retina of the chick during development. Curr Eye Res. 1988;7:503–510. doi: 10.3109/02713688809031804. [DOI] [PubMed] [Google Scholar]
- 77.Tousi F, Hancock WS, Hincapie M. Technologies and strategies for glycoproteomics and glycomics and their application to clinical biomarker research. Anal Methods. 2011;3:20–32. doi: 10.1039/c0ay00413h. [DOI] [PubMed] [Google Scholar]
- 78.Kolarich D, Jensen PH, Altmann F, Packer NH. Determination of site-specific glycan heterogeneity on glycoproteins. Nat Protoc. 2012;7:1285–1298. doi: 10.1038/nprot.2012.062. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


