Abstract
Induced pluripotent stem cells (iPSCs) technology provides a powerful means to generate and regenerate unlimited pluripotent stem cells directly from body tissue cells. Stem cells from apical papilla (SCAP) and Dental pulp stem cells (DPSCs) are present in ‘cell-rich zones’ within the dental pulp region, which are capable of regenerating pulp and dentin tissues in vivo. In this study, we investigated the difference of miRNAs expression in SCAPs and DPSCs before and after the reprogramming. Using miRNA microarray, 134 and 265 differentially expressed miRNAs in DPSCs- and SCAP-iPSCs were up-regulated compared to these before reprogramming. 117 specific miRNAs with enhanced more than 2-fold were identified in both DPSCs- and SCAP-iPSCs. Among the co-regulated miRNAs, miR-19a-3p, miR-92b-3p and miR-130b-3p showed the maximum difference, which had involvement in the cell cycle, TGF beta signaling pathway and epithelial mesenchymal transition. Using qRT-PCR analysis, the expression of miR-19a-3p, miR-92b-3p and miR-130b-3p indicated substantial increases in DPSCs-iPSCs and SCAP-iPSCs. The findings suggest that miRNAs play a part in the difference between DPSCs-iPSCs and DPSCs, as well as between SCAP-iPSCs and SCAP. The variation of miRNA expression in reprogrammed dental-derived pluripotent stem cells revealed different characteristics induced by iPSC generation.
Introduction
In the past ten years, the establishment of induced pluripotent stem cells (iPSCs) has been as one of the breakthroughs in the field of stem cell research [1, 2]. This breakthrough discovery provides a powerful means to generate and regenerate unlimited pluripotent stem cells directly from body tissue cells [3]. Like embryonic stem cells (ESCs), iPSCs share the majority properties with embryonic stem cells, such as morphology, differentiation, DNA methylation, gene expression, unlimited self-renewal ability and potential to generate any differentiated cell type (pluripotency) [4]. As previous studies reported, the self-renewal and pluripotency properties of iPSCs are not only regulated by an array of protein-coding genes but also a group of noncoding microRNA (miRNA) genes [5, 6]. With ongoing advances in miRNA biology, using nonviral vectors, such as RNA and microRNAs (miRNAs), to generate iPSCs has also been revealed [7]. miRNA is ~20–22 nucleotides in length and well known to control extensive cellular functions via affecting the abundance and translation efficiency of cognate mRNA. miRNAs are expressed in a variety of cells, such as embryonic stem cells, iPS cells and somatic cells. A number of co-expressed miRNAs clusters, such as miR-302-367, and miR-290, were disclosed in embryonic stem cells, iPS cells and somatic cells [8].
Stem cells from apical papilla (SCAP) are originated from the developing tissue at the apex of a tooth root termed apical papilla [9]. Dental pulp stem cells (DPSCs) are present in ‘cell-rich zones’ within the dental pulp region [10]. They are capable of regenerating pulp and dentin tissues in vivo. In our previous study, we have reprogrammed the SCAP and DPSCs into to iPS cells [11]. However, the difference of miRNAs expression before and after the reprogramming remain largely unexplored. In this study, chip analysis technology was used to screen the differential miRNAs expression in reprogramming process of human dental iPS cells, thereby providing a basis for further research on the mechanism of miRNA and its target genes in the reprogramming process.
Materials and methods
Isolation, culture and identification of human DPSCs and SCAP in vitro
We collected 2 mandibular third molars from the same patient (age<20) indicated for extraction at department of oral and maxillofacial, first people's Hospital of Yunnan Province. All patients provided written informed consent, and the study was approved by the Ethics Committee of the first people's Hospital of Yunnan Province.
DPSCs and SCAP isolated from third molars (Fig 1) were culture in medium consisting of α-MEM (Gibco/Invitrogen, Grand Island, NY, USA), 10% fetal bovine serum (Thermo Scientific), 1% Glutamax (Gibco/Invitrogen, Grand Island, NY, USA) and 0.5% Penicillin/Streptomycin ((Invitrogen). At P3, DPSCs and SCAP were harvested by trypsinization with 0.05% trypsin (Invitrogen) upon reaching 90% confluence, and re-suspended in DPBS to reach a final cell density of 1.5× 106 cells/ml. An amount of 200ul of cell suspension (1.5×106 cells) was incubated in the dark for 30min at room temperature with fluoro isothycyanate-conjugated antibodies against CD24, CD34, CD45, stro-1 (all from Invitrogen), and phycoerythrin-conjugated antibodies against CD90, CD105, CD146, oct-4 (all from eBioscience) for specific surface antigens analysis by using flow cytometer. All analyses were standardized against negative control cells incubated with Isotype-specific IgG2ak-PE (Invitrogen). The results of flow cytometer were analyzed by using Summit, Version 5.1
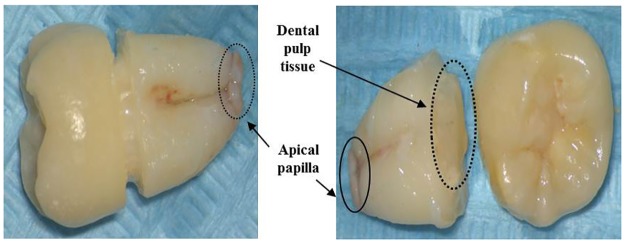
Fig 1. Dental pulp tissue and apical papilla isolated from third molars.
Transduction and reprogramming of DPSCs and SCAP
The transduction was carried out based on the instructions of the CytoTune-iPS Sendai Reprogramming kits. Two days before transfection, DPSCs and SCAP at P3 were seeded separately into wells of 6-well plates with 1 x 105 cells each well. On the transfection day, appropriate volumes of each of the three CytoTune 2.0 Sendai tubes were dissolved in 70uL medium were used for transduction of DPSCs and SCAP and then 130 uL medium was added after one day. On the second day, the medium were replaced by medium included fresh SeV. On the third day, the transfected cells were transferred to the 6-well plates covered with matrix and cultured with reprogramming medium. New DPSCs iPSC and SCAP iPSC colonies emerged in about three weeks. The clone were picked using the "cross method" and transferred into the new plates and cultured with PSC-easy medium included 10μM Y27632 (Selleck, Shanghai, China). H9 (P53) routinely cultured was as a standard control.
miRNA microarray analysis
90% confluent P3 human DPSCs /SCAP and P12 DPSCs/SCAP-iPSCs were collected for the miRNA isolation. Total RNA (including miRNA) was isolated using TRIzol reagent (Invitrogen, Carlsbad, CA, USA) and purified with RNeasy Mini Kit (Qiagen) following the manufacturer’s instructions. The purity and quantity of the isolated RNAs were assessed by 1% formaldehyde-agarose gel electrophoresis and spectrophotometry (NanoDrop ND-1000; Thermo Scientific, Wilmington, DE). The RNAs from P3 human DPSCs /SCAP and P12 DPSCs/SCAP-iPSCs have been respectively mixed and put into microarray analysis. After further processing, the isolated miRNAs were labeled with Hy3™ by using the miRCURY™ Hy3™/Hy5™ Power labeling kit (Exiqon, Vedbaek, Denmark). The Hy3™-labeled miRNAs were then hybridized to individual miRCURYTM LNA microRNA Arrays (v.18.0, Exiqon). Microarray images were obtained by an Axon Genepix 4000B microarray scanner (Axon Instruments, Union City, CA, USA), and processed and analyzed by the accompanying Axon Genepix Pro 6.0 software. Signal values of more than 30 in all samples were extracted for further analysis. After normalization, differentially expressed miRNAs were screened based on the fold change and P-value between cells before and after reprogramming. miRNAs with an adjusted p value less than 0.05 (ANOVA) were identified as differentially expressed miRNAs in these different reprogrammed cells.
Quantitative validation of miRNA using qRT-PCR
Total RNA from DPSCs /SCAP before and after reprogramming were used as template to synthesize cDNA with the One Step PrimeScript miRNA cDNA Synthesis kit (TaKaRa). The qPCR assay was conducted with reagents from the SYBR Green Real-Time PCR kit (TaKaRa), and the qRT-PCR mixture contained SYBR Premix Ex Taq II, forward and reverse primers (Table 1), UnimiR qPCR primer, cDNA, ROX reference dye. The reactions were conducted by using ABIPrism 7000 Sequence Detection System (Applied Biosystems) with initial enzyme activation at 95°C for 30 s, followed by 45 cycles of denaturation at 95°C for 5 s and annealing and extension at 60°C for 34 s. The threshold cycle (Ct) was determined by using the default threshold settings. All experiments were performed in triplicate and repeated three times. The expression level of genes of interest was normalized against housekeeping gene U6. The fold change was calculated by using the equation 2−ΔΔCT method.
Table 1. miRNA-specific primers used in the qRTPCR.
| miRNA | Forward primer (5’-3’) |
|---|---|
| U6 |
F: 5’GCTTCGGCAGCACATATACTAAAAT3’ R: 5’CGCTTCACGAATTTGCGTGTCAT3’ |
| miR-19a-3p |
GSP: 5’GGGGGGGTGTGCAAATCT3' R: 5' GTGCGTGTCGTGGAGTCG3' |
| miR-92b-3p |
GSP: 5’GGTATTGCACTCGTCCCG3’ R: 5'GTGCGTGTCGTGGAGTCG3’ |
| miR-130b-3p |
GSP: 5'GGGCAGTGCAATGATGAAA3' R: 5’GTGCGTGTCGTGGAGTCG3’ |
Bioinformatic prediction and pathway analysis of miRNAs target
The differentially expressed miRNAs targets were determined by using the miRanda (http://mirdb.org/miRDB/), microCosm(http://www.ebi.ac.uk/enright-srv/microcosm/htdocs/targets/v5/) and targetscan (http://www.targetscan.org/) [12]. To determine the biological and functional properties of all the differentially expressed miRNAs target, they were mapped with Gene Ontology Terms (http://geneontology.org/). The target genes of these miRNAs were also mapped to KEGG pathways. The enrichment results are presented in a “bubble plot” using the ggplot2 package [13]. Additionally, the biological functions associated with these networks are also provided.
Results
Primary culture of human DPSCs and SCAP and flow cytometric analyses
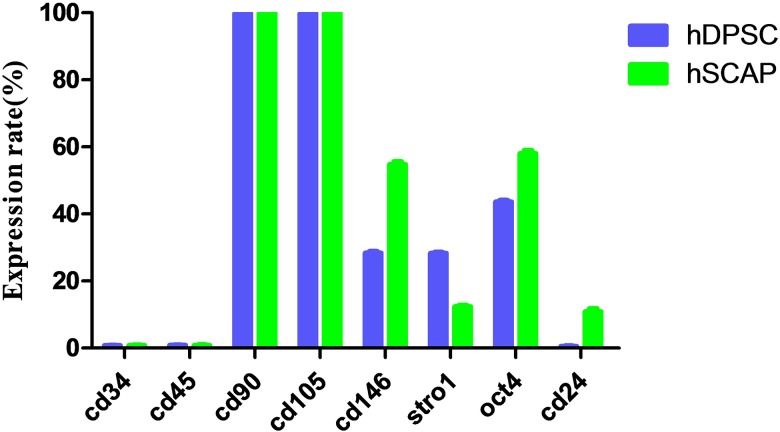
The isolated human DPSCs and SCAP in vitro were cloned after culturing for four days. The flow cytometric analyses showed that DPSCs and SCAP were both negative for the haematopoietic surface markers of CD34 and CD45. On the contrary, DPSCs and SCAP were positive (about 100%) for CD90 and CD105. DPSCs (28.4±0.56) % and SCAP [(54.8±0.96) %] were also shown strong positive for CD146 (Fig 2). At the same time, DPSCs and SCAP were positive for STRO-1[(28.3±0.4) % and (12.4±0.46)%] and OCT-4 [(43.6±0.66)% and (58±1.22)%] respectively (Fig 2). CD24 was only expressed in the SCAP [(10.9±1.06)%] (Fig 2).
Fig 2. The expression rate of surface markers in DPSCs and SCAP.
Generation of DPSCs-iPSCs and SCAP-iPSCs
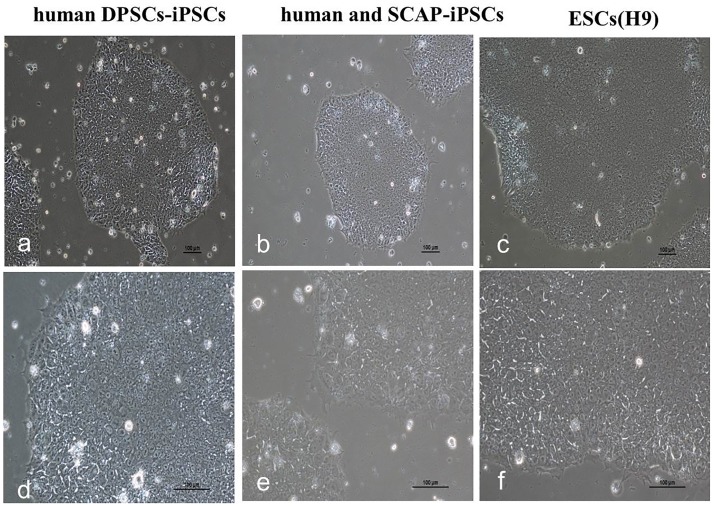
The transfected DPSCs and SCAP were seeded onto Matrigel. Within three to four weeks after transfection, ESC-like colonies emerged on Matrigel (Fig 3a–3e), which were primary iPS cells (P0). The iPS clones were transferred to the new matrix via cross method (P1) and passaged in four to five days. The culture of human DPSCs-iPSCs and SCAP-iPSCs in vitro were as indicated in Fig 3.
Fig 3. The culture of human DPSCs-iPSCs, SCAP-iPSCs and ESCs in vitro.
Differential expression of miRNAs between DPSCs/ SCAP and DPSCs-iPSCs/ SCAP-iPSCs
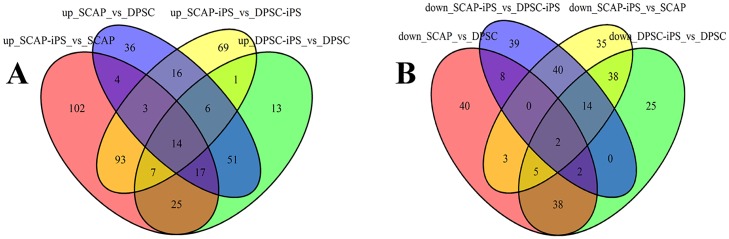
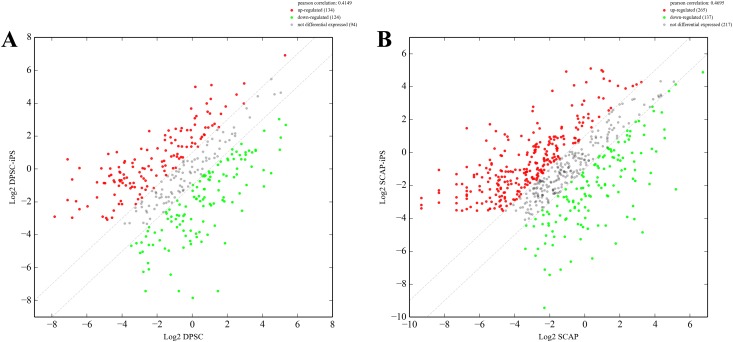
A total of 1201 miRNA genes were all determined by using miRanda, microcosm and targetscan (S1 Table). The venn diagrams of human DPSCs/SCAP-iPSCs showed the differentially expressed miRNA in DPSCs-iPSCs vs. DPSCs, SCAP-iPSCs vs. SCAP, DPSCs vs. SCAP and DPSCs-iPSCs vs. SCAP-iPSCs (Fig 4). Of the miRNA genes analyzed by miRNA microarray, 134 miRNA genes in DPSCs-iPSCs were up-regulated and 124 were down-regulated compared to those in DPSCs (Fig 5A). In SCAP-iPSCs, 265 miRNA genes showed upregulated expression and 137 downregulated expression compared to those in SCAP (Fig 5B). Among these up-regulated miRNA genes, 117 enhanced more than 2-fold were identified in both DPSCs-iPSCs and SCAP-iPSCs. Among these 117 up-regulated miRNA genes, miR-19a-3p, miR-92b-3p and miR-130b-3p showed the maximum difference between reprogrammed iPS cells and DPSCs/SCAP. These results implied that the difference between reprogrammed cells and DPSCs/SCAP might be attributed, at least in part, to differently expressed miRNAs. The following target prediction analysis on miR-19a-3p, miR-92b-3p and miR-130b-3p using bioinformatic neural nets are summarized in Table 2 (S1 Table).
Fig 4. The Venn diagrams of the number of miRNA genes expressed in human DPSCs/SCAP-iPSCs.
A: Up-regulated miRNA genes. B: Down-regulated miRNA genes.
Fig 5. Comparison of the expression levels between DPSCs and DPSCs-iPSCs (A) as well as SCAP and SCAP-iPSCs (B).
The differentially expressed miRNA genes were filtered using P≤0.05 and |log2 ratio|≥1 as a threshold. The red points represent upregulated genes, and the green points indicate downregulated genes. The gray spots represent no significant difference.
Table 2. Putative target genes of the six differentially expressed miRNAs.
| Target gene | Gene description | |
|---|---|---|
| hsa-miR-19a-3p | ACTN1 | actinin, alpha 1 |
| ALPK2 | alpha-kinase 2 | |
| ATP10A | ATPase, class V, type 10A | |
| BCL3 | B-cell CLL/lymphoma 3 | |
| BTG1 | B-cell translocation gene 1, anti-proliferative | |
| CAB39L | calcium binding protein 39-like | |
| CAST | castor zinc finger 1 | |
| DHRS3 | dehydrogenase/reductase (SDR family) member 3 | |
| DLC1 | deleted in liver cancer 1 | |
| DOCK10 | dedicator of cytokinesis 10 | |
| ELK3 | ELK3, ETS-domain protein (SRF accessory protein 2) | |
| FAM114A1 | family with sequence similarity 114, member A1 | |
| FAM69A | family with sequence similarity 69, member A | |
| FBXO8 | F-box protein 8 | |
| FLNC | filamin C, gamma | |
| FOSL1 | FOS-like antigen 1 | |
| FOXF2 | forkhead box F2 | |
| HIPK3 | homeodomain interacting protein kinase 3 | |
| LRIG3 | leucine-rich repeats and immunoglobulin-like domains 3 | |
| LRRTM2 | leucine rich repeat transmembrane neuronal 2 | |
| MAP2K3 | mitogen-activated protein kinase kinase 3 | |
| MAP3K12 | mitogen-activated protein kinase kinase kinase 12 | |
| MICAL2 | microtubule associated monoxygenase, calponin and LIM domain containing 2 | |
| NRP2 | neuropilin 2 | |
| PDE4A | phosphodiesterase 4A, cAMP-specific | |
| PIK3CA | phosphoinositide-3-kinase, catalytic, alpha polypeptide | |
| PNRC1 | proline-rich nuclear receptor coactivator 1 | |
| PON2 | paraoxonase 2 | |
| POSTN | periostin, osteoblast specific factor | |
| PRKAA1 | protein kinase, AMP-activated, alpha 1 catalytic subunit | |
| REEP3 | receptor accessory protein 3 | |
| RHOB | Rho-related BTB domain containing 1 | |
| SH3KBP1 | SH3-domain kinase binding protein 1 | |
| SMARCA2 | SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 | |
| SNX17 | sorting nexin 17 | |
| SPOCK1 | sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 1 | |
| SPRYD3 | SPRY domain containing 3 | |
| ST3GAL5 | ST3 beta-galactoside alpha-2,3-sialyltransferase 5 | |
| THBS1 | thrombospondin 1 | |
| TMEM45A | transmembrane protein 45A | |
| TNFRSF12A | tumor necrosis factor receptor superfamily, member 12A | |
| TNIP1 | TNFAIP3 interacting protein 1 | |
| TOR1B | torsin family 1, member B (torsin B) | |
| WNT1 | wingless-type MMTV integration site family, member 1 | |
| ZFPM2 | zinc finger protein, multitype 2 | |
| ZMYND11 | zinc finger, MYND-type containing 11 | |
| hsa-miR-92b-3p | AATK | apoptosis-associated tyrosine kinase |
| ACAN | aggrecan | |
| ADAMTSL1 | ADAMTS-like 1 | |
| ARHGEF17 | Rho guanine nucleotide exchange factor (GEF) 17 | |
| BAHCC1 | BAH domain and coiled-coil containing 1 | |
| DKK3 | dickkopf homolog 3 (Xenopus laevis) | |
| GATA2 | GATA binding protein 2 | |
| GRIA3 | glutamate receptor, ionotrophic, AMPA 3 | |
| HIPK3 | homeodomain interacting protein kinase 3 | |
| ITGA5 | integrin, alpha 5 (fibronectin receptor, alpha polypeptide) | |
| JPH2 | junctophilin 2 | |
| KIF5B | kinesin family member 5B | |
| KLF2 | Kruppel-like factor 2 (lung) | |
| MYH9 | myosin, heavy chain 9, non-muscle | |
| MYO1B | myosin IB | |
| NKX2-3 | NK2 homeobox 3 | |
| OAZ3 | ornithine decarboxylase antizyme 3 | |
| PAX9 | paired box 9 | |
| POLK | polymerase (DNA directed) kappa | |
| RAB23 | RAB23, member RAS oncogene family | |
| RGS3 | regulator of G-protein signaling 3 | |
| RRBP1 | ribosome binding protein 1 homolog 180kDa | |
| SLC39A6 | solute carrier family 39 (zinc transporter), member 6 | |
| SMAD6 | SMAD family member 6 | |
| hsa-miR-130b-3p | ACVR1 | activin A receptor, type IB |
| APCDD1 | adenomatosis polyposis coli down-regulated 1 | |
| CLIP1 | CAP-GLY domain containing linker protein 1 | |
| COL6A3 | collagen, type VI, alpha 3 | |
| ELK3 | ELK3, ETS-domain protein (SRF accessory protein 2) | |
| FRMD6 | FERM domain containing 6 | |
| INHBB | inhibin, beta B | |
| LYSMD2 | LysM, putative peptidoglycan-binding, domain containing 2 | |
| MAP3K12 | mitogen-activated protein kinase kinase kinase 12 | |
| MLLT6 | myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 6 | |
| NDEL1 | nudE nuclear distribution gene E homolog (A. nidulans)-like 1 | |
| NRP2 | neuropilin 2 | |
| PFKFB3 | 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 3 | |
| PPARG | peroxisome proliferator-activated receptor gamma, coactivator 1 beta | |
| STARD13 | StAR-related lipid transfer (START) domain containing 13 | |
| STX12 | syntaxin 12 | |
| TMEM9B | TMEM9 domain family, member B | |
| WNT1 | wingless-type MMTV integration site family, member 10A |
Analysis of up-regulated miRNAs expressed in DPSCs-iPSCs and SCAP-iPSCs
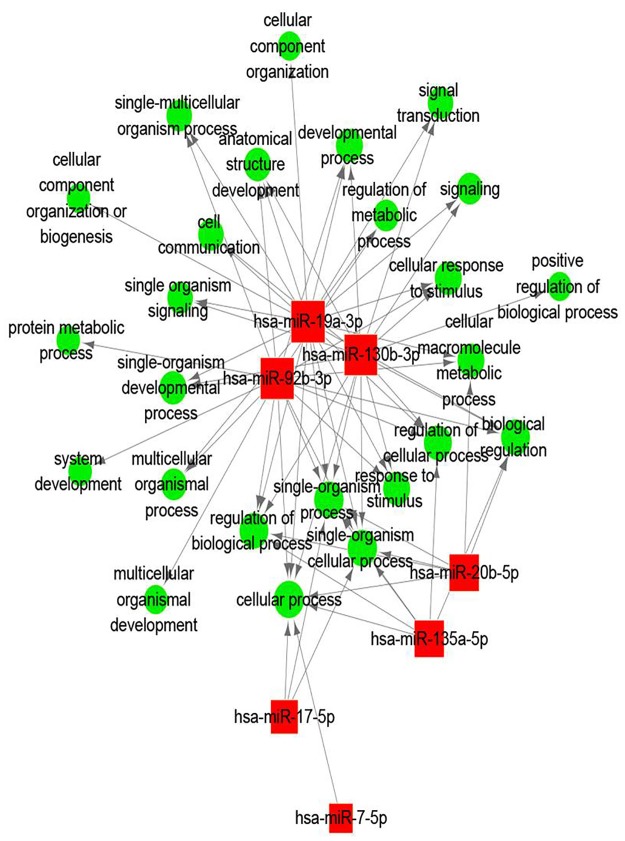
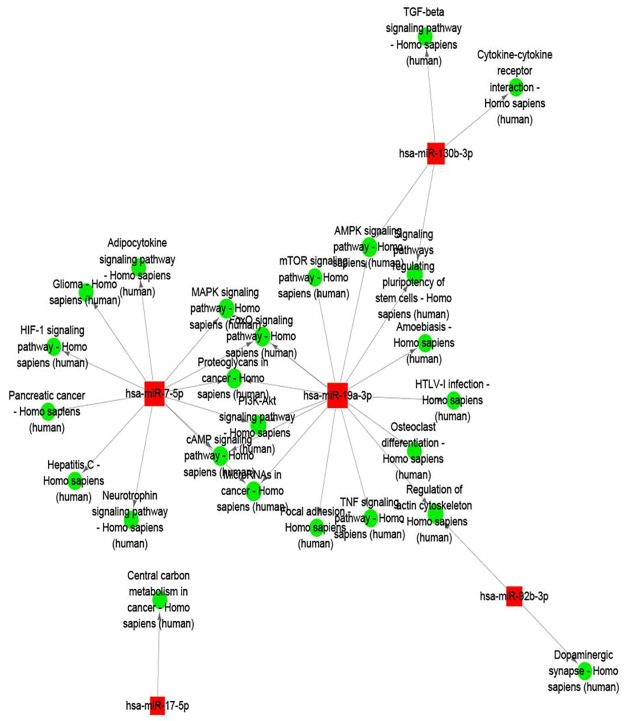
Further bioinformatics analyses are conducive to investigate the roles of the three highly expressed miRNAs in the reprogramming process of human dental iPS cells. Gene Ontology and KEGG pathway enrichment analyses were used to predict the biological functions of the miRNA targets. GO analysis of the miRNA targets showed that miR-19a-3p, miR-92b-3p and miR-130b-3p were mostly involved in the function of cell biological process, metabolic regulation and stimulating reaction (Fig 6 and S2 Table). The biological functions generated from the top 3 up-regulated miRNA demonstrated their involvement in various pathways. The miR-19a-3p, it was related with 15 pathways, such as signaling pathways regulating pluripotency of stem cells, focal adhesion and amoebiasis. And the miR-92b-3p was involved in the regulation of actin cytoskeleton and dopaminergic synapse. As for the miR-130b-3p, it was showed the participate in the cytokine-cytokine receptor interaction, AMPK and TGF-beta signaling pathway and signaling pathways regulating pluripotency of stem cells (Fig 7 and S3 Table). Hence, our prediction that the differentially expressed miRNAs have the capability to target various components in these crucial pathways makes them promising molecular targets for the induction of iPSCs.
Fig 6. The significantly enriched Gene Ontology (GO) terms of the up-regulated expressed miRNAs.
The red nodes represent the target genes of the differentially expressed miRNAs, and the green nodes represent the molecular function of the GO terms.
Fig 7. The predicted target genes involve the KEGG pathway-mediated regulatory network in the reprogramming process of human dental iPS cells.
The red nodes represent the target genes of the differentially expressed miRNAs, and the green nodes represent the involved KEGG pathway.
Validation of the three differentially expressed miRNAs by using qRT-PCR
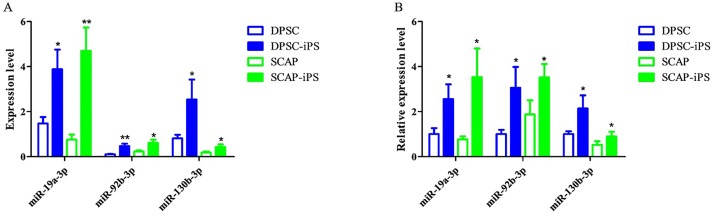
As shown in Fig 8A, the results detected on microarray indicated that the miR-19a-3p, miR-92b-3p and miR-130b-3p displayed substantial increase expression in DPSCs-iPSCs and SCAP-iPSCs. The distinct expressions of the three miRNAs found in DPSCs-iPSCs and SCAP-iPSCs were further confirmed by using qRT-PCR analysis. As a result, they all showed consistent tendency as those detected on microarray (Fig 8B).
Fig 8. Comparison of the levels of three differentially expressed miRNAs detected on microarray (A) and by qRT-PCR(B).
t-test was implemented for comparisons between dental-derived pluripotent stem cells before and after reprogrammed. *, p<0.05; **, p<0.01.
Discussion
Stem cells have the ability to rescue and/or repair injured tissue and could be isolated from the human body. Among these, DPSCs and SCAP are relatively easily obtainable and exhibit high plasticity and multipotential capabilities [14]. A previous had addressed that their isolation, selection, and differentiation are of great importance [15]. Using a Sendai virus vector, we successfully generated the DPSCs-iPSCs and SCAP-iPSCs with typical iPSCs characteristics, fibroblastoid morphology, proliferation, multipotent differentiation capability and the expression of a typical set of haematopoietic surface markers.
It has been clearly revealed that varied types of cell vary not only in the expression of their coding genes, but also in the expression of their non protein-coding portion of the genome [16]. Currently, one of the non protein-coding portion that has received increasing attention is noncoding RNAs (ncRNAs), which is of crucial functional relevance in several mechanisms of gene regulation, both for normal development and physiology, and for human diseases [17]. The most well characterized ncRNA are lncRNAs and miRNAs. As the best-known class of ncRNAs, miRNAs is able to regulate many target genes and control gene expression through translational repression and degradation [18]. In the present study, we compared differences in miRNAs expression between DPSCs/SCAP and DPSCs-iPSCs/SCAP-iPSCs to recognized pluripotent specific miRNAs. 134 up-regulated miRNA genes were identified in DPSCs-iPSCs and 265 in SCAP-iPSCs. Among these up-regulated miRNA genes, 117 miRNAs with more than 2-fold enhancement were found in both DPSCs-iPSCs and SCAP-iPSCs. These results were suggested that the role played by the differently expressed miRNAs in the difference between reprogrammed cells and DPSCs/SCAP. Based on these up-regulated miRNAs, 3 specific miRNAs, namely hsa-miR-19a-3p, hsa-miR-92b-3p and hsa-miR-130b-3p, with the maximum difference between reprogrammed cells and DPSCs/SCAP were entered into further discussion.
The hsa-miR-19a-3p, was reported as a miRNA involved in the leukotriene biosynthesis via targeting 5-Lipoxygenase in a cell type- and stimulus-specific manner [19]. This gene is said to function as vital feedback in signal molecules between irradiated GBM stem-like cells and non-irradiated cells [20]. Interestingly, there are also reports revealing that miR-19a and miR-19b, which are oncogenic in human malignancies, are the key components in human fibroblastic cell reprogramming and enhanced human fibroblast reprogramming, in the presence or absence of cMyc [21]. In this study, miR-19a-3p was highly expressed in two dental stem cells (DPSCs-iPSCs and SCAP-iPSCs), suggesting that hsa-miR-19a-3p may have potent to stimulate induction of iPSCs. Furthermore, the target gene analysis showed that this miRNA was targeted forty genes (Table 2). Among these targeted genes, WNT was relevant to governing transcription of pluripotent genes, self-renewal and differentiation in most of the SCs found in adult tissues [22]. Dissimilar levels of WNT signaling were resulted in distinct lineage-specific differentiation properties in human ESCs [23]. Our further prediction revealed that hsa-miR-19a-3p was involved in signaling pathways regulating pluripotency of stem cells, focal adhesion and amoebiasis. Further study is warrant as for the relationship between hsa-miR-19a-3p and these signaling pathway.
Hsa-miR-92b-3p has been reported to target several important lipogenic gene to regulate the networks of lipid deposition [24]. There are also report indicating that miR-92b-3p is to be specifically associated with neural progenitors [25]. The computational data in our study predicted that hsa-miR-92b-3p is connected to many targeted genes, such as AATK, GATA2, HIPK3, MYO1B, OAZ3 and SMAD6. Further prediction showed that hsa-miR-92b-3p was involved in the regulation of actin cytoskeleton and dopaminergic synapse. This miRNA showed elevated expression in reprogrammed dental cells, implying that miR-92b-3p may be capable of stimulating induction of iPSCs.
hsa-miR-130b-3p has been known to promotes CD133+ liver tumor-initiating cell growth and self-renewal via tumor protein 53-induced nuclear protein 1 [26]. It was also reported that this miRNA was targeted the insulin-like growth factor (IGF-1) in the epithelium [27]. TGF-b signaling reportedly functions in both hES and mES cell self-renewal, and FGF2, a widely used growth factor for ES cell culture, induces TGF-b ligand expression and suppresses BMP-like activities [28]. Our analysis on the hsa-miR-130b-3p showed its involvement in the cytokine-cytokine receptor interaction, AMPK and TGF-b signaling pathway and signaling pathways regulating pluripotency of stem cells, suggesting that hsa-miR-130b-3p may be a potential inducer of iPSCs.
Conclusions
In conclusion, we report differentially expressed miRNAs between reprogrammed cells and DPSCs/SCAP. The specific expression of miRNAs, namely hsa-miR-19a-3p, hsa-miR-92b-3p and hsa-miR-130b-3p, which were prediction to be related to the cell cycle, TGF beta signaling pathway and epithelial mesenchymal transition, may reflect the difference between naturally pluripotent cells and reprogrammed cells. The abilities and mechanism of these specific expression of miRNAs in the reprogramming process of human dental iPS cells warrant further exploration.
Supporting information
(XLSX)
(XLS)
(XLS)
Acknowledgments
This study was supported by National Natural Science Foundation of China (81360161) and Yunnan Provincial Department of Education Science Foundation of China (2015Y153).
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This study was supported by National Natural Science Foundation of China (81360161) and Yunnan Provincial Department of Education Science Foundation of China (2015Y153). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Kazutoshi T, Koji T, Mari O, Megumi N, Tomoko I, Kiichiro T, et al. (2007) Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell 131: 861–872. 10.1016/j.cell.2007.11.019 [DOI] [PubMed] [Google Scholar]
- 2.Yu J, Vodyanik MA, Smugaotto K, Antosiewiczbourget J, Frane JL, Tian S, et al. (2007) Induced pluripotent stem cell lines derived from human somatic cells. Stem Cell Rev 318: 1917–1920. [DOI] [PubMed] [Google Scholar]
- 3.Hu K (2014) All roads lead to induced pluripotent stem cells: the technologies of iPSC generation. Stem Cells Dev 23: 1285–1300. 10.1089/scd.2013.0620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Choi J, Lee S, Mallard W, Clement K, Tagliazucchi GM, Lim H, et al. (2015) A comparison of genetically matched cell lines reveals the equivalence of human iPSCs and ESCs. Nat Biotech 33: 1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Marson A, Levine SS, Cole MF, Frampton GM, Brambrink T, Johnstone S, et al. (2008) Connecting microRNA Genes to the Core Transcriptional Regulatory Circuitry of Embryonic Stem Cells. Cell 134: 521 10.1016/j.cell.2008.07.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Skreka K, Schafferer S, Nat IR, Zywicki M, Salti A, Apostolova G, et al. (2012) Identification of differentially expressed non-coding RNAs in embryonic stem cell neural differentiation. Nucleic Acids Res 40: 6001 10.1093/nar/gks311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Papapetrou EP, Sadelain M (2011) Generation of transgene-free human induced pluripotent stem cells with an excisable single polycistronic vector. Nat Protoc 6: 1251–1273. 10.1038/nprot.2011.374 [DOI] [PubMed] [Google Scholar]
- 8.Sharma A, Wu JC (2013) MicroRNA expression profiling of human-induced pluripotent and embryonic stem cells. Meth Mol Biol 936: 247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sonoyama W, Liu Y, Fang D, Yamaza T, Seo BM, Zhang C, et al. (2006) Mesenchymal Stem Cell-Mediated Functional Tooth Regeneration in Swine. Plos One 1: e79 10.1371/journal.pone.0000079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vasanthan P, Govindasamy V, Gnanasegaran N, Kunasekaran W, Musa S, Kasim NHA (2015) Differential expression of basal microRNAs' patterns in human dental pulp stem cells. J Cell Mol Med 19: 566–580. 10.1111/jcmm.12381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zou XY, Yang HY, Yu Z, Tan XB (2012) Establishment of transgene-free induced pluripotent stem cells reprogrammed from human stem cells of apical papilla for neural differentiation. Stem Cell Res & Ther 3: 43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Balaga O, Friedman Y, Linial M (2012) Toward a combinatorial nature of microRNA regulation in human cells. Nucleic Acids Res 40: 9404–9416. 10.1093/nar/gks759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Walter W, Sánchez-Cabo F, Ricote M (2015) GOplot: an R package for visually combining expression data with functional analysis. Bioinformatics 31: 2912–2914. 10.1093/bioinformatics/btv300 [DOI] [PubMed] [Google Scholar]
- 14.La NM, Paino F, Spina A, Naddeo P, Montella R, Desiderio V, et al. (2014) Dental pulp stem cells: state of the art and suggestions for a true translation of research into therapy. J Dent 42(7):761–8. 10.1016/j.jdent.2014.02.018 [DOI] [PubMed] [Google Scholar]
- 15.Tirino V, Paino F, Rosa AD Papaccio G (2012) Identification, Isolation, Characterization, and Banking of Human Dent. Meth Mol Biol 879(5):443. [DOI] [PubMed] [Google Scholar]
- 16.Barrett JR (2004) Template for toxicants: gene expression varies by cell type. Environ Health Pers 112: A944. [PMC free article] [PubMed] [Google Scholar]
- 17.Misso G, Zarone M R, Grimaldi A, Di MM, Lombardi A, Kawasaki H, et al. (2017). Non Coding RNAs: A New Avenue for the Self-Tailoring of Blood Cancer Treatment. Current Drug Targets, 2016. 18(1):35–55. [DOI] [PubMed] [Google Scholar]
- 18.Hausser J, Zavolan M (2014) Identification and consequences of miRNA-target interactions—beyond repression of gene expression. Nat Rev Gene 15: 599. [DOI] [PubMed] [Google Scholar]
- 19.Busch S, Auth E, Scholl F, Huenecke S, Koehl U, Suess B, et al. (2015) 5-Lipoxygenase Is a Direct Target of miR-19a-3p and miR-125b-5p. J Immunol 194: 1646–1653. 10.4049/jimmunol.1402163 [DOI] [PubMed] [Google Scholar]
- 20.Tokudome T, Sasaki A, Tsuji M, Udaka Y, Oyamada H, Tsuchiya H, et al. (2015) Reduced PTEN expression and overexpression of miR-17-5p, -19a-3p, -19b-3p, -21-5p, -130b-3p, -221-3p and -222-3p by glioblastoma stem-like cells following irradiation. Oncol Lett 10: 2269 10.3892/ol.2015.3594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.He X, Cao Y, Wang L, Han Y, Zhong X, Zhou G, et al. (2014) Human fibroblast reprogramming to pluripotent stem cells regulated by the miR19a/b-PTEN axis. Plos One 9: e95213 10.1371/journal.pone.0095213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wend P, Holland JD, Ziebold U, Birchmeier W (2010) Wnt signaling in stem and cancer stem cells. Semin Cell Dev Biol 25: 254–264. [DOI] [PubMed] [Google Scholar]
- 23.Blauwkamp TA, Nigam S, Ardehali R, Weissman IL, Nusse R (2011) Endogenous Wnt signalling in human embryonic stem cells generates an equilibrium of distinct lineage-specified progenitors. Nat Commun 3: 1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang Z (2015) Identification of Genes Related to Growth and Lipid Deposition from Transcriptome Profiles of Pig Muscle Tissue. Plos One 10: e141138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jönsson ME, Jenny NW, Malin Å, Agnete K, Josephine M, Ludvik BP, et al. (2011) Comprehensive analysis of microRNA expression in regionalized human neural progenitor cells reveals microRNA-10 as a caudalizing factor. Development 142: 3166–3177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ma S, Tang KH, Chan YP, Lee TK, Kwan PS, Castilho A, et al. (2010) miR-130b Promotes CD133(+) liver tumor-initiating cell growth and self-renewal via tumor protein 53-induced nuclear protein 1. Cell Stem Cell 7: 694–707. 10.1016/j.stem.2010.11.010 [DOI] [PubMed] [Google Scholar]
- 27.Li S, Jing G, Xu X, Huang X, Dong L, Jiang D, et al. (2016) miR-130b-3p Modulates Epithelial-Mesenchymal Crosstalk in Lung Fibrosis by Targeting IGF-1. Plos One 11: e150418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Galvin-Burgess KE, Travis ED, Pierson KE, Jay LV (2013) TGF-β-Superfamily Signaling Regulates Embryonic Stem Cell Heterogeneity: Self-Renewal as a Dynamic and Regulated Equilibrium. Stem Cells 31: 48–58. 10.1002/stem.1252 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(XLSX)
(XLS)
(XLS)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.