Abstract
Aims
An intriguing difference between vertebrate skeletal and cardiac muscles is that the lengths of the thin filaments are constant in the former but variable in the latter. The thick filaments have constant lengths in both types of muscles. The contractile behaviour of a muscle is affected by the lengths of both types of filaments as the tension generated during contraction depends on the amount of filament overlap. To understand the behaviour of cardiac muscle, it is important to know the distribution of the thin filament lengths. The previous detailed analysis by Robinson and Winegrad used serial transverse sections to determine the lengths of the thin filaments. However, the precision, set by the 100 nm section thickness, was low. Here, we have used electron tomography to produce 3D images of rat and mouse cardiac muscles in which we can actually see individual thin filaments up to the free ends and see that these free ends have variable locations. For comparison, we also measure the thin filament lengths in skeletal muscle (frog sartorius).
Methods and results
Cardiac papillary muscles were obtained from a rat (Sprague–Dawley) and a mouse (C57/B6). Skeletal muscle (sartorius) was obtained from a frog (Rana pipiens). Longitudinal sections (100 nm thick) were used to produce tilt series and tomograms from which the thin filament paths were traced. Cardiac papillary muscle thin filaments in rat and mouse range from 0.94 to 1.10 µm, with a mean length of 1.04 µm and standard deviation of 0.03 µm. For frog sartorius muscle, the thin filament length was 0.94 µm with standard deviation of 0.01 µm.
Conclusion
Electron tomography of cardiac and skeletal muscles allows direct visualization and high precision measurement of the lengths of thin filaments.
Keywords: Electron microscopy, Contractile function, Contractile apparatus, Myocytes
1. Introduction
In striated muscles, the thick filaments and the A-band have remarkably constant structure.1–4 The thin filaments have constant lengths in skeletal muscle but not in cardiac muscle.4–6 The tension generated during muscular contraction varies as the overlap of the thick and thin filaments7 and hence knowledge of the thin filament lengths and their variability is very important.4–10 Two previous studies have measured thin filament lengths in cardiac muscle by very different methods. Robinson and Winegrad6 used serial transverse sections spanning from the Z-disc to the M-band to measure the thin filament lengths from the number of transverse sections spanned. The precision of the length measurement in this method is low as it is determined by the section thickness ∼100 nm. In this important paper, Robinson and Winegrad showed that thin filaments from relatively compliant atria have a large range of lengths, 0.5 µm, compared with a narrow range, 0.2 µm, in thin filaments from less compliant ventricles. Littlefield and Fowler11 used fluorescence microscopy combined with deconvolution analysis to measure the thin filament lengths. Their method has higher precision, 9 nm, for skeletal muscle compared with 50 nm for cardiac muscle. The obvious method for measuring filament lengths with high precision is by electron microscopy of longitudinal sections. However, direct viewing of electron micrographs has ambiguity of filament superimposition through the section depth, and ambiguity whether the filament end is true or has obliquely exited the section. Ideally, we should measure the lengths from a 3D image where the path of every single thin filament can be seen right up to the filament terminus. This is exactly what electron tomography can provide.12,13 Here, we have produced tomograms of longitudinal sections in mammalian cardiac muscle to view a large region centred at the M-band. We mapped all the thick and thin filaments present within a selected sarcomere region paying special attention to the terminal points (pointed ends) of the thin filaments. Following measurement of the lengths of the sarcomeres from which each tomogram was produced, we determined the lengths of all the thin filaments to within 10 nm. For comparison, the same measurements were also carried out for skeletal muscle in frog.
2. Methods
2.1. Sample preparation
Rat (Sprague–Dawley) papillary muscle was dissected under Krebs with 30 mM BDM, pinned on a Petri dish with a Sylgard gel base, aerated with 95% oxygen/5% carbon dioxide. Then the Krebs/BDM was replaced with Krebs and the aeration continued for half an hour. The muscle was fixed in 3% glutaraldehyde for 1 h, rinsed in Krebs, fixed in aqueous 1% osmium tetroxide, dehydrated in an acetone series, and embedded in Araldite. The same method was used for mouse papillary muscle. A cardiac MyBP-C knockout mouse (C57/B6) was used; we expect the thin filaments to be normal in this muscle.14 The investigation conforms to the Guide for the Care and Use of Laboratory Animals published by the US National Institutes of Health (NIH Publication No. 85-23, revised 1996). Frog sartorius muscle was fixed in 3% glutaraldehyde in situ by exposing muscle on the dissected limb and then processed as above. For each sample, ∼100 nm thin sections were cut with a Reichert Ultracut-E and stained with 2% uranyl acetate and Reynolds lead citrate. At this thickness, there are ∼2.5 unit cells of the ∼40 nm myosin filament lattice within the depth of the section.
2.2. Electron microscopy and tilt series
The electron microscopy and tilt series recording of rat papillary and frog sartorius muscle was done with a JEOL 1200 EX electron microscope. To record the tilt series, the sample was tilted manually from −60° to +60° in 2° steps and images recorded at ×15 000 magnification on a Teitz Fastscan CCD camera with 1024 × 1024 pixels. A tilt/rotate holder was used to obtain dual-axes tilt series. The defocus was set for the first minimum in the Fourier transform to fall at 1/5 nm−1. Hence, we estimate that our filament length measurements have precision better than 10 nm. For all the single-axis tilt series in this study, the tilt axis was along the filaments. For the mouse tomography, the tilt series were recorded using a Philips CM300 FEG electron microscope operated at 300 kV at Florida State University in Tallahassee, FL, and recorded at ×15 000 or ×20 000 magnification on to a Teitz TemCam-F224HD 2k × 2k CCD camera.
2.3. Tomography
Single-axis and dual-axes tilt series for rat papillary and frog sartorius muscle were aligned and back-projected using Imod.15 The mouse tilt series were aligned and back-projected using Protomo software.16 The tracing and modelling of the actin and myosin filaments were done using Imod and the Slicer option which allows tilting of the tomogram to move sets of filaments into the plane of view.
2.4. Measurement of thin filament length
To measure the thin filament lengths, we found it useful to first trace the paths of the thin and thick filaments using the modelling option in Imod. During the modelling, each thin filament was assigned a number which was used to track the filament for subsequent measurements. A 2D image of the model rotated to make the filaments horizontal and where most of the thin filaments ends were visible was exported to ImageJ (http://rsb.info.nih.gov/ij/). The x-coordinate was recorded for each filament. Each filament could be identified and its number determined by the shape of its path or by the path of the surrounding thick or thin filaments. The x-coordinate of the M-band was found from the profile plot of the tomogram sections or projections of a subset. Calibration of the electron microscope images and the tomogram is a vital step for this study, as chemical processing for electron microscopy as well as the electron radiation during the electron microscopy causes shrinkage of the sample by varying amounts.17,18 We used the myosin crossbridge repeat of 43 nm19 to provide internal calibration; this was done from the Fourier transform of selected regions using ImageJ. If the crossbridge repeat is not clear in the Fourier transform, then the width of the A-band, 1.63 µm, can be used for internal calibration.1 The sarcomere lengths for the regions used were measured from the CCD images or by recording at ×20 000 using electron microscope film (Kodak S0-163).
3. Results
We show a schematic view of a striated muscle sarcomere in Figure 1. Although the actin filaments span the width of the Z-disc,20 here we measure the lengths of the thin filaments from the centre of the Z-disc to the free (pointed) ends of the thin filaments (arrowed). The H-zone is the region in the middle of the A-band devoid of actin filaments. It is only clearly visible in electron micrographs of skeletal muscle where the thin filaments have constant lengths. Figure 2 shows snapshots of the tomograms and models produced for rat and mouse cardiac and frog skeletal muscle. The tomograms for this study were derived from ∼100 nm thick sections and hence scanning through the depth of the 3D reconstruction, we could clearly identify at least two layers of the myosin filament lattice. The tomograms were filtered using non-linear anisotropic diffusion methods to improve the visualization of fine structure.21 Distribution histograms of the thin filament length measurements are shown in Figure 3 and the overall results are summarized in Table 1.
Figure 1.
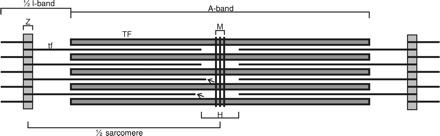
Components of the muscle sarcomere. The A-band comprises thick filaments (TF) and the I-band comprises thin filaments (tf). The Z-disc (Z-line, Z-band, and label Z) forms the sarcomere boundary and is at the centre of the I-band. Actin filaments are tethered at the Z-disc in a square lattice. Actin filaments traverse the full width of the Z-disc,20 but in this study, we measure the length to the centre of the Z-disc. The thin filament lengths from the tomogram were calculated from the half sarcomere length and the separation of the actin filament terminal points (arrows) from the centre of the M-band (M). The H-zone (H) is the central A-band region not overlapped by thin filaments and is typically clearly seen in electron micrographs of skeletal muscle.
Figure 2.
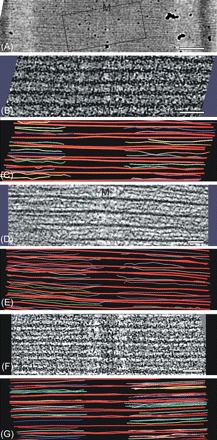
Electron tomography to measure thin filament lengths in rat cardiac, mouse cardiac, and frog skeletal muscle. (A) Electron micrograph of a thin section of rat cardiac (papillary) muscle showing the approximate region used to produce one of the rat muscle tomograms in this study. Using the crossbridge 43 nm periodicity for internal calibration, the sarcomere length was 2.27 µm. M marks the location of the M-band in this figure. (B) An example 2D slice from one of the tomograms of rat papillary muscle. Using Imod, the thick filaments were traced in red and the thin filaments were traced in various colours. The left side and the right side provided two independent sets of data for thin filament measurements. For the filament tracing, the Imod slicer option was used to tilt the tomogram so that particular sets of thin filaments were in the plane of the view. (C) A model produced with Imod of the thin and thick filaments contained in the tomogram for (B). (D) An example 2D slice from the tomogram of mouse papillary muscle. Here, thick and thin filaments are clearer than for the rat tomogram in (B). (E) IMOD model produced from the tomogram of mouse papillary muscle in (D) showing the thick filaments (red) and thin filaments (various colours). (F) An example 2D slice from the tomogram of frog sartorius muscle. (G) Imod model for frog showing thick (red) and thin (various colours) filaments. Scale bars: (A) 0.25 µm, (B, D, F) 100 nm.
Figure 3.
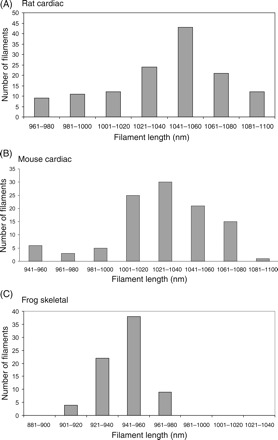
Distribution of thin filament lengths in (A) rat cardiac papillary, (B) mouse cardiac papillary, and (C) frog skeletal sartorius muscles. Rat and mouse thin filaments have a broad distribution of lengths with a mean of 1.04 and 1.03 µm, respectively, and standard deviation of ∼30 nm. In comparison with cardiac thin filaments, the distribution for skeletal muscle thin filaments (C) is very sharp, and within the experimental error, the thin filaments have uniform lengths of 0.94 nm.
Table 1.
Summary of thin filament lengths
| Type of muscle | Filament range (µm) | Filament length (µm) |
|---|---|---|
| Rat cardiac papillary | 0.92–1.10 | 1.04 ± 0.03 (131 filaments) |
| Mouse cardiac papillary | 0.94–1.08 | 1.03 ± 0.03 (103 filaments) |
| Frog skeletal sartorius | 0.90–0.97 | 0.94 ± 0.014 (73 filaments) |
3.1. Rat papillary muscle
Figure 2A shows the region from which one of the dual-tilt axes tilt series for rat papillary muscle was recorded. It is not necessary to produce a tomogram of a whole sarcomere or even of a complete half sarcomere. Instead a generously large ∼1 µm region centred at the M-band that contains all the free (pointed) ends of the thin filaments from both sarcomeres gives best value per tomogram, as we then only need the sarcomere length for the region used to measure the thin filament lengths. A single example slice from the tomogram is shown in Figure 2B. Although this slice is noisy, typical of electron tomograms, it shows thick filaments and fragments of thin filaments. They are seen more clearly if the image is viewed at a glancing angle from the left or right side of the page. Both sets of filaments were modelled by tracing in Imod using the Slicer option that tilts the tomogram to move sets of filaments into the plane of view. Additionally, the tomogram was frequently scanned in Z to observe the emergence and disappearance of filaments, specially noting the terminal points. Although it was not essential, we found it useful to trace the myosin filaments first as this simplified the subsequent tracing of the actin filaments. Part of the modelled tomogram is shown in Figure 2C, with thick red lines depicting the thick filaments and thin multi-coloured lines depicting the thin filaments. The whole model is shown as a movie in the Supplementary material online. In this tomogram, as in the ones from mouse cardiac and frog skeletal, thin filaments occasionally had wavy paths. Lack of straightness is often observed in tomograms.22 The paths of the thin filaments were drawn paying special attention to the terminus. The variable location of the thin filament ends shows directly that thin filaments have variable lengths in cardiac muscle. The results of the measurement are summarized in the graph in Figure 3A and in Table 1. The shape of the distribution is skewed to the smaller lengths. The thin filaments range in length from 0.96 to 1.10 µm and have a mean length of 1.04 µm with a standard deviation of 0.03 µm. We also measured the thin filaments along the wavy paths and this resulted in an increase by ∼10 nm. The overall results in Table 1 are for straight filaments.
3.2. Mouse papillary muscle
A single 2D slice from the tomogram of mouse papillary muscle is shown in Figure 2D. In this case, the favourable orientation of the fibre in the section cut by ultramicrotomy resulted in well-aligned filaments in the tomogram. We see clear thick filaments with two actin filaments between each pair. Part of the modelled region is shown in Figure 2E. The variable position of the thin filament ends is clearly seen. The results are summarized in the distribution histogram in Figure 3B. As in rat papillary, the shape of the distribution is skewed to the smaller lengths. The thin filament lengths are similar to rat papillary muscle; they range from 0.94 to 1.08 µm and have mean length of 1.03 µm with a standard deviation of 0.03 µm. We have also calculated tomograms and filament models for wild-type murine papillary muscle and similar values were obtained (data not shown).
3.3. Frog skeletal muscle
For comparison with cardiac muscle, skeletal muscle (frog sartorius) was also examined. A single slice of the frog tomogram is shown in Figure 2F. The model of the thick and thin filaments is shown in part in Figure 2G and in full in the Supplementary material online. We see directly from the tomogram and the distribution graphs in Figure 3C that the thin filaments lengths are sharply defined. We measured a mean length of 0.94 µm with a very small standard deviation of 14 nm.
4. Discussion
We have used electron tomography to measure the lengths of thin filaments in cardiac muscle with greater precision than was possible in previous studies.6 The 3D images produced here allow us to directly visualize a large region of thin filaments up to the free ends. In rat and mouse ventricular muscle, we find that the mean length of the thin filaments is 1.03 µm with standard deviation of 0.03 µm. Although the thin filament lengths are variable, the distribution graphs in Figure 3 have distinct peaks, showing that there must be some length maintaining mechanism in cardiac muscle in the absence of stoichiometric amounts of nebulin.23 Our comparison with skeletal muscle, frog sartorius, shows that the skeletal thin filaments have a length of 0.94 ± 0.01 µm, and hence have highly uniform lengths.
4.1. Preparation technique
We have used conventional processing methods to prepare our samples for electron microscopy (consisting of glutaraldehyde and osmium fixation, dehydration in acetone series, and embedding in epoxy resin). Similar methods were used in previous length measurement studies.4,6 By using electron tomography, we have been able to measure the lengths with greater precision than was possible before. We have also made some measurements on a frog skeletal muscle (sartorius) processed by rapid-freezing/freeze substitution protocols which resulted in overall superior preservation (data not shown). The thin filaments were slightly longer in the rapid frozen sample with mean length of 0.99 µm. Conventional processing may therefore not be able to preserve the thin filaments perfectly. Hence, it is possible that the measurements for rat and mouse may be longer in situ by ∼0.05 µm. An obvious improvement to the current study would be to study cardiac muscle by rapid freezing protocols.
4.2. Application of electron tomography to measure thin filament length
The attraction of tomography for this study is that we actually see the filaments and hence we can measure their lengths with precision better than 10 nm. We have used both single-axis and dual-axes tomograms, but since we are measuring an essentially linear object, single-axis tilt series with the tilt axis along the filament is sufficient for measurements of the thin filament lengths. For the tomography, our preferred size of sample in the electron microscope was ∼1 µm square centred at the M-band. This includes sufficient actin filament regions on each side and hence gave best value per tomogram. If one used a complete half sarcomere to follow the thin filaments along their entire length, the greater challenge would be tracing the paths through the I-band where the lattice transforms from the square lattice of the Z-band to the hexagonal lattice of the A-band. For simplicity, we have assumed a straight path from the Z-band to the thin filament terminus. Lack of straightness is often observed in electron tomograms22 and this may be the cause for the wavy path of the thin filaments that we observed in these tomograms. We estimate that if the measurement of the filament lengths is done along the contours, it adds ∼10 nm to the filament length within the tomogram region.
In the previous study by Robinson and Winegrad,5,6 the precision of the measurement of thin filament length was set by the section thickness of ∼100 nm. They found a range of 0.9–1.1 µm for rat papillary muscle, almost exactly what we have found (0.96–1.1 µm). The range was much larger for atrial muscle, 0.6–1.1 µm. They concluded that double overlap of the thin filaments at the M-band should exist over a large range of sarcomere lengths. In our tomograms, we only occasionally saw filaments going into the M-band region. Tomography would be excellent for measuring the lengths of thin filaments in atrial muscle and for examining the behaviour of thin filaments if and when they overlap within the M-band. Tomography would also resolve any confusion of thin filaments with M-band components like M-filaments.24
Littlefield and Fowler11 used distribution deconvolution analysis of fluorescence images to measure thin filament lengths with high precision in fast and slow skeletal fibres and cardiomyocytes. It is interesting that thin filaments in fast muscles from different species measured with high 10 nm precision by Littlefield and Fowler (chicken pectoralis major), by Sosa et al.1 (rabbit psoas), and by us (frog sartorius muscles) have nearly the same lengths: 1.003 ± 0.015; 1.11 ± 0.03; 0.99 ± 0.01 µm (fast-freezing) and 0.94 ± 0.01 µm (conventional processing), respectively.
4.3. Regulation of thin filament lengths
The length of actin filaments in muscle is regulated by gain or loss of monomers at the barbed end at the Z-disc or the pointed end (free end) in the A-band and by the interaction of capping proteins, CapZ at the Z-disc, and tropomodulin at the barbed end.25 The giant protein nebulin (500–800 kDa), composed of repeating domains of 35 residues, has a length of ∼1 µm and is found associated with thin filaments in stoichiometric amounts in skeletal muscle.26,27 There is now plenty of evidence that nebulin maintains the length of the thin filaments in skeletal muscle.23,27,28 Until recently, nebulin was not found in cardiac muscle (although the smaller related protein nebulette located at the Z-band was found), hence this was thought to be the cause of the variable thin filament lengths. However, nebulin has now been found in cardiac muscle, although in miniscule amounts.29 Conflicting data were obtained by Witt et al.23 when comparing tissue culture/RNAi knockdown approaches and data in vivo: knockouts for example suggested the assembly of shorter filaments in the absence of nebulin,23 whereas knockdowns reported a dramatic elongation in the presence of reduced nebulin levels.30 With such low amounts, nebulin may be shared between many thin filaments and Horowits31 has proposed a uniform scanning model in which a nebulin molecule tethered at the Z-disc may rotate about its length and hence interact with different thin filaments. Our results allow us to comment on this model. In Horowits’ model, the longest filament length occurs for the most number of filaments, giving a distribution of filament lengths quite different from the near bell-shaped distribution that we have observed here.
4.4. Length–tension relationship in cardiac muscle
The classic length–tension relationship for skeletal muscle under isometric contraction7 has its equivalence in cardiac muscle but which is dominated by the steepness of the ascending limb that arises due to the increasing calcium sensitivity with increasing sarcomere length (Frank-Starling Law) (for review see Allen and Kentish32). Weiwad et al.33 have used Fourier analysis to show the length–tension relationship for a single cardiac myocyte. At intermediate levels of Ca2+, they found a distinct peak separating the ascending and descending limbs at ∼2.4 µm sarcomere length. Comparison with skeletal muscle suggests that thin filaments in cardiac muscle may in fact be longer, ∼1.2 µm, and have more uniform lengths. Our results and those of previous workers5,11 show smaller, ∼1 µm, but variable lengths. Further research is therefore required to resolve this controversy between the measurements of thin filament lengths so far by electron microscopy and the lengths suggested by length–tension experiments.
In conclusion, electron tomography offers a new method to measure thin filament lengths in cardiac muscle with high precision. The distribution graphs that we have determined for cardiac muscle thin filaments give valuable tests for any model of filament length regulation.
Supplementary material
Supplementary material is available at Cardiovascular Research online.
Funding
This research was funded by the British Heart Foundation, Grants PG/06/010/20256 and FS/05/019.
Supplementary Material
Acknowledgements
We are most grateful to the following scientists: Kenneth Taylor for providing access to electron microscope facilities for the tomography of the mouse sample; Rick Moss for the provision of cardiac muscle of a cardiac MyBP-C-ko mouse; Cathy Timson for help with the sample preparations; and Jon Kentish, Steve Marston, and John Squire for help with the cardiac sample preparations and for valuable discussions.
Conflict of interest: none declared.
References
- 1.Sosa H, Popp D, Ouyang G, Huxley HE. Ultrastructure of skeletal muscle fibers studied by a plunge quick freezing method: myofilament lengths. Biophys J. 1994;67:283–292. doi: 10.1016/S0006-3495(94)80479-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Squire JM. Architecture and function in the muscle sarcomere. Curr Opin Struct Biol. 1997;7:247–257. doi: 10.1016/s0959-440x(97)80033-4. [DOI] [PubMed] [Google Scholar]
- 3.Luther PK, Squire JM, Forey PL. Evolution of myosin filament arrangements in vertebrate skeletal muscle. J Morphol. 1996;229:325–335. doi: 10.1002/(SICI)1097-4687(199609)229:3<325::AID-JMOR7>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- 4.Page SG, Huxley HE. Filament lengths in striated muscle. J Cell Biol. 1963;19:369–390. doi: 10.1083/jcb.19.2.369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Robinson TF, Winegrad S. Variation of thin filament length in heart muscles. Nature. 1977;267:74–75. doi: 10.1038/267074a0. [DOI] [PubMed] [Google Scholar]
- 6.Robinson TF, Winegrad S. The measurement and dynamic implications of thin filament lengths in heart muscle. J Physiol. 1979;286:607–619. doi: 10.1113/jphysiol.1979.sp012640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gordon AM, Huxley AF, Julian FJ. The variation in isometric tension with sarcomere length in vertebrate muscle fibres. J Physiol. 1966;184:170–192. doi: 10.1113/jphysiol.1966.sp007909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Huxley HE. Electron microscopic studies on the structure of natural and synthetic protein filaments from striated muscle. J Mol Biol. 1963;7:281–308. doi: 10.1016/s0022-2836(63)80008-x. [DOI] [PubMed] [Google Scholar]
- 9.Traeger L, Goldstein MA. Thin filaments are not of uniform length in rat skeletal muscle. J Cell Biol. 1983;96:100–103. doi: 10.1083/jcb.96.1.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Granzier HL, Akster HA, Ter Keurs HE. Effect of thin filament length on the force-sarcomere length relation of skeletal muscle. Am J Physiol. 1991;260:C1060–C1070. doi: 10.1152/ajpcell.1991.260.5.C1060. [DOI] [PubMed] [Google Scholar]
- 11.Littlefield R, Fowler VM. Measurement of thin filament lengths by distributed deconvolution analysis of fluorescence images. Biophys J. 2002;82:2548–2564. doi: 10.1016/S0006-3495(02)75598-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Frank J, editor. Electron Tomography: Methods for Three-dimensional Visualization of Structures in the Cell. 2nd ed. New York: Springer; 2006. [Google Scholar]
- 13.McIntosh R, Nicastro D, Mastronarde D. New views of cells in 3D: an introduction to electron tomography. Trends Cell Biol. 2005;15:43–51. doi: 10.1016/j.tcb.2004.11.009. [DOI] [PubMed] [Google Scholar]
- 14.Harris SP, Bartley CR, Hacker TA, McDonald KS, Douglas PS, Greaser ML, et al. Hypertrophic cardiomyopathy in cardiac myosin binding protein-C knockout mice. Circ Res. 2002;90:594–601. doi: 10.1161/01.res.0000012222.70819.64. [DOI] [PubMed] [Google Scholar]
- 15.Mastronarde DN. Dual-axis tomography: an approach with alignment methods that preserve resolution. J Struct Biol. 1997;120:343–352. doi: 10.1006/jsbi.1997.3919. [DOI] [PubMed] [Google Scholar]
- 16.Winkler H. 3D reconstruction and processing of volumetric data in cryo-electron tomography. J Struct Biol. 2007;157:126–137. doi: 10.1016/j.jsb.2006.07.014. [DOI] [PubMed] [Google Scholar]
- 17.Luther PK, Lawrence MC, Crowther RA. A method for monitoring the collapse of plastic sections as a function of electron dose. Ultramicroscopy. 1988;24:7–18. doi: 10.1016/0304-3991(88)90322-1. [DOI] [PubMed] [Google Scholar]
- 18.Luther PK. Sample shrinkage and radiation damage of plastic sections. In: Frank J, editor. Electron Tomography: Methods for Three-dimensional Visualization of Structure in the Cell. New York: Springer; 2006. pp. 17–40. [Google Scholar]
- 19.Huxley HE, Brown W. The low-angle x-ray diagram of vertebrate striated muscle and its behaviour during contraction and rigor. J Mol Biol. 1967;30:383–434. doi: 10.1016/s0022-2836(67)80046-9. [DOI] [PubMed] [Google Scholar]
- 20.Luther PK, Padron R, Ritter S, Craig R, Squire JM. Heterogeneity of Z-band structure within a single muscle sarcomere: implications for sarcomere assembly. J Mol Biol. 2003;332:161–169. doi: 10.1016/s0022-2836(03)00883-0. [DOI] [PubMed] [Google Scholar]
- 21.Frangakis AS, Hegerl R. Noise reduction in electron tomographic reconstructions using nonlinear anisotropic diffusion. J Struct Biol. 2001;135:239–250. doi: 10.1006/jsbi.2001.4406. [DOI] [PubMed] [Google Scholar]
- 22.Winkler H, Taylor KA. Three-dimensional distortion correction applied to tomographic reconstructions of sectioned crystals. Ultramicroscopy. 1996;63:125–132. doi: 10.1016/0304-3991(96)00024-1. [DOI] [PubMed] [Google Scholar]
- 23.Witt CC, Burkart C, Labeit D, McNabb M, Wu Y, Granzier H, et al. Nebulin regulates thin filament length, contractility, and Z-disk structure in vivo. Embo J. 2006;25:3843–3855. doi: 10.1038/sj.emboj.7601242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Luther P, Squire J. Three-dimensional structure of the vertebrate muscle M-region. J Mol Biol. 1978;125:313–324. doi: 10.1016/0022-2836(78)90405-9. [DOI] [PubMed] [Google Scholar]
- 25.Fischer RS, Fowler VM. Tropomodulins: life at the slow end. Trends Cell Biol. 2003;13:593–601. doi: 10.1016/j.tcb.2003.09.007. [DOI] [PubMed] [Google Scholar]
- 26.Wang K, Wright J. Architecture of the sarcomere matrix of skeletal muscle: immunoelectron microscopic evidence that suggests a set of parallel inextensible nebulin filaments anchored at the Z line. J Cell Biol. 1988;107:2199–2212. doi: 10.1083/jcb.107.6.2199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kruger M, Wright J, Wang K. Nebulin as a length regulator of thin filaments of vertebrate skeletal muscles: correlation of thin filament length, nebulin size, and epitope profile. J Cell Biol. 1991;115:97–107. doi: 10.1083/jcb.115.1.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McElhinny AS, Kazmierski ST, Labeit S, Gregorio CC. Nebulin: the nebulous, multifunctional giant of striated muscle. Trends Cardiovasc Med. 2003;13:195–201. doi: 10.1016/s1050-1738(03)00076-8. [DOI] [PubMed] [Google Scholar]
- 29.Kazmierski ST, Antin PB, Witt CC, Huebner N, McElhinny AS, Labeit S, et al. The complete mouse nebulin gene sequence and the identification of cardiac nebulin. J Mol Biol. 2003;328:835–846. doi: 10.1016/s0022-2836(03)00348-6. [DOI] [PubMed] [Google Scholar]
- 30.McElhinny AS, Schwach C, Valichnac M, Mount-Patrick S, Gregorio CC. Nebulin regulates the assembly and lengths of the thin filaments in striated muscle. J Cell Biol. 2005;170:947–957. doi: 10.1083/jcb.200502158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Horowits R. Nebulin regulation of actin filament lengths: new angles. Trends Cell Biol. 2006;16:121–124. doi: 10.1016/j.tcb.2006.01.003. [DOI] [PubMed] [Google Scholar]
- 32.Allen DG, Kentish JC. The cellular basis of the length-tension relation in cardiac muscle. J Mol Cell Cardiol. 1985;17:821–840. doi: 10.1016/s0022-2828(85)80097-3. [DOI] [PubMed] [Google Scholar]
- 33.Weiwad WK, Linke WA, Wussling MH. Sarcomere length-tension relationship of rat cardiac myocytes at lengths greater than optimum. J Mol Cell Cardiol. 2000;32:247–259. doi: 10.1006/jmcc.1999.1069. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


