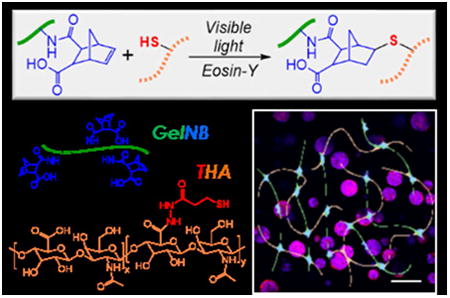
Abstract
Photopolymerized biomimetic hydrogels with adaptable properties have been widely used for cell and tissue engineering applications. As a widely adopted gel crosslinking method, photopolymerization provides experimenters on-demand and spatial-temporal controls in gelation kinetics. Long wavelength ultraviolet (UV) light initiated photopolymerization is among the most popular methods in the fabrication of cell-laden hydrogels owing to its rapid and relatively mild gelation conditions. The use of UV light, however, still causes concerns regarding its potential negative impacts on cells. Alternatively, visible light based photopolymerization can be used to crosslink cell-laden hydrogels. The majority of visible light based gelation schemes involve photoinitiator, co-initiator, and co-monomer. This multi-component initiation system creates added challenges for optimizing hydrogel formulations. Here, we report a co-initiator/co-monomer-free visible light initiated thiol-norbornene photopolymerization scheme to prepare modular biomimetic hydrogels suitable for in situ cell encapsulation. Eosin-Y was used as the sole initiator to initiate modular gelation between synthetic macromers (e.g., thiolated poly(vinyl alcohol) or poly(ethylene glycol)) and functionalized extracellular matrices (ECM), including norbornene-functionalized gelatin (GelNB) and/or thiolated hyaluronic acid (THA). These components are modularly crosslinked to afford bio-inert (i.e., purely synthetic), bioactive (i.e., using gelatin), and biomimetic (i.e., using gelatin and hyaluronic acid) hydrogels. The stiffness of the hydrogels can be easily tuned without affecting the contents of the bioactive components. Furthermore, the use of naturally-derived biomacromolecules (e.g., gelatin and HA) renders these hydrogels susceptible to enzyme-mediated degradation. In addition to demonstrating efficient and tunable visible light mediated gelation, we also utilized this biomimetic modular gelation system to formulate artificial tumor niche and to study the effects of cell density and gel modulus on the formation of pancreatic ductal adenocarcinoma (PDAC) spheroids.
Keywords: Keywords. Hydrogel, thiol-ene, tumor, modular crosslinking, gelatin, hyaluronic acid
Graphical abstract
1. Introduction
Biomimetic hydrogels with adaptable compositions and properties have tremendous potentials in both basic and applied research, including cell and molecular biology, drug delivery, tissue engineering, and regenerative medicine.1-3 From the viewpoint of material science and engineering, the compositions of biomimetic hydrogels must be well defined and highly controllable. From the perspective of basic biological science, the materials and protocols for hydrogel preparation must be readily available and adaptable for different laboratories with various research interests. Most importantly, the components and methods for hydrogel preparation should be highly cytocompatible. Photopolymerization is among the most reliable methods for hydrogel fabrication owing to its ease of preparation, highly controllable gelation kinetics, and predictable degradation (if appropriate degradable motifs are incorporated during macromer synthesis).4-7 Photopolymerized hydrogels have been widely used in biomedical applications, including biosensing and device coating, drug and protein delivery, in vitro cell culture and disease models, stem cell differentiation, and tissue regeneration.1, 2
In principle, any macromers with unsaturated vinyl groups can be crosslinked into hydrogels via photopolymerization. The following three groups of macromers are commonly adopted for fabricating biocompatible photopolymerized hydrogels: (1) Synthetic polymers, such as poly(ethylene glycol) (PEG) and poly(vinyl alcohol) (PVA); (2) Extracellular matrix (ECM)-derived peptides/proteins, including collagen, gelatin, fibronectin, and recombinant proteins, etc.; and (3) Glycosaminoglycans (GAGs), including hyaluronic acid (HA), heparin sulfate, and chondroitin sulfate, etc. The popularity of synthetic polymers stems from their low cost of production, high lot-to-lot consistency in material properties, and high degree of control in chemical modification. On the other hand, the bioactive motifs (e.g., ligands for cell-surface receptors, recognition sequences for protease cleavage) contained in ECM-derived proteins/peptides and GAGs are often utilized to mimic the biochemical milieu of natural tissues. These materials can be used alone or in combinations to form cell-laden hydrogels, as long as they are modified with photocrosslinkable motifs, such as acrylate, thiol, vinyl ether, and norbornene.4-6
We have previously reported the preparation of step-growth modular hydrogels using visible light-mediated thiol-norbornene photopolymerization with eosin-Y as the only photoinitiator.8, 9 The use of visible light negates the potential cellular and/or genetic damages imposed by ultraviolet (UV) light exposure. Furthermore, visible light initiated thiol-norbornene photochemistry eliminates the need for strong base co-initiator (e.g., triethanolamine, TEOA) and co-monomer (e.g., N-vinylpyrrolidone, NVP), two necessary components required in conventional visible light mediated photopolymerization.10-13 The elimination of co-initiator and co-monomer from the pre-polymer solution not only greatly simplifies material preparation, but also prevents potential toxicity of these small molecular weight molecules. Furthermore, the thiol-norbornene hydrogel crosslinking proceeds in a strictly modular and step-growth manner, which produces an idealized network with superior material properties at lower macromer contents.6, 9, 14 Compared with conventional visible light cured chain-growth hydrogels using the tri-initiator system (i.e., eosin-Y, TEOA, and NVP), we have shown that eosin-Y initiated thiol-norbornene gelation (Fig. 1A) is cytocompatible for three-dimensional (3D) culture of human mesenchymal stem cells (MSCs) and pancreatic MIN6 β-cells.9 We have also utilized this unique and simple gelation system to form multi-layer hydrogels,8 microgels,15 and islet surface conformal coating.16 This gelation system was also used to prepare visible light cured thiol-norbornene hydrogels capable of undergoing UV-light-mediated photodegradation.17
Figure 1. Visible light initiated thiol-norbornene reaction for forming orthogonally crosslinked biomimetic hydrogels.
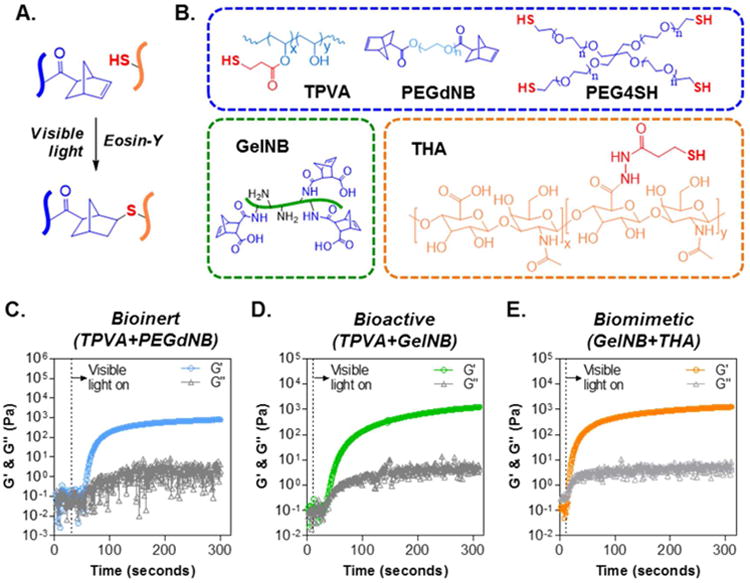
(A) Schematic of thiol-norbornene reaction under visible light exposure and with eosin-Y as the only initiator. (B) Chemical structures of macromers used, including thiolated PVA (TPVA, 6kDa), PEG-di-norbornene (PEGdNB, 10kDa), PEG-tetra-thiol (PEG4SH, 10kDa), gelatin-norbornene (GelNB), and thiolated HA (THA, 250kDa). (C-E) Evolution of storage (G′) and loss (G′) modulus of visible light initiated bio-inert (C), bioactive (D), and biomimetic (E) thiol-norbornene gelation.
While this visible light-mediated thiol-norbornene photochemistry has shown great potential, it has not been designed to impart biomimetic features (except for one example where pendant CRGDS peptide was immobilized in the otherwise inert PEG network).9 Therefore, the primary focus of this contribution is on the development and characterization of step-growth, modular, and biomimetic hydrogels crosslinked from the macromers mentioned above, including synthetic macromers (e.g., thiolated PVA (TPVA), PEG-tetra-thiol (PEG4SH), and PEG-di-norbornene (PEGdNB)), norbornene-functionalized gelatin (GelNB), and thiolated hyaluronic acid (THA). Visible light and eosin-Y mediated photochemistry was used to fabricate hydrogels that are either purely bio-inert (e.g., PEGdNB-TPVA or PEGdNB-PEG4SH gels), bio-active (e.g., TPVA-GelNB or PEG4SH-GelNB), or biomimetic (e.g., GelNB-THA). When appropriately combined, these materials afford a diverse array of tailor-made hydrogels for 3D cell culture. In our previous work, we have explored UV light initiated thiol-norbornene PEG-peptide hydrogels for studying pancreatic ductal adenocarcinoma (PDAC) cell fate and drug responsiveness in 3D.18 Here, we use a PDAC cell line, COLO357, to demonstrate the utility of the visible light polymerized thiol-norbornene biomimetic hydrogels for in vitro study of PDAC cell fate.
2. Experimental Section
2.1 Materials
PEG4SH (MW: 10kDA) and four-arm PEG (MW: 20 kDa) were acquired from JenKem Technology USA. Thiolated hyaluronan (THA, 250 kDa), gelatin, and eosin-Y disodium salt were obtained from ESI Bio, Electron Microscopy Sciences, and MP Biomedical, respectively. Collagenase (300 U/mg) and hyaluronidase (770 U/mg) were purchased from Worthington Biochemical. Linear PEG (MW: 10 kDa), 3-mercaptopropionic acid (MPA) and all other chemicals were purchased from ThermoFisher Scientific unless noted otherwise.
2.2 Macromer synthesis
The following macromers were synthesized in house: TPVA (6 kDa), PEGdNB (10 kDa), and GelNB (type B, 225 Bloom). GelNB synthesis was achieved through reacting gelatin with carbic anhydride as described previously.19, 20 Fluoraldehyde reagent was used to quantified the degree of norbornene functionalization on gelatin. Using unmodified gelatin with known concentrations as standards, the degree of functionalization was determined to be ∼2.22 mM norbornene per 1 wt% of GelNB. PEGNB was synthesized through reacting PEG-OH with 5-norbornene-2-carboxylic acid following an established protocol.6, 14 TPVA was synthesized following an established protocol with modified purification method.21 Briefly, mixture of hydrochloric acid (5 ml, 12 N) and MPA (25 mL) was added slowly into PVA (5 g) pre-dissolved in 50 mL ddH2O at 60°C. After 24 hours of reaction, the product was transferred to a dialysis bag (MWCO: 3600 Da, Spectrum lab) and dialyzed for 3 days in slightly acidic ddH2O (pH 6.5). The product was recovered, lyophilized, and the degree of thiolation was quantified by Ellman's assay.
2.3 Hydrogel crosslinking and rheometry
Hydrogels were fabricated by reacting norbornene moieties of macromers with thiol containing crosslinkers via step-growth photopolymerization.6, 9 Precursor solutions comprised of 0.1 mM EY and desired concentrations of TPVA/PEGNB, TPVA/GelNB, or THA/GelNB were irradiated with visible light (400 – 700 nm, 5 minutes at 70k Lux) using a halogen cold light lamp (AmScope).8, 9 In situ gelation and rheological properties of the hydrogels were characterized with a digital rheometer (Bohlin CVO 100). In situ photorheometry was achieved by polymerizing hydrogel (100 μL) on a quartz plate in a light cure cell with a 25 mm parallel plate geometry. Visible light was directed to the bottom of the quartz plate through a flexible light guide and moduli were recorded in a time-sweep mode using 10 % strain, 1 Hz frequency, and a gap size of 90 μm. The time where storage or shear modulus (G′) exceeded loss modulus (G″) was recorded as the gel point. To obtain moduli of preformed gels with different formulations, 50 μL of pre-polymer solutions were polymerized between glass slides separated by 1mm-thick Teflon spacers. An 8 mm biopsy punch was used to obtain disc-shaped hydrogels for moduli measurements using a parallel plate under strain-sweep mode (0.1% to 5% strain) at 1 Hz frequency.
2.4 Characterization of hydrogel degradation
Visible light polymerized bio-inert TPVA/PEGNB hydrogels were incubated in pH 7.4 PBS and shear moduli of the hydrogels at different time points were used to determine their hydrolytic degradation. Data obtained from rheometry were fitted with pseudo-first order degradation kinetics as reported previously.14, 22 Bioactive (i.e., TPVA/GelNB) and biomimetic (i.e., GelNB/THA) hydrogels were used in proteolytic degradation studies. Briefly, hydrogels were formed and incubated in PBS for 2.5 hours at 37°C. Following measurement of the initial gel mass, hydrogels were transferred to wells containing 0.5 mL of collagenase type I (10U/mL) or hyaluronidase (300U/mL) in PBS and incubated at 37°C. At predetermined time intervals, gels were removed from protease solution, blotted dry, and their mass was recorded. This process was repeated until no remnants of the gel could be seen. Results were presented as percent mass remaining to the initial gel mass before protease treatment.
2.5 Cell culture, encapsulation, and characterizations
PDAC cell line COLO-357 was cultured in high-glucose DMEM supplemented with L-glutamine and sodium pyruvate, 10% fetal bovine serum (FBS), β-mercaptoethanol, and antibiotic-antimycotic. Cells were maintained in a standard cell culture incubator (37°C and 5% CO2). For encapsulation in hydrogels, cells were suspended in pre-polymer solutions composed of eosin-Y, PEG4SH, GelNB, and/or THA at desired concentrations. Cell-laden hydrogels (20 μl/gel) were obtained through 5 minutes of visible light exposure (400-700 nm at 70kLux, equivalent to 10mW/cm2 at 550 nm). Following gel crosslinking, cell-laden hydrogels were maintained in cell culture incubator with media (500 μl per gel) refreshment every two days. To quantify cell metabolic activity, cell-laden hydrogels were incubated in 500μL Alamarblue® reagent (AbD Serotec; 10 % in cell culture medium) for 2.5 hours. 200μl of media was transferred to a 96-well plate for fluorescence quantification (excitation: 560 nm and emission: 590 nm) using a microplate reader.
Live/dead staining was used to evaluate the viability and spheroid size of the encapsulated cells. Briefly, cell-laden hydrogels were stained with live/dead staining kit (Calcein AM stained live cells green and Ethidium homodimer-1 stained dead cells red) for an hour, washed with HBSS for 10 minutes, followed by confocal microscopy imaging (FV1000 Laser Scanning Biological Microscope). Random fields in the hydrogels were selected for scanning and the gel thickness in any image was 100μm (step size of 10μm). The diameter of the spheroids on day 10 was determined using Nikon NIS Elements software. F-actin staining was achieved using Phalloidin. Briefly, cell-laden hydrogels were fixed with 4 % paraformaldehyde for 1 hour at room temperature on an orbital shaker. After cell fixation, hydrogels were washed twice with HBSS (10 minute/wash). Cells were then permeabilized with 0.5 % of TritonX-100 in HBSS for 45 minutes before washing twice with HBSS (10 minutes/wash). Cell-laden gels were incubated in phalloidin solution (at 100 nM) overnight at 4°C. Before imaging, cell nuclei were counter-stained with DAPI for an hour at room temperature and washed three times with HBSS (10 minutes/wash). Hydrogels were imaged with a confocal microscope as described above.
2.6 RNA isolation and gene expression analysis
In preparation for RNA isolation, cell-laden gels and cells recovered from control 2D culture were rapidly frozen using liquid nitrogen and stored in -80°C until use. Collected samples were processed following NucleoSpin® RNA protocols (Clontech). The concentrations of pure RNA were determined using NanoDrop 2000 Spectrophotometer (Thermo Scientific). PrimeScript™ RT Reagent Kit (Clontech) was used to convert RNA into cDNA. Quantitative real time PCR (Applied Biosystems 7500 Fast Real-Time PCR machine) was performed using cDNA and SYBR Premix Ex Taq II kit (Clontech) with appropriate primers (F: forward, R: reverse): GAPDA-F: GAAGGTGAAGGTCGGAGTC; GAPDF-R: GAAGATGGTGATGGGATTTC;23 CTGF-F: AGGAGTGGGTGTGTGACGA; CTGF-R: CCAGGCAGTTGGCTCTAATC;24 SHH-F: GGAAGCAGCCTCCCGATT; SHH-R: CGAGTCCAAGGCACATATCCA;25 MMP14-F: GCAGAAGTTTTACGGCTTGCA, MMP14-R: TCGAACATTGGCCTTGATCTC.26 2-ΔΔct method was used for data analysis where GAPDH was used as the housekeeping gene. Gene expression levels from hydrogel samples were further compared to the 2D samples collected on respective day of culture.
2.7 Statistical analysis
All experiments were conducted independently for three times with a minimum of three samples per conditions. Numerical data were reported as Mean ± SD and analyzed with two-way ANOVA on GraphPad Prism 5 software (p < 0.05 signifies statistical significance).
3. Results
3.1 Modular crosslinking of visible light polymerized thiol-norbornene hydrogels
To obtain visible light-initiated step-polymerized hydrogels with modular properties (Fig. 1A), we utilized a range of bio-inert (TPVA, PEGdNB, or PEG4SH) or bioactive (GelNB or THA) macromers with either thiol or norbornene moiety (Fig. 1B). We named the three classes of step-polymerized thiol-norbornene hydrogels as below: bio-inert (TPVA+PEGdNB), bioactive (TPVA+GelNB), and biomimetic hydrogels (GelNB+THA). In situ photorheometry was used to evaluate gelation efficiency. For both bio-inert (Fig. 1C) and bioactive (Fig. 1D) hydrogels, the gel point (time when G′ surpasses G″) was around 25 seconds. On the other hand, the gel point of biomimetic gelation system was significantly faster (∼5 seconds, Fig. 1E) than both bio-inert and bioactive systems. The gelation of all three groups of gels reached near completion after 5 minutes of visible light exposure.
3.2 Modularly tunable stiffness in visible light cured thiol-norbornene hydrogels
Fig. 2 shows the high tunability of the matrix stiffness in bio-inert, bioactive, and biomimetic thiol-norbornene hydrogels. Fig. 2A demonstrates two levels of control in bio-inert hydrogel crosslinking: the degree of thiol substitution (DS) in TPVA (DS = 26 or 40) and the number of polymerizable moiety or functionality (f) per PEGNB molecule (i.e., f = 2 for PEGdNB and f = 4 for PEG4NB). Regardless of PEGNB functionality, hydrogels were significantly stiffer using TPVA with high DS (i.e., 40). However, the effect of PEGNB functionality on gel crosslinking was more apparent when high DS TPVA was used. Next, we demonstrated the independent control of gel stiffness by varying the content of bio-inert TPVA without affecting the amount of bioactive macromer incorporated (Fig. 2B). At a fixed GelNB content (i.e., 7 wt%), we adjusted the concentration of TPVA to 0.7, 1, and 1.3 wt% so that the stoichiometric ratio of thiol-to-norbornene (RSH/ene) was 0.5, 0.75, and 1, respectively. Not surprisingly, gel modulus increased at increasing RSH/ene, a characteristic trend for orthogonally crosslinked hydrogels.6 Finally, the tunability of gel stiffness in biomimetic hydrogels composed of GelNB and THA was explored (Fig. 2C). Here, we fixed the concentrations of GelNB and THA to 7wt% and 1.1wt% respectively, while adjusting the content of another bio-inert macromer PEG4SH so that RSH/ene was maintained at 0.5, 0.75, or 1. The results are in accordance with that shown in Fig. 2B where an increase in RSH/ene leads to increasing gel stiffness.
Figure 2. Modularly tunable stiffness in visible light cured thiol-norbornene hydrogels.
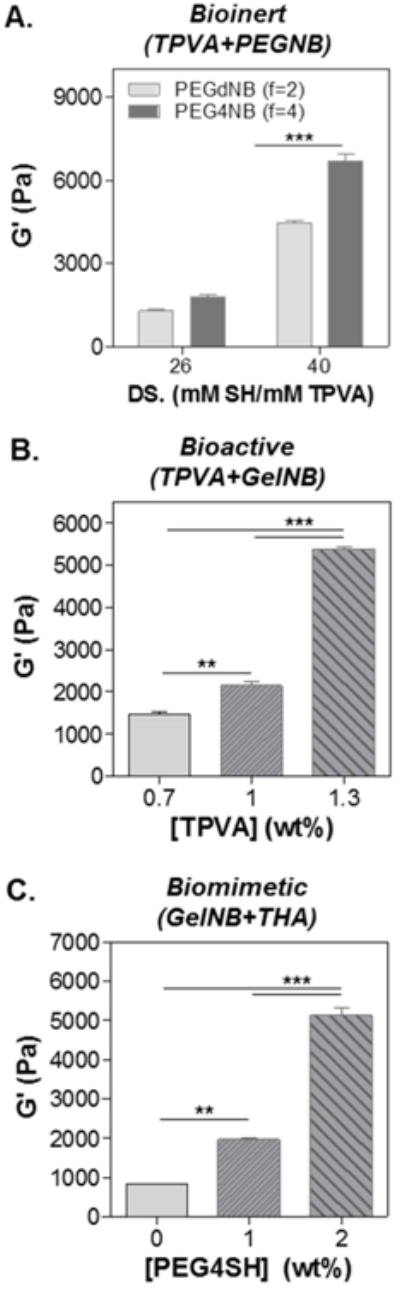
(A) Effect of TPVA degree of substitution (DS) and PEG functionality (PEGdNB or PEG4NB) on shear modulus of bio-inert thiol-norbornene hydrogel. (B) Effect of thiol/ene molar ratio (R[thiol]/[ene]) on shear modulus of bioactive thiol-norbornene hydrogels composed of GelNB (fixed at 7wt%) and TPVADS40 (at 0.7, 1, or 1.3 wt% for R[thiol]/[ene]= 0.5, 0.75, and 1, respectively). (C) Effect of PEG4SH supplement on shear modulus of biomimetic thiol-norbornene hydrogels composed of GelNB (fixed at 7wt%) and THA (fixed at 1.1wt%). PEG4SH = 0, 1.2, 2.4 wt%, corresponding to R[thiol]/[ene]= 0.5, 0.75, and 1.
3.3 Hydrolytic and enzymatic degradation of visible light cured thiol-norbornene biomimetic hydrogels
Next, we examined the degradation of these visible light cured thiol-norbornene hydrogels. In gel formulations containing macromers with ester bonds (e.g., TPVA, PEGdNB, or PEG4NB, Fig. 1B), hydrogels could be degraded via hydrolysis.14, 27 Fig. 3A shows that the shear modulus (G′) of TPVA/PEGNB hydrogels decreased as a function of time, indicative of network cleavage and decreases in gel crosslinking density. Furthermore, the degradation profiles followed pseudo-first order hydrolysis kinetics (Fig. 3A) as reported previously.14, 22, 28 In terms of enzymatic degradation, we prepared hydrogels using GelNB/PEG4SH or GelNB/THA (Fig. 3B) and subjected the gels to hyaluronidase (150 U/mL), which degrades hyaluronic acid but not gelatin. As expected, the mass of hydrogels containing no HA (i.e., GelNB/PEG4SH group) was not affected by hyaluronidase. However, changes in gel mass were noticeable in hydrogels crosslinked by THA. In the first 36 hours of hyaluronidase incubation, the mass of GelNB-THA hydrogels increased substantially, suggestive of increasing water uptake due to partial network cleavage. After 36 hours of hyaluronidase treatment, the mass of HA-containing biomimetic hydrogel (i.e., GelNB/THA) started to decrease until the gels completely dissolved. In addition to hyaluronidase, we also explored collagenase-mediated proteolytic degradation of GelNB/THA hydrogels that were crosslinked with 0, 1, and 2 wt% PEG4SH. The addition of PEG4SH led to gels with the same amount of bioactive components but different stiffness (Fig. 3C. G′ = 1, 2, 5 kPa for soft, medium, and stiff hydrogels, respectively). While all three groups of hydrogels contained the same amount of gelatin (i.e., 7wt% GelNB), the degradation exhibited stiffness-dependent kinetics. Specifically, soft GelNB/THA hydrogels degraded much faster than the stiffer hydrogels. Furthermore, hydrogels crosslinked with 1 or 2 wt% PEG4SH (i.e., medium and stiff hydrogel) gained significant weight during the initial phase of collagenase treatment (i.e., 24 and 48 hours for medium and stiff hydrogels).
Figure 3. Degradation of visible light polymerized thiol-norbornene hydrogels.
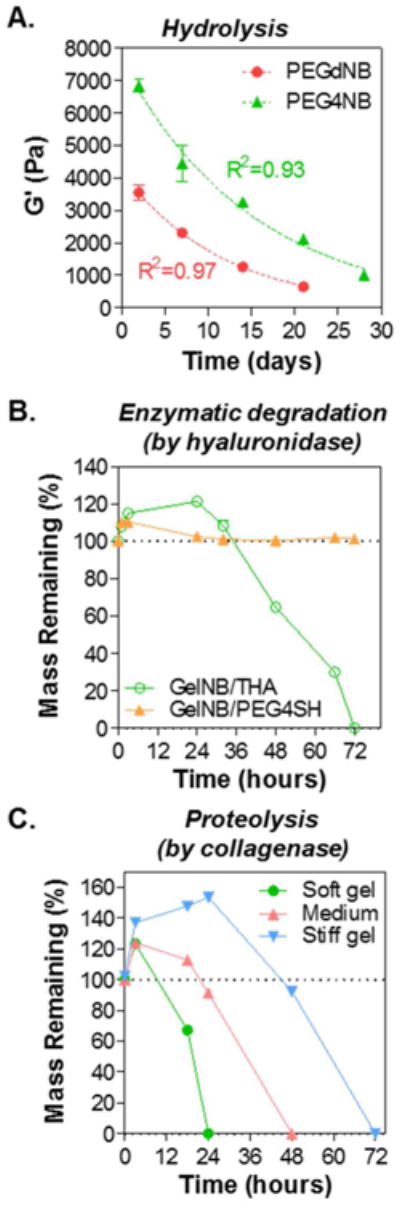
(A) Hydrolysis of bio-inert hydrogels composed of 0.5 wt% of TPVA and 3.6 wt% of PEGdNB or PEG4NB. Time dependent moduli were fitted to pseudo-first order hydrolytic degradation kinetics. (B) Hyaluronidase-mediated proteolytic degradation (at 37°C) of bioactive GelNB/PEG4SH and biomimetic GelNB/THA hydrogels (7wt% GelNB, 2wt% PEG4SH or 0.9wt% THA). (C) Collagenase-mediated proteolytic degradation (at 37°C) of biomimetic hydrogels with different stiffness (Collagenase: 100U/mL. Soft gel: ∼1 kPa, Medium: ∼2 kPa, Stiff gel: ∼ 5 kPa).
3.4 Effect of cell density on the formation of tumor spheroids in biomimetic hydrogels
The cytocompatibility of visible light cured thiol-norbornene hydrogels was evaluated by in situ encapsulation of COLO-357 cells, a PDAC cell line that forms spheroids when cultured in 3D hydrogels.18 Cells were first encapsulated at different cell densities (0.5, 2, 5 ×106 cells/mL) in biomimetic hydrogels composed of GelNB (7wt%) and THA (0.8wt%). The shear moduli of this gel formulation were close to 1kPa (data not shown). Live/Dead staining and confocal images (Fig. 4A, top panel) showed that almost all cells remained alive following encapsulation regardless of cell density. After 10-day in vitro culture, cells formed spheroids with sizes that were inversely proportional to the initial cell density (Fig. 4A, bottom panel). F-actin staining images also showed the inverse relationship between spheroid sizes and cell density at the time of encapsulation (Fig. 4B). Image analysis results showed that cell spheroid sizes were considerably larger when they were encapsulated at lower cell densities (average dia. ∼57 μm, 44 μm, or 21 μm for cells encapsulated at 0.5, 2, or 5×106 cells/mL, respectively. Fig. 4C). Furthermore, cells encapsulated at lower cell density grew into spheroids with higher heterogeneity in sizes (Fig. 4C, Fig. S1). We also quantified metabolic activity of the encapsulated cells over time using Alamarblue® reagent, which measures the reducing activity within the cytosol of living cells. Interestingly, while live/dead images clearly showed that all cells remained alive throughout the 10-day culture, the metabolic activity of the cells increased only in the first three days and dropped gradually from day-3 to day-10, eventually reached similar levels as that in day 1 (Fig. 4D). While cells encapsulated at 0.5×106 cells/mL did not show significantly higher fluorescence than the blank media control, these cells were obviously alive and proliferated into round spheres (Fig. 4A-4C). This was because the Alamarblue® reagent incubation time was fixed at 2.5 hours for all cell densities as longer incubation times would lead to overflow in reading for samples with higher cell density. We also extracted total DNA from cells encapsulated in these gels and the results (Fig. S2) showed that the total DNA contents increased both as a function of time and the encapsulating cell density.
Figure 4. Effect of cell density on the formation of pancreatic cancer cell spheroids.
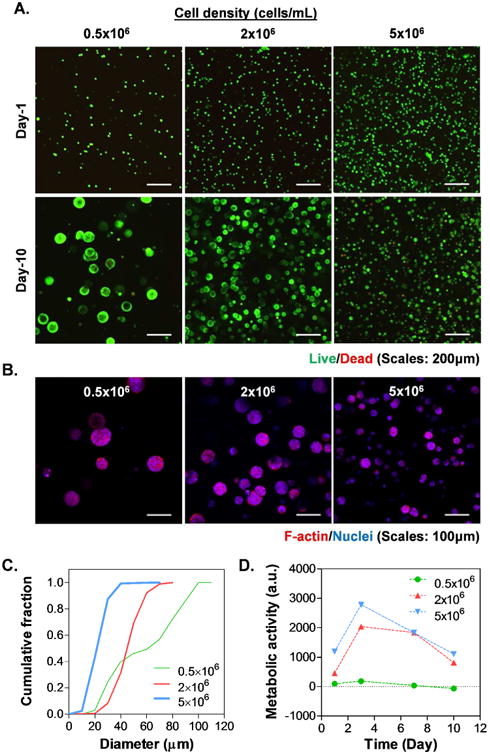
(A) Representative confocal z-stack images of live/dead stained COLO357 cells. (B) Images of F-actin/DAPI staining on day 10. (C) Cumulative distribution of cell spheroid diameter. (D) Cell viability as assessed by Alamarblue® reagent. All gel formulations contained 7 wt% GelNB, 0.9 wt% THA.
3.5 Effect of gel stiffness on the formation of tumor spheroids in biomimetic hydrogels
The impact of matrix stiffness on the formation of tumor spheres was studied using biomimetic hydrogels with fixed gelatin and HA weight percent (7 wt% GelNB and 0.9 wt% THA, respectively) and varied PEG4SH contents (0, 1, 2 wt%). The modulation of PEG4SH content allowed us to control gel stiffness (G′ = 1, 2, and 5kPa for soft, medium, and stiff gels, respectively. Fig. 2C) without altering the concentrations of biomimetic motifs. Since cell density at the time of encapsulation appeared to affect the size of tumor spheroids, we decided to encapsulate the cells at 2×106 cells/mL. Fig. 5A and Fig. 6A showed the live/dead staining and F-actin staining images, respectively. Although all cells formed spheroids regardless of gel modulus, cells encapsulated in softer gels showed higher metabolic activity (Fig. 5B). Spheroid size analysis showed that cells encapsulated in soft gels continued to grow into larger spheroids from day-5 to day-10, whereas the growing of cell spheroids in stiff gels appeared to slow down after day-5 (Fig. 6B, Fig. S3A & S3B). Furthermore, hydrogels with medium stiffness promoted the formation of spheroids with higher heterogeneity in sizes after 10 days of culture (Fig. S3B). The effect of gel stiffness on the total DNA contents from cells encapsulated in these gels were evaluated at all three time points (i.e., day 1, 5, and 10). The results (Fig. S4) showed that the total DNA contents increased as a function of time. However, the DNA contents were lower in cells encapsulated in the stiffer hydrogels than those in the gels with soft and medium stiffness.
Figure 5. Effect of gel stiffness on the formation of pancreatic cancer cell spheroids.
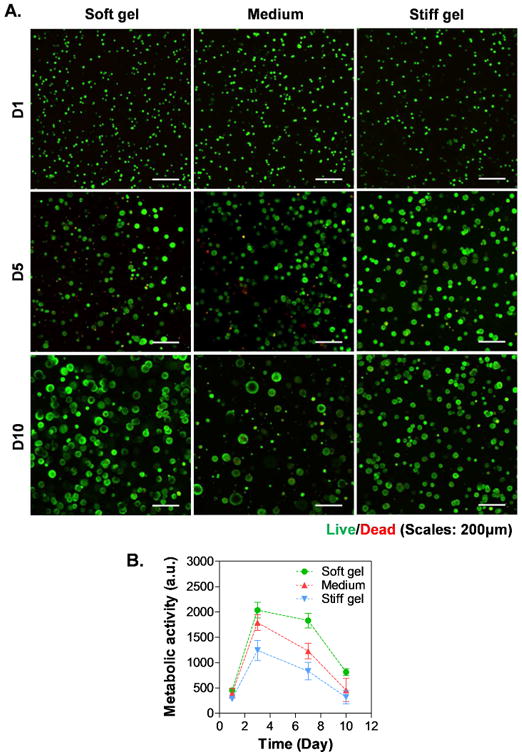
(A) Representative confocal z-stack images of live/dead stained COLO357 cells. (B) Cell metabolic activity as assessed by Alamarblue® reagent. All gel formulations contained 7 wt% GelNB, 0.9 wt% THA, as well as 0, 1 or 2 wt% of PEG4SH (i.e., soft, medium and stiff hydrogel).
Figure 6. Effect of gel stiffness on the sizes of pancreatic cancer cell spheroids.
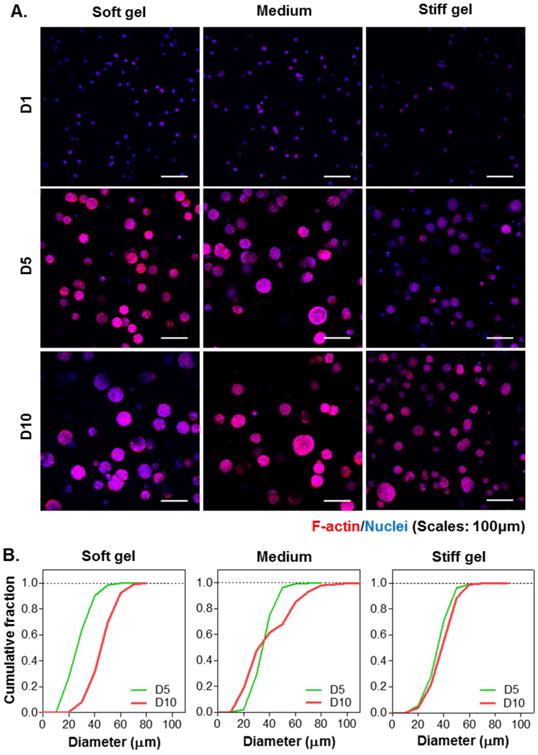
(A) Representative confocal z-stack images of COLO-357 cells stained with F-actin/DAPI. (B) Cumulative distribution of cell spheroid diameter. Gel formulations were identical to that used in Figure 5.
3.6 Effect of gel stiffness on gene expression in tumor spheroids formed in biomimetic hydrogels
Fig. 7 shows mRNA expression of three genes relevant to the growth of tumor spheres. The expression of connective tissue growth factor (CTGF, Fig. 7A) and sonic hedgehog (SHH, Fig. 7B) were evaluated because of their roles in tumor/organ size control, as well as in maintaining stemness in cancer stem cells (CSC).25, 29-32 Moreover, the expression of matrix metalloproteinase 14 (MMP14, Fig. 7C) was examined because of its importance in matrix cleavage during the growth of tumor spheres.18 Data were normalized to an arbitrary value of 1 based on the levels of mRNA expression from cells cultured in soft hydrogels (G′ ∼1 kPa) for the same duration (i.e., 1, 5, or 10 days). At respective day of culture, CTGF mRNA expression levels (Fig. 7A) were generally lower when the cells were cultured in medium (G′ ∼ 3kPa) and stiff (G′ ∼ 5 kPa) gels than in softer gels. However, the influence of stiffness on CTGF mRNA expression seems to be less significant as the cells formed spheroids (i.e., longer culture time) as there were upward trends in CTGF expression in medium and stiff gels. By contrast, SHH expression was generally higher in cells grown for longer time and in stiffer hydrogels (Fig. 7B). Finally, the expression levels of MMP14 were higher in cells encapsulated in stiffer gels than in softer gels (Fig. 7C).
Figure 7. Effect of gel stiffness on mRNA expression in encapsulated cells.
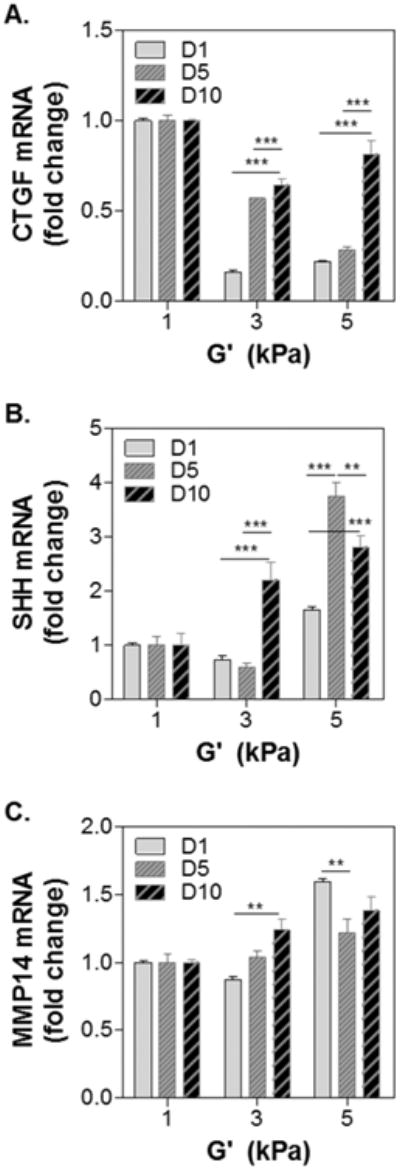
(A) CTGF, (B) SHH, and (C) MMP14. Housekeeping gene: GAPDH. 1-fold: gene expression levels in cells encapsulated in soft (G′ ∼ 1kPa) gels at respective day of culture (i.e., day-1, 5, or 10). Gel formulations were identical to that used in Figure 5. Asterisks: **p<0.01, ***p<0.001.
4. Discussion
Thiol-norbornene PEG-based hydrogels can be rapidly crosslinked by UV light (e.g., λ = 365 nm) irradiation.33 However, the use of UV light poses a concern regarding cancer cell fate, especially for mutation-prone PDAC cells. In this contribution, we report the use of visible light (λ = 400-700 nm) initiated thiol-norbornene biomimetic hydrogels for culturing COLO357 cells, a PDAC cell line that forms spheroids in 3D culture.18, 34 Conventionally, visible light mediated gelation involves the use of (meth)acrylated macromers (e.g., PEGDA or gelatin-methacrylate) coupled with a dual-initiator system (e.g., eosin-Y and TEOA), as well as a co-monomer (e.g., NVP).11 Although careful adjustments of the formulations produce chain-polymerized hydrogels with desired materials properties, some of these required components (e.g., strong base TEOA) may pose undesired side effects to the encapsulated.9 On the other hand, we have reported a visible light initiated thiol-norbornene gelation system using low concentration of eosin-Y (0.1 mM) as the single photoinitiator (Fig. 1A).9 In the prior work, however, we did not explore the effect of co-polymerized bioactive components on cell fate processes. The current contribution fills this technological gap by providing a visible light based step-polymerized gelation strategy with modular controls in matrix properties. Owing to the orthogonal crosslinking nature of thiol-norbornene chemistry, at least two types of macromers must be employed: one with thiol and the other with norbornene moiety.33 Three classes of macromers, including synthetic macromers and derivatives of gelatin and HA, were used to render the resulting hydrogels that are bio-inert (e.g., TPVA+PEGdNB), bioactive (TPVA+GelNB), or biomimetic (GelNB+THA). Most of these macromers are commercially available (e.g., PEGNB, PEGSH, and THA), while others are easily synthesized in aqueous solution with mild reaction conditions (e.g., GelNB). Although either TPVA or multi-arm PEGSH could be used, the former provides a more economical option and a tunable degree of thiolation per macromer that could lead to higher degree of gel crosslinking. The gelation of visible light-based step-polymerized hydrogel system was understandably slower (gel point: ∼ 5 to 25 seconds, Fig. 1C-1E) than its counterpart where UV-light was used (typical gel point: ∼ 1 to 2 seconds). However, the gel points reported in this work were comparable to or even faster than that in our previous study using visible light initiated thiol-norbornene gelation with inert macromers PEG4NB and DTT (gel point ∼ 20 seconds).9 The incorporation of THA induced rapid gelation (Fig. 1E. Gel point ∼ 5 seconds), presumably due to the exceptionally high functionality of THA (DS: ∼216 thiols/THA molecule, per Ellman's assay for thiol quantification).
The ability to control matrix stiffness is becoming a critical consideration when designing hydrogels for 3D cell culture.24, 35 The major benefit of modular crosslinking is that matrix stiffness can be independently controlled without altering the bioactivity of the gel matrix. Here, we show that the stiffness of hydrogels, whether bio-inert, bioactive, or biomimetic, could be readily tuned by controlling various parameters, including the degree of substitution, macromer functionality, or simply the content of bio-inert macromer (Fig. 2). In particular, these manipulations could be achieved without affecting the content of bioactive motifs, which are required for supporting cell survival in 3D but could become a confounding factor when evaluating the effect of matrix stiffness on cell fate.
Predictable or controllable degradation is also critical when engineering matrices for 3D cell culture.36-41 This is because matrix degradation, either through hydrolysis or proteolysis, affects many aspects of cell fate processes, including proliferation, migration, and differentiation. Here, we show that the visible light initiated step-polymerized thiol-norbornene hydrogels exhibit various degrees of degradability, depending on the macromer constituents (Fig. 3). Whereas macromers containing ester bonds are degraded through simple hydrolysis, both gelatin and hyaluronic acid are susceptible to enzyme-specific degradation. Interestingly, the degradation of GelNB-based hydrogels exhibited different modes of degradation, depending on the degree of gel crosslinking. While GelNB hydrogels with lower crosslinking could be readily eroded through enzyme-mediated chain cleavage, highly crosslinked GelNB hydrogels required more time to reach sufficient cleavage and gel mass loss. This degradation behavior is in concord with our previous report using UV-light crosslinked GelNB-PEG hydrogels.19 This phenomenon can potentially affect cells' ability to model their ECM, which may lead to changes in cell spreading and proliferation, as well as alterations in intracellular signaling.
COLO-357 pancreatic cancer cell line was used as a model to illustrate the importance of matrix properties, particularly stiffness, on the growth of tumor spheres. We chose a PDAC cell line because this cancer is associated with marked stroma formation and marked increases in stiffness. We found that the visible light based step-polymerized biomimetic hydrogels were highly cytocompatible for in situ cell encapsulation (Fig. 4). Encapsulated cells were able to proliferate and form spherical cell clusters. Interestingly, cells encapsulated at lower cell density appeared to grow into larger and more heterogeneous spheroids. It is likely that some paracrine effects led to growth inhibition when cells were encapsulated at higher cell density (e.g., 5×106 cells/mL). The relationship of initial cell density and tumor spheroid size was previously observed in breast cancer cells grown in inert PEG-based hydrogels.42, 43 The authors attributed the increased spheroid size at low cell density to enhanced expression of CSC markers for breast cancer cells, such as CD44, ABCG2, and EGFR. Similar phenomenon could contribute to the results observed in Fig. 4. In addition to initial cell density, we also found that softer gels encouraged the formation of larger PDAC cell spheroids (Fig. 6). This was likely due to the fact that hydrogels with lower crosslinking density were less dense and were easily deformed by the growing spheroids. On the other hand, stiff hydrogels were tightly crosslinked and imposed significant strain on the cells, which leads to decrease in spheroid sizes. During the growth of PDAC tumor spheroids, cellular metabolic activity peaked at day-3, but gradually decreased in the next 7 days regardless of initial cell density. Similar reduction in cell metabolic activity was observed when cells were encapsulated at the same cell density but within gels of different moduli (Fig. 5). It is worth noting that Alamarblue® reagent only measures non-specific reduction in cell metabolic activity. Results shown in Fig. 4D and 5B suggest that the metabolism of COLO-357 cells was affected by the formation of multicellular spheroids. The results were also cell-type specific and could be affected by culture media supplements, immobilized biochemical motifs, cell-cell interactions, and biophysical properties of the matrix. The results from additional total DNA quantification (Fig. S2, S4) were largely in agreement with the live/dead staining images (i.e., showing sign of cell proliferation) but not with the metabolic activity assays. The latter highlights the complicated nature of tumor cell metabolism. Although the current data support the notion that matrix properties could affect cancer cell fate processes, future studies are required to elucidate the influence of these matrix properties on tumor cell morphogenesis, including proliferation, invasion, and even drug responsiveness.
The effects of matrix stiffness on PDAC cells were also evaluated using real time PCR of selective genes deemed to be relevant to the growth of tumor spheroids. Note that we normalized the gene expression levels first to GAPDA internal control, then to the expression levels in cells grown in soft hydrogels (G′ ∼ 1kPa). This allowed us to gain insights regarding the differences of cell fate processes on the mRNA level between cells grown in hydrogels with different stiffness. CTGF is an effector gene downstream of the HIPPO pathway, which regulates tissue homeostasis, organ size, tumorigenesis, and even chemoresistance.30, 32 The upregulation of CTGF could be a result of inactivation of the HIPPO pathway, whose activation suppresses tumor progression and metastasis.31 Our results (Fig. 7A) suggest that while cells encapsulated in medium and stiff hydrogels initially had lower CTGF expression than that in cells encapsulated in the soft gels, higher matrix stiffness might suppress HIPPO pathway (and hence promote tumor progression) at later time as the cells gradually formed spheroids. In contrast to CTGF, the expression of SHH was found to be higher in stiff gel matrix. SHH signaling contributes critically to pancreatic desmoplasia and upregulation of SHH has been associated with increased tumor progression.44 A stiffer matrix also promotes expression of MMP-14, especially at later phase of cell growth (day-5 and day-10). We have previously shown that both SHH and MMP14 were upregulated in COLO-357 cells stimulated with transforming growth factor β (TGFβ) and epidermal growth factor (EGF).18 This study shows that, without altering the contents of bioactive components (i.e., gelatin and hyaluronic acid) or soluble factors (e.g., TGFβ and EGF), matrix stiffness alone is sufficient to upregulate SHH and MMP14 expression. The results reported here show vast differences of gene expression between cells encapsulated in soft and stiff hydrogels. For example, there was an upward trend in SHH expression in cells encapsulated in stiffer hydrogels overtime compared to cells grown in softer environment. Although it is not the focus of the current study, these gene expression data suggest the need to design additional experiments for understanding the mechanisms by which matrix properties affect PDAC cell fate. It is also worth noting that the results could be cell-type dependent and future studies should contain other PDAC cells, including cell lines and primary tumor cells. Nonetheless, the work presented in this contribution demonstrates the high tunability of visible light initiated thiol-norbornene biomimetic hydrogels and their potential in cancer cell research.
5. Conclusion
In conclusion, the current study explores the use of visible light polymerized thiol-norbornene biomimetic hydrogels for PDAC cell research. The use of visible light negates potential concerns of UV light induced cellular damage while allowing facile fabrication of biomimetic cell-laden hydrogels using thiol- and norbornene-functionalized macromers. In particular, the gelation of biomimetic hydrogels composed of GelNB and THA was highly efficient and cytocompatible for in situ encapsulation of PDAC cells. We determined that cell density and matrix stiffness are two critical parameters affecting the formation of tumor spheroids. Specifically, there exists an inverse relationship between cell density at the encapsulation and the size of tumor spheroids formed. Furthermore, stiffer matrix upregulated the expression of SHH and MMP-14, suggesting the important influence of matrix stiffness on tumor cell fate. Further work will focus on exploiting this modular and adaptable visible light initiated gelation system for studying cancer cell growth, dormancy, and invasion in 3D.
Supplementary Material
Acknowledgments
This project was funded by the National Cancer Institute of the NIH (R21CA188911) and by the Purdue Research Foundation Doctoral Research Grant (to HS). The authors thank Mr. Brent Hukill for his technical assistance with TPVA/PEGNB hydrogel characterization.
References
- 1.DeForest CA, Anseth KS. Annu Rev Chem Biomol Eng. 2012;3:421–44. doi: 10.1146/annurev-chembioeng-062011-080945. [DOI] [PubMed] [Google Scholar]
- 2.Lin CC. RSC Adv. 2015;5:39844–398583. doi: 10.1039/C5RA05734E. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lutolf MP, Hubbell JA. Nat Biotechnol. 2005;23:47–55. doi: 10.1038/nbt1055. [DOI] [PubMed] [Google Scholar]
- 4.Elbert DL, Hubbell JA. Biomacromolecules. 2001;2:430–441. doi: 10.1021/bm0056299. [DOI] [PubMed] [Google Scholar]
- 5.Salinas CN, Anseth KS. Macromolecules. 2008;41:6019–6026. [Google Scholar]
- 6.Fairbanks BD, Schwartz MP, Halevi AE, Nuttelman CR, Bowman CN, Anseth KS. Adv Mater. 2009;21:5005–5010. doi: 10.1002/adma.200901808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Anderson SB, Lin CC, Kuntzler DV, Anseth KS. Biomaterials. 2011;32:3564–3574. doi: 10.1016/j.biomaterials.2011.01.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shih H, Fraser AK, Lin CC. ACS Appl Mater Interfaces. 2013;5:1673–1680. doi: 10.1021/am302690t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shih H, Lin CC. Macromol Rapid Comm. 2013;34:269–273. doi: 10.1002/marc.201200605. [DOI] [PubMed] [Google Scholar]
- 10.Cruise GM, Hegre OD, Scharp DS, Hubbell JA. Biotechnol Bioeng. 1998;57:655–665. doi: 10.1002/(sici)1097-0290(19980320)57:6<655::aid-bit3>3.0.co;2-k. [DOI] [PubMed] [Google Scholar]
- 11.Bahney CS, Lujan TJ, Hsu CW, Bottlang M, West JL, Johnstone B. Eur Cell Mater. 2011;22:43–55. doi: 10.22203/ecm.v022a04. discussion 55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.West JL, Hubbell JA. Macromolecules. 1999;32:241–244. [Google Scholar]
- 13.Kizilel S, Perez-Luna VH, Teymour F. Langmuir. 2004;20:8652–8658. doi: 10.1021/la0496744. [DOI] [PubMed] [Google Scholar]
- 14.Shih H, Lin CC. Biomacromolecules. 2012;13:2003–2012. doi: 10.1021/bm300752j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fraser AK, Ki CS, Lin CC. Macromol Chem Phys. 2014;215:507–515. [Google Scholar]
- 16.Shih H, Mirmira RG, Lin CC. J Mater Chem B. 2015;3:170–175. doi: 10.1039/C4TB01593B. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ki CS, Shih H, Lin CC. Polymer. 2013;54:2115–2122. doi: 10.1016/j.polymer.2013.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ki CS, Lin TY, Korc M, Lin CC. Biomaterials. 2014;35:9668–9677. doi: 10.1016/j.biomaterials.2014.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Greene T, Lin CC. ACS Biomater Sci Eng. 2015;1:1314–1323. doi: 10.1021/acsbiomaterials.5b00436. [DOI] [PubMed] [Google Scholar]
- 20.Munoz Z, Shih H, Lin CC. Biomaterials Science. 2014;2:1063–1072. doi: 10.1039/c4bm00070f. [DOI] [PubMed] [Google Scholar]
- 21.Dicharry RM, Ye P, Saha G, Waxman E, Asandei AD, Parnas RS. Biomacromolecules. 2006;7:2837–2844. doi: 10.1021/bm060432n. [DOI] [PubMed] [Google Scholar]
- 22.Hao Y, Shih H, Munoz Z, Kemp A, Lin CC. Acta Biomater. 2014;10:104–14. doi: 10.1016/j.actbio.2013.08.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ling H, Sylvestre JR, Jolicoeur P. Oncogene. 2010;29:4543–54. doi: 10.1038/onc.2010.186. [DOI] [PubMed] [Google Scholar]
- 24.Dupont S, Morsut L, Aragona M, Enzo E, Giulitti S, Cordenonsi M, Zanconato F, Le Digabel J, Forcato M, Bicciato S, Elvassore N, Piccolo S. Nature. 2011;474:179–83. doi: 10.1038/nature10137. [DOI] [PubMed] [Google Scholar]
- 25.Jabari S, Meissnitzer M, Quint K, Gahr S, Wissniowski T, Hahn EG, Neureiter D, Ocker M. Int J Oncol. 2009;35:69–80. doi: 10.3892/ijo_00000314. [DOI] [PubMed] [Google Scholar]
- 26.Silveira Correa TC, Massaro RR, Brohem CA, Taboga SR, Lamers ML, Santos MF, Maria-Engler SS. J Cell Biochem. 2010;110:52–61. doi: 10.1002/jcb.22472. [DOI] [PubMed] [Google Scholar]
- 27.Raza A, Lin CC. Macromol Biosci. 2013;13:1048–1058. doi: 10.1002/mabi.201300044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Metters A, Hubbell J. Biomacromolecules. 2005;6:290–301. doi: 10.1021/bm049607o. [DOI] [PubMed] [Google Scholar]
- 29.Lunardi S, Muschel RJ, Brunner TB. Cancer Lett. 2014;343:147–155. doi: 10.1016/j.canlet.2013.09.039. [DOI] [PubMed] [Google Scholar]
- 30.Zhao B, Tumaneng K, Guan KL. Nat Cell Biol. 2011;13:877–83. doi: 10.1038/ncb2303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Serrano I, McDonald PC, Lock F, Muller WJ, Dedhar S. Nat Commun. 2013;4:2976. doi: 10.1038/ncomms3976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kong D, Zhao Y, Men T, Teng CB. J Drug Target. 2015;23:125–33. doi: 10.3109/1061186X.2014.983522. [DOI] [PubMed] [Google Scholar]
- 33.Lin CC, Ki CS, Shih H. J Appl Polym Sci. 2015;132:41563. doi: 10.1002/app.41563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sempere LF, Gunn JR, Korc M. Cancer Biol Ther. 2011;12:198–207. doi: 10.4161/cbt.12.3.15979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yang C, Tibbitt MW, Basta L, Anseth KS. Nat Mater. 2014;13:645–52. doi: 10.1038/nmat3889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hao Y, Lin CC. J Biomed Mater Res A. 2014;102:3813–27. doi: 10.1002/jbm.a.35044. [DOI] [PubMed] [Google Scholar]
- 37.Hudalla GA, Eng TS, Murphy WL. Biomacromolecules. 2008;9:842–849. doi: 10.1021/bm701179s. [DOI] [PubMed] [Google Scholar]
- 38.Khetan S, Guvendiren M, Legant WR, Cohen DM, Chen CS, Burdick JA. Nat Mater. 2013;12:458–465. doi: 10.1038/nmat3586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Levesque SG, Shoichet MS. Bioconjug Chem. 2007;18:874–885. doi: 10.1021/bc0602127. [DOI] [PubMed] [Google Scholar]
- 40.Lutolf MP, Lauer-Fields JL, Schmoekel HG, Metters AT, Weber FE, Fields GB, Hubbell JA. Proc Natl Acad Sci USA. 2003;100:5413–5418. doi: 10.1073/pnas.0737381100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zustiak SP, Leach JB. Biomacromolecules. 2010;11:1348–57. doi: 10.1021/bm100137q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jabbari E, Sarvestani SK, Daneshian L, Moeinzadeh S. PLos ONE. 2015;10:e0132377. doi: 10.1371/journal.pone.0132377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yang X, Sarvestani SK, Moeinzadeh S, He X, Jabbari E. PLos ONE. 2013;8:e59147. doi: 10.1371/journal.pone.0059147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gore J, Korc M. Cancer Cell. 2014;25:711–712. doi: 10.1016/j.ccr.2014.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


