Abstract
Phycobilisomes (PBS) are accessory light harvesting protein complexes that directionally transfer energy towards photosystems. Phycobilisomes are organized in a central core and rods radiating from it. Components of phycobilisomes in Gracilaria chilensis (Gch) are Phycobiliproteins (PBPs), Phycoerythrin (PE), and Phycocyanin (PC) in the rods, while Allophycocyanin (APC) is found in the core, and linker proteins (L). The function of such complexes depends on the structure of each component and their interaction. The core of PBS from cyanobacteria is mainly composed by cylinders of trimers of α and β subunits forming heterodimers of Allophycocyanin, and other components of the core including subunits αII and β18. As for the linkers, Linker core (LC) and Linker core membrane (LCM) are essential for the final emission towards photoreaction centers. Since we have previously focused our studies on the rods of the PBS, in the present article we investigated the components of the core in the phycobilisome from the eukaryotic algae, Gracilaria chilensis and their organization into trimers. Transmission electron microscopy provided the information for a three cylinders core, while the three dimensional structure of Allophycocyanin purified from Gch was determined by X-ray diffraction method and the biological unit was determined as a trimer by size exclusion chromatography. The protein sequences of all the components of the core were obtained by sequencing the corresponding genes and their expression confirmed by transcriptomic analysis. These subunits have seldom been reported in red algae, but not in Gracilaria chilensis. The subunits not present in the crystallographic structure were modeled to build the different composition of trimers. This article proposes structural models for the different types of trimers present in the core of phycobilisomes of Gch as a first step towards the final model for energy transfer in this system.
Introduction
Aquatic organisms, such as cyanobacteria, eukaryotic red alga and cryptophyta, present accessory light harvesting protein complexes called phycobilisomes (PBS) [1]. The structure of PBS allows the energy to be transferred directionally to photosystem II (PSII), thus improving the use of the 400-650nm range of wavelengths. Phycobilisomes are organized in a central core and rods radiating from it. Components of phycobilisomes in Gracilaria chilensis are Phycobiliproteins (PBPs): Phycoerythrin (PE), and Phycocyanin (PC) in the rods, Allophycocyanin (APC) is found in the core, and linker proteins (L) present along the whole system. PBPs are chromophorylated proteins that share a general architecture. They are organized as heterodimers (αβ), which assembly themselves into trimers or hexamers. The subunits of PBPs are mono or multi chromophorylated with linear tetrapyrrols covalently bound to cysteine residues. Efficient energy transfer is achieved through a combination of position and geometry of the chromophores and the protein environment [1–3]. In the present article, we focused on the principal component of the phycobilisomes´ core, the phycobiliprotein Allophycocyanyn or APC. The Allophycocyanin is organized as cylinders, and different numbers of these cylinders have been reported for the core of PBS including bicylindrics, tricylindrics and pentacylindrics, being the tricylindric the most common in cyanobacteria and red alga [4–6]. The tricylindrical core has been proposed to be formed by a foundation of two cylinders associated to membrane and a third cylinder is located at the top of the base forming a pyramid [7]. Four subunits have been identified for APC in the core: α, β, αII and β18, which are codified by apcA, apcB, apcD and apcF genes, respectively [8]. Studies reported on Synechococcus show that subunits αII and β18 are expressed in minor quantities, which make difficult to study the expressed proteins in native conditions [9,10]. Indeed, studies of the same nature in eukaryotic red alga are still missing.
Besides PBPs, the two linkers Core (LC) and core-membrane (LCM) have been identified in the core [1]. The LC of 7.8 kDa is codified by the apcC gene. This linker is associated to APC trimers in cylinders, suggesting a specific role on stability and energy transfer [9,11–13]. The structure of LC has been determined by X-ray diffraction as a complex with APC trimer in Mastigocladus laminosus (PDB ID: IB33) [14]. The LCM, codified by the apcE gene, in a tricylindrical core is a 90kDa chromophorylated linker protein that has been proposed as the final acceptor of the light harvested by the PBS, besides its role in the anchoring of the PBS to the photosynthetic membrane [9,12,13,15,16]. Different domains have been identified in the linker core-membrane; for example, the N-terminal region, called PB domain, is homologous to PBP alpha subunits, and shows 30% similarity with α subunit of APC, and also contains a phycocyanobilin chromophore. This PB domain presents an insertion of 50 residues (PB loop). The PB loop in cyanobacteria has been associated to fluorescence quenching by its interaction with the orange carotenoid protein (OCP) [17], which has not been detected in eukaryotic alga. Zhao and his group [15] produced a recombinant chromophorylated PB domain in absence of lyases, a fact that induced the authors to propose an autocatalytic binding of phycocyanobilin to LCM. Due to their similarity, the PB domain can replace the α subunit and form a PBβ heterodimer, which can associate itself to other αβ heterodimers to form a trimer.
Different trimers composition present in a tricylindrical core have been reported for Synechocystis [18] APC (α3β3), APC_1 (αα3β3Lc), APC_2 (α2αIIβ3Lc) and APC_3 (α2PBβ2β18). Watanabe and Ikiushi [19], in an interesting review proposed also a model for the position of each type of trimer in the three cylinders core, which in Gracilaria chilensis has not been assessed yet.
Gracilaria chilensis (Rhodophyta, Gigartinalis) (G.ch) [20] is one of the economically important red macroalga in the South Pacific. Our research on the structure and function of the PBS in this algae has been focused on understanding how the organization, i.e. composition, structure or component interactions of the PBS provides the frame for the high efficiency of the energy transfer towards PSII. Until now, the structure of PE and PC from Gch has been reported previously by our group [21,22], as well as information about interaction between hexamers in a rod and its role in the function of energy transfer [23]. In this article, we report i) the three cylinders nature of the core of the PBS in Gch, ii) the sequences of the subunits components of the core, iii) the sequence analysis and iv) the X-ray structure for Allophycocyanin from Gch (αβ)3. This structure was used to model the rest of the subunits, which may also form a functional Allophycocyanin, and thereby to build the trimers proposed for the core in cyanobacteria, as our first approach to the analysis of the contribution of both the subunits in the core and LC to the energy transfer pathways in an eukaryotic red alga.
Material and methods
Purification of phycobilisomes
PBS were purified using the condition presented in S1 File, from 200g of intertidal G.ch collected at Coliumo Bay (36° 32’S, 072° 56’ W). The fraction containing complete PBS was identified by its spectroscopic properties (emission at 660nm after excitation at 566nm) and was used for Electron Microscopy.
Electron microscopy: The micrographs were obtained in a Philips Tecnai 12 Bio Twin EM (120kv) at the Pontificia Universidad Católica de Chile facility, using the methodology described [23] for complete phycobilisomes.
The sequences of the subunits
Primers design
The primers were designed by sequence alignment of the genes reported in literature, using the sequences displayed on S2 File. The primers were designed manually considering the conserved sequences, the %GC, Tm and the size of the oligonucleotides. The design was verified with Oligonucleotide PrimerCheck (http://depts.washington.edu/bakerpg/primertemp/primermelttemp.html) and synthesized by IDT. The primers used were:
α subunit Sense: 5’atgagtattattactaaatcaatcgttaatgctgatgcagaagctcgg 3`
Antisense: 3’ttactgcattgcacctaatgtataatcaaaataaaatcct 5’
βsubunit
Sense: 5’atgcaagatgctattacttctgtaattaatgcagctgatgtacaagg 3’
Antisense: 3’ctagcttaaaccagaacaaatataatcaaaatatactccc5’
αII subunit
Sense: 5’atgagcttagttagccaaattattttaaatgcagataatgaattaagat 3’
Antisense: 3’ ttatgacataccttgtattataaaatcaaagtagggttct 5’
β18
Sense: 5’atgcaagacgctattactacaattttaaatcgatatgatttaacaggaaa 3’
Antisense: 3’ ttagatatcttcttcgcttaagtttttaattatatattgaaatggttcaa 5’
Sequencing of the genes codifying for the subunits of Allophycocyanin
RNA was extracted from 0.1g of fresh washed and frozen alga, ground in a mortar with TRI-Reagent (Molecular Research Center) using the protocol recommended by the supplier. cDNA was obtained using Reverse Transcriptase RevertAidTM kit (Fermentas) according to recommendations of suppliers. The cDNA was used for the amplification assays by PCR according to S3 File. The sequences translated to proteins were used for the resolution of the 3D structure of Allophycocyanin and for the molecular modeling of the subunits αII and β18.
The sequence of the linkers associated to the CORE
The PB domain of the LCM:Primers were designed according to the sequences reported for LCM of the red algae Chondrus crispus [24] and Gracilaria tenuistipitata [25]. The sequencing and the amplification methodology were similar to the methodology described above for α and β subunits of APC.
The LC: The sequence of the Linker core of Gracilaria chilensis was obtained from a direct analysis of the transcriptome (AN: SRX1507975) [26].
Purification of Allophycocyanin
APC was obtained as reported in S4 File. The pure protein was characterized by its spectroscopic properties of absorption (λAmax = 651nm) and emission (λEmax = 660nm) and the relationship A651/A280 as purity index. This sample was used for the determination of the oligomerization state of Allophycocyanin, the spectroscopic studies and for crystallization.
Determination of the oligomerization state
50μL of 1 mg/mL of purified APC was separated in a Superdex200 HR/30 column (Amershan Biosciences), equilibrated with 5mM phosphate buffer pH7, and associated to a Merck-Hitachi FPLC with a flux of 0.5ml/min provided by a L6210 Intelligent Pump with a L-4250 UV-Vis Detector. For the calibration, BioRad standards were used. The elution of the proteins was followed at 651 and 280nm.
Protein crystallization
A general screening was performed in the robotic unit at Universidad de Chile. The final crystallization conditions were 2mg/mL protein concentration, 300mM NaCl, 20% v/v PEG 6000, and 100mM MES pH7.
Determination of the three dimensional structure of APC
Crystals were diffracted at Soleil Synchrotron up to 2.3 Å resolution. The phases were obtained by molecular replacement using the data of 1KN1 [27] and the software Phenix [28]. The final model was built in Coot [29] and refined by Phenix Refine [30].
Molecular models for the different trimers present in the core
The trimer for APC was obtained by crystallographic symmetry (P321). The different subunits were modeled with Modeller v 9.1 [31]. The templates were the α subunit of APC from the structure reported here (PDB ID: 5TJF), the PB domain in 4XXI [32], and the β subunit of 5TJF for αII, PB domain and β18, respectively. Trimers APC_1, APC_2, APC_3 were built by substitution of the corresponding subunits followed by a molecular dynamics protocol for steric rearrangement of residues. LC was modelled also with Modeller using the chain O of the reported structure for Mastigoclaudus laminosus (PDB ID: 1B33) [14] as template, and a docking procedure was performed between the LC and (αβ)3 or (αIIα2β3) using ClusPro [33] without restrictions. Each final model was evaluated with PROSA [34] and RAMPAGE [35].
Results and discussion
The phycobilisome's core of G. chilensis is tricylindrical
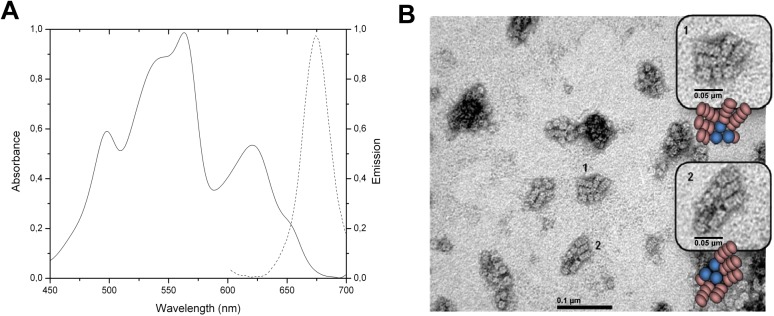
The PBS enriched fractions were analyzed by electron microscopy. The absorption spectrum (Fig 1A) showed the assembly of the PBS. Upon excitation at 566nm corresponding to the absorption maximum of PE (Phycoerythrin). The only emission corresponded to the emission of APC, which means that the assembly was intact. Moreover, the energy has been transferred through the complex to emit at 660nm. The micrograph (TEM) for the PBS is shown in Fig 1B. The image clearly showed three cylinders core of phycobilisomes, which are compatible with the presence of 90kDa LCM, and 5–6 rods formed by the piling of 3–4 hexamers. This is the first time that a photomicrograph of a phycobilisome of Gracilaria chilensis is reported, and at least in shape and organization resembles the PBS from prokaryotes [2].
Fig 1. Characterization of phycobilisomes of Gracilaria chilensis.
A) Absorption(-) and emission(..) spectra. B) Transmission electron micrograph of purified phycobilisomes.The inserts show amplified images. Schematic drawings of PBS are also shown.
Allophycocyanin is mainly present as trimer (αβ)3, despite the transcription of other subunits
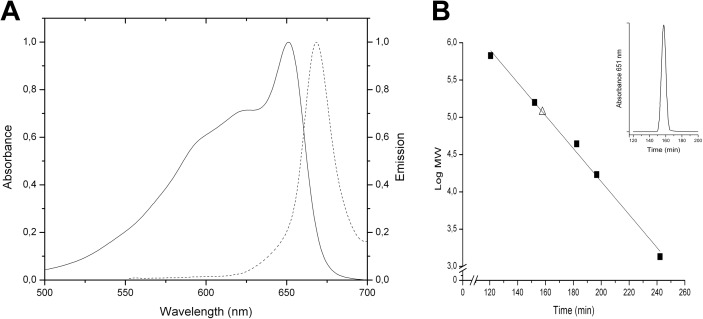
The absorption and emission spectra of the purified samples of Allophycocyanin are shown in Fig 2A. This sample was used to determine the oligomerization state using size exclusion chromatography (Fig 2B).
Fig 2. Spectroscopic characterization and oligomerization state of Allophycocyanin.
A) Absorption (-) and emission (--) spectra of purified Allophycocyanin. B) Molecular sieve chromatogram; the standards and the sample are indicated.
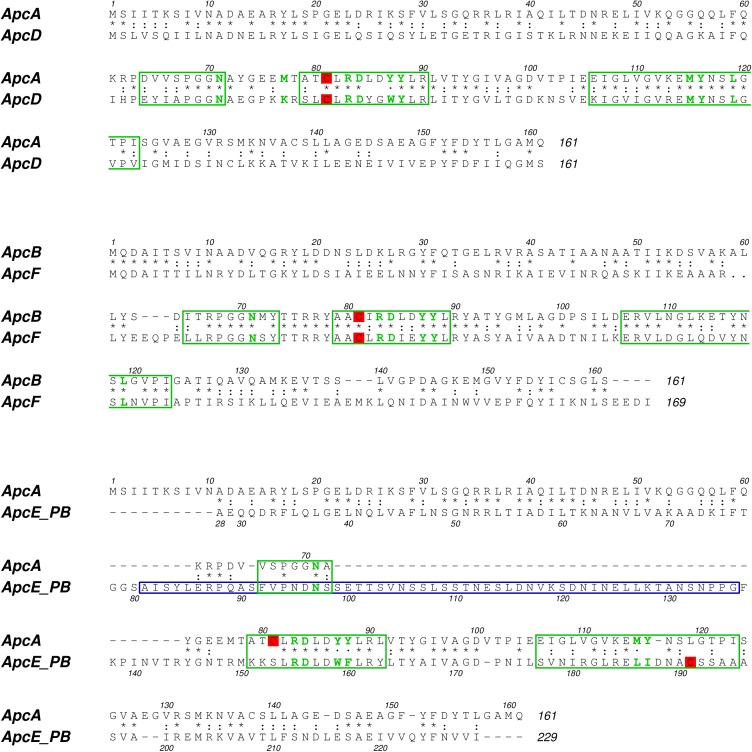
The result showed that Allophycocyanin from Gracilaria chilensis was purified in an oligomeric state (αβ)3. The purified APC was shown to contain only α and β subunits by mass spectrometry experiments. Studies reported in Synechocystis [19] and our results from the sequencing of the genes of the phycobiliproteins in the core of Gracilaria chilensis, revealed that they also contain αII and β18, which are not present in the Allophycocyanin purified. Therefore, in this article the sequences of these two additional subunits are also reported (Fig 3). The sequences for apcA, apcB, apcD and apcF genes (corresponding to α, β, αII and β18 subunits, respectively) are shown in S5 File. Fig 3 shows the amino acid sequence of the α and β subunits corresponding to the apcA and apcB genes, respectively, and comparison with sequences of their homologous subunits αII and β18 (genes apcD and apcF, respectively). The sequence of the PB domain and the LC used for this work are also displayed. These sequences were confirmed by transcriptomics analysis [26].
Fig 3. Sequence comparison between α and αII (ApcA and ApcD), β and β18 (Apcb and ApcF) and α and the PB_domain of the LCM (ApcA and ApcE).
Chromophore binding region are enclosed in green rectangles, and the conserved residues are highlighted in green. The cysteine residues that bind the chromophores are shown in red background. The PB-loop sequence is enclosed in blue lines.
The analysis of the sequences showed that α and αII subunits have 161 residues. They shared 41% identity and the position of the cysteines is conserved (Cys81) in both molecules. β and β18 subunits have 161 and 169 residues, respectively. They share 43% identity and the position of cysteines are also conserved (81 and 82, respectively). The sequence of α subunit and the fragment of the linker core-membrane used for modeling (PB domain) shared 22% identity (Fig 3), with an insertion of 55 amino acids in the PB domain. Cys81 in α subunit is replaced by Ser153 in the sequence alignment, but the chromophore is bound to Cys191 in the sequence of PB domain according to the reported recombinant structure reported (PDB ID: 4XXI) [32].
The X-ray structure of Allophycocyanin form G. chilensis showed a high structural conservation
The purified APC was used for crystallization experiments. The statistics and the refinement data are shown in Table 1. The coordinates have been deposited at the Protein Data Bank under the accession code 5TJF.
Table 1. Information on the data collection and refinement for the determination of the three dimensional structure of Allophycocyanin from Gracilaria chilensis.
| Data collection | Allophycocyanin |
|---|---|
| Space Group | P321 |
| Cell Dimensions | |
| a, b, c (Å) | 97.41, 97.41, 63.83 |
| α, β, γ (°) | 90, 90, 120 |
| Resolution range (Å) | 42.18–2.3 (2.38–2.30) |
| Rmerge | 0.1303 (0.3568) |
| I / σI | 6.18 (2.16) |
| Completeness (%) | 97 (97) |
| Redundancy | 2.9(2.6) |
| Refinement | |
| Resolution (Å) | 2.3 |
| Nº reflections/Nº reflections used | 84722(7757)/29317(2937) |
| Rwork / Rfree | 25.70/28.65 |
| Nº atoms | 3035 |
| Protein | 2422 |
| Ligand/ion | 135 |
| Water | 478 |
| B-factors | 31.75 |
| Protein | 32.01 |
| Ligand | 29.65 |
| Water | 31.04 |
| r.m.s. deviation | |
| Bond lengths (Å) | 0.004 |
| Bond angles (°) | 1.56 |
The values in parenthesis correspond to the last resolution shell.
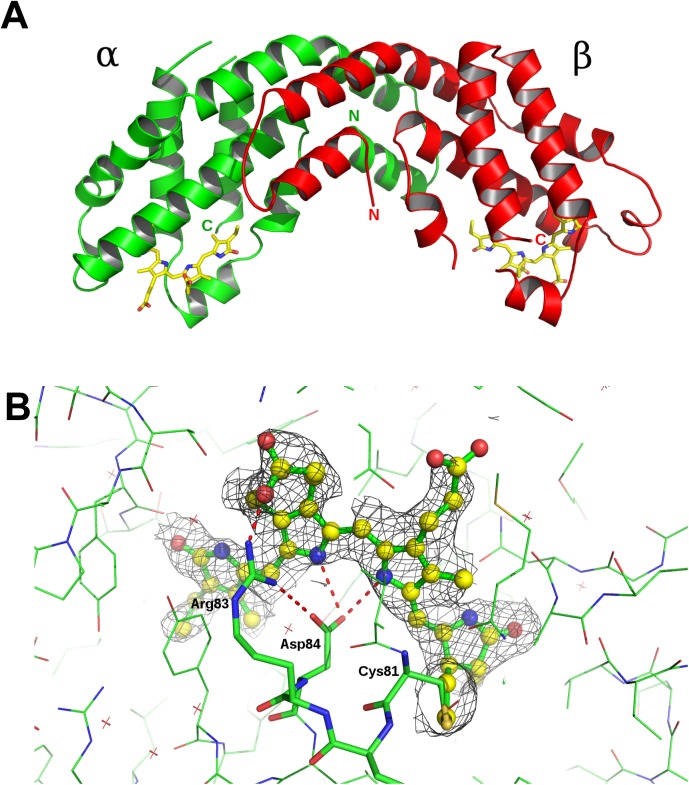
The crystal structure of Allophycocyanin from G. chilensis was determined in the space group P321 with one heterodimer αβ per asymmetric unit using molecular replacement. The final structure was refined to 2.3Å resolution with a final R-work and R-free of 25.7 and 28.69 respectively (Table 1). The resolution allowed the chromophores to be clearly defined in the electron density map (|2Fo-Fc|) (Fig 4B).
Fig 4. Crystallographic structure of Allophycocyanin from Gracilaria chilensis.
A) Ribbon representation of the asymmetric unit, the heterodimer. B) A section of the |2F-Fo| omit electron density map is shown for phycocyanobilin in α subunit. The residues interacting with the chromophore are also shown.
The structure of the (αβ heterodimer is shown in Fig 4A. Indeed, α and β subunits contained one phycocyanobilin bound to cysteine 81 and are stabilized by Asn71, Arg83 and Asp84 in α subunit, and by methyl-Asn71 (Asm71) and Asp84 in β subunit. The biological unit is a trimer of heterodimers as it was determined by size exclusion chromatography. The structure reported in this article is very similar to the structures of APC previously reported, 1B33 that contains the Lc [14] and 1KN1 from the rhodophyta Porphyra yezoensis [27].
The environment of the chromophore phycocyanobilin is highly conserved among different subunits
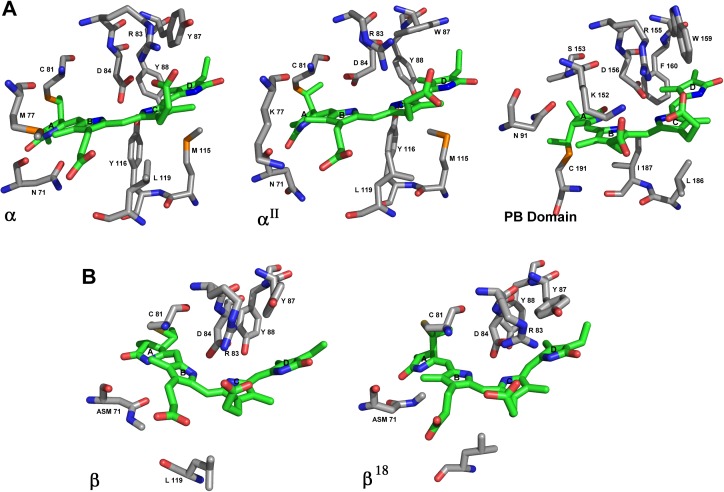
Subunits αII, β18 and PB domain were modeled and analyzed. A comparison among subunits in APC of M. laminosus [14], P. yezoensis [27], G. chilensis (PDB IDs 1B33, 1KN1, and 5TJF (this paper), respectively) and the αII model showed that the residues interacting with the phycocyanobilin chromophore are highly conserved (S1 Table). There is also high degree of conservation for the conformation of the chromophore (ASA) [36, 37], which is confirmed by the distances between rings A and D in the phycocyanobilin (Fig 5A and S1 Table). Instead the PB domain presents a SSA conformation that could account for the different spectroscopic characteristics of the LCM. A comparison between the β and β18 subunit is also shown in Fig 5B. In general, there are not important differences between β and β18 subunits and between α and αII subunits as it was expected by the sequence identity.
Fig 5. Binding sites of phycocynobilin.
A) Sticks representation of binding site of phycocyanobilin in α subunit of 5TJF (this paper), αII and the PB domain in APC_3. The chromophores are represented in green. B) Sticks representation of β subunit in 5TJF and β18 in APC_3. The chromophores are also in green.
The same comparison was performed between α subunit of 5TJF, the structure reported for the PB domain from Nostoc sp. (PDB ID: 4XXI) [32], and the model for the PB domain in Gracilaria chilensis. The PB domain can replace an α subunit in a trimer of heterodimers. Nevertheless, there are obvious differences, such as the insertion of 55 residues between residues 81–136 in the PB domain; 58% of this sequence was recognized as intrinsically disordered by Disprot [38] and also contains 12 Ser, 4 Thr and 1 Tyr, all hydroxylated residues. Four of these residues (Ser98, Ser99, Thr101 and Thr102) were recognized as phosphorylation sites by PROSITE and one of them (Thr101) in a motif recognized by protein kinase C. In prokaryote, this peptide segment has been described as an interaction region for the Orange Carotenoid Protein (OCP), which has not been reported in eukaryotes yet, and also a site for regulation by phosphorylation [39], fact that has not been tested in eukaryotic red algae for potential significance.
Attention is drown to the fact that the LCM and its chromophore have been proposed to be responsible for the final transfer of energy towards chlorophylls in Photosystems. In this regard, it was important to analyze the differences in the binding site of phycocyanobilins in α subunit, the modeled PB domain and the structure reported at the PDB ID: 4XXI [32]. The position of Cys81 in the alignment (Fig 3), which binds the PCB in all the other α subunits, is occupied by Ser153 in the PB domain. The Cys191 is the residue that binds to the phycocyanobilin. In our model, the chromophore was stabilized by Asp156, Arg155, Trp159, and Phe160 (S1 Table). Besides the difference in the cysteine residue, it is possible to see minor differences in the stabilization of the ligand (Fig 5A and S1 Table). Considering the binding site and the conformation of the chromophore, the phycocyanobilin in the PB domain has a SSA conformation instead of ASA for the chromophore in theα subunit [36, 37]. This conformation is the same in the crystal structure of PB domain from Nostoc sp. (PDB ID: 4XXI) [32]. The absorption spectrum, calculated for phycocyanobilin in the model of the PB domain of Gch, using Gaussian 09[40] with a ZINDO-MN semiempirical approach [40], gave the λmaxAbs = 661nm, which agreed with the maximum absorption of the PB domain reported for Nostoc sp. Other relevant difference is that Cys 81 of α subunit binds from the top of PCB, while Cys 165 binds from the bottom in the PB domain (See Fig 5). This difference may be related to the fact that PCB needs a specific cysteine lyase to be bound to Cys 81 in α subunit. However, it has been reported that for LCM, the process of binding the chromophore would be autocatalytic [15]. It has been also reported a mechanism for the addition of PCB to Cys 155 in Phycocyanin in cyanobacteria. It involves the presence of an aspartic acid participating in the formation of an acylimmonium cation on ring A of the phycocyanobilin. After that, the addition reaction to cysteine is produced with the assistance of a tyrosine residue. The authors also report that after the addition, a conformational change is produced in the chromophore. The PB domain model from Gch locates also a tyrosine and an aspartic acid in the vicinity of the phycocyanobilin; however the position of the functional groups seems to be located too far in the binding state. It is possible that before the reaction to occur and the positioning of the chromophores, these residues could be part of the catalysis, and after the binding their location changes to the final position in the molecule.
The crystal structure of Allophycocyanin allowed the building of the different type of trimers present in the core
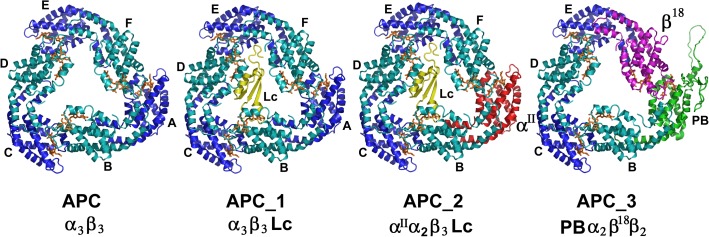
Fig 6 shows the models created for APC, APC_1, APC_2 and APC_3 using the structure of APC reported here, with the corresponding substitution of subunits as is shown.
Fig 6. Structural models proposed for each trimer of Allophycocyanin.
A) Schematic representation of the different composition trimers. B) Structure of APC, APC_1, APC_2, and APC_3.
Each trimer was analyzed considering the number and type of interactions among subunits, the interaction surfaces on each trimer as a whole and considering the substitutions. (The data is shown in S2 Table). A comparison between trimers as follows:
APC/APC_1: The presence of the Lc (Chain G) did not affect the interaction surface (aprox.1450 Å2) between subunits, yet the number of hydrogen bonds between heterodimers decreases. The Lc interacted mainly with the β (chains B, D and F) subunits by hydrogen bonding.
APC_1/APC_2: Both trimers contained the Lc, but APC_2 has the substitution of α by αII (Chain A). The interactions of Lc in both oligomers were very similar with β chains, especially with chain D.
APC/APC_3: The substitution of chain A (α) by PB domain and chain F (β) by β18 produced a trimer that contained these two special subunits in neighbor heterodimers. The αβ heterodimers conserved the interactions (H-bonds/salt bridges), but less salt bridges were observed in heterodimers with the substitutions. The interaction surfaces in both trimers are similar (aprox 1450Å2), except for the lower value measured for the interaction between subunits F and E (1154 Å2)
The structural analysis of the models proposed in this article allowed to arrange them face to face to form hexamers, which interact back to back with other hexamers to organize the cylinders present in the core as it was proposed for Synechococcus [41].
The experimental data available so far, for the subunit composition of trimers proposed in this article, indicated that LCM (PB_domain) and β18 were not in the same heterotrimer with αII [41]. According to spectroscopic data obtained by Kuzminov et al [42], β18 should transfer energy to the LCM, a fact that could be accomplished only if they are in the same trimer but neighboring heterodimers. Finally, they propose that β18 is a mediator in the energy transfer towards the final emitter, with the chromophore present in the LCM (PB_domain).
The modeling of the 55 extra residues (PB_loop) and the analysis of the sequence (Fig 3) indicated a very disordered region that is probably interacting with other proteins. Cross-linking experiments involving cyanobacterial phycobilisomes [43] do not provide information on the possible interactions of this segment of PB domain.
The model for the Linker core fits very well the space in the center of APC; clearly, this small molecule contributes to give asymmetry to allophycocyanin. It has been reported that the LC modifies the spectroscopic properties of the PCB in APC_1 and APC_2. According to our results and those from Reuter et al [14], in the trimeric state, one explanation is the different environment of the chromophore upon binding of LC.
Future perspectives
Our work on the structure of the phycobilisome components of an eukaryotic algae such as Gracilaria chilensis, including the study of the components of its core in this article, pretends to fill a gap produced by the enormous amount of information published in prokaryotes, especially Synechocystis and the small amount of information produced about this system in eukaryotic organisms. From the beginning, the purpose of our study on Phycobilisomes was to understand how the structure of this super complex supported the high efficiency of the energy transfer towards the photosystems. In this article, we are reporting new pieces of the big puzzle.
Supporting information
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
Interface area, numbers of hydrogen bonds and salt bridges between subunits in different trimers.
(DOCX)
Acknowledgments
We would like to thanks to the Soleil Synchrotron and its beamline Proxima 1 for the x-ray data collection. Also, we would like to thanks to the Robotic Unit at Universidad de Chile for the initial screening of crystallization conditions.
Data Availability
All protein structure files are available from the PDB database (accession code 5TJF).
Funding Statement
This work was supported by FONDECYT 113.0256 grant to MB, FONDECYT Doctoral grant 21120260 to JDL, CONICYT scholarship to JDL and CR_Lab Biofísica Molecular 2015-2016. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Glazer AN. Light guides. Directional energy transfer in a photosynthetic antenna. J Biol Chem. 1989;264: 1–4. [PubMed] [Google Scholar]
- 2.MacColl R. Cyanobacterial Phycobilisomes. J Struct Biol. 1998;124: 311–334. doi: 10.1006/jsbi.1998.4062 [DOI] [PubMed] [Google Scholar]
- 3.Adir N. Elucidation of the molecular structures of components of the phycobilisome: reconstructing a giant. Photosynth Res. 2005;85: 15–32. doi: 10.1007/s11120-004-2143-y [DOI] [PubMed] [Google Scholar]
- 4.Sun L, Wang S, Zhao M, Fu X, Gong X, Chen M, et al. Phycobilisomes from Cyanobacteria. Handbook on Cyanobacteria: Biochemistry, Biotechnology and Applications. Nova Science Publishers; 2009. pp. 105–160. [Google Scholar]
- 5.Sidler WA. Phycobilisome and Phycobiliprotein Structures In: Bryant DA, editor. The Molecular Biology of Cyanobacteria. Springer Netherlands; 1994. pp. 139–216. [Google Scholar]
- 6.MacColl R. Allophycocyanin and energy transfer. Biochim Biophys Acta BBA—Bioenerg. 2004;1657: 73–81. [DOI] [PubMed] [Google Scholar]
- 7.Arteni AA, Ajlani G, Boekema EJ. Structural organisation of phycobilisomes from Synechocystis sp. strain PCC6803 and their interaction with the membrane. Biochim Biophys Acta BBA—Bioenerg. 2009;1787: 272–279. [DOI] [PubMed] [Google Scholar]
- 8.Hagopian JC, Reis M, Kitajima JP, Bhattacharya D, Oliveira MC de. Comparative Analysis of the Complete Plastid Genome Sequence of the Red Alga Gracilaria tenuistipitata var. liui Provides Insights into the Evolution of Rhodoplasts and Their Relationship to Other Plastids. J Mol Evol. 2004;59: 464–477. doi: 10.1007/s00239-004-2638-3 [DOI] [PubMed] [Google Scholar]
- 9.Lundell DJ, Glazer AN. Molecular architecture of a light-harvesting antenna. Core substructure in Synechococcus 6301 phycobilisomes: two new allophycocyanin and allophycocyanin B complexes. J Biol Chem. 1983;258: 902–908. [PubMed] [Google Scholar]
- 10.Maxson P, Sauer K, Zhou J, Bryant DA, Glazer AN. Spectroscopic studies of cyanobacterial phycobilisomes lacking core polypeptides. Biochim Biophys Acta BBA—Bioenerg. 1989;977: 40–51. [DOI] [PubMed] [Google Scholar]
- 11.Houmard J, Capuano V, Colombano MV, Coursin T, Marsac NT de. Molecular characterization of the terminal energy acceptor of cyanobacterial phycobilisomes. Proc Natl Acad Sci. 1990;87: 2152–2156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gao X, Wei T-D, Zhang N, Xie B-B, Su H-N, Zhang X-Y, et al. Molecular insights into the terminal energy acceptor in cyanobacterial phycobilisome. Mol Microbiol. 2012;85: 907–915. doi: 10.1111/j.1365-2958.2012.08152.x [DOI] [PubMed] [Google Scholar]
- 13.Mullineaux CW. Phycobilisome-reaction centre interaction in cyanobacteria. Photosynth Res. 2008;95: 175 doi: 10.1007/s11120-007-9249-y [DOI] [PubMed] [Google Scholar]
- 14.Reuter W, Wiegand G, Huber R, Than ME. Structural analysis at 2.2 Å of orthorhombic crystals presents the asymmetry of the allophycocyanin–linker complex, AP LC7. 8, from phycobilisomes of Mastigocladus laminosus. Proc Natl Acad Sci. 1999;96: 1363–1368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhao K-H, Su P, Böhm S, Song B, Zhou M, Bubenzer C, et al. Reconstitution of phycobilisome core–membrane linker, LCM, by autocatalytic chromophore binding to ApcE. Biochim Biophys Acta BBA—Bioenerg. 2005;1706: 81–87. [DOI] [PubMed] [Google Scholar]
- 16.Adir N, Dines M, Klartag M, McGregor A, Melamed-Frank M. Assembly and Disassembly of Phycobilisomes In: Shively JM, editor. Complex Intracellular Structures in Prokaryotes. Springer Berlin Heidelberg; 2006. pp. 47–77. [Google Scholar]
- 17.Jallet D, Gwizdala M, Kirilovsky D. ApcD, ApcF and ApcE are not required for the Orange Carotenoid Protein related phycobilisome fluorescence quenching in the cyanobacterium Synechocystis PCC 6803. Biochim Biophys Acta BBA—Bioenerg. 2012;1817: 1418–1427. [DOI] [PubMed] [Google Scholar]
- 18.Capuano V, Braux AS, Marsac NT de, Houmard J. The “anchor polypeptide” of cyanobacterial phycobilisomes. Molecular characterization of the Synechococcus sp. PCC 6301 apce gene. J Biol Chem. 1991;266: 7239–7247. [PubMed] [Google Scholar]
- 19.Watanabe M, Ikeuchi M. Phycobilisome: architecture of a light-harvesting supercomplex. Photosynth Res. 2013;116: 265–276. doi: 10.1007/s11120-013-9905-3 [DOI] [PubMed] [Google Scholar]
- 20.Bird CJ, McLachlan J, Oliveira EC de. Gracilaria chilensis sp.nov. (Rhodophyta, Gigartinales), from Pacific South America. Can J Bot. 1986;64: 2928–2934. [Google Scholar]
- 21.Contreras-Martel C, Matamala A, Bruna C, Poo-Caamaño G, Almonacid D, Figueroa M, et al. The structure at 2 Å resolution of Phycocyanin from Gracilaria chilensis and the energy transfer network in a PC–PC complex. Biophys Chem. 2007;125: 388–396. doi: 10.1016/j.bpc.2006.09.014 [DOI] [PubMed] [Google Scholar]
- 22.Contreras-Martel C, Martinez-Oyanedel J, Bunster M, Legrand P, Piras C, Vernede X, et al. Crystallization and 2.2 Å resolution structure of R-phycoerythrin from Gracilaria chilensis: a case of perfect hemihedral twinning. Acta Crystallogr D Biol Crystallogr. 2001;57: 52–60. [DOI] [PubMed] [Google Scholar]
- 23.Figueroa M, Martínez-Oyanedel J, Matamala AR, Dagnino-Leone J, Mella C, Fritz R, et al. In silico model of an antenna of a phycobilisome and energy transfer rates determination by theoretical Förster approach. Protein Sci. 2012;21: 1921–1928. doi: 10.1002/pro.2176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Collén J, Porcel B, Carré W, Ball SG, Chaparro C, Tonon T, et al. Genome structure and metabolic features in the red seaweed Chondrus crispus shed light on evolution of the Archaeplastida. Proc Natl Acad Sci. 2013;110: 5247–5252. doi: 10.1073/pnas.1221259110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Guan X, Qin S, Zhao F, Zhang X, Tang X, others. Phycobilisomes linker family in cyanobacterial genomes: divergence and evolution. Int J Biol Sci. 2007;3: 434–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vorphal MA, Gallardo-Escárate C, Valenzuela-Muñoz V, Dagnino-Leone J, Vásquez JA, Martínez-Oyanedel J, et al. De novo transcriptome analysis of the red seaweed Gracilaria chilensis and identification of linkers associated with phycobilisomes. Mar Genomics. 2017;31: 17–19. doi: 10.1016/j.margen.2016.11.001 [DOI] [PubMed] [Google Scholar]
- 27.Liu J-Y, Jiang T, Zhang J-P, Liang D-C. Crystal Structure of Allophycocyanin from Red AlgaePorphyra yezoensis at 2.2-Å Resolution. J Biol Chem. 1999;274: 16945–16952. [DOI] [PubMed] [Google Scholar]
- 28.Adams PD, Afonine PV, Bunkóczi G, Chen VB, Davis IW, Echols N, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66: 213–221. doi: 10.1107/S0907444909052925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60: 2126–2132. doi: 10.1107/S0907444904019158 [DOI] [PubMed] [Google Scholar]
- 30.Afonine PV, Grosse-Kunstleve RW, Echols N, Headd JJ, Moriarty NW, Mustyakimov M, et al. Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallogr D Biol Crystallogr. 2012;68: 352–367. doi: 10.1107/S0907444912001308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Webb B, Sali A. Comparative Protein Structure Modeling Using MODELLER. Current Protocols in Bioinformatics. John Wiley & Sons, Inc.; 2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tang K, Ding W-L, Höppner A, Zhao C, Zhang L, Hontani Y, et al. The terminal phycobilisome emitter, LCM: A light-harvesting pigment with a phytochrome chromophore. Proc Natl Acad Sci. 2015;112: 15880–15885. doi: 10.1073/pnas.1519177113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Comeau SR, Gatchell DW, Vajda S, Camacho CJ. ClusPro: a fully automated algorithm for protein–protein docking. Nucleic Acids Res. 2004;32: W96–W99. doi: 10.1093/nar/gkh354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wiederstein M, Sippl MJ. ProSA-web: interactive web service for the recognition of errors in three-dimensional structures of proteins. Nucleic Acids Res. 2007;35: W407–W410. doi: 10.1093/nar/gkm290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lovell SC, Davis IW, Arendall WB, de Bakker PIW, Word JM, Prisant MG, et al. Structure validation by Cα geometry: ϕ,ψ and Cβ deviation. Proteins Struct Funct Bioinforma. 2003;50: 437–450. [DOI] [PubMed] [Google Scholar]
- 36.Tu P, Yao Y, Li Y, Liu B. Conformational flexibility of phycocyanobilin: Monte-Carlo and DFT study. J Mol Struct THEOCHEM. 2009;894: 9–13. [Google Scholar]
- 37.Göller AH, Strehlow D, Hermann G. Conformational Flexibility of Phycocyanobilin: An AM1 Semiempirical Study. ChemPhysChem. 2001;2: 665–671. doi: 10.1002/1439-7641(20011119)2:11<665::AID-CPHC665>3.0.CO;2-O [DOI] [PubMed] [Google Scholar]
- 38.Piovesan D, Tabaro F, Mičetić I, Necci M, Quaglia F, Oldfield CJ, et al. DisProt 7.0: a major update of the database of disordered proteins. Nucleic Acids Res. 2017;45: D1123–D1124. doi: 10.1093/nar/gkw1279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Harris D, Tal O, Jallet D, Wilson A, Kirilovsky D, Adir N. Orange carotenoid protein burrows into the phycobilisome to provide photoprotection. Proc Natl Acad Sci. 2016;113: E1655–E1662. doi: 10.1073/pnas.1523680113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Frisch MJ, Trucks GW, Schlegel HB, Scuseria GE, Robb MA, Cheeseman JR, et al. Gaussian 09 Revision A.1.
- 41.Tal O, Trabelcy B, Gerchman Y, Adir N. Investigation of Phycobilisome Subunit Interaction Interfaces by Coupled Cross-linking and Mass Spectrometry. J Biol Chem. 2014; jbc.M114.595942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kuzminov FI, Bolychevtseva YV, Elanskaya IV, Karapetyan NV. Effect of APCD and APCF subunits depletion on phycobilisome fluorescence of the cyanobacterium Synechocystis PCC 6803. J Photochem Photobiol B. 2014;133: 153–160. doi: 10.1016/j.jphotobiol.2014.03.012 [DOI] [PubMed] [Google Scholar]
- 43.Chang L, Liu X, Li Y, Liu C-C, Yang F, Zhao J, et al. Structural organization of an intact phycobilisome and its association with photosystem II. Cell Res. 2015;25: 726–737. doi: 10.1038/cr.2015.59 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
Interface area, numbers of hydrogen bonds and salt bridges between subunits in different trimers.
(DOCX)
Data Availability Statement
All protein structure files are available from the PDB database (accession code 5TJF).