Abstract
Objective
NONcNZO10 (NZ10) mice are predisposed to obesity and develop type 2 diabetes (T2D) and hepatic steatosis even when maintained on a control diet (CD) of 6% fat. Studies were designed to determine whether this extreme susceptibility phenotype could be alleviated by diet and if so the molecular targets of diet.
Methods
NZ10 and SWR/J (SWR) control mice were fed a CD or a test diet of high protein and fish oil (HPO) for 19 weeks and then analyzed for steatosis, blood chemistry, hepatic gene and micro-RNA expression.
Results
HPO diet prevented steatosis, significantly increased serum adiponectin and reduced serum cholesterol and triglycerides only in NZ10 mice. The HPO diet repressed hepatic expression of fatty acid metabolic regulators including PPAR-γ, sterol regulatory element-binding protein-c1, peroxisome proliferator-activated receptor gamma co-activator-1, fatty acid synthase, fatty acid binding protein-4, and apolipoprotein A4 genes only in NZ10 mice. Also repressed by a HPO diet were adiponectinR2 receptor, leptin-R, PPAR-α, pyruvate dehydrogenase kinase isoforms 2 and 4, AKT2 and GSK3β. Micro-RNA (miR) arrays identified miRs that were diet and/or genetics regulated. QRTPCR confirmed increased expression of miR-205 and suppression of a series of miRs including miRs-411, 155, 335 and 21 in the NZ10-HPO group, each of which are implicated in the progression of diabetes and/or steatosis. Evidence is presented that miR-205 co-regulates with PPARγ and may regulate fibrosis and EMT during the progression of steatosis in the livers of NZ10-CD mice. The dietary responses of miR-205 are tissue-specific with opposite effects in adipose and liver.
Conclusion
The results confirm that a HPO diet overrides the genetic susceptibility of NZ10 mice and this correlates with the suppression of key genes and perhaps micro-RNAs involved in hyperglycemia, dyslipidemia and inflammation including master PPAR regulators, adiponectin and leptin receptors.
Keywords: obesity, type 2 diabetes, steatosis, NAFLD, microRNA, diet, NoNcNZO10
INTRODUCTION
Non-alcoholic fatty liver disease (NAFLD) is the most common chronic liver disease with prevalence estimated between 20–30% in some European countries and the USA (reviewed in [1, 2]). Obesity and T2D are the most common risk factors and recent studies suggest that genetics plays a significant role in predisposing subjects to NAFLD [2, 3]. Animal and patient studies have also implicated changes of specific miRs associated with NAFLD as well as the transition to non-alcoholic steatohepatitis (NASH) [4–6]. In order to further our understanding of the etiology of NAFLD as well as to develop new treatment strategies it is important to identify susceptibility genes and determine how they interact with diet. The NONcNZO mouse strains were developed at The Jackson Laboratory by introgressing known diabesity QTL from the T2D-prone and obese NZO/HlLt inbred strain into the non-obese and diabetes-resistant NON/Lt strain background [7–9]. The mice provide a model to trace obese and T2D susceptibility conferring loci and resident genes that is not possible in models that use wild type animals or monogenic transgenic (Ob/ob; Db/db) models (reviewed in [10]). NZ10 mice contain 6 such QTL and represent the most severe T2D susceptibility phenotype of the NONcNZO strains although with a relatively mild obesity. The NZ10 phenotype is more similar to human T2D that are the other monogenic or high fat-induced wild type rodent models. Previous studies have described the accumulation of visceral fat, elevated triglycerides and insulin resistance of NZ10 mice after 10 weeks of moderate to high fat feeding [7, 8]. Another studies reported widespread hepatic and pancreatic fatty infiltrates and focal hepatitis of NZ10 mice fed a 6% fat diet [9].
Dietary polyunsaturated fatty acids (PUFA) including omega-3 can regulate hepatic gene expression and have been shown to improve insulin sensitivity, reduce liver triglyceride levels and hepatic steatosis in animal models [11, 12]. Whereas some studies have suggested similar responses in human subjects to dietary fish oil supplements and PUFA (13), these results are not universal, and larger clinical trials have been inconclusive, perhaps because of the multifactorial etiology of the disease and heterogeneity of the subjects (reviewed in [11–13]). Further animal studies to investigate the effects of diet on a complex genetic susceptibility background may help resolve this. The purpose of the present study was to test the effect of a HPO diet on severe hepatic steatosis in NZ10 mice, to quantify key responsive hepatic genes and profile changes of miRs. The results show that HPO has profound effects on the steatosis phenotype with parallel effects on metabolic gene expression and miR profiles.
METHODS
Animals and diets
Four week Male NZ10 and their non-diabetic counterparts SWR/LtJ (SWR) mice were purchased from Jackson Laboratory. After 2 weeks of acclimatization both NZ10 and SWR mice were randomized into 2 groups (12 mice per group) to receive CD or HPO diets for a period of 19 weeks and were maintained in a temperature-controlled facility on a 12-h light-dark cycle. Control Diet (Custom modified Purina LabDiet 5K20) is the diet recommended by Jackson Laboratory to maintain the diabetic phenotype of male NZ10 mice. The High protein fish oil diet (Purina LabDiet 5TWH) was optimized for high protein, fiber and “good” fat (omega-3). The latter diet is modeled on the popularized human South Beach diet that emphasizes high protein, poly/mono-unsaturated fat and low carbohydrate. Feeding for all experimental groups was ad lib. For tissue harvest, mice were fasted for 12-h, sacrificed by cervical dislocation and the liver tissue rapidly dissected and processed for RNA analysis. Blood glucose after 12 hrs of fasting was measured with a glucometer (Freestyle, Abbott Diagnostics). Plasma triglycerides and cholesterol were assayed using Ortho Vitros 250 chemistry analyzer (Ortho, Rochester, NY) on undiluted samples. Total adiponectin levels were measured in the serum using an ELISA kit from Alpco Diagnostics (Salem, NH).
Liver fixing and staining
Livers were harvested at sacrifice and samples of <10 mm thickness placed immediately into 10% phosphate buffered paraformaldehyde at room temperature. After 24h, fixed samples were washed in PBS and incubated overnight in 10% phosphate buffered sucrose. Standard procedures were used for paraffin embedding, washing, dehydrating, cutting sections, staining with haematoxylin and eosin (H&E) and microscopic imaging.
RNA preparation
Total RNA was isolated from liver tissue using TRIzol Reagent (Invitrogen, Carlsbad, CA) according to manufacturer’s instructions. The samples were processed using an RNeasy Mini Kit (Qiagen, Valencia, CA) and digested with RNase free DNase (Qiagen, Valencia, CA). The integrity and size distribution of total RNA was monitored using an Agilent 2100 bioanalyzer (Santa Clara, CA). The concentration of RNA was determined by spectrophotometry, using Nanodrop-1000 (Nanodrop Technologies, Wilmington, DE). Reverse transcription was performed on 200ng/μl of total RNA with High-Capacity cDNA Reverse Transcription kit using random primers (Applied Biosystems, Foster City, CA).
MicroRNA Microarrays
MicroRNA microarrays were done at Ocean Ridge Biosciences (ORB, Palm Beach Gardens, FL) using custom multi-species microarrays containing 697 probes covering 707 mouse mature microRNAs present in Sanger 14.0 miRBase database. The array also contained 892 probes covering 902 human mature microRNAs and 388 probes covering 388 mouse mature microRNAs. The sensitivity of the microarray is such that it could detect as low as 20 amoles of synthetic microRNA being hybridized along with each sample. The microarrays were produced by Microarrays Inc. (Huntsville, Alabama), and consisted of epoxide glass substrates that had been spotted in triplicate with each probe.
Sample Processing
Quality of the total RNA samples was assessed using UV spectrophotometry and agarose gel electrophoresis. The samples were DNAse digested and low-molecular weight (LMW) RNA was isolated by ultrafiltration through YM-100 columns (Millipore) and subsequent purification using the RNeasy MinElute Clean-Up Kit (Qiagen, Valencia, CA). The LMW RNA samples were 3′-end labeled with Oyster-550 fluorescent dye using the Flash Tag RNA labeling Kit (Genisphere, Hatfield, PA). Labeled LMW RNA samples were hybridized to the MicroRNA microarrays according to conditions recommended in the Flash Tag RNA labeling Kit manual. The microarrays were scanned on an Axon Genepix 4000B scanner (Molecular Devices, Sunnyvale, CA) and data was extracted from images using GenePix V4.1 software.
Data Processing
Spot intensities were obtained for the 4860 features on each microarray by subtracting the median local background from the median local foreground for each spot. Detection Thresholds for each array were determined by calculating the 10% trim mean intensity of the negative controls spots and adding 5X the standard deviation of the background (non-spot area). The spot intensities and the Threshold (T) were transformed by taking the log (base 2) of each value. The normalization factor (N) for each microarray was determined by obtaining the 20% trim mean of the mouse probes intensities above threshold in all samples. The log2-transformed spot intensities for all 4860 features were normalized, by subtracting N from each spot intensity, and scaled by adding the grand mean of N across all microarrays. The mean probe intensities for each of the 697 mouse probes on each of the 4 arrays were then determined by averaging the triplicate spot intensities. Spots flagged as poor quality during data extraction were omitted prior to averaging. The 697 mouse non-control log2-transformed, normalized, and averaged probe intensities were filtered to obtain a list of 405 mouse probes showing probe intensity above T in at least one sample.
Microarray Quality Control
Each array contains probes targeting 11 different synthetic miRNAs, each of which is added at a mass of 200 amoles to each RNA sample prior to labeling and hybridization. Sensitivity of the microarray hybridization was confirmed by detection of hybridization signal for all 11 spikes well above the detection threshold. The array also included a set of specificity control probes complementary to three different miRNAs. Each specificity control includes a perfect match, single mismatch, double mismatch, and shuffled version of the probe. Specificity of the hybridization was confirmed by detection of hybridization signal on the microarray for the perfect match probes and not the double mismatch and shuffled version of the probes. Reproducibility of the arrays was determined by monitoring the hybridization intensity for the triplicate mouse spots on each array. The sensitivity, specificity, and reproducibility data for the arrays were compiled into a Quality Control report.
Hierarchical Clustering Of MicroRNA Array Data
Data for the 405 detectable mouse probes were clustered using Cluster 3.0 software [14]. Genes were median centered prior to hierarchical clustering. Hierarchical clustering was conducted using Centered Correlation as the similarity metric and Average Linkage as the clustering method. Intensity scale shown is arbitrary.
Gene Expression Analysis
Gene expression levels were assayed by real-time PCR using the ABI 7900HT thermalcycler using custom assays (Applied Biosystems). The following Assay IDs Mm00456425_m1, Mm01291334_mH, Mm01184030_m1, Mm01184322_m1, Mm01138344_m1, Mm00446681_m1, Mm00443325_m1, Mm00447183_m1, Mm00440939_m1, Mm00662319_m1, Mm00445878_m1, Mm00431814_m1, Mm00440181_m1, Mm01331624_m1, Mm00545827_m1, Mm00444911_m1 were used for Adiponectin, Adiponectin Receptor 1, Adiponectin Receptor 2, Peroxisome Proliferator Activated Receptor Gamma, Sterol Regulatory Element Binding Transcription Factor 1, Pyruvate Dehydrogenase Kinase 2, Pyruvate Dehydrogenase Kinase 4 and Peroxisome and Proliferator Activated Receptor, Gamma, Co-activator 1 Alpha and Peroxisome Proliferator Activated Receptor Alpha, Fatty Acid Synthase, Fatty Acid Binding Protein 4, Apolipoprotein A-IV, Leptin Receptor, Thymoma Viral Proto-Oncogene 1 (AKT1), Thymoma Viral Proto-Oncogene 2 (AKT2), Glycogen Synthase Kinase 3 Beta, genes respectively. Eukaryotic 18S rRNA (Assay ID Hs99999901_s1) was used as an endogenous control for normalizing the data. For gene expression in HepG2 cells the following assay ID’s Hs01088691_m1, Hs00234579_m1, Hs00164004_m1, Hs00900055_m1 were used for SREBF1, MMP9, COL1A1 and VEGFA respectively. All samples were run in duplicates. cDNA was amplified using TaqMan Universal PCR master mix reagent (Applied Biosystems, Foster City, CA) under the following conditions: 20 seconds at 95°C, 40 cycles: 1 second at 95°C and 20 seconds at 60°C. Data was analyzed using software SDS V2.3 from Applied Biosystems (Foster City, CA).
MicroRNA RT-PCR
MiR expression levels were confirmed by real time PCR using a ABI 7900HT thermal cycler with custom assays (Life Technologies, Grand Island, NY). PCR reactions were conducted at 95°C for 10 min and then followed by 40 cycles (15 s at 95°C, 60 s at 60°C). The following Assay IDs 000509, 002238, 002571, 002185, 000397, 002249, 002112, 000413 and 000587 were used for miRNAs -205, 411, 155, 335, 21, 143, 29a, 29b and 29c respectively. snoRNA 202 and 234 (Assay IDs 1232, 1234) were used for miRNA expression normalization.
Western Blot
Cell lysates from transfected HepG2 cells containing equal amounts of protein (30μg) were subjected to SDS-PAGE and western blot as described previously (15). Antibodies were MMP9 (Abcam, MA) SREBP1c (Cell Signaling Technology, MA), Beta-Actin (Chemicon, CA).
Cell Culture and miR-205 transfection
To investigate a role for miR-205 in hepatic steatosis we manipulated miR-205 levels by transfection of precursor (premiR) or scrambled oligonucleotides into human HepG2 cells as described earlier (15). HepG2 cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM) (ATCC) supplemented with 10% fetal bovine serum (FBS). HepG2 cells were grown to 60–70% confluence and 10 nM pre-miR-205 or scrambled miR (Applied Biosystems) were transfected with siPort NeoFx transfection reagent (Applied Biosystems) according to manufacturer’s instructions. After 48-hr of transfection, cells were lysed for western blot and qPCR analysis.
Real Time (RT) PCR
Gene expression levels were assayed by RT-qPCR using an ABI 7900HT thermal cycler with custom assays (8 pooled samples per group) as described previously [15]. Eukaryotic 18S rRNA was used for normalization.
Statistical analysis
Data are expressed as means ± SEM. Differences between two groups were analyzed by Student’s t test and Mann-Whitney U test.
RESULTS AND DISCUSSION
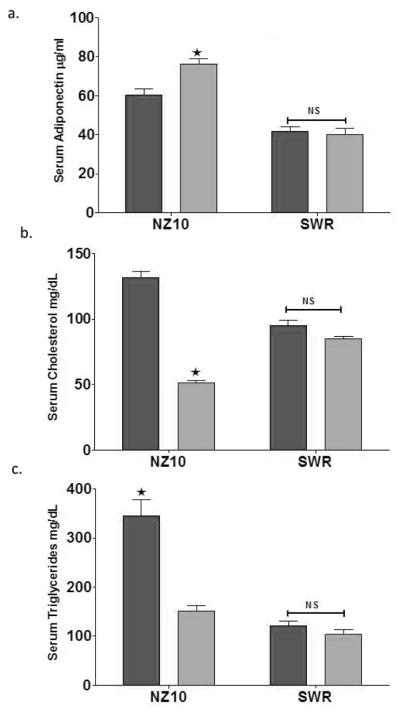
We recently reported that the HPO diet significantly reduced the body weight of NZ10 mice compared with their counterparts on CD or the SWR mice on either diet [15]. Weight gain by SWR mice was not diet-dependent over 25 weeks. NZ10 mice fed CD had increased body fat and displayed hyperinsulinemia and fasting hyperglycemia that was not seen in mice fed HPO or SWR mice fed either diet [15]. These results confirm the development of a diabetogenic phenotype in NZ10 mice fed CD. Adiponectin has anti-inflammatory and insulin-sensitizing properties and low serum adiponectin is predictive of dyslipidemia, independently of insulin sensitivity and visceral obesity [16]. As shown in Fig 1, serum adiponectin was higher in NZ10 mice compared with SWR independently of diet (p<0.01) and the HPO diet mediated an additional significant increase of serum adiponectin in the NZ10 group (1a). The effect of HPO diet on adiponectin in NZ10 mice was paralleled by significantly decreased serum cholesterol (1b) and TG (1c). Serum cholesterol of NZ10 mice fed CD was higher than SWR fed either diet, and serum TG of NZ10 mice fed HPO was decreased to the levels of SWR mice fed either diet. The cholesterol and TG levels of the NZ10-CD group are comparable and greater respectively than those recently reported for these mice (17). Diet did not affect any of these blood parameters in SWR mice. Leiter et al [17] reported that serum adiponectin of insulin-resistant NZ10 mice was the same as insulin-sensitive NZ05 mice after 12 weeks of feeding a 6% fat diet [17]. The authors concluded that serum adiponectin did not discriminate between obese insulin-resistant (NZ10) and obese insulin-sensitive (NZ05) phenotypes in this model. Our results support this conclusion insofar as adiponectin was higher in the NZ10-CD (obese-insulin resistant) group compared with both SWR (lean-insulin sensitive) groups (p<0.001), and therefore not predictive of insulin sensitivity or obesity across strains. Studies of humans have shown that adiponectin levels of subjects with NAFLD were similar to healthy controls when corrected for BMI and low adiponectin appeared only after the progression of NAFLD to NASH [2, 18]. Our results indicate that the HPO diet conferred significantly elevated adiponectin compared with CD in NZ10 mice in parallel with suppressed heaptosteatosis, however our studies do not distinguish between adiposity and insulin sensitivity as regulators of adiponectin production.
Figure 1. Serum lipids and adiponectin.
(a) Total serum adiponectin levels in NZ10 and SWR mice fed the indicated diet for 25 weeks of age. (b and c) Serum cholesterol and triglyceride levels respectively of mice fed CD or SD as indicated for 25 weeks. Dark bars are CD diet; light bars HPO; data are mean ± SEM; * p<0.05; NS is non-significant; n=12.
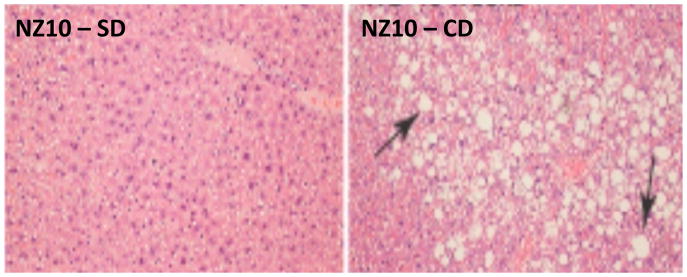
As illustrated in Figure 2, we observed extensive areas of fatty liver and necrosis only in NZ10 mice fed CD confirming previous reports of severe hepatosteatosis and steatohepatitis in this model [8, 9]. Such adipose infiltration of liver was not seen in SWR mice fed either CD or HPO diet (not shown).
Figure 2. H & E staining of NZ10 liver.
Representative fields of liver sections from NZ10 mice fed HPO (left panel) and CD (right panel) at 25 weeks (representative of n=4). Arrows indicate large fat droplets seen in all CD-fed NZ10 livers that were absent in the HPO groups. There was also evidence of cell necrosis in these regions (not shown).
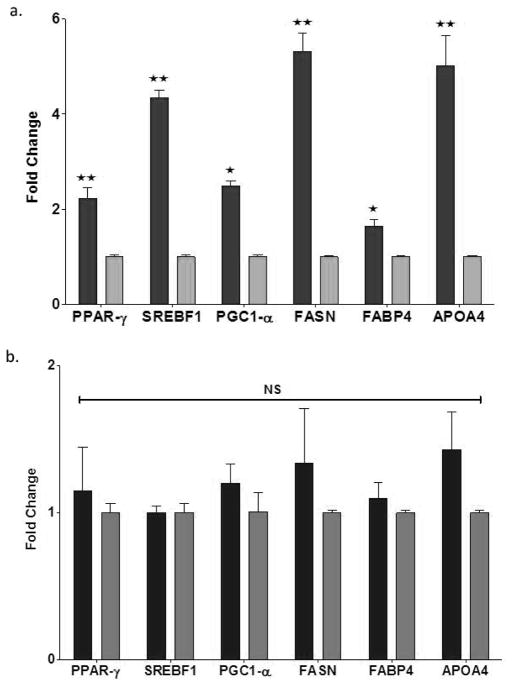
The master transcription factors PPARγ and SREBP1c and associated peroxisome proliferator-activated receptor gamma coactivator 1-alpha (PGC-1α) regulate genes involved in glucose, fatty acid and lipid metabolism, and adipocyte differentiation (reviewed in 19). Hepatic overexpression of these genes associated with NAFLD has been reported in animal models and human subjects [20]. As shown in Figure 3a, livers of NZ10 mice fed CD displayed 2-, 4.5-, and 2–5-fold increased expression levels of PPAR-γ, SREBP-1c and PGC-1α respectively. Consistent with the activation of fatty acid metabolizing pathways in NZ10 by the CD diet, downstream genes including fatty acid synthase (FASN), fatty acid binding protein 4 (FABP4) and apolipoprotein A-4 (APO4) were also significantly higher in the NZ10-CD groups (Fig. 3a) but were not regulated by diet in the SWR group (Fig 3b). In rodents the intestine and liver produce APO4, and the secreted protein regulates hepatic transcellular lipid transport, appetite and satiety and may contribute to NAFLD (21)
Figure 3. Hepatic expression of lipid metabolic pathway genes.
Liver samples from NZ10 (a) and SWR (b) mice at 25 weeks fed CD (dark bars) or HPO (light bars) diets were analyzed for markers of lipid accumulation by q-RTPCR as described in Methods. Data are mean ± SEM * p<0.05; ** p<0.01; NS is non-significant, (n=6).
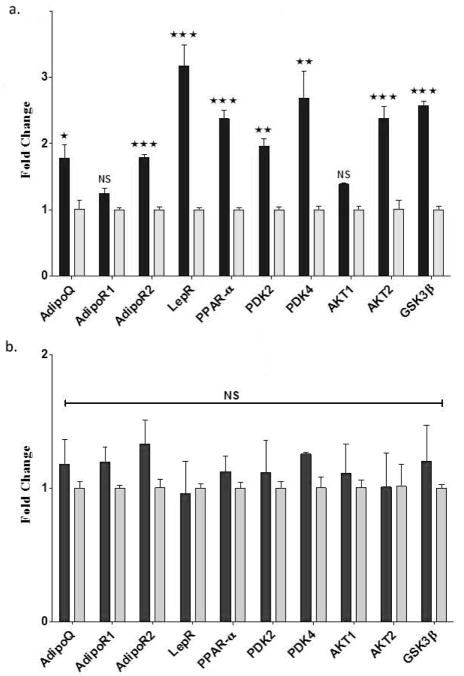
Adiponectin is not expressed in normal liver, however expression has been reported in damaged liver associated with fibrosis and in human subjects with hepatic steatosis (see References in [2]). Consistent with this we found a small but significant increase of adiponectin gene expression in NZ10 mice fed CD relative to HPO or SWR mice fed either diet (Fig. 4a and b). Liver adipocytes, present only in the NZ10-CD group may be responsible for this increased expression of adiponectin. Adiponectin acts via two specific receptors AdipoR1 and R2, expressed primarily in muscle and liver respectively. In diabetic ob/ob mice both AdipoR1 and AdipoR2 receptor expression decreased in liver, muscle and adipose tissue [22] while they remain unchanged in db/db mice [23]. Both receptors were increased in the livers of obesity-prone C57BL/6J, obese-resistant A/J mice or obese (fa/fa) Zucker rats by high-fat feeding [24]. Contradictory results have been reported for AdipoR2 expression in liver of human diabetic subjects [25, 26]. Therefore regulation of these receptors may involve complex interactions between genetic factors, diet, obesity and insulin sensitivity. As shown in Fig 4a, we observed a significant increase of AdipoR2 gene transcripts in the liver of NZ10 mice fed CD and no change of AdipoR1. AdipoR2 gene expression of NZ10 mice fed CD was also higher than that in livers of SWR mice independently of diet (compare Figs 4a and b). The results suggest that the regulation of adiponectin and its receptors is model-dependent. Leptin is the principal adipostatic signaling pathway that controls appetite, food intake, and endocrine function to maintain energy balance in mammals, and leptin receptors (LepR) are present in multiple peripheral and central tissues (reviewed in 27). Leptin acts as a pro-fibrotic cytokine in the liver and is known to contribute to hepatic fibrosis during NAFLD (reviewed in 28, 29). We found a highly significant >3-fold increase in LepR in liver of NZ10 mice fed CD relative to HPO but no diet-related changes in the SWR group (Fig 4a and b). These results suggest indicate diet-mediated regulation of AdipoQ, AdipoR2 and LepR selectively in NZ10 mice in a manner that correlates positively with liver adiposity and perhaps NAFLD
Figure 4. Hepatic expression of adiponectin, leptin and insulin pathways genes.
Liver extracts from NZ10 and SWR mice at 25 weeks fed CD (dark bars) or HPO (light bars) diets were analyzed for the indicated genes by q-RTPCR as described in Methods. Data are mean ± SEM * p<0.05; ** p<0.01; *** p<0.001; NS is non-significant, (n=6).
PPARα is a nuclear receptor transcription factor and a major regulator of lipid metabolism in the liver that is activated under conditions of energy deprivation in response to prolonged fasting. PPARα has anti-inflammatory and insulin-sensitizing properties and promotes FA oxidation by regulating the expression of lipid metabolic genes. Unexpectedly, previous work has shown that PPARα expression is increased in the livers of diabetic ob/ob, db/db, serotonin 5-HT2c receptor mutant mice, obese rats, and high fat-induced wild type mice [30]. Consistent with this, we found that PPARα transcript levels were 6-fold higher in NZ10 compared with SWR mice fed CD (NZ10-CD/SWR-CD = 5.92 ±0.88) and the levels were decreased 2-fold by the HPO diet in NZ10 mice (Fig. 4a). Diet did not significantly effect the expression of PPARα in the SWJ group (Fig. 4b).
The pyruvate dehydrogenase complex (PDC) catalyzes the oxidative decarboxylation of pyruvate, and links glycolysis to the tricarboxylic acid cycle. Suppression of PDC by pyruvate dehydrogenase kinase (PDK), promotes gluconeogenesis and glucose conservation (reviewed in 19). Such suppression is predicted to inhibit fatty acid oxidation, promote ectopic fat accumulation and worsen insulin sensitivity. It has been reported that expression of the major PDK isozymes 2 and 4 is regulated by PPARα ligands, PGC-1α as well as by the availability of plasma free fatty acids [31, 32]. As shown in Figure 4a, and consistent with previous reports with other models, we found that hepatic expression of both PDK2 and PDK4 genes was significantly increased in the livers of NZ10 mice fed the CD diet relative to HPO. Again there were no effects of diet on the expression of these genes in the livers of SWR mice (Fig. 4b). PDK2 and PDK4 gene transcripts were respectively 2.35 ±0.24 and 2.48 ±0.36 fold higher in NZ10-CD versus SWR-CD (both p<0.05; n=6). Finally, two other metabolism/energy transduction-related transcripts, AKT2 and GSK3β were also selectively and significantly increased by CD diet in the livers of NZ10 mice (Fig. 4a). These are insulin/IGF-1 regulated genes involved in glucose metabolism, cell size, growth and survival.
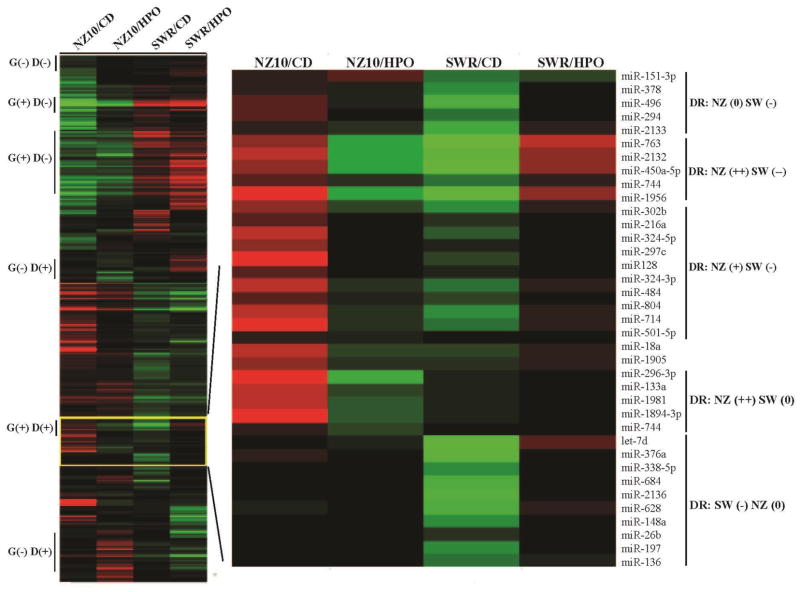
NONcNZ1-10 mice may be useful for identifying susceptibility loci and genetic background-diet interactions [7–9)]. As a first step towards such analyses we implemented micro-RNA arrays using RNA from the livers of NZ10 and SWR strains fed CD or HPO diets as described in Methods. These results are summarized by the heat-maps shown in Figure 5. The vertical bars at left designate regions of the heat-maps containing miRs that are responsive to genetic strain (G) and/or diet (D). MiRs are identified within these regions that remain unchanged by diet or genetic background, (G (−)D(−)); regulated by genetic background alone with minimal influence of diet, (G(+)D(−)); regulated by diet alone independently of genetic background, (G(−)D(+)); or regulated by both genetic background and diet, (G(+)D(+)). MiRs can also be identified that are regulated by diet selectively on the NZ10 or SWR backgrounds. The former are the most likely to contain candidate miRs that confer diabetogenic susceptibility. Figure 5, right panel shows an example set in which differentially expressed miRs of NZ10 and SWR groups are identified as unregulated (0), negatively (−) or positively (+/++) regulated by diet. For example HPO increases miR-763 by >4-fold in NZ10, but decreases the same miR also by >4-fold in SWR mice. Functions of the miRs in the NZ(++)SW(−−) group (Fig 3b) are mostly unknown, but low levels of miR-450a have been linked with hepatocarcinogenesis by enhancing expression of DNA methyltransferase, an activity that correlates negatively hepatocyte proliferation (31, 33). Therefore lower levels of this miR in the NZ10-HPO and SWR-CD groups may support such hepatocyte turnover.
Figure 5. Regulation of miRs by genetic background and diet.
Left panel shows heatmaps comparing miRs of NZ10 and SWR fed CD and HPO as indicated. Analyses were as described in Methods. Vertical bars indicate regions of the heatmap that show regulation or not by genetic background (G) and diet (D). Most of the miRs show some level of regulation by genetic background and/or diet and a smaller number show regulation by diet only on the BZ10 background, (NZ10 D(+). Right panel shows an Expanded region of the yellow-boxed miRs as indicated depicting the individual miRs, labeled at right. DR is diet regulation of grouped miRs, neutral (0), negative (−) or positively (+) regulated by CD versus HPO diet on NZ10 (NZ) or SWR (SW) backgrounds. The trend illustrates up-regulation of miRs by HPO in NZ10 livers and down-regulation of the same miRs in the SWR group.
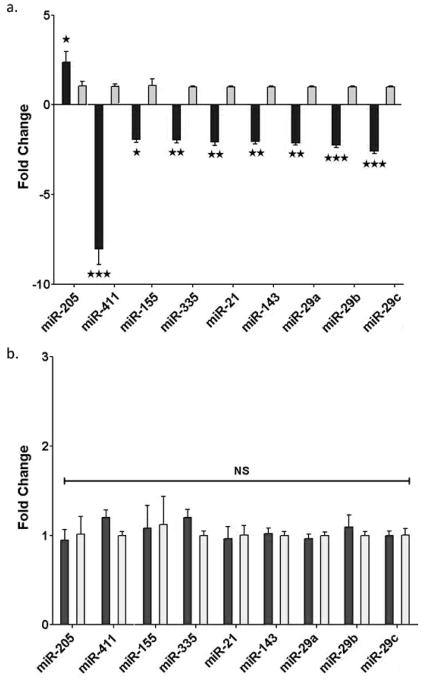
Figure 6 shows a set of miRs that were identified by differentially regulated expression in the arrays and confirmed by rtPCR. The level of miR-205 was increased >2-fold by feeding CD relative to HPO only in NZ10 mice. This is consistent with previous reports including our own that PPAGγ may regulate the expression of miR-205 (15, 34). Nine additional miRs that have been linked with metabolism and T2D and or NAFLD were significantly decreased only in the NZ10-CD group with miR-411 showing the greatest reduction; >8-fold decrease in NZ10-CD liver. MiR-411 targets include Forkhead box O1 (FOXO1), interleukin-18, a pro-inflammatory cytokine, and MAPK pathway regulator sprout homolog 4 (SPRY4) (35–37). It has been reported that mir-411 levels are suppressed by high glucose treatment of endothelial cells suggesting links with diabetes (38). Mir-155 was down-regulated >2-fold only in the NZ10-CD group and has well-defined regulatory roles in inflammation, cholesterol and fatty acid metabolism by targeting the CCAAT/enhancer-binding protein beta (Cebp-β), suppressor of cytokine signaling 1 (Socs1) and the liver X receptor alpha (LXR-α) (39). Mir-155 is thought to have protective roles in NAFLD in part by quenching inflammation (40). The levels are reduced in peripheral blood mononuclear cells of type 2 diabetic patients, consistent with our results (41). MiR-335 has been linked to oxidative stress and inflammatory genes and its levels were report to increase (42) or remain unchanged in T2D (43). Contrary to these reports we found that miR-335 was decreased selectively in the NZ10-CD group (Fig. 6a). The levels of mir-21, considered to be a possible marker of NAFLD (44), were also decreased by >2-fold selectively in the NZ10-CD group. It has been reported that miR-21 regulates triglyceride and cholesterol metabolism by targeting 3-hydroxy-3-methylglutaryl-co-enzyme A reductase (HMGCR) expression and the levels are reduced in sera of NAFLD patients (45–47). We found that miRs-143 and 29a,b,c were each significantly down-regulated in the NZ10-CD group. These results are in contrast to previous reports that these miRs are increased in other diet-induced and genetic rodent models (48, 49). Reported targets for these miRs include oxysterol-binding-protein-related protein-8 (ORP8) for miR-143 (48) and FOXA2 for miR-29 (49).
Figure 6. Hepatic miRNA expression.
Liver RNA samples from NZ10 and SWR mice at 25 weeks fed CD (dark bars) or HPO (light bars) diets were analyzed for miRNA expression by qRT-PCR as described in Methods. MiRs were selected from the microarray panel on the basis of significance and relatedness to diabetes/obesity/liver steatosis. Data are mean ± SEM * p<0.05; ** p<0.01; *** p<0.001; NS is non-significant, (n=6).
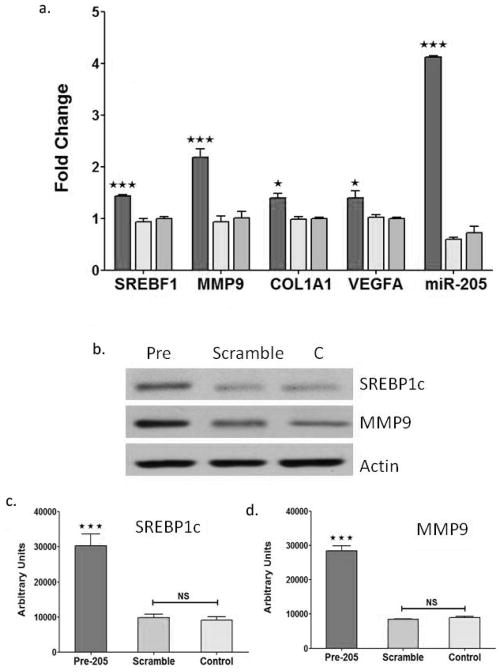
Previous studies have linked miR-205 with epithelial-mesenchyme transition (EMT) and fibrosis (50, 51). Therefore, because both miR-205 and PPARγ levels are increased in livers of NZ10-CD mice, we hypothesized that miR-205 may contribute to the development of fibrosis associated with NAFLD in this model. To test for this we examined the effect of miR-205 overexpression on fibrosis-related and lipid metabolic gene expression by treating human HepG2 hepatocytes with pre-miR-205. As shown in Figure 7, pre-miR-205 but not scrambled sequence ribonucleotides, or controls (transfection mix only) increased expression of each of the target genes at both RNA and protein levels. Fibrosis-associated transcripts of MMP9, ColA1 and VEGF and the sterol regulatory element-binding potein-1 (SREBF1), a key regulator of glucose, fatty acid and lipid metabolism were each significantly increased by pre-miR-205 (Fig. 7a). Western blots confirmed that the protein levels of SREBP1 and MMP9 were also significantly increased by overexpression of miR-205 (Fig. 7b and c). The results are consistent with a pathway whereby enhanced levels of miR-205 in liver of NZ10-CD mice, perhaps linked with elevated PPARγ leads to increased expression of fibrosis and lipid metabolizing genes, and this may contribute to the progression of steatosis, fibrosis and NAFLD in this model. In a recent report we described a pronounced suppression of miR-205 in adipose tissues of NZ10 mice fed CD (52), therefore it should be noted that the responses of miR-205 appear to be tissue-specific with opposite regulation in liver versus adipose. Such an effect may reflect different levels of tissue specific transcription factors and gene expression in the two tissues. Further work is required to identify the targets of miR-205 that confer these phenotypes.
Figure 7. Regulation of fibrosis-related gene expression by pre-miR-205 overexpression in HepG2 cells.
HepG2 hepatocytes were cultured and transfected with pre-miR-205, scrambled sequence miR or no-oligonucleotide (C-control) as described in Methods. (a) qRT-PCR quantified expression of the indicated gene transcripts and miR-205 levels at 48 hrs after transfection. (b) Western blot of HepG2 protein extracts treated as indicated for 48h and probed with SREBP1C, MMP9 and anti-actin antibodies. (c) Quantification of SREBP1 and MMP9 from western blots as in (b) normalized for actin expression. All data are mean ± SEM; * p<0.05; *** p<0.001, NS is non-significant (n=6).
CONCLUSION
Our results confirm the roles of dietary protein and fish oil in protecting against the development of T2D and NAFLD in NZ10 mice. Though preliminary, these findings show that diet rescues the extreme diabetogenic phenotype of these mice coincident with enhanced levels of plasma adiponectin and depressed expression of key genes involved in hepatic lipid metabolism and fibrosis. The studies identify miR sets that are differentially regulated by diet and genetic background, as well as miRs that are diet-responsive only on the NZ10 background some of which may be markers of NAFLD. Further studies using this model may lead to the identification of miRs and genes that mediate predisposition to NAFLD and T2D. This may also assist in the development of new pharmacological therapies for NAFLD.
Acknowledgments
Supported by grants RO1 HL092499 and RO1 HL072924 from the NIH, by a grant from the Florida Heart Research Institute and by a Walter G. Ross Distinguished Chair of Vascular Biology (to K.A.W.).
References
- 1.Tziomalos K, Athyros VG, Karagiannis A. Non-alcoholic fatty liver disease in type 2 diabetes: pathogenesis and treatment options. Curr Vasc Pharmacol. 2012;10:162–72. doi: 10.2174/157016112799305012. [DOI] [PubMed] [Google Scholar]
- 2.Hernaez R. Genetic factors associated with the presence and progression of nonalcoholic fatty liver disease: a narrative review. Gastroenterol Hepatol. 2012;35:32–41. doi: 10.1016/j.gastrohep.2011.08.002. [DOI] [PubMed] [Google Scholar]
- 3.Polyzos SA, Kountouras J, Zavos C, Tsiaousi E. The role of adiponectin in the pathogenesis and treatment of non-alcoholic fatty liver disease. Diabetes Obes Metab. 2010;12:365–83. doi: 10.1111/j.1463-1326.2009.01176.x. [DOI] [PubMed] [Google Scholar]
- 4.Kerr TA, Korenblat KM, Davidson NO. MicroRNAs and liver disease. Transl Res. 2011;157:241–252. doi: 10.1016/j.trsl.2011.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dehwah MAS, Xu A, Huang Q. MicroRNAs and Type 2 Diabetes/Obesity. J Genetics and Genomics. 2012;39:11–18. doi: 10.1016/j.jgg.2011.11.007. [DOI] [PubMed] [Google Scholar]
- 6.Li S, Chen X, Zhang H, Liang X, Xiang Y, Yu C, Zen K, Li Y, Zhang CY. Differential expression of microRNAs in mouse liver under aberrant energy metabolic status. J Lipid Res. 2009;50:1756–65. doi: 10.1194/jlr.M800509-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Leiter EH, Strobel M, O’Neill A, Schultz D, Schile A, Reifsnyder PC. Comparison of Two New Mouse Models of Polygenic Type 2 Diabetes at the Jackson Laboratory, NONcNZO10Lt/J and TALLYHO/JngJ. J Diabetes Res. 2013;2013:165327. doi: 10.1155/2013/165327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cho YR, Kim HJ, Park SY, Ko HJ, Hong EG, Higashimori T, Zhang Z, Jung DY, Ola MS, Lanoue KF, Leiter EH, Kim JK. Hyperglycemia, maturity-onset obesity, and insulin resistance in NONcNZ010/LtJ males, a new mouse model of type 2 diabetes. Am J Physiol Endocrinol Metab. 2007;293:E327–E336. doi: 10.1152/ajpendo.00376.2006. [DOI] [PubMed] [Google Scholar]
- 9.Reifsnyder PC, Leiter EH. Deconstructing and reconstructing obesity-induced diabetes (diabesity) in mice. Diabetes. 2002;51:825–832. doi: 10.2337/diabetes.51.3.825. [DOI] [PubMed] [Google Scholar]
- 10.Nagarajan P, Kumar MJ, Venkatesan R, Majundar SS, Juyal RC. Genetically modified mouse models for the study of nonalcoholic fatty liver disease. World J Gastroenterol. 2012;18:1141–1153. doi: 10.3748/wjg.v18.i11.1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Parker HM, Johnson NA, Burdon CA, Cohn JS, O’Connor HT, George J. Omega-3 supplementation and non-alcoholic fatty liver disease: a systematic review and meta-analysis. J Hepatol. 2012;56:944–951. doi: 10.1016/j.jhep.2011.08.018. [DOI] [PubMed] [Google Scholar]
- 12.Shapiro H, Tehilla M, Attal-Singer J, Bruck R, Luzzatti R, Singer P. The therapeutic potential of long-chain omega-3 fatty acids in nonalcoholic fatty liver disease. Clin Nutr. 2011;30:6–19. doi: 10.1016/j.clnu.2010.06.001. [DOI] [PubMed] [Google Scholar]
- 13.De Wit NJ, Afman LA, Mensink M, Müller M. Phenotyping the effect of diet on non-alcoholic fatty liver disease. J Hepatol. 2012;57:1370–3. doi: 10.1016/j.jhep.2012.07.003. [DOI] [PubMed] [Google Scholar]
- 14.De Hoon M, Imoto JL, Nolan S, Nolan J, Miyano S. Open Source Clustering Software. Bioinformatics. 2004;20:1453–1454. doi: 10.1093/bioinformatics/bth078. [DOI] [PubMed] [Google Scholar]
- 15.Adi NC, Adi JN, Cesar L, Agatston AS, Kurlansky P, Webster KA. Influence of diet on visceral adipose remodeling in NONcNZO10 mice with polygenic susceptibility for type 2 diabetes. Obesity. 2012;20:2142–2146. doi: 10.1038/oby.2012.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Baratta R, Amato S, Degano C, Farina MG, Patanè G, Vigneri R, Frittitta L. Adiponectin relationship with lipid metabolism is independent of body fatmass: evidence from both cross-sectional and intervention studies. J Clin Endocrinol Metab. 2004;89:2665–2671. doi: 10.1210/jc.2003-031777. [DOI] [PubMed] [Google Scholar]
- 17.Leiter EH, Reifsnyder PC, Xiao Q, Mistry J. Adipokine and insulin profiles distinguish diabetogenic and non-diabetogenic obesities in mice. Obesity. 2007;15:1961–1968. doi: 10.1038/oby.2007.234. [DOI] [PubMed] [Google Scholar]
- 18.Jarrar MH, Baranova A, Collantes R, Ranard B, Stepanova M, Bennett C, Fang Y, Elariny H, Goodman Z, Chandhoke V, Younossi ZM. Adipokines and cytokines in non-alcoholic fatty liver disease. Aliment Pharmacol Ther. 2008;27:412–421. doi: 10.1111/j.1365-2036.2007.03586.x. [DOI] [PubMed] [Google Scholar]
- 19.Feige JN, Auwerx J. Transcriptional coregulators in the control of energy homeostasis. Trends Cell Biol. 2007;17:292–301. doi: 10.1016/j.tcb.2007.04.001. [DOI] [PubMed] [Google Scholar]
- 20.Pettinelli P, Videla LA. Up-regulation of PPAR-γ mRNA expression in the liver of obese patients: an additional reinforcing lipogenic mechanism to SREBP-1c induction. J Clin Endocrinol Metab. 2011;96:1424–1430. doi: 10.1210/jc.2010-2129. [DOI] [PubMed] [Google Scholar]
- 21.Wu CL, Zhao SP, Yu BL. Intracellular role of exchangeable apolipoproteins in energy homeostasis, obesity and non-alcoholic fatty liver disease. Biol Rev Camb Philos Soc. 2015;90:367–376. doi: 10.1111/brv.12116. [DOI] [PubMed] [Google Scholar]
- 22.Tsuchida A, Yamauchi T, Ito Y, Hada Y, et al. Insulin/Foxo1 pathway regulates expression levels of adiponectin receptors and adiponectin sensitivity. J Biol Chem. 2004;279:30817–30822. doi: 10.1074/jbc.M402367200. [DOI] [PubMed] [Google Scholar]
- 23.Inukai K, Nakashima Y, Watanabe M, Takata N, et al. Regulation of adiponectin gene expression in diabetic mice. Am J Physiol Endocrinol Metab. 2005;288:E876–E882. doi: 10.1152/ajpendo.00118.2004. [DOI] [PubMed] [Google Scholar]
- 24.Bullen JW, Bluher S, Kelesidis T, Mantzoros CS. Regulation of adiponectin and its receptors in response to development of diet-induced obesity in mice. Am J Physiol Endocrinol Metab. 2006;292:E1079–E-1086. doi: 10.1152/ajpendo.00245.2006. [DOI] [PubMed] [Google Scholar]
- 25.Shimizu A, Takamura T, Matsuzawa N, Nakamura S, Nabemoto S, Takeshita Y, Misu H, Kurita S, Sakurai M, Yokoyama M, Zen Y, Sasaki M, Nakanuma Y, Kaneko S. Regulation of adiponectin receptor expression in human liver and a hepatocyte cell line. Metabolism. 2007;56:1478–85. doi: 10.1016/j.metabol.2007.06.013. [DOI] [PubMed] [Google Scholar]
- 26.Carazo A, León J, Casado J, Gila A, Delgado S, Martín A, Sanjuan L, Caballero T, Muñoz JA, Quiles R, Ruiz-Extremera A, Alcázar LM, Salmerón J. Hepatic expression of adiponectin receptors increases with non-alcoholic fatty liver disease progression in morbid obesity in correlation with glutathione peroxidase-1. Obes Surg. 2011;21:492–500. doi: 10.1007/s11695-010-0353-2. [DOI] [PubMed] [Google Scholar]
- 27.Schwartz GJ, Azzara AV, Heaner MK. Roles for central leptin receptors in the control of meal size. Appetite. 2013;71:466–469. doi: 10.1016/j.appet.2013.04.017. [DOI] [PubMed] [Google Scholar]
- 28.Park PH, Sanz-Garcia C, Nagy LE. Adiponectin as an anti-fibrotic and anti-inflammatory adipokine in the liver. Curr Pathobiol Rep. 2015;3:243–252. doi: 10.1007/s40139-015-0094-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Noureddin M, Rinella ME. Nonalcoholic Fatty liver disease, diabetes, obesity, and hepatocellular carcinoma. Clin Liver Dis. 2015;19:361–79. doi: 10.1016/j.cld.2015.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Patsouris D, Reddy JK, Muller M, Kersten S. Peroxisome proliferator-activated receptor α mediates the effects of high-fat diet on hepatic gene expression. Endocrinology. 2006;147:1508–1516. doi: 10.1210/en.2005-1132. [DOI] [PubMed] [Google Scholar]
- 31.Bajotto G, Murakami T, Nagasaki M, Qin B, Matsuo Y, Maeda K, Ohashi M, Oshida Y, Sato Y, Shimomura Y. Increased expression of hepatic pyruvate dehydrogenase kinases 2 and 4 in young and middle-aged otsuka long-evans tokushima fatty rats: induction by elevated levels of free fatty acids. Metabolism Clinical and Experimental. 2006;55:317–323. doi: 10.1016/j.metabol.2005.09.014. [DOI] [PubMed] [Google Scholar]
- 32.Ma K, Zhang Y, Elam MB, Cook GA, Park EA. Cloning of the rat pyruvate dehydrogenase kinase 4 gene promoter: activation of pyruvate dehydrogenase kinase 4 by the peroxisome proliferator-activated receptor gamma coactivator. J Biol Chem. 2005;280:29525–29532. doi: 10.1074/jbc.M502236200. [DOI] [PubMed] [Google Scholar]
- 33.Weng Z1, Wang D, Zhao W, Song M, You F, Yang L, Chen L. microRNA-450a targets DNA methyltransferase 3a in hepatocellular carcinoma. Exp Ther Med. 2011;2:951–955. doi: 10.3892/etm.2011.288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.John E, Wienecke-Baldacchino A, Liivrand M, Heinäniemi M, Carlberg C, Sinkkonen L. Dataset integration identifies transcriptional regulation of microRNA genes by PPARγ in differentiating mouse 3T3-L1 adipocytes. Nucleic Acids Res. 2012;40:4446–4460. doi: 10.1093/nar/gks025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhao Z, Qin L, Li S. MiR-411 contributes the cell proliferation of lung cancer by targeting FOXO1. Tumor Biol. 2016;37:5551–5560. doi: 10.1007/s13277-015-4425-8. [DOI] [PubMed] [Google Scholar]
- 36.Yamamoto K, Seike M, Takeuchi S, Soeno C, Miyanaga A, Noro R, Minegishi Y1, Kubota K1, Gemma A1. MiR-379/411 cluster regulates IL-18 and contributes to drug resistance in malignant pleural mesothelioma. Oncol Rep. 2014;32:2365–2372. doi: 10.3892/or.2014.3481. [DOI] [PubMed] [Google Scholar]
- 37.Sun M, Huang F, Yu D, Zhang Y, Xu H, Zhang L, Li L, Dong L, Guo L, Wang S. Autoregulatory loop between TGF-β1/miR-411–5p/SPRY4 and MAPK pathway in rhabdomyosarcoma modulates proliferation and differentiation. Cell Death Dis. 2015;20(6):e1859. doi: 10.1038/cddis.2015.225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zitman-Gal T, Green J, Pasmanik-Chor M, Golan E, Bernheim J, Benchetrit S. Vitamin D manipulates miR-181c, miR-20b and miR-15a in human umbilical vein endothelial cells exposed to a diabetic-like environment. Cardiovasc Diabetol. 2014;13:8–18. doi: 10.1186/1475-2840-13-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Khamaneh AM, Alipour MR, Sheikhzadeh Hesari F, Ghadiri Soufi F. A signature of microRNA-155 in the pathogenesis of diabetic complications. J Physiol Biochem. 2015;71:301–9. doi: 10.1007/s13105-015-0413-0. [DOI] [PubMed] [Google Scholar]
- 40.Miller AM1, Gilchrist DS, Nijjar J, Araldi E, Ramirez CM, Lavery CA, Fernández-Hernando C, McInnes IB, Kurowska-Stolarska M. MiR-155 has a protective role in the development of non-alcoholic hepatosteatosis in mice. PLoS One. 2013;21(8):e72324. doi: 10.1371/journal.pone.0072324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Corral-Fernández NE, Salgado-Bustamante M, Martínez-Leija ME, Cortez-Espinosa N, García-Hernández MH, Reynaga-Hernández E, Quezada-Calvillo R, Portales-Pérez DP. Dysregulated miR-155 expression in peripheral blood mononuclear cells from patients with type 2 diabetes. Exp Clin Endocrinol Diabetes. 2013;121:347–353. doi: 10.1055/s-0033-1341516. [DOI] [PubMed] [Google Scholar]
- 42.Esguerra JL, Bolmeson C, Cilio CM, Eliasson L. Differential glucose-regulation of microRNAs in pancreatic islets of non-obese type 2 diabetes model Goto-Kakizaki rat. PLoS One. 2011;6:e18613. doi: 10.1371/journal.pone.0018613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Higuchi C, Nakatsuka A, Eguchi J, Teshigawara S, Kanzaki M, Katayama A, Yamaguchi S, Takahashi N, Murakami K, Ogawa D, Sasaki S, Makino H, Wada J. Identification of circulating miR-101, miR-375 and miR-802 as biomarkers for type 2 diabetes. Metabolism. 2015;64:489–497. doi: 10.1016/j.metabol.2014.12.003. [DOI] [PubMed] [Google Scholar]
- 44.Becker PP, Rau M, Schmitt J, Malsch C, Hammer C, Bantel H, Müllhaupt B, Geier A. Performance of Serum microRNAs -122, -192 and -21 as Biomarkers in Patients with Non-Alcoholic Steatohepatitis. PLoS One. 2015;10:e0142661. doi: 10.1371/journal.pone.0142661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sun C, Huang F, Liu X, Xiao X, Yang M, Hu G, Liu H, Liao L. MiR-21 regulates triglyceride and cholesterol metabolism in non-alcoholic fatty liver disease by targeting HMGCR. Int J Mol Med. 2015;35:847–853. doi: 10.3892/ijmm.2015.2076. [DOI] [PubMed] [Google Scholar]
- 46.Salvoza NC, Klinzing DC, Gopez-Cervantes J, Baclig MO. Association of Circulating Serum miR-34a and miR-122 with Dyslipidemia among Patients with Non-Alcoholic Fatty Liver Disease. PLoS One. 2016;11:e0153497. doi: 10.1371/journal.pone.0153497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Alisi A1, Da Sacco L, Bruscalupi G, Piemonte F, Panera N, De Vito R, Leoni S, Bottazzo GF, Masotti A, Nobili V. Mirnome analysis reveals novel molecular determinants in the pathogenesis of diet-induced nonalcoholic fatty liver disease. Lab Invest. 2011;91:283–293. doi: 10.1038/labinvest.2010.166. [DOI] [PubMed] [Google Scholar]
- 48.Jordan SD1, Krüger M, Willmes DM, Redemann N, Wunderlich FT, Brönneke HS, Merkwirth C, Kashkar H, Olkkonen VM, Böttger T, Braun T, Seibler J, Brüning JC. Obesity-induced overexpression of miRNA-143 inhibits insulin-stimulated AKT activation and impairs glucose metabolism. Nat Cell Biol. 2011;13:434–446. doi: 10.1038/ncb2211. [DOI] [PubMed] [Google Scholar]
- 49.Kurtz CL, Peck BC, Fannin EE, Beysen C, Miao J, Landstreet SR, Ding S, Turaga V, Lund PK, Turner S, Biddinger SB, Vickers KC, Sethupathy P. MicroRNA-29 fine-tunes the expression of key FOXA2-activated lipid metabolism genes and is dysregulated in animal models of insulin resistance and diabetes. Diabetes. 2014;63:3141–3148. doi: 10.2337/db13-1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Igietseme JU, Omosun Y, Stuchlik O, Reed MS, Partin J, He Q, Joseph K, Ellerson D, Bollweg B, George Z, Eko FO, Bandea C, Liu H, Yang G, Shieh WJ, Pohl J, Karem K, Black CM. Role of Epithelial-Mesenchyme Transition in Chlamydia Pathogenesis. PLoS One. 2015;10:e0145198. doi: 10.1371/journal.pone.0145198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cai X, Xia Z, Zhang C, Luo Y, Gao Y, Fan Z, Liu M, Zhang Y. Serum microRNAs levels in primary focal segmental glomerulosclerosis. Pediatr Nephrol. 2013;28:1797–1801. doi: 10.1007/s00467-013-2434-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Adi N, Adi J, Cesar L, Kurlansky P, Agatston A, Webster KA. Role of Micro RNA-205 in Promoting Visceral Adiposity of NZ10 Mice with Polygenic Susceptibility for Type 2 Diabetes. J Diabetes Metab. 2015;6 doi: 10.4172/2155-6156.1000574. [DOI] [PMC free article] [PubMed] [Google Scholar]