Abstract
Major radiations of enigmatic bacteria and archaea with large inventories of uncharacterized proteins are a striking feature of the Tree of Life1,2,3,4,5. The processes that led to functional diversity in these lineages, which may contribute to a host-dependent lifestyle, are poorly understood. Here we show that diversity-generating retroelements (DGRs), which guide site-specific protein hypervariability6,7,8, are prominent features of genomically-reduced organisms from the bacterial candidate phyla radiation (CPR) and yet uncultivated phyla belonging to the DPANN archaeal superphylum. From reconstructed genomes we defined monophyletic bacterial and archaeal DGR lineages that expand known DGR range by 120% and reveal a history of horizontal retroelement transfer. Retroelement-guided diversification is further shown to be active in current CPR and DPANN populations, with an assortment of protein targets potentially involved in attachment, defense, and regulation. Based on observations of DGR abundance, function, and evolutionary history, we find that targeted protein diversification is a pronounced trait of CPR and DPANN phyla compared to other bacterial and archaeal phyla. This diversification mechanism may provide CPR and DPANN organisms a versatile tool that could be used for adaptation to a dynamic, host-dependent, existence.
Diverse environments host archaea and bacteria that define major lineages of predominantly uncultivated organisms: the archaeal superphylum comprising the Diapherotrites, Parvarchaeota, Aenigmarchaeota, Nanoarchaeota, and Nanohaloarchaea (DPANN – which includes the recently discovered Woesearchaeota and Pacearchaeota1,2 phyla as well) and the bacterial candidate phyla radiation (CPR)3,4,5. Members of these lineages have consistently been reported to have small genomes (~0.5 – 1.5 Mbp) and some have been shown to have ultra-small cells9,10,11. Most DPANN and CPR genomes are missing biosynthetic pathways considered vital for autonomous growth, which points to a host-dependent lifestyle12,13. Despite genomic insights, little is known of the mechanisms for genetic diversification that drive either adaptation to environmental stress14 (i.e. nutrient or energy limitation) or interactions with neighboring cells. Based on the recent identification of DGRs in two single-cell DPANN partial genomes from a subsurface environment15, and the established role of DGRs in host-dependent bacteria and their viruses7, we sought to address the hypothesis that organisms with minimal genomes and biosynthetic deficiencies (i.e. for nucleotides, lipids, and amino acids) belonging to CPR and DPANN phyla, are candidates for DGR utility. Diversification mechanisms in CPR and DPANN are not established, but are important, given that these radiations appear to comprise a major fraction of microbial life3,4,5.
Among known biological mechanisms for genetic diversification, DGRs are capable of exploring the highest ceiling on coding sequence variability16. These retroelements are unique in that they promote rapid and targeted mutation of specific genomic loci using a reverse transcriptase (RT) that is predicted to be error-prone. They occur in genomes of bacteriophage, and in bacteria whose lifestyles are typified by parasitism, pathogenesis, and intraspecific competition6,17,18. Recent studies have established various forms of evidence that DGRs offer selective advantages to the host genomes that encode them7,8,17. First, their capacity for targeted mutation allows for diversification in a hypervariable coding scaffold, without altering conserved sequence regions that support fold stability16. Second, the variable proteins whose structures have been experimentally assessed to date appear to contain ligand-binding folds16,19,20. This unity in the structures diversified within an array of different genomes points to a common advantage in expanding ligand specificity for different viral and cellular binding proteins. Finally, DGRs appear to be conserved in multiple strains of both Legionella pneumophila and Treponema denticola and can occur within conjugative elements16,17,21; negative selective pressures are likely to preclude exchange and retention of DGRs across ancestral networks.
The DGR mechanism of mutagenic homing (Figure 1A) deploys RT to target a variable protein for diversification through an RNA intermediate6,7,22. Genes encoding the variable protein contain a variable repeat (VR) in close proximity to an invariant template repeat (TR); RT-induced mutation of TR-RNA adenines and cDNA replacement of VR leads to an extraordinary potential for diversification (>1020 amino acid variants)16. To determine whether DGRs occur in the genomes of CPR and DPANN organisms and to assess their evolutionary importance, we capitalized on the availability of a massive metagenomic dataset2,3,5, containing numerous new draft and complete genomes. Our analyses targeted sequences of size-fractionated bacteria and archaea from an aquifer known to harbor diverse CPR and DPANN phyla. The cells were captured onto sequential 1.2 μm, 0.2 μm and 0.1 μm filters2,3,5. In total, we analyzed 30 metagenomic datasets, as well as six metatranscriptomes. Remarkably, we identified 1,136 non-redundant sequences that encode essential features of a DGR (i.e., an RT gene and a VR/TR pair; Figure 1A), approximately tripling the total number of DGRs that have been previously identified from more than 300 bacterial, archaeal, and viral genomes17.
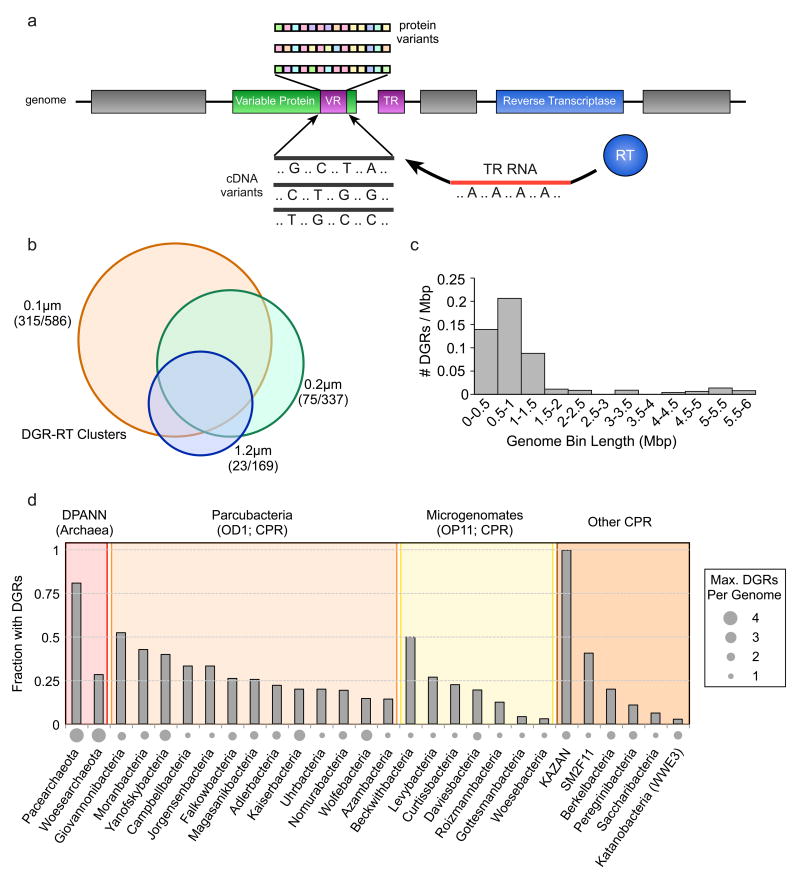
Figure 1. Prevalence of DGRs identified in groundwater metagenomes.
a, Schematic of a genomic diversity-generating retroelement cassette and the mutagenic retrohoming mechanism. b, Clusters of reverse transcriptase protein sequences at >70% global pairwise alignment displayed by filter size, with the numbers of unique/shared RT clusters in parentheses. c, Distribution of DGR occurrence in reconstructed genomes, given as a fraction of 1 Mbp for each discrete interval. d, Incidence of DGRs in the archaeal DPANN superphyla, Parcubacteria (OD1) and Microgenomates (OP11), and other CPR phyla.
We determined the number of distinct DGR sequence types and examined the frequency of these sequences in the genomes of cells separated by size filtration. DGR-like RTs were grouped based on >70% amino-acid identity, generating 699 protein clusters. Only 23 clusters are unique to samples from the largest size fraction (Figure 1B); conversely, 75 are unique to the mid-size fraction, and 315 to the small size fraction. Furthermore, we identified 63 distinctive DGRs affiliated with genomes that were previously linked to cells examined by cryo-electron tomography, which passed through a 0.2 μm filter11. Importantly, out of the 542 genomes in which we identified DGRs, only 19 are linked to non-CPR/DPANN phyla highlighting a low frequency in organisms predicted to have larger cell sizes. Based on these results we conclude that DGRs are enriched in the genomes of ultra-small bacterial and archaeal cells, providing an entirely new niche association for this adaptive mechanism.
To estimate the genomic incidence of targeted protein diversification in these samples, we focused on DGR-containing sequences in 542 high-quality draft genomes; i.e., reconstructed bacterial and archaeal genomes, including 530 containing >75% of a set of universal single copy genes2,23. Notably, a prevalence of DGRs is observed in reconstructed genomes of 0.5 – 1 Mbp (Figure 1C), consistent with the hypothesis that DGRs are common to genomically-reduced phyla. Moreover, these retroelements are overrepresented (found in > 25% of genomes) in the genomes of DPANN archaea (i.e. Pacearchaeota and Woesearchaeota), Parcubacteria (i.e. Campbellbacteria, Falkowbacteria, Giovannonibateria, Jorgensenbacteria, Magasanikbacteria, Moranbacteria, and Yanofskibacteria), and Microgenomates (i.e. Beckwithbacteria and Levybacteria), while likewise abundant in other CPR phyla (Figure 1D) – providing an extraordinary association of DGRs with CPR and DPANN phyla. Among the genomes that contain a DGR, 26% of archaeal genomes and 19% of bacterial genomes encode multiple distinct cassettes. The high incidence of DGRs encoded in reduced genomes raises the question of whether mutagenic homing is a broadly available evolutionary tool among CPR and DPANN phyla (i.e. facilitating genetic adaptation to various selective pressures), and whether DGRs remain active in extant populations.
To determine whether variable proteins have been recently diversified, we assessed the pattern of heterogeneity in reads mapped to VR coding regions of DGR protein targets. The VRs were aligned with corresponding bases in cognate TRs to determine the proportion of adenine-specific mismatches leading to non-synonymous substitutions. Here, a stringent approach excluded sequences with non-synonymous changes in the VR loci that were not aligned with a TR adenine position. Overall, we detected 132 DGR sequences with pronounced levels of adenine-specific mutagenesis, unlikely to arise due to stochastic mutation (resulting in >3 non-synonymous substitutions per VR). This finding provides evidence that proteins in a subset of the recovered genomes were undergoing diversification leading up to sampling. These results are consistent with prior evidence that DGR-RT misincorporates bases in VR that correspond to TR adenines7 (Supplementary Figure S1). Closer examination of DGRs in these genomes identified 34 sequences capable of forming stem-loops in loci proximal to their VRs (Supplementary Table S1); such cis-acting elements are important for DGR function24. We then searched for analogs of Avd, a low molecular weight accessory protein (14.7 kDa) required for mutagenic homing in other DGR systems7. This analysis uncovered a group of 72 DGRs encoded alongside conserved, small proteins (average 9.57 kDa) predicted to function analogously to Avd. Taken together, these findings suggest that the intact DGRs we identified have a capacity for driving adenine mutagenesis.
To further determine if these DGRs were active contemporaneous with sampling, we analyzed six metatranscriptomes, identifying DGRs that aligned with at least one metatranscriptomic read. DGRs from Parcubacteria and Woesearchaeota appear to be disproportionately expressed, with between 13 and 94 metatranscriptomic reads that were mapped to one or more DGR feature(s) (Supplementary Table S2). In contrast to the high transcriptional levels observed for DGRs, few to no reads were mapped to protein-coding genes as putative mRNA; the majority of total metatranscriptomic reads were aligned to rRNA, tRNA, and tmRNA. In the case of each DGR cassette, reads appear to map primarily, but not exclusively, to TR regions. Strikingly, in some instances, metatranscriptomic read mapping was comparable (i.e., within 10x) between TR-RNA and other highly expressed regions (i.e. tmRNA) on the same contig (Supplementary Figure S2). These observations point to DGR expression of a stable RNA molecule in organisms affiliated with both CPR and DPANN, in a manner that is consistent with TR-specific expression in DGR systems of Trichodesmium erythraeum18. Taken together with evidence for adenine-mutagenesis, these findings point to a functional subset of DGRs in groundwater-associated genomes, and by their established mechanism, to protein diversification in CPR and DPANN phyla.
We next analyzed the phylogeny of RT proteins to determine whether the newly identified DGRs are closely related to known DGRs, or if the elements described in this study represent novel lineages. We find that the majority of newly discovered bacterial RTs appear only distantly related to those from known bacterial DGRs. DGR-RTs from CPR genomes almost exclusively form novel lineages (Figure 2A). Moreover, RTs identified in genomes of DPANN phyla form a monophyletic archaeal clade along with previously described DGR-RTs found in single-cell DPANN genomes15. Whereas representatives from archaea and CPR form separate DGR-RT lineages, we observe paraphyletic patterns at higher taxonomic resolution (Figure 2A). Notably, closely related RTs are shared between Woesearchaeota and Pacearchaetoa, and separately, groups of similar RTs appear to link Parcubacteria and Microgenomates with members of Berkelbacteria, Peregrinibacteria, and Saccharibacteria. Moreover, these DGR groups have highly similar TR nucleotide sequences, offering independent evidence of exchange by horizontal gene transfer (HGT).
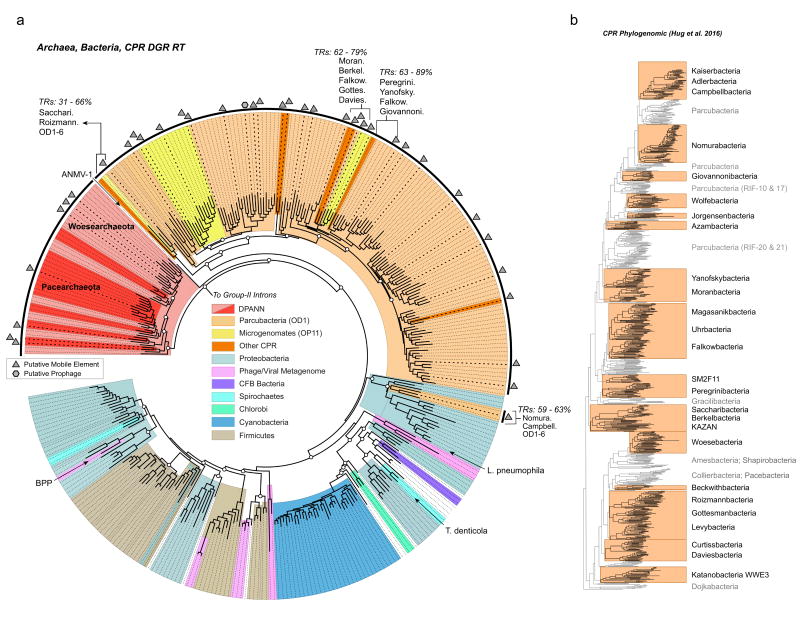
Figure 2. Phylogeny of diversity-generating retroelements and radiation of novel lineages.
a, Phylogeny and taxonomic association of diversifiers. Sequences obtained in this study are highlighted in black on the outer edge of the tree. Shaded slices indicate either candidate superphyla comprising groundwater organisms, or bacterial phyla and phage with previously sequenced RTs. The tree was constructed with 346 sequences and 261 alignment positions. Archaeal sequences from the DPANN superphylum are indicated in either dark red (Pacearchaeota), or light red (Woesearchaeota) shaded slices. Paraphyletic groups of species with closely related RTs are indicated with the associated range of pairwise TR sequence similarity. Symbols (hexagons and triangles) highlight DGRs identified in this study, which are found in close proximity to putative prophage or mobile elements (i.e. transposons or conjugative elements). Diversifiers that have been previously examined are: BPP, a Bordetella pertussis phage; T. denticola, Treponema denticola; L. pneumophila, Legionella pneumophila strain Corby; and ANMV-1, a marine virus of an uncultivated archaeal host. White circles indicate bootstrap values >70% for basal nodes. The scale indicates amino acid substitutions per site. b, Phylogenomic tree of CPR organisms, highlighting major lineages that contain at least one DGR. The phylogenomic tree was originally presented by Hug et al. (2016).
To further investigate whether RTs were subject to HGT amongst CPR and DPANN organisms, we inspected genes in proximity to DGRs (i.e., +/− 10 kbp of an RT gene) for characteristics of prophage (e.g., viral proteins, terminase, integrase), or other mobile elements, such as transposons or conjugative elements. This search revealed numerous DGRs that occur in close proximity to at least one transposase gene, whereas only a single DGR could be linked to a recognizable prophage-like region (Figure 2A). The apparent capacity for horizontal transfer on mobile elements suggests that DGRs offer selective advantages in these bacteria and archaea. We additionally sought to examine monophyletic lineages from a previously constructed phylogenomic tree of CPR representatives4 for examples of conserved DGR. Groups of related DGR RTs, and separate VP groups, appear to be conserved across the Yanofskybacteria clade of CPR phyla (Supplementary Figure S3). Retention across a broad evolutionary distance suggests that DGRs offer advantages to the genomes that encode them. It is also worth noting that, since the CPR phylogenomic tree was constructed including partial genomes, we likely have a limited view of DGR retention for certain representatives (Supplementary Figure S4).
Prior to this study, DGRs were known to occur in bacterial genomes from many phyla7,17 belonging to various microbiomes, but were rare in the genomes of most cultured isolates25,26,27 Our findings are in stark contrast, revealing an extraordinary radiation of novel DGR clades from newly described bacteria belonging mostly to candidate phyla (Figure 2A). Further, these elements are enriched in numerous genomes of CPR bacteria (especially Parcubacteria at >37% incidence). Importantly, DGRs were discovered across most of the major CPR lineages (Figure 2B), and not solely within a closely related subset of organisms. The findings presented herein expand upon prior evidence of two archaeal genomes that encode DGRs, revealing a multitude of diversifiers in other representatives of DPANN archaea (especially Pacearchaeota at >80% incidence, and Woesearchaeota at >28% incidence). When compared with other microbial DGRs from an array of different environments, the groundwater-associated representatives in this study account for 55% of cumulative branch length, or apparent diversity (i.e., substitutions per site), on a non-redundant tree of all representatives (Figure 2A). Remarkably, while DGRs account for an estimated 3% of RTs in previously sequenced bacterial genomes28, they make up 57% of recognizable RTs in the analyzed metagenomes. These findings highlight DGRs as a prominent genetic feature for CPR and DPANN phyla, and reveal an exemplary biome wherein DGRs have evolved to become prominent and active agents of adaptation.
The phylogenetic distribution, horizontal exchange, and retained function of DGRs discovered here, considered alongside their established roles in cellular interaction, raise the question of whether these retroelements are recruited preferentially for adaptive evolution of proteins that enable symbiosis. To address this question we performed functional predictions for DGR variable proteins. To estimate their functional richness, we clustered variable proteins, and separately extracted VR domains (i.e., with >30% intra-cluster similarity; Supplementary Figure S5). We identified 396 variable protein clusters and 284 VR-domain clusters. The most common protein domain annotations include a conserved domain of unknown function (DUF1566), AAA+ related ATPase, and C-type lectin domain (Supplementary Figure S5). Notably, the majority of VRs are located in the C-terminus of their variable protein, which is consistent with VR placement in previously characterized DGRs7,16. Moreover, C-terminal VR domains appear to be predominately localized to C-type lectin (CLec-like) domains and DUF1566 domains, which were recently shown to have CLec-folds29. Amongst an assortment of DUF1566 domains linked to putative transmembrane proteins, we identified putative pilin structures, lipoproteins, fimbrial protein FimH, and a rearrangement hotspot (rhs) toxin, suggesting broad involvement of these proteins in cell attachment and defense (Supplementary Table S3). While most DGR variable proteins are putative single domain proteins, less than 350 amino acids in length (Supplementary Figure S6), an unexpected array of mutidomain architectures is also observed.
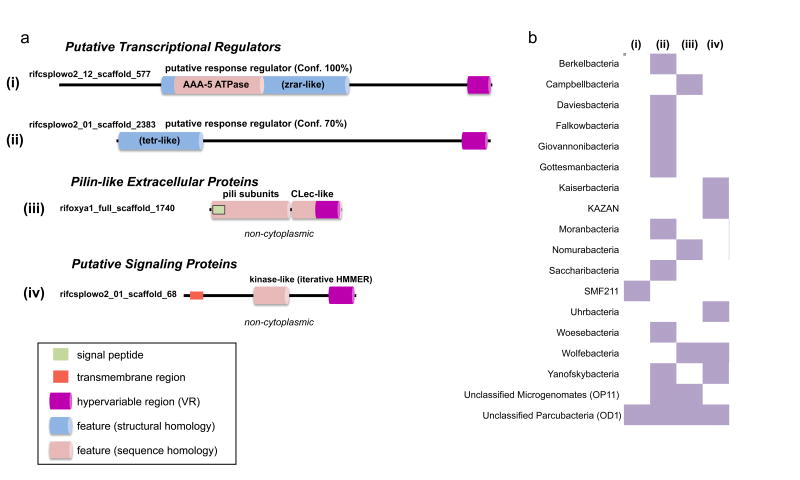
Through further analysis of the draft genomes from candidate phyla, we identified additional clusters of variable proteins whose domain architectures are associated with transcriptional regulation, cell-cell attachment, and signal transduction (Figure 3A). Analyses of homology identified variable proteins, which are common to multiple CPR lineages, containing AAA+ ATPase modules, pilin-like N-terminal regions fused to C-terminal CLec-like ligand-binding domains, and putative kinase-like regions. Moreover, the ATPase domains in CPR variable proteins each belong to the AAA-5 subgroup of eukaryotic-like midasins (Supplementary Table S4). Distinct classes of these variable proteins are associated with genomes representing a range of candidate phyla (Figure 3B). Whether chaperones, cell-cell attachment proteins, or signaling proteins, each of the otherwise distinct variable protein classes are likely to function as ligand-binding receptors. The general role of ligand binding has been described as a unifying attribute of both signaling and regulatory proteins serving core cellular functions in prokaryotes30. Our findings of HGT and adenine-specific mutations of the VR point to the selective advantage of DGRs in CPR organisms, which is perhaps related to the utility in diversifying modular ligand-binding domains – driving expansion of substrate specificity for regulation, signaling, and attachment.
Figure 3.
Putative functional classes of DGR variable proteins. a, Functional annotations for variable proteins identified in at least two distinct candidate phyla. Conserved domains and features based on structural homology are shown. Phyre2 confidence values are given for predictions and their closest known structures. b, Distribution of common variable proteins found in candidate phyla, labeled i – iv, based on the examples in (a).
Presumably then, variable protein genes that offer selective advantages to their genomes would be conserved in both DGR and non-DGR loci. To address this hypothesis, we identified examples of variable protein paralogues occurring both within DGR cassettes and in the absence of proximal DGR features (Supplementary Figure S7 and Supplementary Figure S8). Finding homologous variable protein genes in both DGR and non-DGR loci of a genome suggests diversification might be followed by preservation of a particular variant gene. In addition to potential advantages linked to specific cellular functions, DGRs also offer more general benefit in conferring genetic variability for minimal genomes. Targeted and localized mutagenesis could provide benefit to organisms with minimal genomes that cannot otherwise accommodate extensive variant repertoires.
Myriad selective pressures on CPR or DPANN organisms can impose a need for genetic hypervariability, and interactions with neighboring cells are likely to act as such evolutionary pressures. Based on our results, we conclude that DGRs are prevalent in ultra-small, genomically-reduced cells belonging to both CPR bacteria and DPANN archaea. This prevalence provides an indication of selective pressures that transcend the ancient divergence of CPR bacteria and DPANN archaea. As explanation, we hypothesize an enhanced utility of DGR-mediated diversification that emerges from selective pressure, balancing minimal genome size against the need for dynamic response to manage host association. Furthermore, the capacity of DGRs for accelerated protein evolution suggests a need on the part of some CPR and DPANN to rapidly evolve their symbiotic associations, which in turn suggests they may sometimes exploit intercellular associations to receive greater benefit than their host – perhaps shifting between mutualism, predation, and parasitism.
Methods
Study site and sampling
This study used data from several previously-described samples2,3,5. In brief, sampling was conducted within an unconfined aquifer at the Rifle Integrated Field Research Challenge (IFRC) site, which is adjacent to the Colorado River, near Rifle, Colorado, USA (39° 31′ 44.69″ N, 107° 46′ 19.71″ W). Groundwater samples were collected from three different field experiments: 6 sampling time points across the duration of acetate amendment, A-F; 4 sampling time points across the duration oxygen injection A-D; and 2 sampling time points from natural high and low oxygen conditions in the groundwater, driven by fluctuations in the water table at the site. Aquifer well CD-01 was monitored as part of a 95-day acetate amendment experiment during which acetate was added to the aquifer between August 25th and December 12th, 2011 as previously described2. Following this experiment, aquifer well CD-01 was monitored as part of a 132-day oxygen injection experiment where oxygen-saturated water was injected into the aquifer from August 2nd 2012 to December 12th 2012.
Aquifer well FP-101 was sampled during two specific time points characterized by high and low oxygen in the groundwater. All groundwater samples were collected from 5 m below the ground surface and cells were collected on serial 1.2, 0.2 and 0.1μm filters (Supor disc filters; Pall Corporation) towards differential sampling of small-celled organisms. Following groundwater sampling, filters were immediately frozen prior to DNA extraction, either on dry ice, or in liquid nitrogen.
Metagenomic and metatranscriptomic sequencing
As described previously2,3,5, genomic DNA of groundwater organisms was extracted from ~1.5 g filter samples, using a PowerSoil DNA Isolation Kit (MO-BIO). Filters were cut into strips, which were then vortexed in PowerBead solution, before and after an interval of flash freezing and thawing. Following thawing, the solution was incubated for 30 minutes at 65°C while shaking. DNA was then eluted and concentrated by sodium acetate and ethanol precipitation in glycogen, followed by resuspension in 50 μl of PowerSoil elution buffer. Sequencing was performed at the Joint Genome Institute, using the Illumina HiSeq 2000 platform to generate 2×150 paired-end reads.
As reported previously3, prior to RNA extraction, genomic DNA removal and cleaning was done using RNase-Free DNase Set kit (Qiagen) and Mini RNeasy kit (Qiagen). Next, RNA extractions were performed using Invitrogen TRIzol Reagent (Invitrogen). Sample aliquots were analyzed before sequencing using the Agilent 2100 Bioanalyzer to assess the quality of purified RNA. The cDNA sequence preparation was performed using a SOLiD Total RNA-Seq kit (Applied Biosystems). Samples were sequenced on the SOLiD 5500XL platform, at the DOE Environmental Molecular Sciences Laboratory, a facility of the Pacific Northwest National Laboratory. Initial genome sequence mapping was conducted using LifeScope software (version 2.5; SOLiD) with default parameters; additional metatranscriptomic read mapping details are given below.
Assembly, annotation, and binning of metagenomic sequences
This study involved the analysis of sequences that were previously preprocessed, assembled, and binned2,3,5. Briefly, sequencing reads were filtered for quality using Sickle software version 1.33 (https://github.com/najoshi/sickle), with default parameters. Next, sequences were assembled with IDBA_UD, using default parameters for paired-end reads31. Only assembled scaffolds exceeding 5kb were used for downstream annotation and binning steps. Open reading frame (ORF) annotation was performed using Prodigal32 with the metagenome mode setting. In the present study, functional annotations for genes were determined using hmmsearch v3.133 against an in-house HMM database constructed based on KEGG orthology, while sequences for DGR variable proteins and putative reverse transcriptases were also compared with the Uniprot database using pHMMER33. Here, variable proteins were also analyzed for homology to known protein structures using Phyre234. Read mapping was conducted using Bowtie235. Previously, scaffolds were binned to specific organisms by using coverage across the samples, phylogenetic identity, and GC content, both automatically with the ABAWACA algorithm3 and manually using ggKbase (http://ggkbase.berkeley.edu/). ABAWACA is an algorithm that generates genome bins based on scaffold taxonomic affiliations, time-series abundance patterns, and nucleotide frequencies (https://github.com/CK7/abawaca). Genome bins generated by ABAWACA were manually inspected within ggKbase. Binning purity was confirmed using an Emergent Self-Organizing Map (ESOM)36,37. Each bin was previously assigned a genome phylogeny if it met the following criteria: 1) determined to be high quality2,3,5 (i.e. scaffolds could not be further separated into distinct bins); 2) contains at least 75% of the conserved, single-copy genes found broadly across bacterial genomes, or separately across archaeal genomes38. For draft genomes that contain less than 75% of conserved single copy genes (12 genomes examined in this study), taxonomy was determined from ribosomal protein and 16S rRNA phylogeny as previously described2.
Identification, annotation, and clustering analysis of DGR Features
The following methods were carried out in this study. Several genomic features that are encoded by all DGRs – namely a reverse transcriptase gene and VR/TR pairs – can be used as diagnostic indicators for in silico identification of these retroelements7,28. To this end, we used a consensus sequence of aligned DGR-RT protein sequences from Treponema denticola, Bordetella pertussis phage, Legionella pneumophila, archaeal virus ANMV-1, and uncultivated nanoarchaeota, to conduct a tblastn search for RT-like hits in assembled scaffolds from groundwater metagenomes (Supplementary Figure S9). Next, a custom python script was used to identify near-repeats within 10 kb of the putative RT gene, by applying a sliding window (200 bp windows; 50 bp step) blastall search with the following parameters: -word_size 8 -reward 1 -penalty -1 -evalue 1e-5 -gapopen 6 -gapextend 6 -perc_identity 50. This output was filtered for near repeats: >60bp pairs, which contain >5 adenine-specific mismatches and no more than one non-adenine mismatch (i.e. putative VR/TR pairs). Given that adenines of AAY codons are selectively targeted by DGRs16, ORFs were only identified as variable protein genes where the majority of TR adenines correspond to the first two positions of a given codon in VR.
Putative stem-loop encoding regions (100bp downstream of VR) were extracted from DGR cassette nucleotide sequences, and regions capable of forming a stem-loop were identified using MFold39. Translated reverse transcriptase sequences were clustered using CD-HIT40 with a global alignment (identity) threshold of 70%. Variable protein sequences were clustered using H-CD-HIT with three iterative rounds of clustering and identity thresholds at 90%, 60%, and 30%. VR domains, including 50 flanking amino acids, were extracted from the variable protein sequences and separately clustered using H-CD-HIT with the same three-iteration identity settings as above.
Clustering was also conducted to assess DGR occurrence in genomes by putative cell size. DGR-like RTs were clustered using H-CD-HIT40 as above, resulting in 699 protein clusters, which were then inspected for representatives from individual metagenomic libraries for each filter pore size (i.e., 1.2 μm, 0.2 μm, & 0.1 μm). It should be noted that the cells in the smallest size fraction passed through a filter commonly used for filter-sterilization. Larger organisms (e.g., Spirochaetes), and viruses, are able to pass through 0.2 μm filters, thus highlighting the need to carefully assess the phylogenetic affiliation of DGR-containing contigs linked to smaller filter fractions. To address the concern that filtration is an imperfect method for size exclusion, we examined genomes from one sample that was previously used for cryo-electron tomography to quantify the size of cells belonging to CPR bacteria Parcubacteria (OD1 superphylum) and Microgenomates (OP11 superphylum)11.
DGR readmapping and metatranscriptomic analysis
We analyzed the variability of TR adenine sites leading to non-synonymous versus synonymous substitutions, by assessing reads mapped to the corresponding VR sequence on assembled scaffolds. First, assembly errors were inspected for each scaffold using stringent criteria for each basecall: any regions that were not supported with paired reads with at most one mismatch were replaced with Ns; errors are re-assembled using stringent mapping for one read in a pair; scaffolds were split if insert (Ns) coverage is zero. For the adenine-variability analysis, only uninterrupted VR and TR sequences were used (i.e. without Ns). The scaffold sequence of aligned TR and VR regions was inspected in-frame with respect to the variable protein-encoding gene’s stop codon. Reads mapped to each VR codon were analyzed for non-synonymous or synonymous substitutions at TR adenines. To compare with variability that could have resulted from stochastic mutation, non-synonymous and synonymous substitutions were also tabulated for non-adenine positions in TR.
Stringent metatranscriptomic read mapping to DGR regions was performed using Bowtie235, whereby only matching metatranscriptomic reads, with at most a single mismatch, were mapped to the scaffold. Regions without an apparent ORF, but with >10 reads mapped, were searched against the rFAM database41, to identify potential RNA-encoding sections of DGR-containing scaffolds. We determined metatranscriptomic read coverage for DGR features separately (variable protein; RT; VR; TR), in addition to calculating coverage for the whole DGR cassette.
Phylogenetic analyses
To compare DGR representatives that were derived from groundwater metagenomes with DGR-like RTs from other bacterial and archaeal genomes, we sought a phylogenetic reconstruction for RT protein sequences. A consensus sequence from an alignment of previously studied DGRs15,17 was used to search the NCBI-nr protein database for additional DGR-like hits (blastp; e-value <10−20). Next, the returned hits were individually used towards blast searches against NCBI-nr, to obtain up to 20 top hits for each DGR-like representative (e-value <10−20). Before alignment and tree construction, we performed clustering on the redundant list of RT sequences using CD-Hit46 with an intracluster global alignment threshold of 90% and default parameters. Representatives that were assessed as DGR-like (i.e. included in the DGR-specific RT tree) exhibited a monophyletic association with known DGR representatives, branching separately from Group-II intron-associated RTs, as previously shown15,42. Groundwater-associated and other DGR-like representatives were aligned to a hidden markov model for the reverse transcriptase protein family (PF00078) using HMMalign43. A phylogenetic tree of the RT alignment was constructed using FastTree2 with the WAG model and CAT approximation.
Variable protein domains, including midasin-like ATPase, and separately, DUF1566-like, were aligned using ClustalW and manually inspected to remove ambiguously aligned sites. Trees for variable protein clusters were constructed in Geneious v 8.1.4 (Biomatters), using PhyML44 with the model LG + G, while branch support was determined with 100 bootstrap replicates. All trees for this study were visualized using FigTree (v1.4.2).
Data Availability
The data supporting the results of this study are available within the paper and its Supplementary Information and Supplementary Data Files. Assembled metagenomic sequences are available in the NCBI BioProject database under the accession numbers PRJNA268032, PRJNA273161, PRJNA288027, and KY476664-KY476802.
Supplementary Material
Acknowledgments
This research was funded by National Science Foundation grant OCE-1046144 to D.L.V., NIH grant R01 AI096838 to J.F.M and P.G, and by the US Department of Energy (DOE), Office of Science, Office of Biological and Environmental Research under award number DE-AC02-05CH11231 (Sustainable Systems Scientific Focus Area; Lawrence Berkley National Laboratory operated by the University of California) and award number DE-SC0004918 (Systems Biology Knowledge Base Focus Area). Sequencing was performed at the US DOE Joint Genome Institute, a DOE Office of Science User Facility, supported under contract DE-AC02-05CH11231. Metatranscriptomes were sequenced at the DOE-supported Environmental Molecular Sciences Laboratory at Pacific Northwest National Laboratory. B.G.P was supported by a postdoctoral fellowship from the Center for Dark Energy Biosphere Investigations (C-DEBI). D.B. was supported by a long-term EMBO fellowship. We are grateful to Karthik Antharaman for assistance with genome binning, Andrea Singh and Christopher T. Brown, who aided in examining CPR and DPANN genomes, and Cara Magnabosco for offering insights on phylogenetic reconstruction. This is C-DEBI contribution number 361.
Footnotes
Accession Codes:
Assembled metagenomic sequences are available in the NCBI BioProject database under the accession numbers PRJNA268032, PRJNA273161, PRJNA288027, and KY476664-KY476802.
Contributions:
B.G.P and D.L.V. developed the project. B.G.P., D.B., C.J.C., B.C.T., and J.F.B. performed reassembly, read mapping, and annotation of the metagenomic and metatranscriptomics datasets. B.G.P., D.B., C.J.C., E.C., D.A., S.H., P.G., J.F.M., J.F.B., and D.L.V conducted bioinformatic analyses on DGR sequences. B.G.P., D.B., C.J.C., J.F.B., and D.L.V. wrote the manuscript.
Competing financial interests:
JFM is a cofounder, equity holder, and chair of the scientific advisory board of AvidBiotics Inc., a biotherapeutics company in San Francisco. No other authors declare competing financial interests.
Contributor Information
Blair G. Paul, Marine Science Institute, University of California, Santa Barbara, California, 93106, USA
David Burstein, Department of Earth and Planetary Science, University of California, Berkeley, California 94720, USA.
Cindy J. Castelle, Department of Earth and Planetary Science, University of California, Berkeley, California 94720, USA
Sumit Handa, Department of Chemistry and Biochemistry, UC San Diego, La Jolla, CA, 92093, USA.
Diego Arambula, Department of Microbiology, Immunology and Molecular Genetics, University of California, Los Angeles, California, 90095, USA.
Elizabeth Czornyj, Department of Microbiology, Immunology and Molecular Genetics, University of California, Los Angeles, California, 90095, USA.
Brian C. Thomas, Department of Earth and Planetary Science, University of California, Berkeley, California 94720, USA
Partho Ghosh, Department of Chemistry and Biochemistry, UC San Diego, La Jolla, CA, 92093, USA.
Jeffery F. Miller, Department of Microbiology, Immunology and Molecular Genetics, University of California, Los Angeles, California, 90095, USA
Jillian F. Banfield, Department of Earth and Planetary Science, University of California, Berkeley, California 94720, USA
David L. Valentine, Marine Science Institute, University of California, Santa Barbara, California, 93106, USA.
References
- 1.Rinke C, et al. Insights into the phylogeny and coding potential of microbial dark matter. Nature. 2013;499:431–437. doi: 10.1038/nature12352. [DOI] [PubMed] [Google Scholar]
- 2.Castelle CJ, et al. Genomic expansion of domain archaea highlights roles for organisms from new phyla in anaerobic carbon cycling. Curr Biol. 2015;25:690–701. doi: 10.1016/j.cub.2015.01.014. [DOI] [PubMed] [Google Scholar]
- 3.Brown CT, et al. Unusual biology across a group comprising more than 15% of domain Bacteria. Nature. 2015;523:208–211. doi: 10.1038/nature14486. [DOI] [PubMed] [Google Scholar]
- 4.Hug LA, et al. A new view of The Tree of Life. Nat Microbiol. 2016;1:16048. doi: 10.1038/nmicrobiol.2016.48. [DOI] [PubMed] [Google Scholar]
- 5.Anantharaman K, et al. Thousands of microbial genomes shed light on interconnected biogeochemical processes in an aquifer system. Nat Commun. 2016;7 doi: 10.1038/ncomms13219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liu M, et al. Reverse transcriptase-mediated tropism switching in Bordetella bacteriophage. Science. 2002;295:2091–2094. doi: 10.1126/science.1067467. [DOI] [PubMed] [Google Scholar]
- 7.Doulatov S, et al. Tropism switching in Bordetella bacteriophage defines a family of diversity-generating retroelements. Nature. 2004;431:476–481. doi: 10.1038/nature02833. [DOI] [PubMed] [Google Scholar]
- 8.Guo H, Arambula D, Ghosh P, Miller JF. Diversity-generating retroelements in phage and bacterial genomes. Microbiol Spectr. 2014;2 doi: 10.1128/microbiolspec.MDNA3-0029-2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Comolli LR, Baker BJ, Downing KH, Siegerist CE, Banfield JF. Three-dimensional analysis of the structure and ecology of a novel, ultra-small archaeon. The ISME Journal. 2009;3:159–167. doi: 10.1038/ismej.2008.99. [DOI] [PubMed] [Google Scholar]
- 10.Baker BJ, et al. Enigmatic, ultrasmall, uncultivated Archaea. Proc Natl Acad Sci USA. 2010;107:8806–8811. doi: 10.1073/pnas.0914470107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Luef B, et al. Diverse uncultivated ultra-small bacterial cells in groundwater. Nat commun. 2015;6 doi: 10.1038/ncomms7372. [DOI] [PubMed] [Google Scholar]
- 12.Gong J, Qing Y, Guo X, Warren A. Candidatus Sonnebornia yantaiensis, a member of candidate division OD1, as intracellular bacteria of the ciliated protist Paramecium bursaria (Ciliophora, Oligohymenophorea) Syst Appl Microbiol. 2014;37:35–41. doi: 10.1016/j.syapm.2013.08.007. [DOI] [PubMed] [Google Scholar]
- 13.Nelson WC, Stegen JC. The reduced genomes of Parcubacteria (OD1) contain signatures of a symbiotic lifestyle. Front Microbiol. 2015;6:713. doi: 10.3389/fmicb.2015.00713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Valentine DL. Adaptations to energy stress dictate the ecology and evolution of the Archaea. Nat Rev Microbiol. 2007;5:316–323. doi: 10.1038/nrmicro1619. [DOI] [PubMed] [Google Scholar]
- 15.Paul BG, et al. Targetted diversity generation by intraterrestrial archaea and archaeal viruses. Nat Commun. 2015;6 doi: 10.1038/ncomms7585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Le Coq J, Ghosh P. Conservation of the C-type lectin fold for massive sequence variation in a Treponema diversity-generating retroelement. Proc Natl Acad Sci USA. 2011;108:14649–14653. doi: 10.1073/pnas.1105613108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Arambula D, et al. Surface display of a massively variable lipoprotein by a Legionella diversity-generating retroelement. Proc Natl Acad Sci USA. 2013;110:8212–8217. doi: 10.1073/pnas.1301366110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pfreundt U, Kopf M, Belkin N, Berman-Frank I, Hess WR. The primary transcriptome of the marine diazotroph Trichodesmium erythraeum IMS101. Scientific Reports. 2014;4:6187. doi: 10.1038/srep06187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Miller JL, et al. Selective ligand recognition by a diversity-generating retroelement variable protein. PLoS biology. 2008;6:e131. doi: 10.1371/journal.pbio.0060131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Handa S, Paul BG, Valentine DL, Miller JF, Ghosh P. Conservation of the C-type lectin fold for accommodating massive sequence variation in archaeal diversity-generating retroelements. BMC Struct Biol. 2016;16 doi: 10.1186/s12900-016-0064-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nimkulrat S, et al. Genomic and Metagenomic Analysis of Diversity-Generating Retroelements Associated with Treponema denticola. Front Microbiol. 2016;7 doi: 10.3389/fmicb.2016.00852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Guo H, et al. Diversity-generating retroelement homing regenerates target sequences for repeated rounds of codon rewriting and protein diversification. Mol Cell. 2008;31:813–823. doi: 10.1016/j.molcel.2008.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sharon I, et al. Time series community genomics analysis reveals rapid shifts in bacterial species, strains, and phage during infant gut colonization. Genome Res. 2013;23:111–120. doi: 10.1101/gr.142315.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Guo H, et al. Target Site Recognition by a Diversity-Generating Retroelement. Plos Genetics. 2011;7:e1002414. doi: 10.1371/journal.pgen.1002414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Minot S, Grunberg S, Wu GD, Lewis JD, Bushman FD. Hypervariable loci in the human gut virome. Proc Natl Acad Sci USA. 2012;109:3962–3966. doi: 10.1073/pnas.1119061109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schillinger T, Lisfi M, Chi J, Cullum J, Zingler N. Analysis of a comprehensive dataset of diversity generating retroelements generated by the program DiGReF. BMC Genomics. 2012;13:430. doi: 10.1186/1471-2164-13-430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ye Y. Identification of diversity-generating retroelements in human microbiomes. Int J Mol Sci. 2014;15:14234–14246. doi: 10.3390/ijms150814234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zimmerly S, Wu L. An Unexplored Diversity of Reverse Transcriptases in Bacteria. Microbiol Spectr. 2015;3 doi: 10.1128/microbiolspec.MDNA3-0058-2014. [DOI] [PubMed] [Google Scholar]
- 29.Xu Q, et al. A distinct type of pilus from the human microbiome. Cell. 2016;165:690–703. doi: 10.1016/j.cell.2016.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Anantharaman V, Koonin EV, Aravind L. Regulatory potential, phyletic distribution and evolution of ancient, intracellular small-molecule-binding domains. J Mol Biol. 2001;307:1271–1292. doi: 10.1006/jmbi.2001.4508. [DOI] [PubMed] [Google Scholar]
- 31.Peng Y, Leung HC, Yiu SM, Chin FY. IDBA-UD: a de novo assembler for single-cell and metagenomic sequencing data with highly uneven depth. Bioinformatics. 2012;28:1420–1428. doi: 10.1093/bioinformatics/bts174. [DOI] [PubMed] [Google Scholar]
- 32.Finn RD, Clements J, Eddy SR. HMMER web server: interactive sequence similarity searching. Nucleic Acids Res. 2011;39:W29–W37. doi: 10.1093/nar/gkr367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hyatt D, et al. Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC bioinformatics. 2010;11:1. doi: 10.1186/1471-2105-11-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kelley LA, Sternberg MJ. Protein structure prediction on the Web: a case study using the Phyre server. Nat Protoc. 2009;4:363–371. doi: 10.1038/nprot.2009.2. [DOI] [PubMed] [Google Scholar]
- 35.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nature methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ultsch A, Moerchen F. Technology Report. Vol. 46. Depatrment of Mathematics and Computer Science, University of Marburg; Germany: 2005. ESOM-maps: Tools for Clustering. Visualization, and Classification with Emergent SOM. [Google Scholar]
- 37.Dick GJ, et al. Community-wide analysis of microbial genome sequence signatures. Genome biology. 2009;10:1–16. doi: 10.1186/gb-2009-10-8-r85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Raes J, Korbel JO, Lercher MJ, Von Mering C, Bork P. Prediction of effective genome size in metagenomic samples. Genome biology. 2007;8:R10. doi: 10.1186/gb-2007-8-1-r10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucl Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li W, Godzik A. Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics. 2006;22:1658–1659. doi: 10.1093/bioinformatics/btl158. [DOI] [PubMed] [Google Scholar]
- 41.Burge SW, et al. Rfam 11.0: 10 years of RNA families. Nucleic Acids Res. 2013;41:D226–D232. doi: 10.1093/nar/gks1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Simon DM, Zimmerly S. A diversity of uncharacterized reverse transcriptases in bacteria. Nucleic Acids Res. 2008;36:7219–7229. doi: 10.1093/nar/gkn867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Eddy S. Profile hidden Markov models. Bioinformatics. 1998;14:755–763. doi: 10.1093/bioinformatics/14.9.755. [DOI] [PubMed] [Google Scholar]
- 44.Guindon S, et al. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol. 2010;59:307–321. doi: 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data supporting the results of this study are available within the paper and its Supplementary Information and Supplementary Data Files. Assembled metagenomic sequences are available in the NCBI BioProject database under the accession numbers PRJNA268032, PRJNA273161, PRJNA288027, and KY476664-KY476802.