Abstract
Antimicrobial agents are used in cattle production systems for the prevention and control of bacterial associated diseases. A consequence of their use is the potential development of antimicrobial resistance (AMR). Enterococcus faecium and Enterococcus faecalis that are resistant to antimicrobials are of increased concern to public health officials throughout the world as they may compromise the ability of various treatment regimens to control disease and infection in human medicine. Australia is a major exporter of beef; however it does not have an ongoing surveillance system for AMR in cattle or foods derived from these animals. This study examined 910 beef cattle, 290 dairy cattle and 300 veal calf faecal samples collected at slaughter for the presence of enterococci. Enterococcus were isolated from 805 (88.5%) beef cattle faeces, 244 (84.1%) dairy cattle faeces and 247 (82.3%) veal calf faeces with a total of 800 enterococci subsequently selected for AMR testing. The results of AMR testing identified high levels of resistance to antimicrobials that are not critically or highly important to human medicine with resistance to flavomycin (80.2%) and lincomycin (85.4–94.2%) routinely observed. Conversely, resistance to antibiotics considered critically or highly important to human medicine such as tigecycline, daptomycin, vancomycin and linezolid was not present in this study. There is minimal evidence that Australian cattle production practices are responsible for disproportionate contributions to AMR development and in general resistance to antimicrobials of critical and high importance in human medicine was low regardless of the isolate source. The low level of antimicrobial resistance in Enterococcus from Australian cattle is likely to result from comprehensive controls around the use of antimicrobials in food-production animals in Australia. Nevertheless, continued monitoring of the effects of all antimicrobial use is required to support Australia’s reputation as a supplier of safe and healthy food.
Introduction
Australia produces approximately 4% of the world’s beef supply yet exports more than 70% of production making it the world’s largest beef exporter in 2015 [1]. Antimicrobial agents are used in cattle production systems for the prevention and control of bacterial associated diseases. As many classes of antimicrobials are approved for and may be used in cattle production systems there is the potential for antimicrobial resistance (AMR) to develop in bacteria, including zoonotic pathogens which can be transferred to the human population via the food chain or by direct exposure to animals [2, 3]. Novel resistance phenotypes continue to emerge in zoonotic foodborne pathogens and commensal bacteria isolated from food production animals [4, 5]. In particular, Enterococcus faecalis and Enterococcus faecium have become of increasing importance over recent decades because of life-threatening hospital-acquired infections [6]. Consequently, understanding, assessing and mitigating the risks of non-human use of antimicrobials on human health outcomes remains a high priority. The World Health Organisation (WHO) has developed and maintains criteria and ranks antimicrobials based on their importance to human medicine [7]. Such information will help regulators and stakeholders identify appropriate antimicrobials for use in food animal production systems [8].
A number of countries have established AMR surveillance programs in place. Whilst the main focus of these programs is AMR in bacteria from humans, there is considerable and increasing emphasis on assessing AMR in bacteria from animals during production and from foods at retail. Multi-focus surveillance programs enable trends in AMR development to be further evaluated with respect to production practices and animal type and are particularly useful in addressing concerns from regulators about the overall impact of antimicrobial use. Countries without sophisticated multi-focus surveillance programs instead rely on relatively short-term intensive surveys to achieve the same result. The aim of this study was to conduct a short-term surveillance program to determine the prevalence and AMR status of Enterococcus isolates from Australian cattle populations.
Materials and methods
Sample collection
Faecal samples were collected from Australian cattle at slaughter as previously described [9]. Briefly, samples were collected from three animal groups: beef cattle, dairy cattle, and veal calves. Animals were defined as veal if their carcass weight was no more than 150kg. A total of 910, 290 and 300 samples were collected from beef cattle, dairy cattle and veal calves, respectively. Samples were collected across two sampling windows with sampling occurring once per abattoir in each window. Abattoirs were expected to collect up to a maximum of 40 samples per sampling day therefore all samples were expected to be collected a minimum of 12 minutes apart. Faecal samples were collected post-evisceration by cutting the intestine 15–30 cm from the rectal end and squeezing at least 40 g of material into a sterile jar. Samples were kept chilled and returned to the laboratory by overnight courier for processing.
Isolation of Enterococcus
The presence of Enterococcus was determined by enriching 1 g of faeces in 10 ml of BBL Enterococcosel Broth (BD, Maryland, USA) for 18–24 h at 35 ± 2°C. Enriched broths were then plated onto BBL Enterococcosel Agar (BD) and incubated for 18–24 h at 35 ± 2°C. Translucent colonies with brownish-black to black zones were then streaked onto Sheep Blood Agar (SBA) and incubated for 18–24 h at 35 ± 2°C. Isolates were confirmed as Enterococcus spp. by PCR [10]. A species specific PCR was then used to identify E. faecalis and E. faecium strains [11]. Further speciation was not performed and the remaining isolates were labelled Enterococcus spp.
Phenotypic detection of antimicrobial resistance
The AMR phenotype of isolates was initially determined using the broth microdilution method and the Sensititre apparatus. A total of 800 Enterococcus isolates comprising all E. faecium and E. faecalis and a selection of Enterococcus spp. Isolates from across the three animal groups were selected for AMR testing. Custom susceptibility panels for Enterococcus (AUSVP2; TREK Diagnostic Systems, UK) were used to test all isolates. The dilution ranges and breakpoints for each antimicrobial are shown in Table 1. Interpretation of the MIC values was based on CLSI interpretive criteria when available; otherwise EUCAST and NARMS values were used. Isolates that exceeded the MIC value of the susceptible breakpoint were reported as non-susceptible. E. faecalis ATCC 29212 was used as the control strain.
Table 1. Dilution ranges and breakpoints for antimicrobial susceptibility testing.
| Antimicrobial | Range | Breakpoint* |
|---|---|---|
| Ampicillin | 0.5–16 | ≥16 |
| Chloramphenicol | 2–32 | ≥32 |
| Daptomycin | 0.125–4 | ≥8 |
| Erythromycin | 0.25–8 | ≥8 |
| Flavomycin | 1–32 | ≥32 |
| Gentamicin | 32–1024 | ≥512 |
| Kanamycin | 128–1024 | ≥1024 |
| Lincomycin | 1–32 | ≥8 |
| Linezolid | 0.5–8 | ≥8 |
| Penicillin | 0.5–16 | ≥16 |
| Streptomycin | 256–1024 | ≥1024 |
| Teicoplanin | 0.125–4 | 2 |
| Tetracycline | 2–16 | ≥16 |
| Tigecycline | 0.016–0.5 | ≥0.5 |
| Vancomycin | 0.25–32 | ≥32 |
| Virginiamycin | 1–32 | >8 |
* CLSI breakpoints were adopted for ampicillin, daptomycin, erythromycin, linezolid, penicillin and vancomycin. NARMS breakpoints were used for chloramphenicol, flavomycin, gentamicin, kanamycin, lincomycin, streptomycin, tetracycline, tigecycline with EUCAST breakpoints used for teicoplanin and virginiamycin.
Isolates that demonstrated resistance to antimicrobials (e.g daptomycin and tigecycline) of critical or high importance to human medicine in the Sensititre testing process were further evaluated using M.I.C. Evaluator strips (Oxoid, UK). Susceptibility testing was conducted as per the manufacturer’s recommendations with each isolate suspended in cation adjusted Mueller-Hinton broth at 0.5 MacFarland standard. Each isolate was subsequently spread plated onto Mueller-Hinton agar and overlaid with the appropriate MIC Evaluator strip. The MIC or zone of clearance was measured after 24 hours incubation at 37°C.
Genotypic detection of antimicrobial resistance
Isolates that demonstrated resistance to daptomycin and tigecycline were tested for the presence of AMR genes or SNPs that have previously been shown to be associated with resistance to the aforementioned antimicrobials [12–14]. The primers, cycling conditions and expected product sizes are shown in Table 2. Detection of SNPs in liaR, liaS and rpsJ was conducted by Sanger sequencing of PCR products (AGRF, Brisbane) and subsequent analysis in Vector NTi (Life Technologies, Australia).
Table 2. Primers, cycling conditions and expected product sizes of Enterococcus AMR gene PCRs.
| Resistance to: | Oligo (5’–3’) | Cycling conditions | Products size (bp) | Reference |
|---|---|---|---|---|
| Daptomycin | liaR-F:GGTCCGATCATCCACATCTA liaR-R:CCGTTTAGGCGTTTCATCAT |
30s 94°C, 30s 60°C, 30s 72°C x 30 | 553 | This study |
| liaS-F:AAAGTCATTGGTGGGGAGAA liaS-R:GACTGGGAAGCGTTGATGAT |
30s 94°C, 30s 60°C, 30s 72°C x 30 | 526 | ||
| Tigecycline | rpsJ-F:AGAGGTTGCGACACGCCCGG rpsJ-R:TCTACAACAGTTACTGGAAT |
30s 94°C, 30s 60°C, 30s 72°C x 30 | 525 | [13] |
Results
Prevalence and identity
In total, 1500 faecal samples comprising 910 beef cattle faeces, 290 dairy cattle faeces and 300 veal calf faeces were tested for the presence of Enterococcus. Enterococcus were isolated from 805 (88.5%) beef cattle faeces, 244 (84.1%) dairy cattle faeces and 247 (82.3%) veal calf faeces. Species specific PCR determined that 96 (6.4%) of faecal samples yielded E. faecalis and 120 (8.0%) yielded E. faecium. Veal samples (14.3%) were significantly (p = 0.05) more likely to contain E. faecalis than dairy (3.1%) or beef (4.8%) samples. No significant differences in prevalence were observed between the three animal groups for E. faecium.
Antimicrobial susceptibility testing
Sensititre evaluation
A total of 800 Enterococcus isolates comprising 96 E. faecalis, 120 E. faecium, and 584 Enterococcus spp. were submitted for AMR analysis using the Sensititre test system. The distribution of MICs for each antimicrobial and species group is shown in Table 3. Breakpoints are not available for unspeciated Enterococcus isolates and therefore resistance data is only shown for E. faecium and E. faecalis. Streptogramin MIC values for E. faecalis are not presented as this species is intrinsically resistant. Similarly, flavomycin MIC values for E. faecium are not shown as they are inherently nonsusceptible. Irrespective of animal group and species, resistance to flavomycin (77.3–88.9%) and lincomycin (77.8–100.0%) was common. There was a strong association between daptomycin resistant E. faecalis and veal calves, however this was not considered to be statistically significant (p = 0.05). Resistance to tetracycline (2.3–13.0%) and erythromycin (0.0–13.6%) were observed in the majority of the three animal groups except for erythromycin resistance in E. faecium from veal calves. Furthermore, tigecycline resistance was only observed in E. faecium and E. faecalis from grass-fed animals, and whilst tetracycline resistance in E. faecalis was more common in grain-fed isolates, the opposite relationship existed in E. faecium with tetracycline resistance only detected in isolates from grass-fed animals.
Table 3. Distribution of MICs and occurrence of resistance among Enterococcus isolates using the Sensititre test system.
| Class | Antimicrobial | Species | N = | % Resistant | 95% CI | Antimicrobial concentration (μg/ml) | |||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0.015 | 0.03 | 0.06 | 0.12 | 0.25 | 0.5 | 1 | 2 | 4 | 8 | 16 | 32 | 64 | 128 | 256 | 512 | 1024 | >1024 | ||||||
| Aminoglycoside | Gentamicin | Enterococcus faecalis | 96 | 0.0 | 0.00–3.77 | 100.0 | |||||||||||||||||
| Enterococcus faecium | 120 | 0.0 | 0.00–3.03 | 99.2 | 0.8 | ||||||||||||||||||
| Enterococcus spp | 584 | NA | NA | 99.7 | 0.3 | ||||||||||||||||||
| Kanamycin | Enterococcus faecalis | 96 | 1.0 | 0.03–5.67 | 92.7 | 5.2 | 1.0 | 1.0 | |||||||||||||||
| Enterococcus faecium | 120 | 0.8 | 0.02–4.56 | 99.2 | 0.8 | ||||||||||||||||||
| Enterococcus spp | 584 | NA | NA | 95.0 | 3.8 | 0.3 | 0.3 | 0.5 | |||||||||||||||
| Streptomycin | Enterococcus faecalis | 96 | 1.0 | 0.03–5.67 | 99.0 | 1.0 | |||||||||||||||||
| Enterococcus faecium | 120 | 0.0 | 0.00–3.03 | 100.0 | |||||||||||||||||||
| Enterococcus spp | 584 | NA | NA | 99.1 | 0.9 | ||||||||||||||||||
| Glycopeptides | Teicoplanin | Enterococcus faecalis | 96 | 0.0 | 0.00–3.77 | 30.2 | 46.9 | 17.7 | 5.2 | ||||||||||||||
| Enterococcus faecium | 120 | 0.0 | 0.00–3.03 | 42.5 | 40.0 | 14.2 | 3.3 | ||||||||||||||||
| Enterococcus spp | 584 | NA | NA | 33.2 | 41.1 | 19.7 | 6.0 | ||||||||||||||||
| Vancomycin | Enterococcus faecalis | 96 | 0.0 | 0.00–3.77 | 1.0 | 40.6 | 34.4 | 11.5 | 10.4 | 2.1 | |||||||||||||
| Enterococcus faecium | 120 | 0.0 | 0.00–3.03 | 46.7 | 22.5 | 15.0 | 15.0 | 0.8 | |||||||||||||||
| Enterococcus spp | 584 | NA | NA | 0.5 | 41.6 | 28.3 | 13.4 | 12.3 | 3.9 | ||||||||||||||
| Glycylcycline | Tigecycline | Enterococcus faecalis | 96 | 2.1 | 0.25–7.32 | 2.1 | 45.8 | 41.7 | 3.1 | 5.2 | 2.1 | ||||||||||||
| Enterococcus faecium | 120 | 2.5 | 0.52–7.13 | 43.3 | 44.2 | 5.8 | 4.2 | 2.5 | |||||||||||||||
| Enterococcus spp | 584 | NA | NA | 2.1 | 45.0 | 37.7 | 8.0 | 4.3 | 2.7 | 0.2 | |||||||||||||
| Lincosamide | Lincomycin | Enterococcus faecalis | 96 | 85.4 | 76.74–91.79 | 10.4 | 1.0 | 3.1 | 8.3 | 25.0 | 34.4 | 17.7 | |||||||||||
| Enterococcus faecium | 120 | 94.2 | 88.35–97.62 | 4.2 | 0.8 | 0.8 | 2.5 | 27.5 | 50.0 | 14.2 | |||||||||||||
| Enterococcus spp | 584 | NA | NA | 12.3 | 1.9 | 2.1 | 6.2 | 30.7 | 37.5 | 9.4 | |||||||||||||
| Lipopeptide | Daptomycin | Enterococcus faecalis | 96 | 9.4 | 4.38–17.05 | 75.0 | 15.6 | 9.4 | |||||||||||||||
| Enterococcus faecium | 120 | 2.5 | 0.52–7.13 | 5.8 | 45.0 | 46.7 | 2.5 | ||||||||||||||||
| Enterococcus spp | 584 | NA | NA | 0.2 | 3.3 | 24.8 | 40.6 | 24.5 | 6.7 | ||||||||||||||
| Macrolide | Erythromycin | Enterococcus faecalis | 96 | 10.4 | 5.11–18.32 | 33.3 | 15.6 | 14.6 | 20.8 | 5.2 | 1.0 | 9.4 | |||||||||||
| Enterococcus faecium | 120 | 8.3 | 4.07–14.79 | 53.3 | 13.3 | 12.5 | 9.2 | 3.3 | 4.2 | 4.2 | |||||||||||||
| Enterococcus spp | 584 | NA | NA | 45.9 | 13.2 | 11.0 | 17.6 | 6.3 | 1.2 | 4.8 | |||||||||||||
| Oxazolidinones | Linezolid | Enterococcus faecalis | 96 | 0.0 | 0.00–3.77 | 6.3 | 87.5 | 6.3 | |||||||||||||||
| Enterococcus faecium | 120 | 0.0 | 0.00–3.03 | 5.8 | 87.5 | 6.7 | |||||||||||||||||
| Enterococcus spp | 584 | NA | NA | 0.3 | 5.0 | 88.2 | 6.5 | ||||||||||||||||
| Penicillins | Ampicillin | Enterococcus faecalis | 96 | 0.0 | 0.00–3.77 | 44.8 | 52.1 | 3.1 | |||||||||||||||
| Enterococcus faecium | 120 | 0.0 | 0.00–3.03 | 30.0 | 60.0 | 10.0 | |||||||||||||||||
| Enterococcus spp | 584 | NA | NA | 45.9 | 29.5 | 22.3 | 2.4 | ||||||||||||||||
| Penicillin | Enterococcus faecalis | 96 | 0.0 | 0.00–3.77 | 13.5 | 28.1 | 26.0 | 31.3 | 1.0 | ||||||||||||||
| Enterococcus faecium | 120 | 0.0 | 0.00–3.03 | 12.5 | 34.2 | 30.8 | 20.8 | 1.7 | |||||||||||||||
| Enterococcus spp | 584 | NA | NA | 19.3 | 27.1 | 22.8 | 21.2 | 9.2 | 0.2 | 0.2 | |||||||||||||
| Phenicol | Chloramphenicol | Enterococcus faecalis | 96 | 0.0 | 0.00–3.77 | 17.7 | 82.3 | ||||||||||||||||
| Enterococcus faecium | 120 | 0.0 | 0.00–3.03 | 1.7 | 51.7 | 40.8 | 5.8 | ||||||||||||||||
| Enterococcus spp | 584 | NA | NA | 0.3 | 38.4 | 59.4 | 1.9 | ||||||||||||||||
| Phosphoglycolipid | Flavomycin# | Enterococcus faecalis | 96 | 80.2 | 70.83–87.64 | 15.6 | 1.0 | 2.1 | 1.0 | 2.1 | 78.1 | ||||||||||||
| Enterococcus faecium | NA | NA | NA | ||||||||||||||||||||
| Enterococcus spp | 584 | NA | NA | 11.8 | 1.0 | 0.7 | 1.0 | 0.7 | 2.1 | 82.7 | |||||||||||||
| Streptogramins | Virginiamycin* | Enterococcus faecalis | NA | NA | NA | ||||||||||||||||||
| Enterococcus faecium | 120 | 0.0 | 0.00–3.03 | 85.0 | 5.8 | 9.2 | |||||||||||||||||
| Enterococcus spp | 584 | NA | NA | 86.6 | 6.8 | 6.0 | 0.2 | 0.3 | |||||||||||||||
| Tetracycline | Tetracycline | Enterococcus faecalis | 96 | 7.3 | 2.98–14.45 | 86.5 | 2.1 | 4.2 | 3.1 | 4.2 | |||||||||||||
| Enterococcus faecium | 120 | 11.7 | 6.53–18.80 | 78.3 | 8.3 | 1.7 | 1.7 | 10.0 | |||||||||||||||
| Enterococcus spp | 584 | NA | NA | 83.2 | 3.9 | 2.1 | 1.9 | 8.9 | |||||||||||||||
* Enterococcus faecalis isolates are intrinsically resistant to streptogramins
# Enterococcus faecium isolates are inherently nonsuceptible to flavomycin.
Solid vertical lines indicate breakpoints for resistance. The white fields indicate the dilution range tested for each antimicrobial. Values in the shaded area indicate MIC values greater than the highest concentration tested.
Additional phenotypic AMR testing
Initial evaluation of AMR in the Enterococcus isolates identified resistance to antimicrobials of human clinical significance. In particular, resistance to daptomycin and tigecycline was noted. Resistance to these antimicrobials was higher than anticipated and therefore was further investigated. Testing of daptomycin and tigecycline was completed in duplicate on each of the isolates that had previously demonstrated resistance to these antimicrobials in the Sensititre system, using the M.I.C. Evaluator system. MICs for tigecycline were all below the clinical breakpoint and therefore all isolates should be considered susceptible to tigecycline. The three E. faecium and two E. faecalis isolates all had an MIC of 0.12 μg/mL and are consistent with wild-type strains. None of the Enterococcus spp. isolates had MICs greater than the clinical breakpoint used for E. faecium and E. faecalis. Similarly, the three E. faecium and nine E. faecalis isolates previously identified as resistant to daptomycin all had MICs below the clinical breakpoint on re-testing. The E. faecium isolates all had MICs of 2 μg/mL whereas the E. faecalis isolates ranged from 0.25 to 2 μg/mL. One Enterococcus spp. isolate had an elevated MIC of 8 μg/mL, however the MICs of all remaining isolates were below the clinical breakpoint.
Genotypic investigation of AMR
In an attempt to corroborate the findings of the additional daptomycin and tigecycline AMR testing, all isolates including those not identified as E. faecalis or E. faecium, exhibiting MICs greater than the clinical breakpoints for daptomycin or tigecycline were tested by PCR for a range of genetic markers known to be associated with resistance to these antimicrobials. In total, 42 daptomycin resistant isolates and 22 tigecycline resistant isolates were tested further. For tigecycline resistant isolates, fragments of rpsJ were amplified, sequenced and analysed for a SNP that encodes a predicted amino acid change of Asp60 to Tyr. Fragments of rpsJ were amplified from all three E. faecium isolates and from 14 (82.4%) of 17 Enterococcus spp. isolates. Fragments of rpsJ were not amplified in either of the two E. faecalis isolates. Analysis of the 17 rpsJ fragments determined that none of the isolates harboured the SNP that has been shown to be associated with reduced susceptibility to tigecycline. Daptomycin resistant isolates were tested for the presence of SNPs in liaR and liaS. PCR products for liaR or liaS were generated from eight (88.9%) of nine E. faecalis isolates and all three E. faecium isolates but was found in only two (6.7%) of 30 Enterococcus spp. isolates. E. faecalis strains were most likely to harbour liaR on its own whereas E. faecium were more likely to harbour liaS. One E. faecalis and one E. faecium isolate were shown to contain both liaR and liaS. Sequencing of the PCR fragments determined that none of the isolates possessed the Thr120 to Ala SNP in liaR or the Trp73 to Cys SNP in liaS.
Re-categorisation of daptomycin and tigecycline results
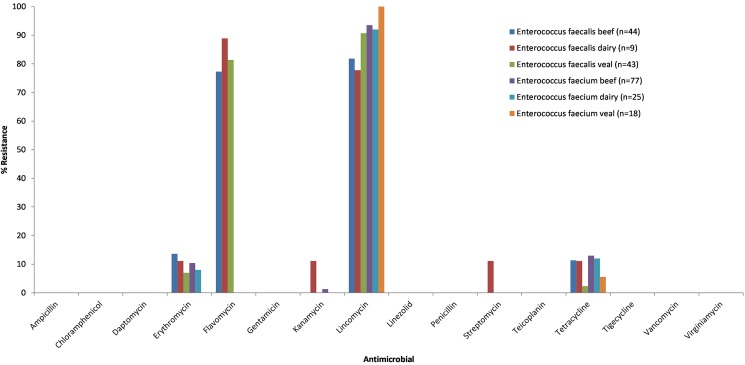
The inability to reproduce the findings of the primary phenotypic antimicrobial testing conducted using the Sensititre test system with custom AMR plates and the absence of the identification of AMR-linked genetic markers suggests that the original phenotypic assessment for resistance to daptomycin and tigecycline is incorrect and likely comprised of major errors. The major error rates for E. faecalis isolates against daptomycin and tigecycline were 9.4% and 2.1%, respectively. Whilst the major error rates for E. faecium isolates against daptomycin and tigecycline were low at 2.5% for both. The acceptance of major errors in the assessment of the three antimicrobials results in the re-categorisation of the results for the E. faecalis and E. faecium isolates in question. The revised results are shown in Table 4 and the distribution of AMR in E. faecalis and E. faecium from the three animal groups shown in Fig 1.
Table 4. Revised MICs and occurrence of resistance among Enterococcus isolates following additional phenotypic and genotypic assessment.
| Class | Antimicrobial | Species | N = | % Resistant | 95% CI | Antimicrobial concentration (μg/ml) | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0.015 | 0.03 | 0.06 | 0.12 | 0.25 | 0.5 | 1 | 2 | 4 | 8 | 16 | ||||||
| Glycylcycline | Tigecycline | Enterococcus faecalis | 96 | 0.0 | 0.00–3.77 | 2.1 | 45.8 | 41.7 | 5.2 | 5.2 | ||||||
| Enterococcus faecium | 120 | 0.0 | 0.00–3.03 | 43.3 | 44.2 | 5.8 | 4.2 | |||||||||
| Enterococcus spp | 584 | NA | NA | 2.1 | 45.0 | 37.7 | 8.0 | 4.3 | ||||||||
| Lipopeptide | Daptomycin | Enterococcus faecalis | 96 | 9.4 | 0.00–3.77 | 2.1 | 4.2 | 2.1 | 76.0 | 15.6 | ||||||
| Enterococcus faecium | 120 | 2.5 | 0.00–3.03 | 5.8 | 47.5 | 46.7 | ||||||||||
| Enterococcus spp | 584 | NA | NA | 0.2 | 0.7 | 5.3 | 25.5 | 40.9 | 27.2 | 0.2 | ||||||
Solid vertical lines indicate breakpoints for resistance. The white fields indicate the dilution range tested for each antimicrobial.
Fig 1. Prevalence of AMR in Enterococcus faecalis and faecium isolates from beef cattle, dairy cattle and veal calf faecal samples.
Antimicrobial resistance profiles
Resistance to three or more classes of antimicrobial, which we defined as multidrug resistance (MDR), was observed in 18 (8.3%) of all E. faecium and E. faecalis isolates. Table 5 shows the resistance profiles for each E. faecium and E. faecalis. MDR was observed in six (5.0%) E. faecium and 12 (12.5%) E. faecalis isolates. Resistance to four or more antimicrobial classes was less commonly observed with only four (3.3%) E. faecium isolates falling into this category. Antimicrobial resistance profiles of E. faecium were dominated by resistance to lincomycin (78.3%). FLV-LIN was the most common resistance profile associated with E. faecalis isolates with 53 (55.2%) of 96 isolates harbouring this combination. The main MDR profiles for E. faecium and E. faecalis were ERY-LIN-TET (3.3%) and ERY-FLV-LIN (5.2%), respectively.
Table 5. Antimicrobial resistance profiles of Enterococcus faecium and faecalis isolates from beef cattle, dairy cattle and veal calf faecal samples.
| Antimicrobial resistance profile* | E. faecium (N = 120) | E. faecalis (N = 96) |
|---|---|---|
| ALL SENSITIVE | 6 | 1 |
| FLV | 13 | |
| LIN | 95 | 17 |
| TET | 1 | |
| ERY LIN | 5 | |
| FLV LIN | 53 | |
| LIN TET | 8 | |
| ERY FLV LIN | 5 | |
| ERY LIN TET | 4 | 1 |
| ERY KAN LIN | 1 | |
| FLV LIN TET | 2 | |
| ERY FLV LIN TET | 3 | |
| ERY FLV KAN LIN STR TET | 1 |
* FLV–flavomycin, LIN–lincomycin, TET–tetracycline, ERY–erythromycin, KAN–kanamycin, STR—streptomycin
Discussion
Bacteria that are resistant to antimicrobials are of increased concern to public health officials throughout the world as they may compromise the ability of treatment regimens to address disease and infection in humans. Knowledge and understanding of both the types of AMR present in food production animals and the type of antimicrobials being used is key to determining the risk that AMR bacteria in the food chain pose to human health. Australia currently does not have a nationally coordinated program for the ongoing surveillance and analysis of AMR bacteria in animals or bacteria in food derived from animals. Consequently it relies heavily on testing of human and animal clinical isolates as well as infrequent surveys of isolates from animals or from food of animal origin to understand AMR development and trends.
Enterococci are ubiquitous bacteria that demonstrate intrinsic resistance to a number of first-line antimicrobial agents and have also demonstrated capacity to rapidly acquire resistance to antimicrobials including quinolones, macrolides, tetracyclines, streptogramins and glycopeptides [15]. They are also frequently associated with mobile genetic elements harbouring AMR genes and have the potential for resistance to virtually all antimicrobials of importance to human medicine [16]. The importance of enterococci as the third most commonly isolated nosocomial pathogen [17] and the clear relationship between exposure to parental antimicrobials and the development of resistance [18] warrants their ongoing inclusion in any human, animal or food AMR surveillance program. E. faecalis and E. faecium were recovered from 6.4% and 8.0% of samples in this survey and although they are the two enterococcal species most associated with human infections, monitoring of additional species of enterococci other than E. faecalis and E. faecium is useful as it may provide insights to trends of MIC’s which may be of concern to the more clinically relevant species. From a human clinical perspective, resistance in E. faecalis and E. faecium to ampicillin, vancomycin, linezolid, daptomycin and tigecycline are the key concerns. Resistance to other older antimicrobials such as lincomycin, flavomycin, tetracycline and erythromycin are seldom considered, as either resistance is common or the antimicrobials are seldom used in human medicine [18]. The findings of this study reinforce this segregation of concern with increased levels of resistance to lincomycin, flavomycin (E. faecalis only), tetracycline and erythromycin observed in E. faecium and E. faecalis isolates from all animal groups. Furthermore, whilst the highest levels of resistance in this study were to lincomycin and flavomycin, these results are consistent with published studies on enterococci from cattle [19] and likely stem from intrinsic resistance as opposed to the development of resistance resulting from widespread use of these antimicrobials.
Conversely, resistance to antimicrobials of critical and high importance to human medicine is of much greater concern to the ongoing treatment strategies for enterococcal infections. Resistance to ampicillin, linezolid and vancomycin was not observed in this study in any E. faecium or E. faecalis isolates. This is significant as ampicillin remains the preferred therapy for uncomplicated enterococcal infections. Similarly, the absence of vancomycin resistant enterococcus assists in maintaining optimal treatment options. Resistance to daptomycin and tigecycline initially observed with the Sensititre system could not be confirmed using gradient diffusion techniques. Publications detailing genes conferring resistance to these antimicrobials in Enterococcus isolates are extremely limited, however whole genome analysis of strains demonstrating reduced susceptibility have identified a number of single nucleotide polymorphisms (SNPs) present in those isolates when compared with wild-type populations [12–14, 20]. Importantly, recent studies have demonstrated that rpsJ mutations may also be present in tigecycline susceptible isolates and perhaps represent just a small component of a complex interplay between numerous resistance mechanisms required to overcome the selective pressure of tigecycline [21]. Investigation of the SNPs in the liaFSR regulon and rpsJ determined that the isolates in this study share sequence homology with wild-type isolates and do not contain these known AMR associated mutations. When combined with the agar dilution results the original Sensititre test results are believed to be incorrect and the overall data set has been modified to reflect these findings. As a consequence, this study reports that resistance to the critical or high importance antimicrobials linezolid, daptomycin, tigecycline and vancomycin was not detected in enterococcal isolates from Australian cattle regardless of the production system.
The generation of discordant AMR results after testing with multiple phenotypic test systems is concerning though not confined to this study alone. Several studies have detailed discrepancies in essential agreement and categorical agreement between test systems when single antimicrobial / bacteria combinations are considered. The United States Food and Drug Administration will approve the marketing of AMR tests system provided that very major errors (false-negatives) and major errors (false-positives) do not exceed 1.5% and 3% respectively and essential MIC agreement within one doubling MIC dilution of >90% occurs between the test system and the reference CLSI method [22]. This study has identified major errors with daptomycin and tigecycline, however only the combination of daptomycin with E. faecalis strains exceed the allowable 3% major error rate. The in vitro evaluation of daptomycin and tigecycline resistance has been shown to be highly dependent on the culture conditions used and may provide an explanation for the higher than acceptable major error rate observed in this study [23–25].
Overall, the results corroborate previous Australian based animal and retail food surveys that have shown a low level of AMR, relatively small proportions of MDR and most importantly the maintenance of susceptibility to most antimicrobials of critical and high importance to human health [9, 26–29]. Importantly, it would appear that the production practices in Australian cattle populations are not generating pools of resistance that are likely to result in the inability to treat human infections caused by enterococci. Nevertheless, it is necessary to maintain strict guidelines and controls around the use of antimicrobials in food-production animals in Australia and monitoring the effects of all antimicrobial use is required to support Australia’s reputation as a supplier of safe and healthy food.
Supporting information
(XLSX)
Acknowledgments
The authors thank Dr Darren Trott, Dr Sam Abraham, Dr John Turnidge and Dr Stephen Page for their assistance with the design of the study and analysis of results. We gratefully acknowledge funding from Meat & Livestock Australia and the Commonwealth Scientific and Industrial Research Organisation (CSIRO). We also acknowledge the significant contribution of samples from processing establishments, without which this project would not have been possible.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
The authors received funding from Meat & Livestock Australia (MLA) and the Commonwealth Scientific and Industrial Research Organisation (CSIRO) for this work.
References
- 1.Meat & Livestock Australia. Fast Facts 2016; Australia's beef industry. 2016. https://www.mla.com.au/globalassets/mla-corporate/prices—markets/documents/trends—analysis/fast-facts—maps/mla_beef-fast-facts-2016.pdf Available at: https://www.mla.com.au/globalassets/mla-corporate/prices—markets/documents/trends—analysis/fast-facts—maps/mla_beef-fast-facts-2016.pdf. Accessed 27 December 2016.
- 2.Collignon P, Angulo FJ. Fluoroquinolone-resistant Escherichia coli: food for thought. J Infect Dis. 2006;194(1):8–10. doi: 10.1086/504922 [DOI] [PubMed] [Google Scholar]
- 3.Heuer OE, Hammerum AM, Collignon P, Wegener HC. Human health hazard from antimicrobial-resistant enterococci in animals and food. Clinical infectious diseases: an official publication of the Infectious Diseases Society of America. 2006;43(7):911–6. [DOI] [PubMed] [Google Scholar]
- 4.Szmolka A, Nagy B. Multidrug resistant commensal Escherichia coli in animals and its impact for public health. Front Microbiol. 2013;4:258 doi: 10.3389/fmicb.2013.00258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Walsh C, Fanning S. Antimicrobial resistance in foodborne pathogens—a cause for concern? Curr Drug Targets. 2008;9(9):808–15. [DOI] [PubMed] [Google Scholar]
- 6.Hao H, Sander P, Iqbal Z, Wang Y, Cheng G, Yuan Z. The Risk of Some Veterinary Antimicrobial Agents on Public Health Associated with Antimicrobial Resistance and their Molecular Basis. Front Microbiol. 2016;7:1626 doi: 10.3389/fmicb.2016.01626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.World Health Organisation. Critically important antimicrobials for human medicine—3rd revision World Health Organization, 2011. Available at: http://apps.who.int/iris/bitstream/10665/77376/1/9789241504485_eng.pdf. Accessed 27 December 2016. [Google Scholar]
- 8.Collignon P, Powers JH, Chiller TM, Aidara-Kane A, Aarestrup FM. World Health Organization Ranking of Antimicrobials According to Their Importance in Human Medicine: A Critical Step for Developing Risk Management Strategies for the Use of Antimicrobials in Food Production Animals. Clinical Infectious Diseases. 2009;49(1):132–41. doi: 10.1086/599374 [DOI] [PubMed] [Google Scholar]
- 9.Barlow RS, McMillan KE, Duffy LL, Fegan N, Jordan D, Mellor GE. Prevalence and Antimicrobial Resistance of Salmonella and Escherichia coli from Australian Cattle Populations at Slaughter. J Food Prot. 2015;78(5):912–20. doi: 10.4315/0362-028X.JFP-14-476 [DOI] [PubMed] [Google Scholar]
- 10.Ke DB, Picard FJ, Martineau F, Menard C, Roy PH, Ouellette M, et al. Development of a PCR assay for rapid detection of Enterococci. J Clin Microbiol. 1999;37(11):3497–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dutkamalen S, Evers S, Courvalin P. Detection of Glycopeptide Resistance Genotypes and Identification to the Species Level of Clinically Relevant Enterococci by Pcr. Journal of Clinical Microbiology. 1995;33(1):24–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Arias CA, Panesso D, McGrath DM, Qin X, Mojica MF, Miller C, et al. Genetic Basis for In Vivo Daptomycin Resistance in Enterococci. New Engl J Med. 2011;365(10):892–900. doi: 10.1056/NEJMoa1011138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cattoir V, Isnard C, Cosquer T, Odhiambo A, Bucquet F, Guerin F, et al. Genomic Analysis of Reduced Susceptibility to Tigecycline in Enterococcus faecium. Antimicrobial Agents and Chemotherapy. 2015;59(1):239–44. doi: 10.1128/AAC.04174-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Diaz L, Tran TT, Munita JM, Miller WR, Rincon S, Carvajal LP, et al. Whole-Genome Analyses of Enterococcus faecium Isolates with Diverse Daptomycin MICs. Antimicrobial Agents and Chemotherapy. 2014;58(8):4527–34. doi: 10.1128/AAC.02686-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hammerum AM. Enterococci of animal origin and their significance for public health. Clinical Microbiology and Infection. 2012;18(7):619–25. doi: 10.1111/j.1469-0691.2012.03829.x [DOI] [PubMed] [Google Scholar]
- 16.Ramos S, Igrejas G, Capelo-Martinez JL, Poeta P. Antibiotic resistance and mechanisms implicated in fecal enterococci recovered from pigs, cattle and sheep in a Portuguese slaughterhouse. Ann Microbiol. 2012;62(4):1485–94. [Google Scholar]
- 17.Hidron AI, Edwards JR, Patel J, Horan TC, Sievert DM, Pollock DA, et al. Antimicrobial-Resistant Pathogens Associated With Healthcare-Associated Infections: Annual Summary of Data Reported to the National Healthcare Safety Network at the Centers for Disease Control and Prevention, 2006–2007. Infect Cont Hosp Ep. 2008;29(11):996–1011. [DOI] [PubMed] [Google Scholar]
- 18.Hollenbeck BL, Rice LB. Intrinsic and acquired resistance mechanisms in Enterococcus. Virulence. 2012;3(5):421–33. doi: 10.4161/viru.21282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jackson CR, Lombard JE, Dargatz DA, Fedorka-Cray PJ. Prevalence, species distribution and antimicrobial resistance of enterococci isolated from US dairy cattle. Lett Appl Microbiol. 2011;52(1):41–8. doi: 10.1111/j.1472-765X.2010.02964.x [DOI] [PubMed] [Google Scholar]
- 20.Aarestrup FM. Association between the consumption of antimicrobial agents in animal husbandry and the occurrence of resistant bacteria among food animals. Int J Antimicrob Agents. 1999;12(4):279–85. [DOI] [PubMed] [Google Scholar]
- 21.Fiedler S, Bender JK, Klare I, Halbedel S, Grohmann E, Szewzyk U, et al. Tigecycline resistance in clinical isolates of Enterococcus faecium is mediated by an upregulation of plasmid-encoded tetracycline determinants tet(L) and tet(M). J Antimicrob Chemother. 2016;71(4):871–81. doi: 10.1093/jac/dkv420 [DOI] [PubMed] [Google Scholar]
- 22.Jorgensen JH, Ferraro MJ. Antimicrobial Susceptibility Testing: A Review of General Principles and Contemporary Practices. Clinical Infectious Diseases. 2009;49(11):1749–55. doi: 10.1086/647952 [DOI] [PubMed] [Google Scholar]
- 23.Butaye P, Devriese LA, Haesebrouck F. Effects of different test conditions on MICs of food animal growth-promoting antibacterial agents for enterococci. Journal of Clinical Microbiology. 1998;36(7):1907–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kelesidis T, Humphries R, Uslan DZ, Pegues DA. Daptomycin Nonsusceptible Enterococci: An Emerging Challenge for Clinicians. Clinical Infectious Diseases. 2011;52(2):228–34. doi: 10.1093/cid/ciq113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rathe M, Kristensen L, Ellermann-Eriksen S, Thomsen MK, Schumacher H. Vancomycin-resistant Enterococcus spp.: validation of susceptibility testing and in vitro activity of vancomycin, linezolid, tigecycline and daptomycin. Apmis. 2010;118(1):66–73. doi: 10.1111/j.1600-0463.2009.02559.x [DOI] [PubMed] [Google Scholar]
- 26.Abraham S, Groves MD, Trott DJ, Chapman TA, Turner B, Hornitzky M, et al. Salmonella enterica isolated from infections in Australian livestock remain susceptible to critical antimicrobials. International Journal of Antimicrobial Agents. 2014;43(2):126–30. doi: 10.1016/j.ijantimicag.2013.10.014 [DOI] [PubMed] [Google Scholar]
- 27.Barlow RS, Gobius KS. Pilot survey for antimicrobial resistant (AMR) bacteria in Australian food. Cannon Hill, Queensland: 2008 2008. Available at: http://foodregulation.gov.au/internet/fr/publishing.nsf/Content/784616537FC3F1C3CA25806A007C5D5C/$File/pilot%20survey%20AMR%20part%201.pdf. Accessed 27 December 2016.
- 28.Barton MD, Pratt R, Hart WS. Antibiotic resistance in animals. Communicable diseases intelligence. 2003;27 Suppl:S121–6. [DOI] [PubMed] [Google Scholar]
- 29.DAFF. Pilot surveillance program for antimicrobial resistance in bacteria of animal origin Australian Government Department of Agriculture, Fisheries and Forestry; 2007. Available at: http://www.agriculture.gov.au/animal/health/amr/antimicrobial-resistance-bacteria-animal-origin. Accessed 27 December 2016. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(XLSX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.