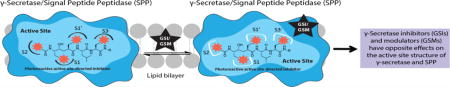
Abstract
γ-Secretase inhibitors (GSIs) and modulators (GSMs) are at the frontline of cancer and Alzheimer’s disease research, respectively. While both are therapeutically promising, not much is known about their interactions with proteins other than γ-secretase. Signal peptide peptidase (SPP), like γ-secretase, is a multispan transmembrane aspartyl protease that catalyzes regulated intramembrane proteolysis. We used active site-directed photophore walking probes to study the effects of different GSIs and GSMs on the active sites of γ-secretase and SPP and found that nontransition state GSIs inhibit labeling of γ-secretase by activity-based probes but enhance labeling of SPP. The opposite is true of GSMs, which have little effect on the labeling of γ-secretase but diminish labeling of SPP. These results demonstrate that GSIs and GSMs are altering the structure of not only γ-secretase but also SPP, leading to potential changes in enzyme activity and specificity that may impact the clinical outcomes of these molecules.
Graphical abstract
Signal peptide peptidase (SPP) and γ-secretase are aspartyl proteases that catalyze regulated intramembrane proteolysis, a process that controls the activity and function of membrane proteins in all living systems studied to date.1 SPP proteolyzes signal peptides in their transmembrane region after they have been cleaved off from the protein by signal peptidase. The signal peptide fragments released by SPP are required for important cellular events such as immune surveillance by HLA-E epitopes and HCV polyprotein cleavage critical for viral lifecycle.2,3 γ-Secretase has over 90 reported substrates, of which Notch and APP are the most heavily studied due to the roles they play in cancer and Alzheimer’s disease (AD).4 However, how many of these substrates are processed under physiological conditions needs to be further investigated.5 SPP and γ-secretase are multispan transmembrane (TM) proteins that share the same YD/GXGD catalytic motif6 but are structurally and functionally distinct (Figure 1). The key differences between the two proteins are, first, γ-secretase is a multisubunit protein complex that includes Aph1, Nct, Pen2, and Presenilin (PS).7–10 PS, the catalytic subunit of the complex,11–13 can be found as either the PS1 or PS2 homologue, which is present in mutually exclusive γ-secretase complexes.14 Unlike γ-secretase, SPP appears to function alone without the participation of other protein cofactors.15 Second, SPP and PS have opposite orientations within the membrane.15–17 As a consequence, γ-secretase cleaves type 1 TM substrates while the substrates of SPP are type 2. Third, endogenous PS is expressed as a full-length protein that is activated by endoproteolytic cleavage during protein maturation.8,18 Endoproteolysis results in the formation of two fragments, PS amino-terminal fragment (PS-NTF), and PS carboxy-terminal fragment (PS-CTF), which remain associated within active γ-secretase complexes. Conversely, SPP functions as a full-length protein.15,19,20 There is evidence to suggest that SPP forms dimers and tetramers within the cell.19–21
Figure 1.
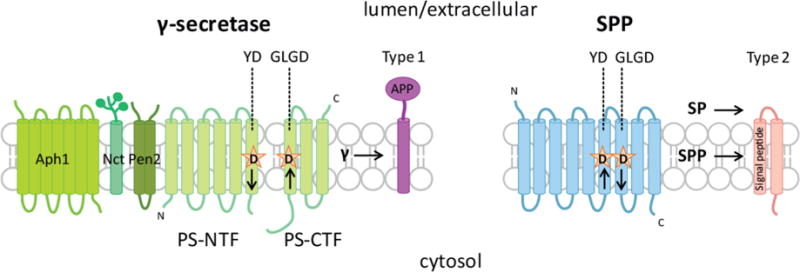
Structural and functional similarities/differences between SPP and γ-secretase. SPP and γ-secretase are similar in that they are both multipass transmembrane enzymes that share the YD/GXGD catalytic motif and cleave their respective substrates in the transmembrane region. SPP and PS, the catalytic subunit of γ-secretase, both transverse the membrane 9 times. The differences between the enzymes include their limited sequence homology, their inverse orientation in the membrane, and their specificity for either type 1 or type 2 substrates. Additionally, SPP functions without protein cofactors while PS requires at least 3 additional proteins (Aph1, Nct, and Pen2) for γ-secretase activity.
Although PS and SPP have opposite topologies with limited sequence homology, they share pharmacological characteristics: transition-state γ-secretase inhibitor L-685,458 (L458) inhibits SPP activity,22,23 and L458-based photoaffinity probes L-852,646 (L646) and CBAP-BPyne specifically label SPP.22,24,25 These data suggest structural similarity within the active sites of SPP and γ-secretase. However, because there is no crystal structure of either SPP or γ-secretase, scant information exists regarding the active site architecture of these enzymes. Moreover, a comparison study exploring the effect of γ-secretase inhibitors (GSIs) and modulators (GSMs), which are being developed for cancer and AD treatment, on the active sites of SPP and γ-secretase has not been reported. We use a chemical biology approach to probe the active sites of PS1 and SPP. Particularly, we employ active site-directed photoprobes, in a method called photophore walking, to target the various subpockets of the enzyme active site (Figure 2A).5,26–29 Comparison of the photolabeling profiles of PS1 and SPP demonstrated that the active sites of these proteins are similar, yet some differences are apparent in specific active site subpockets. Furthermore, we used the photophore walking approach to determine the effects of GSIs and GSMs on the active site conformations of γ-secretase and SPP, which is an important step toward understanding off-target effects of GSI and GSM treatment. In addition, we provide confirming evidence that active SPP exists as a stable homodimer.
Figure 2.
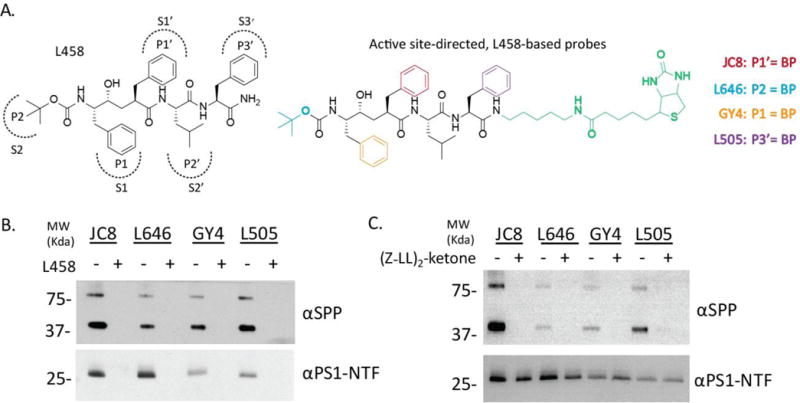
L458-based photoreactive probes specifically label PS1 and SPP. (A) Structures of L458 and photoreactive probes JC8, L646, GY4, and L505. L458 side chain residues (P and P′ sites) interact with corresponding subpockets of γ-secretase and SPP (S and S′ sites). Photoreactive probes JC8, L646, GY4, and L505 have an L458 backbone (black), a biotin moiety (green), and a cross-linkable benzophenone (BP). Each probe has a BP incorporated into a different site on the L458 backbone. The location of the BP is illustrated by the color scheme. For example, JC8 has a BP at the P1′ site, in place of the red benzene ring. JC8, L646, GY4, and L505 label S1′, S2, S1, and S3′ subsites of the enzymes, respectively. (B) HeLa membranes were labeled with 20 nM of photoprobes JC8, L646, GY4, or L505 in the presence of 0.25% CHAPSO, and either with (+) or without (−) 2 μM L458. Samples were run on SDS-PAGE and analyzed by Western blot. Anti-SPP and PS1-NTF antibodies were used to detect SPP (upper panel) and PS1-NTF (lower panel). (C) Same as B, except 2 μM (Z-LL)2-ketone was used to block the labeling of SPP (upper panel) and PS1-NTF (lower panel).
RESULTS AND DISCUSSION
PS1 and SPP active site conformations are similar yet distinct
L458 has been a valuable tool in the study of γ-secretase.30 We incorporated a photoreactive benzophenone moiety into different side chain positions of L458 (P and P′ sites) to generate a series of photoprobes. These photoprobes, JC8, L646, GY4, and L505, photo-cross-link to S1′, S2, S1, or S3′ subsites within the γ-secretase active site, respectively (Figure 2A).12,26,27,31 A biotin linker was attached for isolation of the labeled species. This photophore walking approach facilitates the comparison of active site conformational changes of target enzymes, as the photo-cross-linking efficiency is determined by the distance and orientation between the contact residues of the enzyme active site and the benzophenone moiety of the photoprobes. By comparing the photo-cross-linking efficiency of each probe to PS1 and SPP, we can gain insight into the conformational differences between the active sites of these enzymes.
Total membrane isolated from HeLa cells was used as the source of endogenous γ-secretase and SPP. First, we confirmed that JC8, L646, GY4, and L505 photolabeled PS1-NTF and this labeling was specific since an excess amount of L458 blocked the labeling (Figure 2B, lower panel). Furthermore, the photolabeling profile of PS1-NTF revealed that JC8 labeled PS1-NTF with the strongest intensity followed by L646. GY4 and L505, on the other hand, were both weaker photoprobes for PS1-NTF (Figure 2B, lower panel). SPP was photo-cross-linked with the same set of photoprobes, and the labeling was specific, as excess L458 was able to block the photoinsertion of the probes (Figure 2B, upper panel). Additionally we used (Z-LL)2-ketone, a peptidomimetic SPP inhibitor (Figure 2C) with an in vitro IC50 of 50 nM,32 to confirm that SPP is being specifically labeled and inhibited in our system. As expected, excess (Z-LL)2-ketone blocked the photolabeling of SPP completely (Figure 2C, upper panel) but had little effect on the photolabeling of PS1-NTF (Figure 2C, lower panel).
These results indicate that the photolabeling profile of SPP is different from that of PS1. Although JC8 labeled both PS1 and SPP with the strongest intensity, L646 was a much weaker photoprobe for SPP. Instead, L505 photolabeled SPP with similar efficiency to JC8. GY4 was a weaker photoprobe for both PS1 and SPP (Figure 2B). Comparison of the PS1 and SPP photolabeling profiles reveals that while two probes label the enzymes with similar efficiency (JC8 and GY4), the other two probes exhibit opposite labeling efficiencies (L646 and L505), suggesting that γ-secretase and SPP have similar architecture of the S1′ and S1 subpockets, whereas the S2 and S3′ subpockets are organized differently. In conclusion, the overall structure of the PS1 and SPP active sites is similar since both enzymes can be specifically labeled by all four active site-directed photoprobes, but the structure of the active site subsites differs between the two enzymes, exemplified by the difference in labeling efficiency among photoprobes. How variation of these subsites contributes to the activity and specificity of γ-secretase and SPP remains to be investigated.
Active SPP exists predominantly as a homodimer
Initial characterization of SPP showed that endogenous SPP is a monomer.15,22 Further studies by Nyborg et al. demonstrated that SPP is a homodimer that is SDS-stable and partially heat-labile.19,20 However, only the monomeric form of SPP was labeled with the active site-directed photoprobe III-63.33,34 Additionally, endogenous DDM-solubilized human and drosophila SPP formed higher molecular weight complexes around 180−200 kDa.21 We utilized a photolabeling approach to elucidate whether endogenous active SPP is a homodimer or monomer. We synthesized JC10, a photoprobe structurally similar to JC8 except with the addition of a disulfide bond in the biotin linker that can be eluted from the streptavidin matrix with the reducing agent TCEP (Figure 3A).35 Our data showed that the majority of SPP was monomeric when heat was used to elute SPP from JC8 (Figure 3B, first panel), with some residual SPP dimer still detected. However, in the absence of heat, JC10 photolabeled SPP was predominantly a homodimer (Figure 3B, second panel). Similar to the SPP labeled with JC8 (Figures 2B, C and 3B), most of the SPP dimer was converted into the monomer when heat was applied (Figure 3B, third panel). This confirmed the finding that dimer SPP is heat-labile and provided further evidence that endogenous, active SPP exists as a homodimer. To detect predominantly SPP dimer, we eluted proteins with Laemmli Sample Buffer + 2 mM biotin at 70 °C. Under these conditions, SPP does not dissociate into its monomeric form. As a result, we show only SPP dimer in subsequent experiments.
Figure 3.
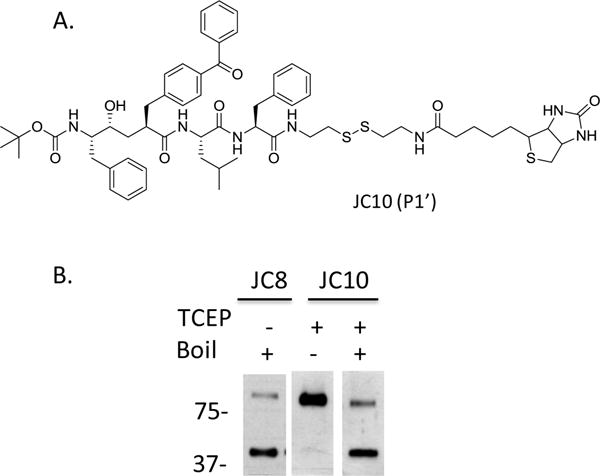
Endogenous, active SPP is a homodimer. (A) Structure of JC10. (B) JC8 and JC10 were used to photolabel SPP. TCEP and/or heat were used to elute photolabeled proteins.
GSIs and GSMs have opposite effects on the active site conformational changes of PS1 and SPP
We have previously used photoprobes to detect γ-secretase active site conformational changes induced by the binding of GSIs and GSMs.26,27,29 However, the effect of GSIs and GSMs on the active site conformation of SPP has not been reported.
We investigated the effects of four GSIs and two GSMs on the active site conformations of γ-secretase and SPP (Figure 4). GSIs inhibit production of Notch intracellular domain (NICD) and all Aβ species including Aβ40 and Aβ42 while GSMs have little impact on NICD production and selectively inhibit production of Aβ42.5 BMS-708163, commonly known as avagacestat, was discordantly reported as both a Notch-sparing GSI36 and a nonselective GSI,37,38 that completed phase II clinical trials but did not proceed any further. LY450139, also known as semagacestat, advanced into a Phase III clinical trial but was terminated prematurely due to side effects potentially stemming from Notch-associated toxicity.39 Compound X (cpd X) and GSI-34 are both GSIs.40,41 E2012 is an imidazole-derived GSM which has been shown to bind SPP,42 and GSM-616 is an acetic acid GSM43 that binds SPP.29
Figure 4.
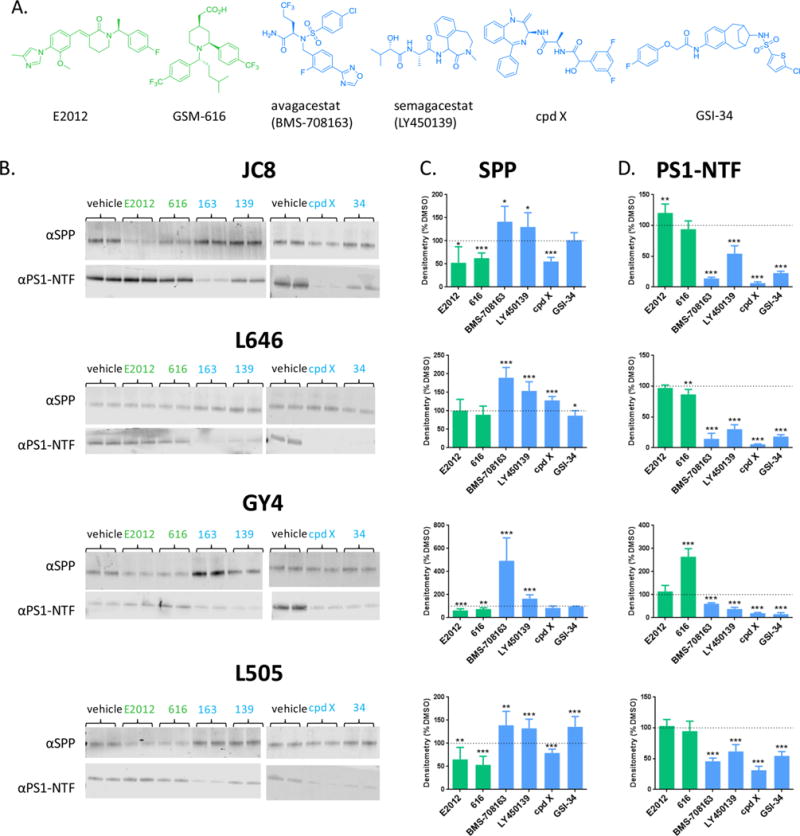
GSIs and GSMs have opposite effects on the photolabeling profiles of γ-secretase and SPP. (A) Structures of E2012, GSM-616, BMS-708163, LY450139, cpd X, and GSI-34. (B) Photoprobes JC8, L646, GY4, and L505 were incubated with HeLa membrane in 0.25% CHAPSO in the presence of 25 μM GSMs E2012/616 (green) or 10 μM GSIs 708163/139/cpd X/GSI-34 (blue). Samples were run on SDS-PAGE and analyzed by Western blot. Anti-SPP and PS1-NTF antibodies were used to detect SPP (upper panel) and PS1-NTF (lower panel). (C) Densitometry quantification of SPP labeling. GSMs are graphed in green, and GSIs are graphed in blue. (D) Same as C, except PS1-NTF. ns P > 0.05; *P ≤ 0.05; **P ≤ 0.01; ***P ≤ 0.001.
We tested the effects of BMS-708163, LY450139, cpd X and GSI-34 on the photolabeling of PS1 and SPP by active site-directed probes. The effect of these GSIs was assessed by comparison to vehicle control. As anticipated, these GSIs inhibited the photolabeling of PS1-NTF (Figure 4D). Surprisingly, they selectively enhanced the labeling of SPP (Figure 4C). These data suggest that the GSIs tested have opposite effects on the active site conformations of SPP and γ-secretase. The increase in SPP labeling in the presence of GSIs may be due to a direct interaction between the GSI and SPP, leading to a change in SPP active site conformation that improves labeling with the active site-directed probe. Alternatively, the increase in labeling may be due to increased availability of the active site-directed probe as a result of a reduction in the number of active γ-secretase complexes available for binding. The latter hypothesis does not require direct binding between GSIs and SPP, and is based on the data that shows a reduction in PS1 labeling in the presence of GSIs (Figure 4B and C), which may suggest that the active site-directed probes that are not engaged in labeling PS1 are labeling SPP. While both hypotheses are feasible explanations for the increase in SPP labeling in the presence of GSIs, the data support the “direct labeling” hypothesis for the following reasons: 1. In the presence of GSIs, SPP labeling is enhanced for some, but not all, active site-directed probes. If the increase in SPP labeling were a result of an increase in probe availability, all active site-directed probes would be expected to label SPP more robustly, but we do not observe this. 2. Fuwa et al. found that a compound E-based probe, which is identical to cpd X with the exception of a single hydroxyl group, specifically labels SPP, showing direct binding between this GSI and SPP.44 For these reasons it is likely that the GSIs studied here are directly binding SPP. We also tested the effects of E2012 and GSM-616 on the photolabeling of PS1 and SPP. Although these GSMs have been shown to modulate γ-secretase activity,29,42 they had little effect on the active site labeling of PS1-NTF (with the exception of the S1 subsite for GSM-616), suggesting that these compounds affect γ-secretase activity without drastically altering the active site conformation (Figure 4D). More interestingly, these GSMs partially reduced the active site labeling of SPP by all photoprobes except L646 (Figure 4C), suggesting that both of these structurally distinct GSMs affect the same subpockets of the SPP active site. Additionally, we and others have reported that GSM-1, which is a close homologue of GSM-616, and GSM E2012, directly bind SPP.29,42
The combined data show that while GSIs inhibit labeling of PS1 and have no effect on or enhance labeling of SPP, the opposite is true of GSMs, which inhibit labeling of SPP and have little to no effect on labeling of PS1. A clear exception is the pronounced increase in GY4 labeling of PS1 in the presence of GSM-616 (Figure 4D), which was previously reported.29 The trend, therefore, is that GSIs and GSMs have opposite effects on the photolabeling profiles of γ-secretase and SPP (Figure 5). The data suggest that not only GSMs, as previously reported, but also GSIs directly bind to SPP, potentially leading to the observed conformational change in its active site. Consequently, GSIs in clinical trials for cancer and GSMs developed for AD treatment may lead to undesirable effects associated with concomitant changes in SPP structure. This possibility is worth studying as SPP is essential in eukaryotes45–47 and a change in its activity and specificity may affect the therapeutic windows of GSIs and GSMs.
Figure 5.
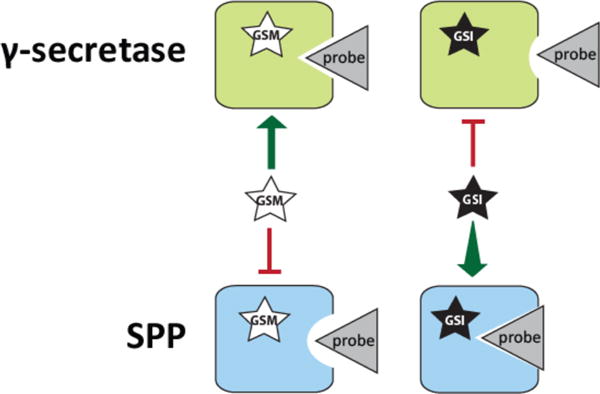
Model for the change in active site conformation of γ-secretase and SPP that occurs upon binding by GSIs and GSMs. We propose that the GSIs and GSMs studied here allosterically bind to γ-secretase and SPP, causing a conformational change in the active sites of the enzymes. Surprisingly, the induced conformational change is opposite for the two enzymes, as evidenced by their binding to active site-directed probes. Specifically, GSIs cause decreased binding between γ-secretase and probe while increasing binding between SPP and probe. GSMs cause little change in binding between γ-secretase and probe but reduce binding between SPP and probe. This suggests a model in which GSIs cause the active site of γ-secretase to assume a “closed” conformation but have the reverse impact on the active site structure of SPP.
CONCLUSION
Determining allosteric site-induced conformational changes in the active sites of enzymes for which the crystal structures have not been solved has been a big challenge. To address this, we developed and applied the photophore walking technique for probing the active sites of two such enzymes, γ-secretase and SPP. We found that while the S1′ and S1 subpockets of both enzymes are similar, the S2 and S3′ subpockets are structurally distinct. The strong overall similarity in the active sites of these two enzymes that cleave entirely different substrates may mean that the active site does not play an important role in determining substrate specificity. On the flip side, some substrate specificity may be conferred by the subtle differences in two of the four active site subpockets. It is possible that the conserved structure of the S1′ and S1 subsites is a result of the importance of these subsites in catalysis. We were surprised to find that the active sites of γ-secretase and SPP are differently affected by allosteric GSIs and GSMs. Despite the strong structural homology of the active sites of these enzymes, GSIs had a diametrical impact on the conformations of the two enzyme active sites, as gauged by interaction with active site-directed probes. Although enzyme binding to active site-directed probes is not a direct proxy for activity, there is a high likelihood that if the active site is changing, so is enzyme activity/specificity. This has important ramifications for drug development, as changes in SPP activity may occur during GSI/GSM treatment.
METHODS
HeLa cell membrane preparation and chemical compounds
HeLa membrane fraction was isolated from HeLa-S3 cells (National Cell Culture Center) as previously described.12,27 Synthesis of L458, cpd X, GSI-34, L646, GY4, JC8, L505 and (Z-LL)2-ketone were described previously.12,30–32,35,48 LY450139, BMS-708163, E2012 and GSM-616 were kindly provided by Dr. Douglas Johnson from Pfizer.
Peptide synthesis and anti-SPP antibody production
SPP peptide corresponding to the N-terminal of human SPP (MDSALSDPHNGSAEAC) was synthesized with an automated solid phase peptide synthesizer (ProteinTech) using Fmoc chemistry. The purified peptide was conjugated to maleimide functionalized keyhole limpet hemocyanin (KLH) according to the manufacturer’s instructions (Pierce). This antigen was used to immunize rabbits for polyclonal antibody production (Covance). Serum was collected and tested for SPP detection using Western blot analysis.
Photolabeling of γ-secretase and SPP.12,24,28,29,37
Total HeLa cell membrane was preincubated in the presence of 0.25% CHAPSO with or without inhibitors at 37 °C for 30 min. Then, 20 nM of photoactive probes was added to the mixture and incubated for an additional 1 h at 37 °C. The reaction mixtures were irradiated at 350 nm for 45 min and solubilized with RIPA buffer (50 mM Tris base, pH 8.0, 150 mM NaCl, 0.1% SDS, 1% Nonidet P-40, 0.5% deoxycholate) for 1 h. Photo-cross-linked proteins in the soluble fraction were pulled-down with Streptavidin Plus UltraLink Resin (Pierce) overnight at 4 °C. The resin was washed with RIPA buffer and proteins were eluted with Laemmli Sample Buffer (32.9 mM Tris-HCl, pH 6.8, 1% SDS, 13% (w/v) glycerol, 0.005% bromophenol blue) at 95 °C (Figure 2 and 3), 25 mM TCEP in PBS (Figure 3) or 2 mM biotin in Laemmli Sample Buffer at 70 °C (Figure 4). Eluted proteins were resolved on SDS-PAGE and transferred to PVDF (Millipore). PS1-NTF, PS1-CTF and SPP were detected with anti-PS1-NTF (gift from Dr. Min Xu), anti-PS1-CTF (Millipore) or anti-SPP antibody.
Acknowledgments
We thank D. Johnson from Pfizer for providing GSIs and GSMs and D. Iaea for critically reading the manuscript.
Funding
This work is supported by NIH Grant R01AG026660 (Y.M.L.), R01NS076117 (Y.M.L.), Alzheimer Association IIRG-12-242137 (Y.M.L.), the MetLife Foundation (Y.M.L.), the JPB Foundation (Y.M.L.), William H. Goodwin and Alice Goodwin, the Commonwealth Foundation for Cancer Research, the Experimental Therapeutics Center of MSKCC, and the William Randolph Hearst Fund in Experimental Therapeutics.
ABBREVIATIONS
- GSI
γ-secretase inhibitor
- GSM
γ-secretase modulator
- SPP
signal peptide peptidase
- PS
presenilin
- Aβ
amyloid beta
Footnotes
Notes
The authors declare no competing financial interest.
References
- 1.Brown MS, Ye J, Rawson RB, Goldstein JL. Regulated intramembrane proteolysis: a control mechanism conserved from bacteria to humans. Cell. 2000;100:391–398. doi: 10.1016/s0092-8674(00)80675-3. [DOI] [PubMed] [Google Scholar]
- 2.Lemberg MK, Bland FA, Weihofen A, Braud VM, Martoglio B. Intramembrane proteolysis of signal peptides: an essential step in the generation of HLA-E epitopes. J Immunol. 2001;167:6441–6446. doi: 10.4049/jimmunol.167.11.6441. [DOI] [PubMed] [Google Scholar]
- 3.McLauchlan J, Lemberg MK, Hope G, Martoglio B. Intramembrane proteolysis promotes trafficking of hepatitis C virus core protein to lipid droplets. EMBO J. 2002;21:3980–3988. doi: 10.1093/emboj/cdf414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Haapasalo A, Kovacs DM. The many substrates of presenilin/gamma-secretase. J Alzheimers Dis. 2011;25:3–28. doi: 10.3233/JAD-2011-101065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Crump CJ, Johnson DS, Li YM. Development and mechanism of gamma-secretase modulators for Alzheimer’s disease. Biochemistry. 2013;52:3197–3216. doi: 10.1021/bi400377p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fluhrer R, Steiner H, Haass C. Intramembrane proteolysis by signal peptide peptidases: a comparative discussion of GXGD-type aspartyl proteases. J Biol Chem. 2009;284:13975–13979. doi: 10.1074/jbc.R800040200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.De Strooper B. Aph-1, Pen-2, and Nicastrin with Presenilin generate an active gamma-Secretase complex. Neuron. 2003;38:9–12. doi: 10.1016/s0896-6273(03)00205-8. [DOI] [PubMed] [Google Scholar]
- 8.Takasugi N, Tomita T, Hayashi I, Tsuruoka M, Niimura M, Takahashi Y, Thinakaran G, Iwatsubo T. The role of presenilin cofactors in the gamma-secretase complex. Nature. 2003;422:438–441. doi: 10.1038/nature01506. [DOI] [PubMed] [Google Scholar]
- 9.Edbauer D, Winkler E, Regula JT, Pesold B, Steiner H, Haass C. Reconstitution of gamma-secretase activity. Nat Cell Biol. 2003;5:486–488. doi: 10.1038/ncb960. [DOI] [PubMed] [Google Scholar]
- 10.Gertsik N, Chiu D, Li YM. Complex regulation of gamma-secretase: from obligatory to modulatory subunits. Front Aging Neurosci. 2014;6:342. doi: 10.3389/fnagi.2014.00342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wolfe MS, Xia W, Ostaszewski BL, Diehl TS, Kimberly WT, Selkoe DJ. Two transmembrane aspartates in presenilin-1 required for presenilin endoproteolysis and gamma-secretase activity. Nature. 1999;398:513–517. doi: 10.1038/19077. [DOI] [PubMed] [Google Scholar]
- 12.Li YM, Xu M, Lai MT, Huang Q, Castro JL, DiMuzio-Mower J, Harrison T, Lellis C, Nadin A, Neduvelil JG, Register RB, Sardana MK, Shearman MS, Smith AL, Shi XP, Yin KC, Shafer JA, Gardell SJ. Photoactivated gamma-secretase inhibitors directed to the active site covalently label presenilin 1. Nature. 2000;405:689–694. doi: 10.1038/35015085. [DOI] [PubMed] [Google Scholar]
- 13.Ahn K, Shelton CC, Tian Y, Zhang X, Gilchrist ML, Sisodia SS, Li YM. Activation and intrinsic γ-secretase activity of presenilin 1. Proc Natl Acad Sci U S A. 2010;107:21435–21440. doi: 10.1073/pnas.1013246107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lai MT, Chen E, Crouthamel MC, DiMuzio-Mower J, Xu M, Huang Q, Price E, Register RB, Shi XP, Donoviel DB, Bernstein A, Hazuda D, Gardell SJ, Li YM. Presenilin-1 and presenilin-2 exhibit distinct yet overlapping gamma-secretase activities. J Biol Chem. 2003;278:22475–22481. doi: 10.1074/jbc.M300974200. [DOI] [PubMed] [Google Scholar]
- 15.Weihofen A, Binns K, Lemberg MK, Ashman K, Martoglio B. Identification of signal peptide peptidase, a presenilin-type aspartic protease. Science. 2002;296:2215–2218. doi: 10.1126/science.1070925. [DOI] [PubMed] [Google Scholar]
- 16.Nyborg AC, Jansen K, Ladd TB, Fauq A, Golde TE. A signal peptide peptidase (SPP) reporter activity assay based on the cleavage of type II membrane protein substrates provides further evidence for an inverted orientation of the SPP active site relative to presenilin. J Biol Chem. 2004;279:43148–43156. doi: 10.1074/jbc.M405879200. [DOI] [PubMed] [Google Scholar]
- 17.Friedmann E, Lemberg MK, Weihofen A, Dev KK, Dengler U, Rovelli G, Martoglio B. Consensus analysis of signal peptide peptidase and homologous human aspartic proteases reveals opposite topology of catalytic domains compared with presenilins. J Biol Chem. 2004;279:50790–50798. doi: 10.1074/jbc.M407898200. [DOI] [PubMed] [Google Scholar]
- 18.Thinakaran G, Borchelt DR, Lee MK, Slunt HH, Spitzer L, Kim G, Ratovitsky T, Davenport F, Nordstedt C, Seeger M, Hardy J, Levey AI, Gandy SE, Jenkins NA, Copeland NG, Price DL, Sisodia SS. Endoproteolysis of presenilin 1 and accumulation of processed derivatives in vivo. Neuron. 1996;17:181–190. doi: 10.1016/s0896-6273(00)80291-3. [DOI] [PubMed] [Google Scholar]
- 19.Nyborg AC, Kornilova AY, Jansen K, Ladd TB, Wolfe MS, Golde TE. Signal peptide peptidase forms a homodimer that is labeled by an active site-directed gamma-secretase inhibitor. J Biol Chem. 2004;279:15153–15160. doi: 10.1074/jbc.M309305200. [DOI] [PubMed] [Google Scholar]
- 20.Nyborg AC, Herl L, Berezovska O, Thomas AV, Ladd TB, Jansen K, Hyman BT, Golde TE. Signal peptide peptidase (SPP) dimer formation as assessed by fluorescence lifetime imaging microscopy (FLIM) in intact cells. Mol Neurodegener. 2006;1:16. doi: 10.1186/1750-1326-1-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Miyashita H, Maruyama Y, Isshiki H, Osawa S, Ogura T, Mio K, Sato C, Tomita T, Iwatsubo T. Three-dimensional structure of the signal peptide peptidase. J Biol Chem. 2011;286:26188–26197. doi: 10.1074/jbc.M111.260273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Weihofen A, Lemberg MK, Friedmann E, Rueeger H, Schmitz A, Paganetti P, Rovelli G, Martoglio B. Targeting presenilin-type aspartic protease signal peptide peptidase with gamma-secretase inhibitors. J Biol Chem. 2003;278:16528–16533. doi: 10.1074/jbc.M301372200. [DOI] [PubMed] [Google Scholar]
- 23.Iben LG, Olson RE, Balanda LA, Jayachandra S, Robertson BJ, Hay V, Corradi J, Prasad CV, Zaczek R, Albright CF, Toyn JH. Signal peptide peptidase and gamma-secretase share equivalent inhibitor binding pharmacology. J Biol Chem. 2007;282:36829–36836. doi: 10.1074/jbc.M707002200. [DOI] [PubMed] [Google Scholar]
- 24.Gertsik N, Ballard TE, Am Ende CW, Johnson DS, Li YM. Development of CBAP-BPyne, a probe for gamma-secretase and presenilinase. MedChemComm. 2014;5:338–341. doi: 10.1039/C3MD00281K. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Crump CJ, am Ende CW, Ballard TE, Pozdnyakov N, Pettersson M, Chau DM, Bales KR, Li YM, Johnson DS. Development of clickable active site-directed photoaffinity probes for gamma-secretase. Bioorg Med Chem Lett. 2012;22:2997–3000. doi: 10.1016/j.bmcl.2012.02.027. *Co-corresponding. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shelton CC, Zhu L, Chau D, Yang L, Wang R, Djaballah H, Zheng H, Li YM. Modulation of gamma-secretase specificity using small molecule allosteric inhibitors. Proc Natl Acad Sci U S A. 2009;106:20228–20233. doi: 10.1073/pnas.0910757106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tian Y, Bassit B, Chau D, Li YM. An APP inhibitory domain containing the Flemish mutation residue modulates gamma-secretase activity for Abeta production. Nat Struct Mol Biol. 2010;17:151–158. doi: 10.1038/nsmb.1743. [DOI] [PubMed] [Google Scholar]
- 28.Chau DM, Crump CJ, Villa JC, Scheinberg DA, Li YM. Familial Alzheimer Disease Presenilin-1 Mutations Alter the Active Site Conformation of gamma-secretase. J Biol Chem. 2012;287:17288–17296. doi: 10.1074/jbc.M111.300483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Crump CJ, Fish BA, Castro SV, Chau DM, Gertsik N, Ahn K, Stiff C, Pozdnyakov N, Bales KR, Johnson DS, Li YM. Piperidine acetic acid based gamma-secretase modulators directly bind to Presenilin-1. ACS Chem Neurosci. 2011;2:705–710. doi: 10.1021/cn200098p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shearman MS, Beher D, Clarke EE, Lewis HD, Harrison T, Hunt P, Nadin A, Smith AL, Stevenson G, Castro JL. L-685,458, an aspartyl protease transition state mimic, is a potent inhibitor of amyloid beta-protein precursor gamma-secretase activity. Biochemistry. 2000;39:8698–8704. doi: 10.1021/bi0005456. [DOI] [PubMed] [Google Scholar]
- 31.Yang G, Yin YI, Chun J, Shelton CC, Ouerfelli O, Li YM. Stereo-controlled synthesis of novel photoreactive gamma-secretase inhibitors. Bioorg Med Chem Lett. 2009;19:922–925. doi: 10.1016/j.bmcl.2008.11.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Weihofen A, Lemberg MK, Ploegh HL, Bogyo M, Martoglio B. Release of signal peptide fragments into the cytosol requires cleavage in the transmembrane region by a protease activity that is specifically blocked by a novel cysteine protease inhibitor. J Biol Chem. 2000;275:30951–30956. doi: 10.1074/jbc.M005980200. [DOI] [PubMed] [Google Scholar]
- 33.Sato T, Nyborg AC, Iwata N, Diehl TS, Saido TC, Golde TE, Wolfe MS. Signal peptide peptidase: biochemical properties and modulation by nonsteroidal antiinflammatory drugs. Biochemistry. 2006;45:8649–8656. doi: 10.1021/bi060597g. [DOI] [PubMed] [Google Scholar]
- 34.Sato T, Ananda K, Cheng CI, Suh EJ, Narayanan S, Wolfe MS. Distinct pharmacological effects of inhibitors of signal peptide peptidase and gamma-secretase. J Biol Chem. 2008;283:33287–33295. doi: 10.1074/jbc.M805670200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chun J, Yin YI, Yang G, Tarassishin L, Li YM. Stereoselective Synthesis of Photoreactive Peptidomimetic gamma-Secretase Inhibitors. J Org Chem. 2004;69:7344–7347. doi: 10.1021/jo0486948. [DOI] [PubMed] [Google Scholar]
- 36.Gillman KW, Starrett JE, Parker MF, Xie K, Bronson JJ, Marcin LR, McElhone KE, Bergstrom CP, Mate RA, Williams R, Meredith JE, Burton CR, Barten DM, Toyn JH, Roberts SB, Lentz KA, Houston JG, Zaczek R, Albright CF, Decicco CP, Macor JE, Olson RE. Discovery and Evaluation of BMS-708163, a Potent, Selective and Orally Bioavailable γ-Secretase Inhibitor. ACS Med Chem Lett. 2010;1:120–124. doi: 10.1021/ml1000239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Crump CJ, Castro SV, Wang F, Pozdnyakov N, Ballard TE, Sisodia SS, Bales KR, Johnson DS, Li YM. BMS-708,163 Targets Presenilin and Lacks Notch-Sparing Activity. Biochemistry. 2012;51:7209–7211. doi: 10.1021/bi301137h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chavez-Gutierrez L, Bammens L, Benilova I, Vandersteen A, Benurwar M, Borgers M, Lismont S, Zhou L, Van Cleynenbreugel S, Esselmann H, Wiltfang J, Serneels L, Karran E, Gijsen H, Schymkowitz J, Rousseau F, Broersen K, De Strooper B. The mechanism of gamma-Secretase dysfunction in familial Alzheimer disease. EMBO J. 2012;31:2261–2274. doi: 10.1038/emboj.2012.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Doody RS, Raman R, Farlow M, Iwatsubo T, Vellas B, Joffe S, Kieburtz K, He F, Sun X, Thomas RG, Aisen PS, Alzheimer’s Disease Cooperative Study. Steering C, Siemers E, Sethuraman G, Mohs R, Semagacestat Study, G A phase 3 trial of semagacestat for treatment of Alzheimer’s disease. N Engl J Med. 2013;369:341–350. doi: 10.1056/NEJMoa1210951. [DOI] [PubMed] [Google Scholar]
- 40.Placanica L, Chien JW, Li YM. Characterization of an atypical gamma-secretase complex from hematopoietic origin. Biochemistry. 2010;49:2796–2804. doi: 10.1021/bi901388t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yang T, Arslanova D, Xu X, Li YM, Xia W. In vivo manifestation of Notch related phenotypes in zebrafish treated with Alzheimer’s amyloid reducing gamma-secretase inhibitors. J Neurochem. 2010;113:1200–1209. doi: 10.1111/j.1471-4159.2010.06681.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pozdnyakov N, Murrey HE, Crump CJ, Pettersson M, Ballard TE, am Ende CW, Ahn K, Li YM, Bales KR, Johnson DS. γ-Secretase Modulator (GSM) Photoaffinity Probes Reveal Distinct Allosteric Binding Sites on Presenilin. J Biol Chem. 2013;288:9710–9720. doi: 10.1074/jbc.M112.398602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Crump CJ, Johnson DS, Li YM. Development and Mechanism of γ-Secretase Modulators for Alzheimer’s Disease. Biochemistry. 2013;52:3197–3216. doi: 10.1021/bi400377p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fuwa H, Takahashi Y, Konno Y, Watanabe N, Miyashita H, Sasaki M, Natsugari H, Kan T, Fukuyama T, Tomita T, Iwatsubo T. Divergent synthesis of multifunctional molecular probes to elucidate the enzyme specificity of dipeptidic gamma-secretase inhibitors. ACS Chem Biol. 2007;2:408–418. doi: 10.1021/cb700073y. [DOI] [PubMed] [Google Scholar]
- 45.Grigorenko AP, Moliaka YK, Soto MC, Mello CC, Rogaev EI. The Caenorhabditis elegans IMPAS gene, imp-2, is essential for development and is functionally distinct from related presenilins. Proc Natl Acad Sci U S A. 2004;101:14955–14960. doi: 10.1073/pnas.0406462101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Krawitz P, Haffner C, Fluhrer R, Steiner H, Schmid B, Haass C. Differential localization and identification of a critical aspartate suggest non-redundant proteolytic functions of the presenilin homologues SPPL2b and SPPL3. J Biol Chem. 2005;280:39515–39523. doi: 10.1074/jbc.M501645200. [DOI] [PubMed] [Google Scholar]
- 47.Casso DJ, Tanda S, Biehs B, Martoglio B, Kornberg TB. Drosophila signal peptide peptidase is an essential protease for larval development. Genetics. 2005;170:139–148. doi: 10.1534/genetics.104.039933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Seiffert D, Bradley JD, Rominger CM, Rominger DH, Yang F, Meredith JE, Jr, Wang Q, Roach AH, Thompson LA, Spitz SM, Higaki JN, Prakash SR, Combs AP, Copeland RA, Arneric SP, Hartig PR, Robertson DW, Cordell B, Stern AM, Olson RE, Zaczek R. Presenilin-1 and -2 are molecular targets for gamma-secretase inhibitors. J Biol Chem. 2000;275:34086–34091. doi: 10.1074/jbc.M005430200. [DOI] [PubMed] [Google Scholar]


