Abstract
Phylogenetic comparative methods have become standard for investigating evolutionary hypotheses, including in studies of human evolution. While these methods account for the non-independence of trait data due to phylogeny, they often fail to consider intraspecific variation, which may lead to biased or erroneous results. We assessed the degree to which intraspecific variation impacts the results of comparative analyses by investigating the “social brain” hypothesis, which has provided a framework for explaining complex cognition and large brains in humans. This hypothesis suggests that group life imposes a cognitive challenge, with species living in larger social groups having comparably larger neocortex ratios than those living in smaller groups. Primates, however, vary considerably in group size within species, a fact that has been ignored in previous analyses. When within-species variation in group size is high, the common practice of using a mean value to represent the species may be inappropriate. We conducted regression and resampling analyses to ascertain whether the relationship between neocortex ratio and group size across primate species persists after controlling for within-species variation in group size. We found that in a sample of 23 primates, 70% of the variation in group size was due to between-species variation. Controlling for within-species variation in group size did not affect the results of phylogenetic analyses, which continued to show a positive relationship between neocortex ratio and group size. Analyses restricted to non-monogamous primates revealed considerable intraspecific variation in group size, but the positive association between neocortex ratio and group size remained even after controlling for within-species variation in group size. Our findings suggest that the relationship between neocortex size and group size in primates is robust. In addition, our methods and associated computer code provide a way to assess and account for intraspecific variation in other comparative analyses of primate evolution.
Keywords: Sociality, Phylogenetic regression, Measurement error, Neocortex
1. Introduction
Comparative analyses have played an important role in studies of primate and human evolution (Harvey and Pagel,1991; Nunn, 2011). These analyses examine interspecific variation between different traits to infer adaptation, using species as the units of analysis. Because species are products of the evolutionary process and share traits via common descent, however, they cannot be considered as independent data points for statistical purposes (Felsenstein, 1985). As a consequence, evolutionary biologists have developed several procedures to control for phylogenetic non-independence (Nunn, 2011; Garamszegi, 2014a). The development and adoption of these techniques have increased the impact of comparative methods, but they make several important assumptions. One relatively neglected assumption concerns intraspecific variation. Specifically, most comparative methods assume that the entire range of variation in a trait of interest can be represented by a single data point, the species average (Garamszegi and Møller, 2010). Thus, variation around that value—due to measurement error, differences in sample size, or biologically relevant within-species variation—is typically ignored (Garamszegi, 2014b).
Ignoring within-species variation may lead to biases and spurious results in phylogenetic comparative analyses. For example, simulations revealed that failure to account for intraspecific variation can lead to a high Type I error rate (Harmon and Losos, 2005; Felsenstein, 2008; Silvestro et al., 2015). In cases where within-species variation exceeds between-species variation, it may be inappropriate to use mean values to represent an entire species in interspecific comparisons. This is of particular importance in studies of primates, given the considerable variability exhibited within species. This has long been noted as a concern in comparative studies of primates. For example, Strier (2003: 5) stated that, “the compression of intraspecific variation in any behavioral trait into a single, species-specific value precludes interspecific comparisons…”. Similarly, Struhsaker (2000: 119) warned that there is a “need to better understand interpopulational and intraspecific variation […]. Until this level of variation is understood and taken into consideration, broad interspecific comparisons and generalizations are misleading, if not counterproductive, in furthering the field of behavioral ecology”.
In addition, relying on small sample sizes may lead to poor quality data for traits that show high within-species variation (Garamszegi and Møller, 2010). Attention to the quality of data is a growing concern in comparative studies (Borries et al., 2013; Patterson et al., 2014). Sample size and variability are also salient issues in the study of hominin evolution and taxonomy. A small number of specimens, often quite variable due to biases in preservation (Behrensmeyer and Kidwell, 1985), may serve as the basis for inferences for an entire population (Wood and Lonergan, 2008). It is therefore crucial to interpret the hominin fossil record with methods that account for small sample size and accurately assess variation both within- and between-species.
Some comparative methods provide a way to incorporate intraspecific variation into analyses (Martins and Hansen, 1997; Ives et al., 2007; Felsenstein, 2008; Revell and Reynolds, 2012; Garamszegi, 2014b), but such analyses are rarely carried out in practice (Garamszegi and Møller, 2010). In many cases, data on intraspecific variation are unavailable. Even when such data are available, few studies have attempted to quantify the amounts of interspecific and intraspecific variation in traits being compared. The default procedure assumes that no within-species variation exists. The effect of ignoring intraspecific variation in comparative analyses has been studied for only a few systems, predominantly relying on simulated rather than actual data (Harmon and Losos, 2005; Ives et al., 2007; Felsenstein, 2008; Hansen and Bartoszek, 2012). Researchers have only recently begun to assess intraspecific variation in a comparative context (e.g., Kamilar and Baden, 2014; Pap et al., 2015).
Group size in primates provides an example of the issues raised above and is the focus of this paper. Group size is a commonly employed variable in comparative studies of primates and other animals, as this trait is hypothesized to play a central role in the evolution of feeding ecology (Wrangham et al., 1993; Janson and Goldsmith, 1995), social relationships (Sterck et al., 1997; Lehmann et al., 2007), parasitism (Nunn et al., 2003; Rifkin et al., 2012), predator defense (Janson and Goldsmith, 1995; Hill and Lee, 1998), and complex cognition and large brains (Deaner et al., 2000; MacLean et al., 2013). Group size features prominently in tests of the social intelligence and social brain hypotheses, which suggest that, among primates, a selective premium is placed on the cognitive abilities of individuals who must manage relationships with multiple conspecifics in large groups (Chance and Mead, 1953/1988; Jolly, 1966; Humphrey, 1976; Byrne and Whiten, 1988; Dunbar, 1992). Support for the social brain hypothesis comes from several studies showing that neocortex size is positively correlated with group size across primate species (Sawaguchi and Kudo, 1990; Dunbar, 1992; Sawaguchi, 1992; Dunbar and Shultz, 2007). Understanding the evolution of large brains and complex cognition is a major question in the study of human evolution; thus, understanding the nuances of the analyses that underlie the social brain hypothesis is critically important for evolutionary anthropology.
The aforementioned studies used single values of group size to represent an entire species. This practice assumes that negligible variation exists in group size, yet this trait is known to vary considerably within primate species (Struhsaker, 2000; Mitani et al., 2012; Patterson et al., 2014; Strier et al., 2014). In chimpanzees, for example, communities range in size from approximately 20 to 200 individuals (e.g., Wilson et al., 2014), yet in one widely cited paper analyzing the relationship between neocortex size and group size, this variability is collapsed into a single value, 53.5 (Dunbar, 1992). While a species mean may accurately reflect the situation in solitary and socially monogamous primates where the only source of variation in “group” size is due to offspring being born and dispersing at maturity, a species mean is likely to conceal biologically meaningful variation for primates living in larger social groups (Kamilar and Baden, 2014; Patterson et al., 2014).
In this paper, we investigated the degree to which intraspecific variation in group size influences comparative tests of the social brain hypothesis in primates. First, we examine whether between-species variation in group size exceeds the variation found within species. To do this, we estimate within-species “repeatability” in group size in terms of the amount of interspecific variability in traits being analyzed in comparative studies relative to the total amount of variation in the data (Sokal and Rohlf, 1981; Nakagawa and Schielzeth, 2010). If repeatability is high (i.e., approximates 1), then most of the variation is due to differences between species. In contrast, when repeatability is low, considerable variation exists within species. When between-species repeatability is low, controlling for intraspecific variation becomes particularly important for comparative studies (Harmon and Losos, 2005).
Next, we implemented a regression method to control for intraspecific variability in the estimates of group size when analyzing the relationship between group size and neocortex ratio (Ives et al., 2007). Using a generalized linear model, this method uses as input the means for each species and the variance or standard errors around the mean. It then estimates parameters of the linear model, taking intraspecific variation into account.
Finally, we employed a resampling procedure to assess how different estimates of group size affect the relationship between neocortex ratio and group size. For this, we treated each within-species group size value as a potentially true value, using resampled values iteratively in statistical tests analyzing the association between neocortex ratio and group size. If within-species variation has negligible effects on the results of the comparative analysis, different group size values should produce little variance in the parameter estimates of different regression models. We also compared results from this analysis to a test of phylogenetic uncertainty, or error due to different probable phylogenies, as this is another source of variation that is more commonly assessed in comparative analyses (Pagel and Lutzoni, 2002; Arnold et al., 2010).
2. Materials and methods
2.1. Data collection
We collected brain size and group size values from the published literature for the same 26 monkey and ape species included in Dunbar’s (1992) sample. Following Dunbar, we used the brain size data published by Stephan et al. (1981). Dunbar found the strongest correlation between group size and neocortex ratio, although other measures, including absolute neocortex volume, showed statistically significant positive relationships with group size. Continued debate exists about which brain variables are the best metrics to assay cognition for comparative research (Deaner et al., 2000; Healy and Rowe, 2007). Here, we followed Dunbar and colleagues by using the neocortex ratio, which is the volume of the neocortex divided by the rest of the brain, excluding the neocortex. Few samples of the neocortex sizes of individuals of different primate species exist, and lacking estimates of within species variation in this measure, we focus only on variation in group size. We note, however, that the procedures that we implement could also be applied to analyses where both predictor and response variables vary within and between species, and the effect of measurement error (i.e., within-species variance) is different when present on the predictor or the response (Hansen and Bartoszek, 2012).
We employed data on group size from Patterson et al. (2014). These authors searched the primary literature for estimates of group size from studies in which subjects were habituated to human presence and where all individuals or their age-sex classes were known. Adopting these criteria may underestimate true variation within species; however, this procedure ensures the quality of the data and accuracy of group size estimates (Patterson et al., 2014). We found group size estimates for 23 species (range = 3–18 groups per species, median = 10 groups per species). For two taxa in our sample, Alouatta and Cebus, Stephan et al. (1981) only specified the genus, with no species identified for these data on brain size. For our analyses, we used group sizes for Alouatta palliata and Cebus capucinus.
2.2. Statistical analyses
2.2.1. Repeatability
We calculated repeatability as the ratio of between-species variance relative to total variance (i.e., within-plus between-species variance). Group size values were log10-transformed to meet the assumptions of the repeatability analyses for normally-distributed data (Nakagawa and Schielzeth, 2010). We conducted a linear mixed-effect model (LMM) to obtain variance components of repeatability (Nakagawa and Schielzeth, 2010). The first analysis employed the “lme4” package (Bates et al., 2013) in R (R Core Team, 2013), with log group size as the response variable and species identity as a random factor. We then conducted a permutation test to determine whether the observed repeatability differed from a null model in which we reshuffled the group size data 1000 times and calculated repeatability on each dataset. We compared our observed value to the tail of the distribution (see Supplementary Online Material [SOM] 1). To determine confidence intervals, we applied a parametric bootstrap in which data were simulated based on the estimated model parameters, and then the same model was fitted to estimate repeatability (sensu Efron and Tibshirani, 1993). This procedure was iterated 1000 times, and the range of simulated repeatabilities was used to produce a confidence interval (2.5% and 97.5% quantiles).
Not all primates are expected to vary in group size to the same degree, and including species with different social systems may skew repeatability values. As mentioned previously, solitary and monogamous species have low within-species variability due to their small groups. Including these species in an analysis of repeatability will bias the result toward a high repeatability, i.e., higher between- relative to within-species variation, and may not be representative of within-species variation in primate group size. We therefore conducted a second repeatability analysis in which we excluded monogamous species.
2.2.2. Phylogenetic regression models
We calculated species-specific trait values by averaging untransformed group size values, and conducted a phylogenetic generalized least squares (PGLS) regression analysis to assess the relationship between log neocortex ratio and log group size without accounting for within-species variance. We allowed the phylogenetic scaling factor (λ) to take a value at the maximum likelihood (Freckleton, 2009). To conduct this analysis, we used the ‘caper,’ ‘ape,’ and ‘nlme’ packages in R (Paradis et al., 2004; Orme et al., 2013; Pinheiro et al., 2013; R Core Team, 2013). For all analyses assessing the relationship between group size and neocortex ratio, we defined neocortex ratio as the predictor variable and group size as the response variable, following previous tests of the social brain hypothesis (Dunbar, 1992; Dunbar and Shultz, 2007). As with repeatability analyses, we conducted two sets of regression analyses, the first including all 23 species and the second consisting of 17 group-living species that exclude socially monogamous ones.
2.2.3. Controlling for within-species variation
We created measurement error models that adjust for within-species variation in group size through an error component of the PGLS model residuals. For this, we counted the number of samples of group size for each species and calculated their standard errors and variances. We log-transformed the variances and the means using the procedures described in Ives et al. (2007). The transformations assume that the values follow a log-normal distribution and are derived from the probability density function of that distribution (see also Quan and Zhang, 2003). Conducting such transformations is recommended for the statistical tests and also removes scale-dependence.
We compared the outputs of the measurement error model (i.e., model fit and parameter estimates) to the transformed PGLS model that only considers phylogeny and assumes that traits do not vary within species. Because the data are transformed, the parameters of the models controlling for measurement error are compared to models that used transformed data without controlling for measurement error, but are not comparable to the parameters from the original, untransformed PGLS model. We conducted analyses using the “MERegPHYSIG” package (Ives et al., 2007) in MATLAB. We conducted two sets of regression analyses, the first including all 23 species and the second consisting of 17 group-living species that exclude monogamous species.
2.2.4. Within-species resampling
The measurement error method treats within-species variation as an error component in the linear model and attempts to recover the true mean values of the traits and the subsequent parameter estimates of the model. It is possible, however, that a single mean value is not an appropriate metric for a trait that is highly variable, and any single datum is just as accurate as any other for describing a species’ trait. For certain traits, different populations of a single species may have very similar values, whereas for other traits, different populations of a single species may have wildly different values. A mean value may be appropriate for the former, but inappropriate for the latter.
With these concerns in mind, we implemented a resampling procedure as another way to incorporate within-species variation into comparative analyses. Here, we randomly chose one group size datum for each species from the available measurements and accepted this as the species-specific estimate (Mönkkönen and Martin, 2000). This resampling technique is similar to randomization procedures used by paleoanthropologists (e.g., Lockwood et al., 1996). This method does not account for the downward shift (attenuation bias) of the parameter estimates (e.g., slope) when within-species variation is taken into account (Freckleton, 2011; Garamszegi, 2014b). Nevertheless, this analysis is useful for assessing uncertainty in parameter estimates and visualizing the effect of within-species variation. Using this resampling scheme, we created 1000 species-specific datasets that were subsequently analyzed by PGLS to investigate the relationship between group size and neocortex ratio. Some iterations encountered optimization errors, which relate to calculating the maximum likelihood of λ. Such optimization errors (approximately 15) were ignored, and resampling continued until we had produced 1000 models. To make inferences across these models, we determined the 95% confidence intervals of the derived intercepts and slopes and plotted the results. As with previous analyses, we conducted two sets of regression analyses, the first including all 23 species and the second consisting of 17 non-monogamous species.
2.2.5. Phylogenetic uncertainty
Another potential source of error in comparative analyses is uncertainty in the phylogenetic relationships of species in the sample. Controlling for the evolutionary relationships of species addresses the problem of non-independence of data points. However, phylogenies themselves are not without error (Lutzoni et al., 2001; Pagel and Lutzoni, 2002; Arnold et al., 2010). We evaluated the effect of phylogenetic uncertainty by conducting a series of analyses where the evolutionary tree varies and the trait value of group size is held constant (Garamszegi and Mundry, 2014). We compared the results of these analyses to those derived from the resampling method examining the influence of within-species variation to assess which source of variation had a greater effect, trait uncertainty or phylogenetic uncertainty.
We assessed the influence of uncertainty in phylogenetic relationships by using 1000 different phylogenetic trees from 10kTrees version 3, with the Wilson and Reeder (2005) taxonomy (Arnold et al., 2010). These trees were created using Bayesian phylogenetic methods, with trees thus sampled in proportion to their probability (Arnold et al., 2010). We then examined the relationship between neocortex ratio and group size using PGLS separately for each tree, resulting in 1000 PGLS models. For each model, we allowed the phylogenetic scaling factor (λ) to take a value at its maximum likelihood (Freckleton, 2009). To make inferences from the large number of models, we applied model averaging that takes into account the different degree of fit of the models to estimate regression slopes and intercepts (Garamszegi and Mundry, 2014). We also evaluated the regression outcomes graphically. We conducted the above analyses for all 23 species. For the subset of non-monogamous species, λ was close to 0, however. The impact of phylogeny on the regression models was negligible (see below), and thus, we did not investigate the effects of phylogenetic uncertainty for the non-monogamous subset of species.
To facilitate the application of these methods to other datasets, we provide the R code [SOM 1] and associated data files [SOM 2–3] for others to use. In addition, many of the analyses employed here are available as R code with instructions online: http://www.mpcm-evolution.org/ (Garamszegi, 2014a).
3. Results
3.1. Repeatability
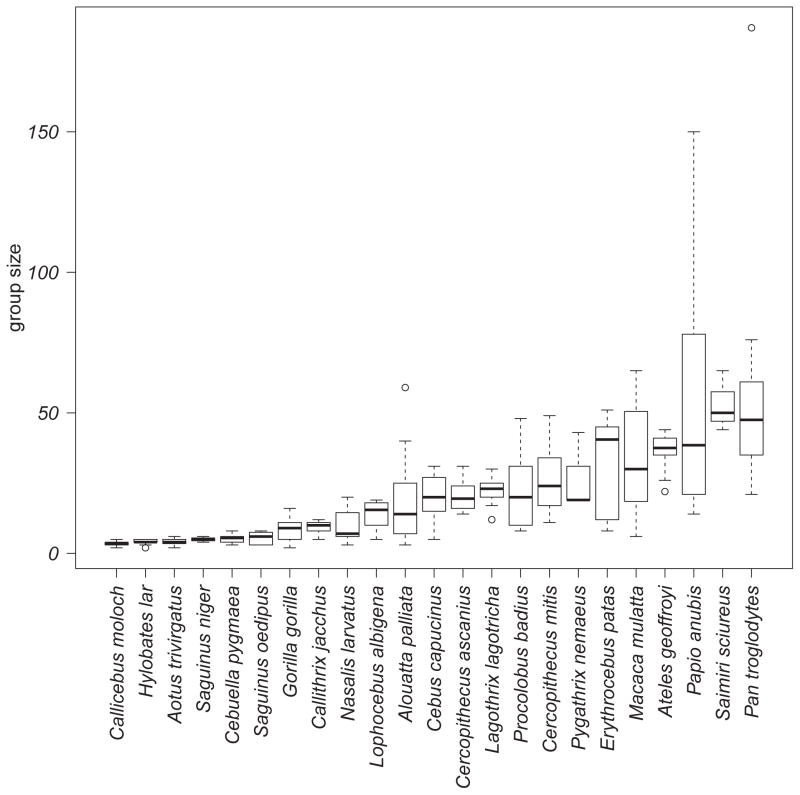
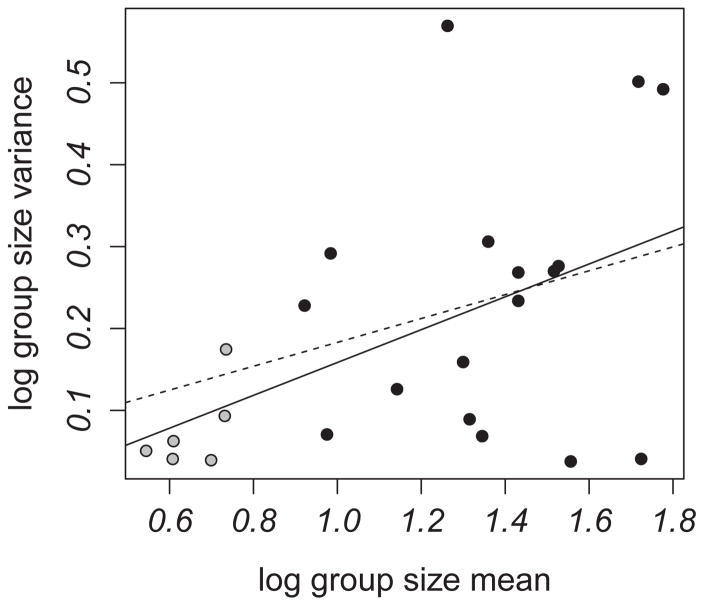
Group size was variable within and between species (Fig. 1). Across all 23 primate species, between-species repeatability of group size can be considered moderate or high (linear mixed effect model [LMM] with parametric bootstrapping: R = 0.733, p < 0.0001, 95% CI = 0.565–0.825). Variance appeared to increase with increasing group size even after logarithmic transformation (Fig. 2). We assessed the relationship between mean values and variance in group size and found a weak but significant positive association, with larger groups exhibiting greater variance than smaller groups (PGLS regression: slope = 0.200 ± 0.077, adjusted R2 = 0.207, p = 0.017).
Figure 1.
Within- and between-species variation in group size across sample of 23 primates. The boxplot shows the median and spread of data: the box represents the 25th and 75th percentiles, whiskers represent minimums and maximums relative to the interquartile range, and circles represent outliers.
Figure 2.
The relationship between average primate group size and its variance. Gray dots = monogamous species, black dots = non-monogamous species, solid line = significant relationship for all 23 species, dotted line = non-significant relationship between group size and variance among non-monogamous species.
The relationship between the mean and variance in group size seemed to be driven partly by monogamous species that live in small groups and exhibit low variance. When monogamous species were excluded, we found no relationship between log-transformed mean group size and its variance (Fig. 2; PGLS regression: slope = 0.146 ± 0.159, adjusted R2 = −0.010, p = 0.374). We also conducted repeatability analyses after excluding monogamous species and found that between-species repeatability of group size was lower (LMM and parametric bootstrapping: R = 0.477, p < 0.0001, 95% CI = 0.275–0.628). Thus, greater within-species variability in group size exists in non-monogamous primates than in the wider primate sample; estimates of within- and between-species variation were similar in the non-monogamous species.
3.2. Phylogenetic regressions with and without controlling for intraspecific variation
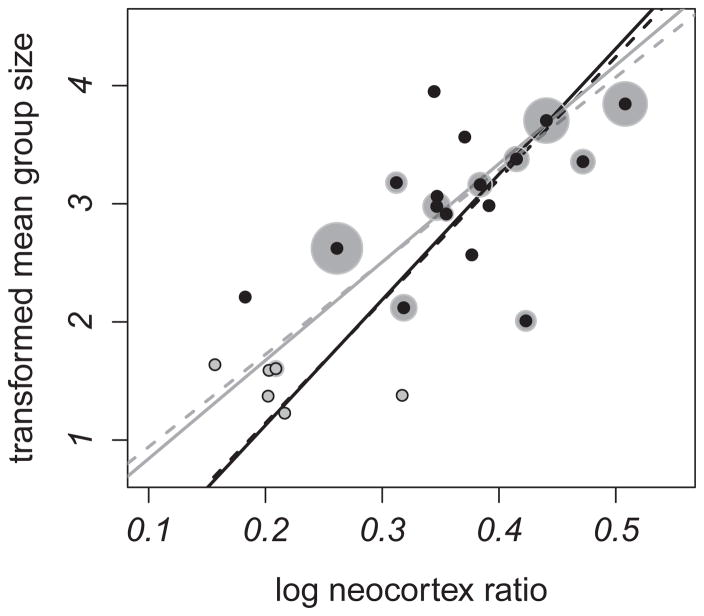
Using our group size estimates for all 23 primate species (Patterson et al., 2014), we found a strong association between mean group size and neocortex ratio using standard phylogenetic comparative methods that did not control for intraspecific variation (Fig. 3; untransformed PGLS regression: slope = 4.081 ± 0.735, t = 5.551, R2 = 0.575, p < 0.001, λ = 0.647). These results are thus similar to those from previous analyses (Dunbar, 1992; Dunbar and Shultz, 2007). When monogamous species were excluded, a significant association between mean group size and neocortex ratio remained (untransformed PGLS regression: slope = 1.818 ± 0.734, t = 2.475, R2 = 0.243, p = 0.026, λ = 0.001).
Figure 3.
The relationship between neocortex ratio versus mean group size. Gray dots = monogamous species, black dots = non-monogamous species, light gray circles = group size transformed variance, black lines = all 23 species, gray lines = non-monogamous species only, solid lines = transformed PGLS regression with no measurement error, dashed lines = Ives et al. (2007) max. likelihood model.
In analyses that controlled for within-species variation, we again recovered a positive relationship between neocortex ratio and group size. The slopes derived from the models correcting for measurement error did not differ substantially from the model that did not incorporate measurement error (Table 1). Within the subset of non-monogamous species, controlling for measurement error recovered a positive relationship, and the slopes derived from the models correcting for measurement error did not differ substantially from the equivalent model that ignored measurement error (Fig. 3). There was, however, additional noise, as there were moderate standard errors around the slope (Table 1).
Table 1.
Coefficients and standard errors of models examining the relationship between neocortex ratio and group size.a
| All species
|
Group-living (monogamous excluded)
|
|||
|---|---|---|---|---|
| Slope | Intercept | Slope | Intercept | |
| Without considering within-species variance | 10.625 | −1.00 | 8.323 | 0.010 |
| With approx. variance (estimated generalized least-square) | 10.625 ± 1.634 | −1.013 ± 0.672 | 8.323 ± 2.277 | −0.003 ± 0.947 |
| With approx. variance (maximum likelihood) | 10.332 ± 1.549 | −0.923 ± 0.622 | 7.822 ± 2.026 | 0.160 ± 0.829 |
Values are shown for models ignoring variance (top model) and those incorporating within-species variance (lower two models, based on Ives et al., 2007).
3.3. Within-species resampling
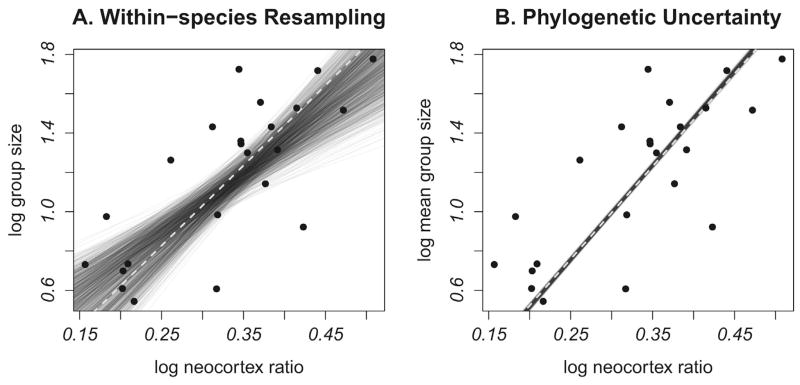
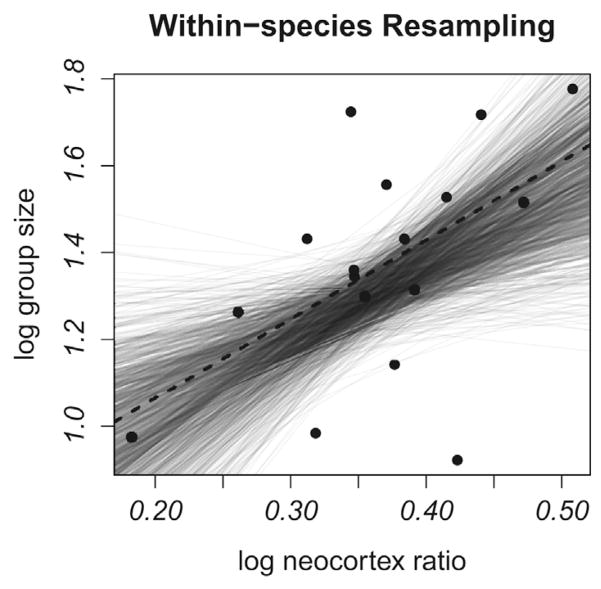
How consistent is the relationship between group size and neocortex ratio using different measures of group size for each species? The resampling analysis produced a positive association between neocortex ratio and brain size (median slope = 3.314, 95% CI=2.058–4.864;median intercept =0.050,95% CI =−0.531–0.434), with modest noise (Fig. 4a). In some cases, the slope was shallow, but the 95% confidence interval did not include zero.
Figure 4.
Evaluating the effects of within species variation in group size and phylogenetic uncertainty. (A) Within species variation in group size. The relationship between group size and neocortex ratio is shown after resampling species-specific group size values using a consensus tree (1000 regressions). (B) Phylogenetic uncertainty. The relationship between group size and neocortex ratio is shown using 1000 different probable phylogenies with mean group size values. Gray lines = individual regressions, dashed lines = regression based on mean group size values and the consensus tree.
The resampling analysis that included only non-monogamous species yielded similar results, although we documented considerable noise (Fig. 5). Regressions produced slopes that approached zero, and on rare occasions regressions produced negative slopes, but the 95% confidence interval excluded zero (median slope = 1.801, 95% CI = 0.269–3.456; median intercept = 0.650, 95% CI = 0.518–1.20). Taken together, these results reveal that, despite considerable within-species variation in group size, a positive correlation between group size and neocortex ratio persists.
Figure 5.
The relationship between group size and neocortex ratio in a sample of 17 non-monogamous primate species. Results of resampling species-specific group size values with a consensus tree (1000 regressions) are shown. Gray lines = individual regressions, dashed line = regression based on mean group size values and the consensus tree.
3.4. Phylogenetic uncertainty
Phylogenetic uncertainty had weaker effects on the relationship between neocortex ratio and group size. Across all 23 species, model averaging indicated that the average slope was 4.148 with a 95% confidence interval of 2.697–5.599. The 95% confidence interval represented a wider range of slopes than actually occurred, as it is an “unconditional” confidence interval calculated based on the model averaged standard errors (Buckland et al., 1997; Burnham and Anderson, 2002; Mazerolle, 2013). Overall, the 1000 regressions using different trees consistently produced similar findings (Fig. 4b).
4. Discussion
In addition to phylogenetic non-independence, within-species variation may influence the outcome of comparative analyses and should be considered in studies that investigate correlated trait evolution (Garamszegi, 2014b). Across a sample of primates, populations of the same species display variable group sizes. Substantially more variation, however, occurs between- rather than within-species. Specifically, we found that about 70% of the total variation was explained by interspecific variation. Controlling for within-species variation had little effect on the relationship between group size and neocortex ratio given this moderate repeatability. This finding is consistent with the results of a simulation study that found within-species variation has weak effects on the results of comparative analyses when at least 60% of the total variation in traits is explained by variation between species (Harmon and Losos, 2005).
In our analyses, incorporating within-species variation in group size had a negligible effect on regression models that investigated the relationship between neocortex ratio and group size. Similar results were derived from the resampling method. Resampling based on data from different populations created variable slope estimates, although these analyses recovered the positive relationship. The variability in slope estimates created by within-species variation was greater than that produced by phylogenetic uncertainty among the primate species in these analyses.
Some species vary more than others in group size. For example, solitary and monogamous species live in small groups, which vary little in size. Thus, these species are expected to have consistently small groups with little within-species variation. Group size is mediated by the mating and social system of the species: in polygynous and polygynandrous mating systems, sizeable influxes of males and/or females can occur over time. Monogamous species, however, do not generally allow immigrants to join their group, and their offspring disperse upon reaching maturity.
When we excluded monogamous species from the analysis, only 50% of the total variance in group size was explained by between-species differences. The lower within-species repeatability is likely driven by the exclusion of species that rarely vary, given the nature of pair bonded social systems. Using models that controlled for within-species variance recovered a positive association and produced similar slopes, although accounting for measurement error introduced considerable uncertainty. The resampling analysis that included only non-monogamous group-living primates revealed that slopes from regressions of group size on neocortex ratio ranged from 0.25 to 3.5. However, the slopes from models that controlled for within-species variance did not differ significantly from the slopes in models that ignored within-species variance.
The finding that neocortex ratio is positively associated with group size after controlling for intraspecific variation in the latter trait in a sample of non-monogamous species is perhaps surprising. Our sample, however, was small, consisting of only 17 species. This small sample size may explain why, despite showing lower between-species repeatability than in the full sample, within-species variation failed to blur the relationship between brain size and group size. Additional analyses incorporating a larger sample of primates may shed additional light on whether within-species variation in traits influences the results of this and other comparative analyses.
One limitation of our study is that we only incorporated within-species variation in the response variable, as multiple measures of the neocortex size of individuals in different primate species are not available. This could further underestimate the true effect of within-species variation. Indeed, studies that have investigated brain size within and between species have documented considerable intraspecific variation (Sherwood et al., 2004). Ideally, analyses should incorporate variance in both the predictor and response variables. Similarly, alternative statistical approaches, such as those incorporating Bayesian probability and model selection, may add additional rigor to comparative studies, especially when data on intraspecific variation in the traits of interest are available (Revell and Reynolds, 2012).
We applied a diversity of approaches to investigate whether within-species variation influences the outcome of comparative studies, which has long been raised as a concern in studies of primate comparative biology (Struhsaker, 2000; Strier, 2003; Nunn, 2011). These approaches could be used in other comparative studies in evolutionary anthropology and human evolution. For example, these analyses could be added to the repertoire of paleoanthropologists, who already use randomization and resampling procedures to make conclusions utilizing small samples or single specimens (e.g., Lockwood, 1999; Cardini and Elton, 2007; Cofran and DeSilva, 2015) and are increasingly incorporating phylogenetic comparative methods in their research (e.g., Pontzer et al., 2014; Gonzales et al., 2015; Pampush, 2015; Russo, 2016).
We focused on group size, which is a key variable in comparative studies inferring the evolution of large brains (Sawaguchi and Kudo, 1990; Dunbar, 1992; Sawaguchi, 1992; Dunbar and Shultz, 2007). Understanding encephalization in primates and humans is a major question in evolutionary anthropology, and the social brain hypothesis has provided a powerful framework for understanding brain evolution. Our findings indicate that the wide range of group size values present in several primate species does not erase the association between group size and neocortex ratio across primates. This association is recovered despite a high degree of within-species variation in group size among non-monogamous primates. Thus, among monkeys and apes, species that tend to live in larger groups also tend to have relatively larger neocortices, affirming the relationship between brain size and sociality in primates. This lends support to the hypothesis that sociality, not just ecology, has been an important factor in the evolution of brain size and cognitive abilities among primates, including humans.
Given the potential impact of intraspecific variation, researchers who have previously conducted comparative analyses utilizing species averages of group size may be tempted to breathe a sigh of relief. However, our results should not imply that within-species variation can be ignored. Whether between-species variation exceeds within-species variation will be influenced by the taxa and traits under consideration. Hence, when the data exist, analyses such as those presented here should be performed. In future studies, when repeatability is low, methods incorporating within-species variation should be employed. Including within-species variation in phylogenetic comparative analyses can reduce biases created by ignoring known variance and increase confidence about parameter estimates. Additional research investigating other sources of error that can arise in comparative studies is required and will ultimately strengthen the conclusions from these analyses. Finally, additional data on neocortex size and comparative analyses of brain structures, group size, and other variables, including ecological traits, will further elucidate the forces associated with the evolution of large brains.
Supplementary Material
Acknowledgments
We thank the Center for Statistical Consultation and Research at the University of Michigan for help with R code and methodological advice, especially Yanming Li, Breanna Miller, and Kerby Shedden. For training in phylogenetic comparative methods, we thank the AnthroTree Workshop that is supported by the National Science Foundation (BCS-0923791) and the National Evolutionary Synthesis Center (NSF grant EF-0905606). For helpful comments on an earlier draft of the manuscript, we thank S. Elton, P.C. Lee, one anonymous reviewer, and an Associate Editor of Journal of Human Evolution. A.A. Sandel was supported by the National Science Foundation Graduate Research Fellowship under Grant No. F031543. J.A. Miller was supported by a department grant from the National Science Foundation (DGE-0801634). J.C. Mitani is currently supported by National Institutes of Health RO1AG049395. C.L. Nunn was supported by Duke University and the National Science Foundation (BCS-1355902). S.K. Patterson was supported by the National Science Foundation Graduate Research Fellowship under Grant No. DGE-1311230. L.Z. Garamszegi was supported by funds from the Spanish Government within the frame of the “Plan Nacional” program (ref. no. CGL2012-38262 and CGL2012-40026-C02-01) and the National Research, Development and Innovation Office, Hungary (NKFIH, K-115970).
Footnotes
Supplementary online material related to this article can be found at http://dx.doi.org/10.1016/j.jhevol.2016.03.007.
References
- Arnold C, Matthews LJ, Nunn CL. The 10kTrees website: a new online resource for primate phylogeny. Evol Anthropol. 2010;19:114–118. [Google Scholar]
- Bates D, Maechler M, Bolker B, Walker S. R package version 1.0-5. 2013. lme4: Linear mixed-effects models using Eigen and S4. [Google Scholar]
- Behrensmeyer AK, Kidwell SM. Taphonomy’s contributions to paleobiology. Paleobiology. 1985;11:105–119. [Google Scholar]
- Borries C, Gordon AD, Koenig A. Beware of primate life history data: a plea for data standards and a repository. PLoS One. 2013;8:e67200. doi: 10.1371/journal.pone.0067200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckland ST, Burnham KP, Augustin NH. Model selection: an integral part of inference. Biometrics. 1997;53:603–618. [Google Scholar]
- Burnham KP, Anderson DR. Model selection and multimodel inference: a practical information-theoretic approach. 2. Springer; New York: 2002. [Google Scholar]
- Byrne RW, Whiten AW. Machiavellian intelligence: social expertise and the evolution of intellect in monkeys, apes, and humans. Clarendon Press; Oxford: 1988. [Google Scholar]
- Cardini A, Elton S. Sample size and sampling error in geometric morphometric studies of size and shape. Zoomorphology. 2007;126:121–134. [Google Scholar]
- Chance MRA, Mead AP. Social behaviour and primate evolution. In: Byrne RW, Whiten AW, editors. Machiavellian intelligence: social expertise and the evolution of intellect in monkeys, apes, and humans. Clarendon Press; Oxford: 1953/1988. pp. 34–49. [Google Scholar]
- Cofran Z, DeSilva JM. A neonatal perspective on Homo erectus brain growth. J Hum Evol. 2015;81:41–47. doi: 10.1016/j.jhevol.2015.02.011. [DOI] [PubMed] [Google Scholar]
- Deaner RO, Nunn CL, van Schaik CP. Comparative tests of primate cognition: Different scaling methods produce different results. Brain Behav and Evol. 2000;55:44–52. doi: 10.1159/000006641. [DOI] [PubMed] [Google Scholar]
- Dunbar RIM. Neocortex size as a constraint on group size in primates. J Hum Evol. 1992;22:469–493. [Google Scholar]
- Dunbar RIM, Shultz S. Evolution in the social brain. Science. 2007;317:1344–1347. doi: 10.1126/science.1145463. [DOI] [PubMed] [Google Scholar]
- Efron B, Tibshirani RJ. An introduction to the bootstrap. monographs on statistics and applied probability 57. Chapman & Hall; New York: 1993. [Google Scholar]
- Felsenstein J. Phylogenies and the comparative method. Amer Nat. 1985;125:1–15. [Google Scholar]
- Felsenstein J. Comparative methods with sampling error and within-species variation: contrasts revisited and revised. Amer Nat. 2008;171:713–725. doi: 10.1086/587525. [DOI] [PubMed] [Google Scholar]
- Freckleton RP. The seven deadly sins of comparative analysis. J Evol Bio. 2009;22:1367–1375. doi: 10.1111/j.1420-9101.2009.01757.x. [DOI] [PubMed] [Google Scholar]
- Freckleton RP. Dealing with collinearity in behavioural and ecological data: model averaging and the problems of measurement error. Behav and Brain Sci. 2011;65:91–101. [Google Scholar]
- Garamszegi LZ. Modern phylogenetic comparative methods and their application in evolutionary biology. Springer-Verlag; Berlin: 2014a. [Google Scholar]
- Garamszegi LZ. Uncertainties due to within-species variation in comparative studies: measurement errors and statistical weights. In: Garamszegi LZ, editor. Modern phylogenetic comparative methods and their application in evolutionary biology. Springer-Verlag; Berlin: 2014b. pp. 157–199. [Google Scholar]
- Garamszegi LZ, Møller AP. Effects of sample size and intraspecific variation in phylogenetic comparative studies: a meta-analytic review. Biol Rev. 2010;85:797–805. doi: 10.1111/j.1469-185X.2010.00126.x. [DOI] [PubMed] [Google Scholar]
- Garamszegi LZ, Mundry R. Multimodel-inference in comparative analyses. In: Garamszegi LZ, editor. Modern phylogenetic comparative methods and their application in evolutionary biology. Springer-Verlag; Berlin: 2014. [Google Scholar]
- Gonzales LA, Benefit BR, McCrossin ML, Spoor F. Cerebral complexity preceded enlarged brain size and reduced olfactory bulbs in Old World monkeys. Nature Comm. 2015;6:7580. doi: 10.1038/ncomms8580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen TF, Bartoszek K. Interpreting the evolutionary regression: The Interplay between observational and biological errors in phylogenetic comparative studies. Syst Biol. 2012;61:413–425. doi: 10.1093/sysbio/syr122. [DOI] [PubMed] [Google Scholar]
- Harmon LJ, Losos JB. The effect of intraspecific sample size on type I and type II error rates in comparative studies. Evolution. 2005;59:2705–2710. [PubMed] [Google Scholar]
- Harvey P, Pagel M. The comparative method in evolutionary biology. Oxford University Press; Oxford: 1991. [Google Scholar]
- Healy SD, Rowe C. A critique of comparative studies of brain size. Proc R Soc B. 2007;274:453–464. doi: 10.1098/rspb.2006.3748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill RA, Lee PC. Predation risk as an influence on group size in cercopithecoid primates: implications for social structure. J Zool. 1998;245:447–456. [Google Scholar]
- Humphrey NK. The social function of intellect. In: Bateson P, Hinde R, editors. Growing points in ethology. Cambridge University Press; Cambridge: 1976. pp. 303–317. [Google Scholar]
- Ives AR, Midford PE, Garland T. Within-species variation and measurement error in phylogenetic comparative methods. Syst Biol. 2007;56:252–270. doi: 10.1080/10635150701313830. [DOI] [PubMed] [Google Scholar]
- Janson CH, Goldsmith ML. Predicting group size in primates: foraging costs and predation risks. Behav Ecol. 1995;6:326–336. [Google Scholar]
- Jolly A. Lemur social behavior and primate intelligence. Science. 1966;153:501–506. doi: 10.1126/science.153.3735.501. [DOI] [PubMed] [Google Scholar]
- Kamilar JM, Baden AL. What drives flexibility in primate social organization? Behav Ecol Sociobiol. 2014;68:1677–1692. [Google Scholar]
- Lehmann J, Korstjens AH, Dunbar RIM. Group size, grooming and social cohesion in primates. Anim Behav. 2007;74:1617–1629. [Google Scholar]
- Lockwood CA. Sexual dimorphism in the face of Australopithecus africanus. Amer J Phys Anth. 1999;108:97–127. doi: 10.1002/(SICI)1096-8644(199901)108:1<97::AID-AJPA6>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- Lockwood CA, Richmond BG, Jungers WL. Randomization procedures and sexual dimorphism in Australopithecus afarensis. J Hum Evol. 1996;31:537–548. [Google Scholar]
- Lutzoni F, Pagel M, Reeb V. Major fungal lineages are derived from lichen symbiotic ancestors. Nature. 2001;411:937–940. doi: 10.1038/35082053. [DOI] [PubMed] [Google Scholar]
- MacLean EL, Sandel AA, Bray J, Oldenkamp RE, Reddy RB, Hare BA. Group size predicts social but not nonsocial cognition in lemurs. PLoS One. 2013;8:e66359. doi: 10.1371/journal.pone.0066359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martins EP, Hansen TF. Phylogenies and the comparative method: a general approach to incorporating phylogenetic information into the analysis of interspecific data. Amer Nat. 1997;149:646–667. [Google Scholar]
- Mazerolle MJ. AICcmodavg: model selection and multimodel inference based on (Q)AIC(c) 2013. R package version 1.35. [Google Scholar]
- Mitani JC, Call J, Kappeler PM, Palombit RA, Silk JB. The evolution of primate societies. University of Chicago Press; Chicago: 2012. [Google Scholar]
- Mönkkönen M, Martin TE. Sensitivity of comparative analyses to population variation in trait values: clutch size and cavity excavation tendencies. J Avian Biol. 2000;31:576–579. [Google Scholar]
- Nakagawa S, Schielzeth H. Repeatability for Gaussian and non-Gaussian data: a practical guide for biologists. Biol Rev. 2010;85:935–956. doi: 10.1111/j.1469-185X.2010.00141.x. [DOI] [PubMed] [Google Scholar]
- Nunn CL. The comparative approach in evolutionary anthropology and biology. University of Chicago Press; Chicago: 2011. [Google Scholar]
- Nunn CL, Altizer S, Jones KE, Sechrest W. Comparative tests of parasite species richness in primates. Amer Nat. 2003;162:597–614. doi: 10.1086/378721. [DOI] [PubMed] [Google Scholar]
- Orme D, Freckleton RP, Thomas G, Petzoldt T, Fritz S, Isaac N, Pearse W. caper: comparative analyses of phylogenetics and evolution in R. 2013 R package version 0.5.2. [Google Scholar]
- Pagel M, Lutzoni F. Accounting for phylogenetic uncertainty in comparative studies of evolution and adaptation. In: Lässig M, Valleriani A, editors. Biological evolution and statistical physics. Springer; Berlin Heidelberg: 2002. pp. 148–161. [Google Scholar]
- Pampush JD. Selection played a role in the evolution of the human chin. J Hum Evol. 2015;82:127–136. doi: 10.1016/j.jhevol.2015.02.005. [DOI] [PubMed] [Google Scholar]
- Pap PL, Osváth G, Sándor K, Vincze O, Bărbos L, Marton A, Nudds RL, Vágási CI. Interspecific variation in the structural properties of flight feathers in birds indicates adaptation to flight requirements and habitat. Funct Ecol. 2015;29:746–757. [Google Scholar]
- Paradis E, Claude J, Strimmer K. APE: analyses of phylogenetics and evolution in R language. Bioinformatics. 2004;20:289–290. doi: 10.1093/bioinformatics/btg412. [DOI] [PubMed] [Google Scholar]
- Patterson SK, Sandel AA, Miller JA, Mitani JC. Data quality and the comparative method: the case of primate group size. Int J Primatol. 2014;35:990–1003. [Google Scholar]
- Pinheiro J, Bates D, DebRoy S, Sarkar D R Core Team. nlme: Linear and Nonlinear Mixed Effects Models. 2013. [Google Scholar]
- Pontzer H, Raichlen DA, Gordon AD, Schroepfer-Walker KK, Hare B, O’Neill MC, Muldoon KM, Dunsworth HM, Wood BM, Isler K, Burkart J, Irwin M, Shumaker RW, Lonsdorf EV, Ross SR. Primate energy expenditure and life history. Proc Natl Acad Sci USA. 2014;111:1433–1437. doi: 10.1073/pnas.1316940111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quan H, Zhang J. Estimate of standard deviation for a log-transformed variable using arithmetic means and standard deviations. Stat Med. 2003;22:2723–2736. doi: 10.1002/sim.1525. [DOI] [PubMed] [Google Scholar]
- R Core Team. R: A language and environment for statistical computing. Vienna, Austria: 2013. [Google Scholar]
- Revell LJ, Reynolds RG. A new Bayesian method for fitting evolutionary models to comparative data with intraspecific variation. Evolution. 2012;66:2697–2707. doi: 10.1111/j.1558-5646.2012.01645.x. [DOI] [PubMed] [Google Scholar]
- Rifkin JL, Nunn CL, Garamszegi LZ. Do animals living in larger groups experience greater parasitism? A meta-analysis. Amer Nat. 2012;180:70–82. doi: 10.1086/666081. [DOI] [PubMed] [Google Scholar]
- Russo GA. Comparative sacral morphology and the reconstructed tail lengths of five extinct primates: Proconsul heseloni, Epipliopithecus vindobonensis, Archaeolemur edwardsi, Megaladapis grandidieri, and Palaeopropithecus kelyus. J Hum Evol. 2016;90:135–162. doi: 10.1016/j.jhevol.2015.10.007. [DOI] [PubMed] [Google Scholar]
- Sawaguchi T. The size of the neocortex in relation to ecology and social structure in monkeys and apes. Folia Primatol. 1992;58:131–145. [PubMed] [Google Scholar]
- Sawaguchi T, Kudo H. Neocortical development and social structure in primates. Primates. 1990;31:283–289. [Google Scholar]
- Sherwood CC, Cranfield MR, Mehlman PT, Lilly AA, Garbe JAL, Whittier CA, Nutter FB, Rein TR, Bruner HJ, Holloway RL, Tang CY, Naidich TP, Delman BN, Steklis HD, Erwin JM, Hof PR. Brain structure variation in great apes, with attention to the mountain gorilla (Gorilla beringei beringei) Am J Primatol. 2004;63:149–164. doi: 10.1002/ajp.20048. [DOI] [PubMed] [Google Scholar]
- Silvestro D, Kostikova A, Litsios G, Pearman PB, Salamin N. Measurement errors should always be incorporated in phylogenetic comparative analysis. Methods Ecol Evol. 2015;6:340–346. [Google Scholar]
- Sokal RR, Rohlf FJ. Biometry: the principles and practice of statistics in biological research. 2. W.H. Freeman & Company; New York: 1981. [Google Scholar]
- Stephan H, Frahm H, Baron G. New and revised data on volumes of brain structures in insectivores and primates. Folia Primatol. 1981;35:1–29. doi: 10.1159/000155963. [DOI] [PubMed] [Google Scholar]
- Sterck EH, Watts DP, van Schaik CP. The evolution of female social relationships in nonhuman primates. Behav Ecol Sociobiol. 1997;41:291–309. [Google Scholar]
- Strier KB. Primatology comes of age: 2002 AAPA Luncheon address. Amer J Phys Anthropol. 2003;122:2–13. doi: 10.1002/ajpa.10383. [DOI] [PubMed] [Google Scholar]
- Strier KB, Lee PC, Ives AR. Behavioral flexibility and the evolution of primate social states. PLoS One. 2014;9:e114099. doi: 10.1371/journal.pone.0114099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Struhsaker TT. Variation in adult sex ratios of red colobus monkey social groups: implications for interspecific comparisons. In: Kappeler PM, editor. Primate Males. Cambridge University Press; Cambridge: 2000. pp. 108–119. [Google Scholar]
- Wilson DE, Reeder DM. Mammal species of the world: a taxonomic and geographic reference. 3. Johns Hopkins University Press; Baltimore: 2005. [Google Scholar]
- Wilson ML, Boesch C, Fruth B, Furuichi T, Gilby IC, Hashimoto C, Hobaiter CL, Hohmann G, Itoh N, Koops K, Lloyd JN, Matsuzawa T, Mitani JC, Mjungu DC, Morgan D, Muller MN, Mundry R, Nakamura M, Pruetz J, Pusey AE, Riedel J, Sanz C, Schel AM, Simmons N, Waller M, Watts DP, White F, Wittig RM, Zuberbuhler K, Wrangham RW. Lethal aggression in Pan is better explained by adaptive strategies than human impacts. Nature. 2014;513:414–417. doi: 10.1038/nature13727. [DOI] [PubMed] [Google Scholar]
- Wood B, Lonergan N. The hominin fossil record: taxa, grades and clades. J Anat. 2008;212:354–376. doi: 10.1111/j.1469-7580.2008.00871.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrangham RW, Gittleman JL, Chapman CA. Constraints on group size in primates and carnivores: population density and day-range as assays of exploitation competition. Behav Ecol Sociobiol. 1993;32:199–209. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.