Abstract
Inherited arrhythmia disorders are a group of potentially lethal diseases that remain diagnostic and management challenges. While the genetic basis for many of these disorders is well known, the pathogenicity of individual mutations and the resulting clinical outcomes are difficult to predict. Treatment options remain imperfect, and optimizing therapy for individual patients can be difficult. Recent advances in the derivation of induced pluripotent stem cells (iPSCs) from patients and creation of genetically engineered human models using CRISPR/Cas9 has the potential to dramatically advance translational arrhythmia research. In this review, we discuss the current state of modeling inherited arrhythmia disorders using human iPSC-derived cardiomyocytes. We also discuss current limitations and areas for further study.
Keywords: Arrhythmia, iPSCs, Sudden cardiac death, long QT syndrome, arrhythmogenic cardiomyopathy, catecholaminergic polymorphic ventricular tachycardia, brugada syndrome
Introduction
Even before the first recordings of the ECG by Willem Einthoven in the early 1900s the notion that the heart was coordinated by electrical currents was well-recognized. Indeed the first case of the inherited arrhythmia syndrome long QT syndrome (LQTS) was described in 1856 when a deaf girl died after her teacher yelled at her in a family with a previous child who had died after a fright1,2. However it was not until the late 1990s when the first genes for congenital LQTS were cloned by Keating and colleagues, leading to the broad understanding that inherited arrhythmia disorders (IADs) were often caused by mutations in ion channels or related proteins3–5. Since then, more than 15 genes have been found to cause LQTS6. In addition to LQTS several other IADs have been described, including arrhythmogenic cardiomyopathy (ACM)7, catecholaminergic polymorphic ventricular tachycardia (CPVT)8 and Brudaga syndrome (BrS)9. Each of these disorders is caused by mutations in an ever-growing number of genes (Table 1)10–12.
Table 1.
The Genetics of Inherited Arrhythmia Disorders
| Disorder | Causative genes | Cellular Phenotype | Clinical Phenotype |
|---|---|---|---|
| Long QT Syndrome | KCNQ1 (LQT1), KCNE1 | Decreased IKs | TdP |
| KCNH2 (LQT2), KCNE2 | Decreased IKr | TdP | |
| SCN5A (LQT3), SCN4B, SNTA1 | Increased INa | TdP, Bradycardia | |
| ANK2 | Decreased Na/K-ATP activity | Bradycardia, conduction block, TdP | |
| KCNJ2 (Andersen-Tawil Syndrome) | Decreased IK1 | Facial anomalies, periodic paralysis, stress-induced VT | |
| CACNA1C (Timothy Syndrome) | Decreased ICa | TdP, Autism, syndactyly | |
| CAV3 | Decreased IKr | TdP | |
| CALM1, CALM2 | Altered calcium handling | VT | |
|
| |||
| CPVT | RYR2 | Increased calcium leak, altered calcium handling | Stress and exercise-induced VT/VF |
| CASQ2 | Decreased SR calcium buffering | Stress and exercise-induced VT/VF | |
| CALM1 | Abnormal Ca2+ signaling | Stress and exercise-induced VT/VF/Long QT | |
| Triadin | Altered release of Ca2+ | Stress and exercise-induced VT/VF | |
|
| |||
| Brugada | SCN5A, SCN10A, SCN1B, SCN2B, SCN3B, GPD1L, MOG1, SLMAP, PKP2, HEY2 | Decreased INa+ | ECG changes, VT/VF at rest or with fever |
| CACNA1C, CACNB2, CACNA2D1 | Decreased ICa2+ | VT/VF at rest | |
| HCN4, KCNE3, KCNE5, KCND3, ABCC9, KCNJ8, KCNH2, PKP2 | Increased IK+;Ito | VT/VF | |
|
| |||
| ACM | PKP2, DSG2, DSC2, DSP, JUP | Decreased INa and gap junctions; apoptosis | Fibrosis, cardiac dysfunction, VT |
Tdp, Torsades de Pointes; VT, ventricular tachycardia, VF, ventricular fibrillation
Key questions that arise when evaluating a patient with a potential inherited arrhythmia disorder are: Is the mutation truly pathogenic? What is the patient’s risk for catastrophic events, including sudden cardiac death (SCD)? What is the optimal therapy for the patient? What are the implications of the patient’s diagnosis for family members? Unfortunately, these questions can be difficult to answer prospectively. Obtaining DNA sequencing information can be helpful to establish a diagnosis and assess family members, but often does not help to stratify risk or direct optimal therapy. One major reason is that these disorders can have variable penetrance or expressivity ranging from no overt clinical manifestation to mild ECG abnormalities to frequent life-threatening arrhythmias and SCD13. The genetic or cellular mechanisms that underlie variable expressivity for cardiac channelopathies remain poorly understood, making it difficult to predict clinical outcomes based solely on the genetic abnormality present.
Model systems have been essential for understanding how mutations in IAD genes cause arrhythmias. Expression of mutant genes in non-cardiac expression systems has allowed the characterization of the effect of mutations on individual channel function14,15. These analyses have been augmented by structural homology modeling16 and by computational models that integrate individual channel properties into virtual APs16,17. Genetically modified mouse, and more recently rabbit and pig, models have been useful to assess the effect of mutations within intact animals18–20. While these methods can provide evidence about the pathogenicity and mechanism of action of particular mutations, there is uncertainty in their ability to predict clinical outcomes21. Lack of appropriate physiological context in heterologous systems22, non-comprehensive protein structural information23, or differences in ion channel expression24 and cardiac electrophysiological properties25 between animal models and humans all limit the ability of traditional disease models to predict human clinical phenotype, no less predict inter-individual variation that arises from the interaction of the mutation with genetic modifiers.
Recently, Yamanaka and colleagues developed a groundbreaking platform for human disease modeling26,27 which promises to enhance mechanistic studies and, potentially, provide individualized information on risk and optimal therapy. Using a cocktail of reprogramming factors, this group showed that adult somatic cells such as skin fibroblasts could be converted into pluripotent stem cells (“induced pluripotent stem cells”, iPSCs), which can then be directed to differentiate into other cell types including cardiomyocytes. Since its original description, generation of iPSCs from a wide range of tissue sources (skin; blood; urine) has become a routine procedure. Because iPSCs carry the genetic material of the cell donor, human iPSCs (hiPSCs) derived from patients yield patient-specific disease models, including models of cardiomyopathies28–30 and IADs (Table 2; Figure 1). Theoretically, these patient-specific models should be able to recapitulate genetically determined phenotypic differences between patients, although empiric validation of this concept is sparse31,32. It is this area where iPSC technology is uniquely poised to provide an entirely new method to study both monogenetic and complex polygenetic diseases.
Table 2.
iPSC-CM Models of Inherited Arrhythmia Disorders.
| IAD model | Gene Mutation | Experimental Methods | Findings | Ref. |
|---|---|---|---|---|
| LQT1 | KCNQ1 (R190Q) | Patch-clamp | Prolongation of AP, Inappropriate AP adaptation at higher pacing frequencies | 62 |
| LQT2 | KCNH2 (A614V) | Patch-clamp, MEA | Prolongation of AP and induction of EADs; Decreased IKr; potential improvement with pinacidil | 48 |
| LQT2 | KCNH2 (G1681A) | Voltage-clamp | Comparison of symptomatic and asymptomatic carrier; AP prolongation and EADs; stronger phenotype in symptomatic carrier | 61 |
| LQT2 | KCNH2 (R176W) | Voltage-clamp, MEA | AP prolongation; differences in severity between voltage-clamp and MEA recordings | 59 |
| BrS/LQT3 | E1784K | Patch-clamp, MEA | LQT3 phenotype, but with SCN3B knockdown, BrS phenotype unmasked | 46 |
| LQT8 | CACNA1C (G1216A) | Patch-clamp, calcium imaging | AP prolongation; Slower contraction rates in mutant cells; prolonged Ca2+ transients; Possible therapeutic effect of rosovitine | 73 |
| CPVT | RYR2 (F243I) | Voltage-clamp, calcium imaging | Increased DADs with isoproterenol; increased spontaneous calcium sparks | 54 |
| CPVT | RYR (E2311N) | Voltage-clamp, calcium imaging | Increased DADs; increased spontaneous calcium sparks; normalization with CaMKii inhibition | 84 |
| CPVT | RYR (P2328S) | Voltage-clamp, calcium imaging | Altered calcium signaling; DADs and EADs at baseline and with isoproterenol | 85 |
| CPVT | RYR2 (S406L) | Voltage-clamp, calcium imaging | Increased DADs with isoproterenol; improvement with dantrolene | 55 |
| CPVT | RYR2 (L3741P) | Calcium imaging, MEA | Increased DADs; increased spontaneous calcium sparks; reversal of phenotype with flecainide | 57 |
| CPVT | CASQ2 (D307H) RYR2 (R420Q) |
Voltage-clamp calcium imaging | Increased DADs with isoproterenol; increased spontaneous calcium sparks | 56 |
| ACM | PKP2 (L614P) | Patch-clamp, calcium imaging, IF, EM | Reduction of both PKP2 and plakoglobin by immunofluorescence | 90 |
| ACM | PKP2 G784AfsA PKP2 K628Rfs |
IF, EM | 89 | |
| ACM | PKP2 A324fs | MEA, IF, EM | 91 | |
| BrS | Unknown SCN5A 1795insD |
Patch-clamp | No definitive changes in INa, Ito or ICa,L consistent with BrS | 99 |
| BrS/LQT3 | SCN5A (1795insD) | Patch-clamp | Decreased INa density and larger late current | 44 |
AP, action potential; EADs, early after depolarizations; DADs, delayed after depolarizations; MEA, multiple electrode array. IF, immunofluorescence. EM, electron microscopy.
homozygous mutation.
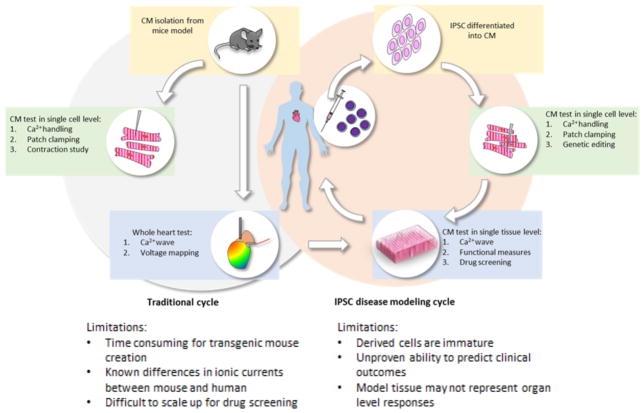
Figure 1.
Flowcharts for traditional IAD analysis with murine models as compared to those using hiPSC-CMs
Electrophysiologic Characteristics of hiPSC-CMs as Compared to Adult Cardiomyocytes
Several protocols for directed cardiac differentiation exist in the literature, often based on timed inhibition of GSK3β and WNT signaling. These protocols for generation of hiPSC-derived cardiomyocytes (hiPSC-CMs) are robust, straightforward, and highly efficient33,34, often yielding cultures in which 70–90% of cells express the cardiomyocyte marker TNNT2. Without positive or negative selection, 50–70% of these hiPSC-CMs are ventricular or ventricular-like cells, with the remaining cells being either atrial or nodal subtypes35. Methods for directed differentiation and selection of atrial36, ventricular37, and even cardiac purkinje38, subtypes of iPSC-CMs have been described, but these have not been applied in most reported studies. Therefore cellular heterogeneity must be considered when interpreting data from hiPSC-CM experiments.
Using single-cell patch clamp, a great deal of work has already been done to define the electrophysiologic properties of hiPSC-CMs. In general, hiPSC-CMs have less mature physiology than adult CMs from human patients or animal models. The resting membrane potential (RMP) of hiPSC-CMs is more depolarized (−50 to −75 mV) than adult CMs (−82 to 87 mV; in this review CM is used to indicate cardiomyocytes obtained from animals or humans), consistent with a more immature phenotype39. This elevated RMP is due to reduced expression of the constitutive inward potassium current IK1 in iPSC-CMs compared to mature CMs (0.9 pA/pF39 versus 3.6 pA/pF40). Forced expression of KCNJ2 (also known as Kir2.1), which underlies IK1, normalized RMP, improved upstroke velocity, and reduced the percentage of spontaneously beating CMs in human embryonic stem-cell derived cardiomyocytes (hESC-CMs)41. These data correlate with comparative gene expression studies of long-term cultures of hiPSC-CMs and adult CMs, which demonstrate a gradual increase in expression of KCNJ2 as well as a relative decrease in the amount of the pacemaker channel HCN4, responsible for a slow depolarizing current that contributes to increased automaticity of iPSC-CMs42.
The expression and relative levels of ion channel proteins and their associated subunits play a major role in shaping the cardiac action potential (AP) and calcium oscillation43. The cardiac sodium channel, which produces the cardiac Na+ current (INa) is responsible for the AP upstroke in chamber (atrial or ventricular) CMs. INa has relatively comparable biophysical properties in hiPSC-CMs compared to isolated adult CMs, including voltage-dependent activation and inactivation characteristics. However, hiPSC-CM peak currents were considerably larger (approximately −216 pA/pF compared to −20 pA/pf)39,44,45. Interestingly, while the upstroke velocities (dV/dtmax) of hiPSC-CMs are generally slower than adult CMs (9 to 40 V/s compared to 215 to 234 V/s) the sodium channel density does not appear to be the limiting factor. Rather, reduced upstroke velocity likely reflects partial sodium channel inactivation secondary to the relatively more depolarized RMP. Consistent with this hypothesis, quiescent hiPSC-CMs exhibited both more depolarized RMP and increased up-stroke velocity compared to their spontaneously beating counterparts, consistent with relatively higher maturity of quiescent compared to spontaneously beating hiPSC-CMs44. Despite the reduced AP upstroke velocity in hiPSC-CMs, patient-derived hiPSC-CMs harboring SCN5A or SCN3B mutations in large part recapitulated the abnormalities observed in CMs derived from transgenic mice with similar genetic anomalies44,46.
The L-type (ICa,L) calcium current, the major calcium current expressed in the mammalian heart, is present in hiPSC-CMs. However, its biophysical properties are more comparable to those observed in adult atrial rather than ventricular CMs39,47: where the V1/2 (The voltage where 50% of the channels are active or inactive) of activation and inactivation are much lower (left shift) in hiPSC-CMs than native ventricular ventricular myocytes The pharmacology of the ICa,L in hiPSC-CMs appears similar to adult CMs, as application of nifedipine shortens the AP without significant effects on upstroke velocities39,48.
Although hiPSC-CMs recapitulate many features of ICa,L, these cells lack T-tubules, an extensive network of plasma membrane invaginations that are key components of calcium release units of CMs. T-tubules, a hallmark of mature ventricular CMs, permit rapid propagation of APs into the center of cardiomyocytes, thereby coordinating calcium release and contraction despite the large size of these cells. The calcium release unit is composed of “dyads”, where junctional domains of the sarcoplasmic reticulum (jSR) are closely juxtaposed with T-tubules. As a result, ryanodine receptor 2 (RYR2), the major CM intracellular calcium release channel located on jSR, is positioned close to L-type calcium channels, located on T-tubules, thereby facilitating cardiomyocyte calcium-induced calcium release. hiPSC-CMs, like embryonic and neonatal CMs (and mature rodent atrial CMs)49,50, lack T-tubules51. In these cells, RYR2 within SR continues to align with sarcomere Z-lines, as it does in mature CMs, but L-type calcium channel is limited to the cell periphery, resulting in sequential and less synchronous calcium release50. In addition, inositol-3-phosphate receptor contributes to a much greater proportion of calcium transients that in adult CMs, another feature consistent with immaturity.52,53 Despite these differences in calcium handling between hiPSC-CMs and adult ventricular CMs, hiPSC-CMs have been productively used to model diseases such as CPVT that are caused by mutations that affect calcium handling54–57 (Table 2).
There are multiple potassium currents that contribute to the cardiac AP (please see the review by Nerbonne and Kass58 for a more comprehensive analysis). The delayed rectifier potassium current that is responsible for repolarization consists primarily of rapid (IKr) and slow (IKs) components, in addition to the aforementioned IK1 that contributes to RMP as well as the terminal phase of the AP. Both IKr and IKs are present in hiPSC-CMs, and these share similar biophysical properties and expression with human CMs39,59. The expression of IKr is comparable between hiPSC-CMs and native ventricular CMs with similar current densities and voltages of activation and inactivation48,60. Functional expression of IKr has also been demonstrated by AP prolongation and early afterdepolarizations (EADs) with the potassium channel E4031 48,61. In contrast the current density for IKs appears to be much more variable, as two different studies have reported vastly different average current densities varying between 0.31 pA/pF39 to as high as 2.5 pA/pF62. In either case, the current density for IKs is significantly higher in hiPSC-CMs than what has been reported for native left ventricular human CMs where the average current density is approximately 0.18 pA/pF63. One possibility for this variability may be the relative expression of KCNE1, which encodes an IKS subunit and has been demonstrated to have a significant effect on IKs current density in hESC-CMs64.
Overall, in several respects hiPSC-CMs are more similar to immature neonatal myocytes than adult CMs. While some aspects of hiPSC-CM channel expression and physiology are well developed, others (RMP and IK1 expression; INa upstroke velocity; calcium release kinetics and calcium release unit ultrastructure) are not under current culture conditions. Three dimensional tissue engineering, mechanical loading, modulation of substrate stiffness, phasic electrical stimulation, hormonal treatment (e.g. thyroid hormone), and long term culture have all been used to enhance hiPSC-CM maturity, with some success65–72. Further progress will require improved understanding of the natural mechanisms that regulate CM maturity. Caution must be applied when using hiPSC-CMs to model IADs, particularly those involving physiologic parameters that are divergent between hiPSC-CMs and mature CMs. Nevertheless, hiPSC-CMs have proven to be useful IAD models (see next section), just as genetically engineered rodents have been imperfect but valuable models for understanding IAD pathogenesis.
Use of hiPSC-derived Cardiomyocytes to Model Inherited Arrhythmia Disorders
Long QT Syndrome
One of the first examples of using patient-specific hiPSCs to model human disease was for LQTS48,62,73, IADs caused by mutations that prolong repolarization by either decreasing repolarizing inward currents or by increasing depolarizing outward currents. In these studies, fibroblasts isolated from patients with clinical and genetic evidence for congenital LQTS were reprogrammed into hiPSCs. Subsequent differentiation into cardiomyocytes (hiPSC-CMs) then provided a platform to study LQTS mutations in an appropriate cellular context as compared to more traditional heterologous expression systems such as HEK cells.
Several groups have demonstrated expected electrophysiologic abnormalities in hiPSC-CM LQTS models, including LQT148,62, LQT2 74, LQT344 and LQT873. These studies have focused on the phenotypes of single cells or small clusters of cells, as measured by patch-clamp recordings. Extracellular field potential duration, recorded by multiple electrode arrays, or whole cell calcium transients, recorded using fluorescent calcium sensitive dyes, have been used as higher-throughput assays75. Collectively these studies showed that LQTS hiPSC-CMs exhibit increased AP duration (APD), an in vitro correlate of prolongation of the QT interval. Voltage-clamp studies of hiPSC-CMs developed from LQTS patients exhibited prolonged APDs and frequent early after-depolarizations (EADs), cellular abnormalities hypothesized to initiate the hallmark arrhythmia of LQTS, Torsades de Pointes 48. That hiPSC-CMs accurately reproduce known electrophysiologic abnormalities provides evidence supporting the use of hiPSC-CMs to model LQTS and by extension other forms of IADs.
hiPSC-CM models can provide new insights into patient disease pathogenesis. For example, hiPSC-CMs from patients with complex or previously unreported mutations, which are difficult to study by more traditional methods, have been informative. Kass, Keller, and colleagues, systematically dissected the relative importance of abnormal sodium and potassium currents in iPSC-CMs from a patient with variants in both SCN5A (LQT3) and KCNH2 (LQT2)76. The authors elegantly demonstrated by patch-clamp recordings that the observed clinical phenotype of severe QT prolongation is likely dominated by dysfunction in the late sodium current without significant contribution from the more common variant in KCNH2. It is this type of analysis which clearly demonstrates the promise of patient-specific hiPSC-CMs as a platform to determine the potential pathogenicity of specific mutations or variants of unknown significance (VUS) as compared to traditional methods77.
Another important aspect of hiPSC-CMs is the ability to test drug responses in vitro. The potential for a nearly unlimited supply of hiPSC-CMs to screen for novel drugs or test the side-effects of existing compounds has garnered significant interest from the drug development industry78. Use of human PSC-CMs is one of the three pillars of the Comprehensive In vitro Pro-arrhythmia Assay (CiPA) initiative, intended to become a mainstay for regulatory evaluation of drugs for their potential to prolong the QT interval and thereby potentially cause lethal arrhythmias79. hiPSC-CMs accurately modeled the protective effect of beta-blockers, an established therapy for LQTS62. Several groups have used hiPSC-CMs to show that already-approved medications have efficacy in single-cell assays, suggesting the possibility that they could be repurposed as novel therapies for inherited arrhythmia syndromes. Yazawa and colleagues developed a model of LQT8 (Timothy syndrome) and demonstrated that the cyclin-dependent kinase inhibitor roscovitine (Selciclib) could partially rescue the abnormal phenotypes observed in Timothy syndrome hiPSC-CMs73. High-throughput platforms to measure AP duration and other electrophysiologic parameters for drug screening have been reported80. Whether these methods have the phenotype sensitivity and/or specificity to screen for novel therapeutics is yet to be determined.
Catecholaminergic Polymorphic Ventricular Tachycardia (CPVT)
CPVT is characterized by recurrent ventricular or atrial arrhythmias during times of stress or exercise8. While most CPVT patients suffer from RYR2 mutations, pathologic variants have also been reported in calsequestrin (CASQ2)81, calmodulin (CaM) genes82 and triadin (Trd)83. These mutations generally increase SR calcium release through RYR2, resulting in elevated intracellular calcium that can drive the sodium calcium exchanger NCX1 in reverse10. This generates a depolarizing current that trigger EADs or DADs, which are thought to underlie the polymorphic or bidirectional ventricular tachycardia that are hallmarks of the disease.
To date, there are five hiPSC-CM models of RYR2 mutations have been described54,55,57,84,85. These cover three of the four regions of RYR2 where the vast majority of pathogenic mutations have been described8. Using primarily voltage-clamp and calcium imaging of small cell clusters, it has been demonstrated that CPVT hiPSC-CMs have an increased frequency of atypical calcium transients, spontaneous calcium release events (Ca2+ sparks), and single-cell voltage membrane depolarization abnormalities, resulting in EADs and DADs54,55,57,84. Consistent with the clinical phenotype of arrhythmia precipitated by catecholaminergic stimulation, these abnormalities are exacerbated by administration of the beta-adrenergic agonist isoproterenol. Several groups have also demonstrated that the duration of spontaneous Ca2+ sparks are prolonged in CPVT-hiPSC-CMs compared to controls57,84. Preininger et. al further investigated calcium homeostasis in CPVT-derived hiPSC-CMs by using tetracaine to inhibit RYR2 Ca2+ release from the sacroplasmic reticulum (SR) and demonstrated increased diastolic Ca2+ leak as well as lower SR Ca2+ loading after tonic stimulation55. These data suggest that enhanced diastolic calcium release (Ca2+ leak) and possibly impaired SR calcium loading may be the mechanisms for the observed abnormalities in CPVT-hiPSC-CMs, rather than SR Ca2+ overload which has been proposed as one mechanism in studies of isolated CMs from transgenic mice86.
hiPSC-CM models of CPVT have been used to test new therapeutic strategies. Dantrolene is used in malignant hyperthermia to stabilize the closed state of the skeletal ryanodine receptor (RYR1). In a single-cell CPVT model, dantrolene was effective in suppressing excessive calcium sparks, EADs, and triggered beats caused by RYR2 mutation55,84. Although CPVT occurs under conditions of high adrenergic stimulation, the downstream targets of adrenergic signaling that interact with RYR2 mutation to induce arrhythmogenesis are not known. Treatment of hiPSC-CM and mouse models of CPVT with inhibitors of CaMKII, a major kinase activated downstream of the β-adrenergic receptor, blocked arrhythmic phenotypes84,87, suggesting that CaMKII phosphorylation of downstream target(s) is central to disease pathogenesis.
These data suggest that hiPSC-CMs recapitulate cardinal features of CPVT that make them useful to investigate arrhythmia mechanisms and to expedite drug development.
Arrhythmogenic Cardiomyopathy (ACM)
ACM, also known as arrhythmogenic right ventricular cardiomyopathy, is a set of disorders caused by mutation of desmosome genes. Through uncertain mechanisms, ACM causes cardiomyocyte loss, cardiac fibrosis, and fatty infiltration, resulting in cardiac dilatation, dysfunction, and arrhythmia. Although cardiac dysfunction and fibrosis undoubtedly contribute to arrhythmias in this disease, some patients present with arrhythmia early in the disease that is out of proportion to the extent of cardiac involvement, suggesting primary arrhythmogenic mechanisms88.
There are several reported hiPSC-CM models of ACM89–92. These models recapitulated the desmosomal abnormalities that are central to the disease89–92. ACM hiPSC-CMs had increased propensity to accumulation of lipid droplets under adipogenic culture conditions89–92. While interesting, it is important to note that ACM myocardium is infiltrated by bona fide adipocytes, whereas lipid-laden CMs are not a frequently noted feature of clinical ACM. These metabolic derangements were linked to increased hiPSC-CM apoptosis89,91,92. There have only been two studies that have examined the characteristics of ACM hiPSC-CMs direct relevant to conduction and arrhythmia. One study reported no electrophysiologic abnormalities or differences in response to calcium channel blockade (nifedipine) or adrenergic stimulation (isoproterenol)90. A second study found that field potential rise time was prolonged in ACM hiPSC-CMs, suggesting reduced conduction or excitability91. hiPSC-CMs also exhibited decreased Cx43 gap junction density and wider desmosomes, which interestingly was correlated with the extent of intracellular lipid accumulation. The interaction between desmosomes, metabolism, and conduction are intriguing, but more research will be necessary to determine if hiPSC-CMs can serve as effective models for arrhythmia in ACM.
Brugada Syndrome (BrS)
BrS is an IAD characterized by stereotypical electrocardiogram findings without significant structural heart disease and is associated with a high incidence of SCD, especially in males93,94. The gene most frequently mutated in BrS is SCN5A, encoding the cardiac sodium channel, but SCN5A mutations only account for about 20% of all BrS patients12. Interestingly, SCN5A mutations also cause LQT3, and some specific “overlap” mutations have been implicated in both BrS and LQT3. It is generally accepted that depolarization abnormalities, especially in the right ventricular outflow tract, underlie the pathophysiology of BrS. One of the challenges in diagnosis is that a number of asymptomatic patients present only with incidental ECG changes95 but the first symptoms are often SCD or aborted cardiac arrest96. While various diagnostic and clinical criteria have been studied, few parameters have been found that reliably identify patients at greatest risk.
Davis and colleagues44 studied a patient-derived hIPSC-CMs with heterozygous SCN5A 1795insD mutation, an overlap mutation associated with both BrS and LQT397,98. By patch clamp, these cells had decreased INa density (BrS phenotype) as well as increased late sodium current (LQT3 phenotype), consistent with CMs from mice with the equivalent mutation that were studied in parallel44. However, a second study of hiPSC-CMs from three patients with clinically well-defined BrS but with undefined genetics as well as hiPSC-CMs from a patient with 1795insD did not reach the same conclusions99. Using very-detailed single-cell electrophysiologic analysis, the authors did not find the characteristic abnormalities in INa typically observed in CMs harboring pathogenic BrS mutations. They also did not detect any consistent changes in the other currents proposed to contribute to BrS (Ito or ICa,L)95. A third study of hiPSC-CMs from a patient with E1784K SCN5A mutation and a mixed LQT3 and BrS phenotype found that the hiPSC-CMs manifested electrophysiological abnormalities consistent with LQT3 but not BrS46. SCN3B, the embryonic beta subunit of the cardiac sodium channel, was found to be expressed in hiPSC-CMs at higher levels than in mature CMs. Knockdown of SCN3B unmasked the BrS phenotype. Thus gene expression changes related to maturity impact phenotypic expression of the mutant genotype in hiPSC-CMs compared to adult CMs and perhaps also accounts for the adult onset of BrS.
Genome editing in disease modeling
Designer meganucleases such as TALENs and in particular CRISPR/Cas9 have enabled precise genome editing with minimal off-target mutagenesis100. These nucleases can be used to inactivate genes with targeted insertions or deletions, or make precise changes through homology-directed repair (HDR). Genome editing is an essential additional to the hiPSC-based disease modeling toolbox for three reasons. First, the ability to study different mutations in the same genetic background reduces the potential variability in phenotype that might be introduced by epigenetic differences101 or unknown genetic modifiers102,103. By maintaining a well-defined genetic background, the introduction of isogenic mutations can also improve the specificity of the quantifiable phenotypes attributable to particular mutations and potentially reveal novel relevant mechanisms of disease104,105. Second, the reciprocal approach is to use genome editing to correct the known mutations from patient-derived iPSC lines to create the proper controls for comparison, rather than using unrelated samples or unaffected family members106. This method can also provide information about the role of the genetic background or the contribution of a single mutation in complex multi-gene disorders44. Finally, genome editing will likely be a powerful tool to explore complex signaling pathways and the contributions of individual signaling events on cellular phenotypes.
Prediction of clinical phenotypes
As reviewed above, it is clear that hiPSC-CMs have applications for studying disease mechanisms and for identifying potential therapies. The potential for making patient-specific models that capture the differences between patients is perhaps the most exciting and disruptive feature of this technology, but at this point there is scant empiric evidence that hiPSC-CMs reproduce phenotypic differences between patients32. Proof-of-concept will require correlating phenotypes across a panel of patients to their cellular equivalents, measured on the hiPSC-CMs derived from these patients. Some studies have done this anecdotally. For example, some CPVT patients respond to flecainide but others do not107. Preininger and colleagues identify a patient with CPVT that responds to flecainide and then demonstrate the abnormalities at a single-cell level in calcium handling including increased spark duration and frequency with isoproterenol administration is greatly mitigated by pre-incubation with flecainide57. However, systematic studies of many patients are needed to determine in practice how well hiPSC-CM models capture inter-individual differences, a prerequisite if information from these models will be practically useful to prospectively guide clinical decision making.
Single Cell Versus Tissue Level Phenotypes
To date, hiPSC-CM IAD models have focused almost exclusively on the cellular phenotypes that arise from mutation of IAD genes. These phenotypes have primarily been studied in individual cells or small islands of cells. However, in the heart CMs are integrated into a highly connected network of cells, and this electrical coupling of excitable cells profoundly influences expressed phenotypes. When individual cells are electrically coupled, aberrant activity of a single cell cell is buffered by dissipation of its electrical activity across the remaining cells. This “source-sink mismatch” can suppress asynchronous aberrant activity, such as EADs and DADs, so that single cell phenotypes may not be the same as phenotypes recorded from cells integrated into tissues.
Clinical arrhythmias are not cellular phenotypes, but rather are the emergent properties of tissues. In many cases the link between cellular phenotypes, such as EADs/DADs, and clinically relevant arrhythmias such as ventricular tachycardia remain cryptic108. To accurately model IADs, it will be necessary to assemble cardiomyocytes into anisotropic sheets that are rhythmically paced in defined geometries. Achieving this goal will require combining patient-derived or genetically engineered iPSC-CMs with bioengineering to assemble patterned, two- and three- dimensional tissues. Examples of these approaches have been used to model cardiomyopathies29,30 and are being extended to arrhythmias. Incorporation of optogenetic techniques109 will permit spatially precise optical stimulation and recording from the engineered tissues110. Combining iPSC-CMs with bioengineered cardiac tissues and optogenetics will certainly enhance IAD models and make it more likely that these models will be useful for predicting clinical outcomes.
Future Perspectives
In a short period of time, IAD modeling with hiPSC-CMs has advanced rapidly. Despite challenges and limitations arising from the immaturity of iPSC-CMs, pioneering studies summarized in this review have established the utility of hiPSC-CMs to faithfully recapitulate core features of IADs and to advance the current state of knowledge by revealing new disease mechanisms or facilitating drug discovery. Ongoing work will enhance hiPSC-CM models by enhancing cellular maturity and by yielding pure preparations of relevant hiPSC-CM subtypes. Expanding the use of genome editing will allow a greater range of clinically relevant mutations to be studied, and will allow for rigorous experimental designs that appropriately control for genetic background. Incorporation of tissue engineering and optogenetics will establish tissue level disease models, which are necessary to understand how molecular defects cause tissue level arrhythmia. A major area for future work will be to establish the extent to which hiPSC-CM models will be able to fulfill their promise of providing individualized disease models to stratify patient risk, guide therapy, and facilitate genetic counseling for family members. Undoubtedly, fulfilling this vision will require successful development of methods to enhance iPSC-CM maturation and integrate them into tissue level models.
Acknowledgments
WTP was supported by grants from the American Heart Association (16CSA28750006) and the National Institutes of Health (R01HL128694), and by charitable donations from the Department of Cardiology of Boston Children’s Hospital.
Bibliography
- 1.Jervell A, Lange-Nielsen F. Congenital deaf-mutism, functional heart disease with prolongation of the Q-T interval and sudden death. Am Heart J. 1957;54:59–68. doi: 10.1016/0002-8703(57)90079-0. [DOI] [PubMed] [Google Scholar]
- 2.Tranebjaerg L, Bathen J, Tyson J, Bitner-Glindzicz M. Jervell and Lange-Nielsen syndrome: a Norwegian perspective. Am J Med Genet. 1999;89:137–146. [PubMed] [Google Scholar]
- 3.Curran ME, Splawski I, Timothy KW, Vincent GM, Green ED, Keating MT. A molecular basis for cardiac arrhythmia: HERG mutations cause long QT syndrome. Cell. 1995;80:795–803. doi: 10.1016/0092-8674(95)90358-5. [DOI] [PubMed] [Google Scholar]
- 4.Sanguinetti MC, Jiang C, Curran ME, Keating MT. A mechanistic link between an inherited and an acquird cardiac arrthytmia: HERG encodes the IKr potassium channel. Cell. 1995;81:299–307. doi: 10.1016/0092-8674(95)90340-2. [DOI] [PubMed] [Google Scholar]
- 5.Wang Q, Curran ME, Splawski I, Burn TC, Millholland JM, VanRaay TJ, et al. Positional cloning of a novel potassium channel gene: KVLQT1 mutations cause cardiac arrhythmias. Nat Genet. 1996;12:17–23. doi: 10.1038/ng0196-17. [DOI] [PubMed] [Google Scholar]
- 6.Nakano Y, Shimizu W. Genetics of long-QT syndrome. J Hum Genet. 2016;61:51–55. doi: 10.1038/jhg.2015.74. [DOI] [PubMed] [Google Scholar]
- 7.Marcus FI, Edson S, Towbin JA. Genetics of arrhythmogenic right ventricular cardiomyopathy: a practical guide for physicians. J Am Coll Cardiol. 2013;61:1945–1948. doi: 10.1016/j.jacc.2013.01.073. [DOI] [PubMed] [Google Scholar]
- 8.Venetucci L, Denegri M, Napolitano C, Priori SG. Inherited calcium channelopathies in the pathophysiology of arrhythmias. Nat Rev Cardiol. 2012;9:561–575. doi: 10.1038/nrcardio.2012.93. [DOI] [PubMed] [Google Scholar]
- 9.Refaat MM, Hotait M, Scheinman M. Brugada Syndrome. Card Electrophysiol Clin. 2016;8:239–245. doi: 10.1016/j.ccep.2015.10.036. [DOI] [PubMed] [Google Scholar]
- 10.Bezzina CR, Lahrouchi N, Priori SG. Genetics of sudden cardiac death. Circ Res. 2015;116:1919–1936. doi: 10.1161/CIRCRESAHA.116.304030. [DOI] [PubMed] [Google Scholar]
- 11.Tester DJ, Ackerman MJ. Genetics of long QT syndrome. Methodist Debakey Cardiovasc J. 2014;10:29–33. doi: 10.14797/mdcj-10-1-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Watanabe H, Minamino T. Genetics of Brugada syndrome. J Hum Genet. 2015;61:57–60. doi: 10.1038/jhg.2015.97. [DOI] [PubMed] [Google Scholar]
- 13.Giudicessi JR, Ackerman MJ. Determinants of incomplete penetrance and variable expressivity in heritable cardiac arrhythmia syndromes. Transl Res. 2013;161:1–14. doi: 10.1016/j.trsl.2012.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Anderson CL, Kuzmicki CE, Childs RR, Hintz CJ, Delisle BP, January CT. Large-scale mutational analysis of Kv11.1 reveals molecular insights into type 2 long QT syndrome. Nat Commun. 2014;5:5535. doi: 10.1038/ncomms6535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhao Y-T, Valdivia CR, Gurrola GB, Powers PP, Willis BC, Moss RL, et al. Arrhythmogenesis in a catecholaminergic polymorphic ventricular tachycardia mutation that depresses ryanodine receptor function. [accessed 11/1/2016];Proc Natl Acad Sci U S A. 2015 112:E1669–77. doi: 10.1073/pnas.1419795112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Romero L, Trenor B, Yang P-C, Saiz J, Clancy CE. In silico screening of the impact of hERG channel kinetic abnormalities on channel block and susceptibility to acquired long QT syndrome. J Mol Cell Cardiol. 2015;87:271–282. doi: 10.1016/j.yjmcc.2015.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yang P-C, El-Bizri N, Romero L, Giles WR, Rajamani S, Belardinelli L, et al. A computational model predicts adjunctive pharmacotherapy for cardiac safety via selective inhibition of the late cardiac Na current. J Mol Cell Cardiol. 2016;99:151–161. doi: 10.1016/j.yjmcc.2016.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Garcia-Gras E, Lombardi R, Giocondo MJ, Willerson JT, Schneider MD, Khoury DS, et al. Suppression of canonical Wnt/beta-catenin signaling by nuclear plakoglobin recapitulates phenotype of arrhythmogenic right ventricular cardiomyopathy. J Clin Invest. 2006;116:2012–2021. doi: 10.1172/JCI27751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Brunner M, Peng X, Liu GX, Ren X-Q, Ziv O, Choi B-R, et al. Mechanisms of cardiac arrhythmias and sudden death in transgenic rabbits with long QT syndrome. J Clin Invest. 2008;118:2246–2259. doi: 10.1172/JCI33578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rizzi N, Liu N, Napolitano C, Nori A, Turcato F, Colombi B, et al. Unexpected Structural and Functional Consequences of the R33Q Homozygous Mutation in Cardiac Calsequestrin: A Complex Arrhythmogenic Cascade in a Knock In Mouse Model. Circ Res. 2008;103:298–306. doi: 10.1161/CIRCRESAHA.108.171660. [DOI] [PubMed] [Google Scholar]
- 21.Schwartz PJ. Introduction to the arrhythmogenic disorders of genetic origin series. Circ Arrhythm Electrophysiol. 2012;5:604–605. doi: 10.1161/CIRCEP.112.971846. [DOI] [PubMed] [Google Scholar]
- 22.Maltsev VA, Kyle JW, Undrovinas A. Late Na+ current produced by human cardiac Na+ channel isoform Nav1.5 is modulated by its beta1 subunit. J Physiol Sci. 2009;59:217–225. doi: 10.1007/s12576-009-0029-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ahuja S, Mukund S, Deng L, Khakh K, Chang E, Ho H, et al. Structural basis of Nav1.7 inhibition by an isoform-selective small-molecule antagonist. Science. 2015;350:aac5464. doi: 10.1126/science.aac5464. [DOI] [PubMed] [Google Scholar]
- 24.Jost N, Virág L, Comtois P, Ordög B, Szuts V, Seprényi G, et al. Ionic mechanisms limiting cardiac repolarization reserve in humans compared to dogs. J Physiol. 2013;591:4189–4206. doi: 10.1113/jphysiol.2013.261198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nerbonne JM. Studying cardiac arrhythmias in the mouse--a reasonable model for probing mechanisms? Trends Cardiovasc Med. 2004;14:83–93. doi: 10.1016/j.tcm.2003.12.006. [DOI] [PubMed] [Google Scholar]
- 26.Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 27.Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, et al. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 28.Lan F, Lee AS, Liang P, Sanchez-Freire V, Nguyen PK, Wang L, et al. Abnormal calcium handling properties underlie familial hypertrophic cardiomyopathy pathology in patient-specific induced pluripotent stem cells. Cell Stem Cell. 2013;12:101–113. doi: 10.1016/j.stem.2012.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang G, McCain ML, Yang L, He A, Pasqualini FS, Agarwal A, et al. Modeling the mitochondrial cardiomyopathy of Barth syndrome with induced pluripotent stem cell and heart-on-chip technologies. Nat Med. 2014;20:616–623. doi: 10.1038/nm.3545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hinson JT, Chopra A, Nafissi N, Polacheck WJ, Benson CC, Swist S, et al. HEART DISEASE. Titin mutations in iPS cells define sarcomere insufficiency as a cause of dilated cardiomyopathy. Science. 2015;349:982–986. doi: 10.1126/science.aaa5458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sharma A, Marceau C, Hamaguchi R, Burridge PW, Rajarajan K, Churko JM, et al. Human induced pluripotent stem cell-derived cardiomyocytes as an in vitro model for coxsackievirus B3-induced myocarditis and antiviral drug screening platform. Circ Res. 2014;115:556–566. doi: 10.1161/CIRCRESAHA.115.303810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Burridge PW, Li YF, Matsa E, Wu H, Ong S-G, Sharma A, et al. Human induced pluripotent stem cell-derived cardiomyocytes recapitulate the predilection of breast cancer patients to doxorubicin-induced cardiotoxicity. Nat Med. 2016;22:547–556. doi: 10.1038/nm.4087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lian X, Zhang J, Azarin SM, Zhu K, Hazeltine LB, Bao X, et al. Directed cardiomyocyte differentiation from human pluripotent stem cells by modulating Wnt/beta-catenin signaling under fully defined conditions. Nat Protoc. 2013;8:162–175. doi: 10.1038/nprot.2012.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Burridge PW, Matsa E, Shukla P, Lin ZC, Churko JM, Ebert AD, et al. Chemically defined generation of human cardiomyocytes. Nat Methods. 2014;11:855–860. doi: 10.1038/nmeth.2999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang J, Wilson GF, Soerens AG, Koonce CH, Yu J, Palecek SP, et al. Functional cardiomyocytes derived from human induced pluripotent stem cells. [Last accessed 11/1/2016];Circ Res. 2009 104:e30–41. doi: 10.1161/CIRCRESAHA.108.192237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Devalla HD, Schwach V, Ford JW, Milnes JT, El-Haou S, Jackson C, et al. Atrial-like cardiomyocytes from human pluripotent stem cells are a robust preclinical model for assessing atrial-selective pharmacology. EMBO Mol Med. 2015;7:394–410. doi: 10.15252/emmm.201404757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bizy A, Guerrero-Serna G, Hu B, Ponce-Balbuena D, Willis BC, Zarzoso M, et al. Myosin light chain 2-based selection of human iPSC-derived early ventricular cardiac myocytes. Stem Cell Res. 2013;11:1335–1347. doi: 10.1016/j.scr.2013.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tsai SY, Maass K, Lu J, Fishman GI, Chen S, Evans T. Efficient Generation of Cardiac Purkinje Cells from ESCs by Activating cAMP Signaling. Stem Cell Reports. 2015;4:1089–1102. doi: 10.1016/j.stemcr.2015.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ma J, Guo L, Fiene SJ, Anson BD, Thomson JA, Kamp TJ, et al. High purity human-induced pluripotent stem cell-derived cardiomyocytes: electrophysiological properties of action potentials and ionic currents. Am J Physiol Heart Circ Physiol. 2011;301:H2006–17. doi: 10.1152/ajpheart.00694.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Magyar J, Iost N, Kortvely A, Banyasz T, Virag L, Szigligeti P, et al. Effects of endothelin-1 on calcium and potassium currents in undiseased human ventricular myocytes. Pflugers Arch. 2000;441:144–149. doi: 10.1007/s004240000400. [DOI] [PubMed] [Google Scholar]
- 41.Lieu DK, Fu JD, Chiamvimonvat N, Tung KC, McNerney GP, Huser T, et al. Mechanism-based facilitated maturation of human pluripotent stem cell-derived cardiomyocytes. Circ Arrhythm Electrophysiol. 2013;6:191–201. doi: 10.1161/CIRCEP.111.973420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yang X, Pabon L, Murry CE. Engineering adolescence: maturation of human pluripotent stem cell-derived cardiomyocytes. Circ Res. 2014;114:511–523. doi: 10.1161/CIRCRESAHA.114.300558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Grant AO. Cardiac ion channels. Circ Arrhythm Electrophysiol. 2009;2:185–194. doi: 10.1161/CIRCEP.108.789081. [DOI] [PubMed] [Google Scholar]
- 44.Davis RP, Casini S, van den Berg CW, Hoekstra M, Remme CA, Dambrot C, et al. Cardiomyocytes derived from pluripotent stem cells recapitulate electrophysiological characteristics of an overlap syndrome of cardiac sodium channel disease. Circulation. 2012;125:3079–3091. doi: 10.1161/CIRCULATIONAHA.111.066092. [DOI] [PubMed] [Google Scholar]
- 45.Sakakibara Y, Furukawa T, Singer DH, Jia H, Backer CL, Arentzen CE, et al. Sodium current in isolated human ventricular myocytes. Am J Physiol. 1993;265:H1301–9. doi: 10.1152/ajpheart.1993.265.4.H1301. [DOI] [PubMed] [Google Scholar]
- 46.Okata S, Yuasa S, Suzuki T, Ito S, Makita N, Yoshida T, et al. Embryonic type Na+ channel beta-subunit, SCN3B masks the disease phenotype of Brugada syndrome. Sci Rep. 2016;6:34198. doi: 10.1038/srep34198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mewes T, Ravens U. L-type calcium currents of human myocytes from ventricle of non-failing and failing hearts and from atrium. J Mol Cell Cardiol. 1994;26:1307–1320. doi: 10.1006/jmcc.1994.1149. [DOI] [PubMed] [Google Scholar]
- 48.Itzhaki I, Maizels L, Huber I, Zwi-Dantsis L, Caspi O, Winterstern A, et al. Modelling the long QT syndrome with induced pluripotent stem cells. Nature. 2011;471:225–229. doi: 10.1038/nature09747. [DOI] [PubMed] [Google Scholar]
- 49.Bootman MD, Smyrnias I, Thul R, Coombes S, Roderick HL. Atrial cardiomyocyte calcium signalling. Biochim Biophys Acta. 2011;1813:922–934. doi: 10.1016/j.bbamcr.2011.01.030. [DOI] [PubMed] [Google Scholar]
- 50.Trafford AW, Clarke JD, Richards MA, Eisner DA, Dibb KM. Calcium signalling microdomains and the t-tubular system in atrial mycoytes: potential roles in cardiac disease and arrhythmias. Cardiovasc Res. 2013;98:192–203. doi: 10.1093/cvr/cvt018. [DOI] [PubMed] [Google Scholar]
- 51.Lieu DK, Liu J, Siu C-W, McNerney GP, Tse H-F, Abu-Khalil A, et al. Absence of transverse tubules contributes to non-uniform Ca(2+) wavefronts in mouse and human embryonic stem cell-derived cardiomyocytes. Stem Cells Dev. 2009;18:1493–1500. doi: 10.1089/scd.2009.0052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Knollmann BC. Induced Pluripotent Stem Cell-Derived Cardiomyocytes: Boutique Science or Valuable Arrhythmia Model? Circ Res. 2013;112:969–976. doi: 10.1161/CIRCRESAHA.112.300567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Itzhaki I, Rapoport S, Huber I, Mizrahi I, Zwi-Dantsis L, Arbel G, et al. Calcium handling in human induced pluripotent stem cell derived cardiomyocytes. [Last accessed 11/1/2016];PLoS One. 2011 6:e18037. doi: 10.1371/journal.pone.0018037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Fatima A, Xu G, Shao K, Papadopoulos S, Lehmann M, Arnáiz-Cot JJ, et al. In vitro modeling of ryanodine receptor 2 dysfunction using human induced pluripotent stem cells. Cell Physiol Biochem. 2011;28:579–592. doi: 10.1159/000335753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jung CB, Moretti A, Mederos y Schnitzler M, Iop L, Storch U, Bellin M, et al. Dantrolene rescues arrhythmogenic RYR2 defect in a patient-specific stem cell model of catecholaminergic polymorphic ventricular tachycardia. EMBO Mol Med. 2012;4:180–191. doi: 10.1002/emmm.201100194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Novak A, Barad L, Zeevi-Levin N, Shick R, Shtrichman R, Lorber A, et al. Cardiomyocytes generated from CPVTD307H patients are arrhythmogenic in response to β-adrenergic stimulation. J Cell Mol Med. 2012;16:468–482. doi: 10.1111/j.1582-4934.2011.01476.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Preininger MK, Jha R, Maxwell JT, Wu Q, Singh M, Wang B, et al. A human pluripotent stem cell model of catecholaminergic polymorphic ventricular tachycardia recapitulates patient-specific drug responses. Dis Model Mech. 2016;9:927–939. doi: 10.1242/dmm.026823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Nerbonne JM, Kass RS. Molecular physiology of cardiac repolarization. Physiol Rev. 2005;85:1205–1253. doi: 10.1152/physrev.00002.2005. [DOI] [PubMed] [Google Scholar]
- 59.Lahti AL, Kujala VJ, Chapman H, Koivisto AP, Pekkanen-Mattila M, Kerkela E, et al. Model for long QT syndrome type 2 using human iPS cells demonstrates arrhythmogenic characteristics in cell culture. Dis Model Mech. 2012;5:220–230. doi: 10.1242/dmm.008409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jost N, Acsai K, Horváth B, Bányász T, Baczkó I, Bitay M, et al. Contribution of I Kr and I K1 to ventricular repolarization in canine and human myocytes: is there any influence of action potential duration? Basic Res Cardiol. 2009;104:33–41. doi: 10.1007/s00395-008-0730-3. [DOI] [PubMed] [Google Scholar]
- 61.Matsa E, Rajamohan D, Dick E, Young L, Mellor I, Staniforth A, et al. Drug evaluation in cardiomyocytes derived from human induced pluripotent stem cells carrying a long QT syndrome type 2 mutation. Eur Heart J. 2011;32:952–962. doi: 10.1093/eurheartj/ehr073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Moretti A, Bellin M, Welling A, Jung CB, Lam JT, Bott-Flügel L, et al. Patient-specific induced pluripotent stem-cell models for long-QT syndrome. N Engl J Med. 2010;363:1397–1409. doi: 10.1056/NEJMoa0908679. [DOI] [PubMed] [Google Scholar]
- 63.Iost N, Virág L, Opincariu M, Szécsi J, Varró A, Papp JG. Delayed rectifier potassium current in undiseased human ventricular myocytes. Cardiovasc Res. 1998;40:508–515. doi: 10.1016/s0008-6363(98)00204-1. [DOI] [PubMed] [Google Scholar]
- 64.Wang K, Terrenoire C, Sampson KJ, Iyer V, Osteen JD, Lu J, et al. Biophysical properties of slow potassium channels in human embryonic stem cell derived cardiomyocytes implicate subunit stoichiometry. J Physiol. 2011;589:6093–6104. doi: 10.1113/jphysiol.2011.220863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tulloch NL, Muskheli V, Razumova MV, Korte FS, Regnier M, Hauch KD, et al. Growth of engineered human myocardium with mechanical loading and vascular coculture. Circ Res. 2011;109:47–59. doi: 10.1161/CIRCRESAHA.110.237206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Jacot JG, McCulloch AD, Omens JH. Substrate stiffness affects the functional maturation of neonatal rat ventricular myocytes. Biophys J. 2008;95:3479–3487. doi: 10.1529/biophysj.107.124545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhang D, Shadrin IY, Lam J, Xian H-Q, Snodgrass HR, Bursac N. Tissue-engineered cardiac patch for advanced functional maturation of human ESC-derived cardiomyocytes. Biomaterials. 2013;34:5813–5820. doi: 10.1016/j.biomaterials.2013.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Nunes SS, Miklas JW, Liu J, Aschar-Sobbi R, Xiao Y, Zhang B, et al. Biowire: a platform for maturation of human pluripotent stem cell-derived cardiomyocytes. Nat Methods. 2013;10:781–787. doi: 10.1038/nmeth.2524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Mihic A, Li J, Miyagi Y, Gagliardi M, Li S-H, Zu J, et al. The effect of cyclic stretch on maturation and 3D tissue formation of human embryonic stem cell-derived cardiomyocytes. Biomaterials. 2014;35:2798–2808. doi: 10.1016/j.biomaterials.2013.12.052. [DOI] [PubMed] [Google Scholar]
- 70.Chattergoon NN, Giraud GD, Louey S, Stork P, Fowden AL, Thornburg KL. Thyroid hormone drives fetal cardiomyocyte maturation. FASEB J. 2012;26:397–408. doi: 10.1096/fj.10-179895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lundy SD, Zhu W-Z, Regnier M, Laflamme MA. Structural and functional maturation of cardiomyocytes derived from human pluripotent stem cells. Stem Cells Dev. 2013;22:1991–2002. doi: 10.1089/scd.2012.0490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ruan J-L, Tulloch NL, Razumova MV, Saiget M, Muskheli V, Pabon L, et al. Mechanical Stress Conditioning and Electrical Stimulation Promote Contractility and Force Maturation of Induced Pluripotent Stem Cell-Derived Human Cardiac Tissue. Circulation. 2016 doi: 10.1161/CIRCULATIONAHA.114.014998. [Internet] Available from: http://dx.doi.org/10.1161/CIRCULATIONAHA.114.014998. [DOI] [PMC free article] [PubMed]
- 73.Yazawa M, Hsueh B, Jia X, Pasca AM, Bernstein JA, Hallmayer J, et al. Using induced pluripotent stem cells to investigate cardiac phenotypes in Timothy syndrome. Nature. 2011;471:230–234. doi: 10.1038/nature09855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wang Y, Liang P, Lan F, Wu H, Lisowski L, Gu M, et al. Genome editing of isogenic human induced pluripotent stem cells recapitulates long QT phenotype for drug testing. J Am Coll Cardiol. 2014;64:451–459. doi: 10.1016/j.jacc.2014.04.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Denning C, Borgdorff V, Crutchley J, Firth KSA, George V, Kalra S, et al. Cardiomyocytes from human pluripotent stem cells: From laboratory curiosity to industrial biomedical platform. Biochim Biophys Acta. 2016;1863:1728–1748. doi: 10.1016/j.bbamcr.2015.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Terrenoire C, Wang K, Tung KW, Chung WK, Pass RH, Lu JT, et al. Induced pluripotent stem cells used to reveal drug actions in a long QT syndrome family with complex genetics. J Gen Physiol. 2013;141:61–72. doi: 10.1085/jgp.201210899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Steffensen AB, Refaat MM, David JP, Mujezinovic A, Calloe K, Wojciak J, et al. High incidence of functional ion-channel abnormalities in a consecutive Long QT cohort with novel missense genetic variants of unknown significance. Sci Rep. 2015;5:10009. doi: 10.1038/srep10009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Roden DM. Drug-induced prolongation of the QT interval. N Engl J Med. 2004;350:1013–1022. doi: 10.1056/NEJMra032426. [DOI] [PubMed] [Google Scholar]
- 79.Cavero I, Holzgrefe H. Comprehensive in vitro Proarrhythmia Assay, a novel in vitro/in silico paradigm to detect ventricular proarrhythmic liability: a visionary 21st century initiative. Expert Opin Drug Saf. 2014;13:745–758. doi: 10.1517/14740338.2014.915311. [DOI] [PubMed] [Google Scholar]
- 80.Klimas A, Ambrosi CM, Yu J, Williams JC, Bien H, Entcheva E. OptoDyCE as an automated system for high-throughput all-optical dynamic cardiac electrophysiology. Nat Commun. 2016;7:11542. doi: 10.1038/ncomms11542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Postma AV, Denjoy I, Hoorntje TM, Lupoglazoff J-M, Da Costa A, Sebillon P, et al. Absence of calsequestrin 2 causes severe forms of catecholaminergic polymorphic ventricular tachycardia. [Last accessed 11/1/2016];Circ Res. 2002 91:e21–6. doi: 10.1161/01.res.0000038886.18992.6b. http://circres.ahajournals.org/cgi/pmidlookup?view=long&pmid=12386154. [DOI] [PubMed] [Google Scholar]
- 82.Gomez-Hurtado N, Boczek NJ, Kryshtal DO, Johnson CN, Sun J, Nitu FR, et al. Novel CPVT-Associated Calmodulin Mutation in CALM3 (CALM3-A103V) Activates Arrhythmogenic Ca Waves and Sparks. Circ Arrhythm Electrophysiol. 2016:9. doi: 10.1161/CIRCEP.116.004161. Internet. Available from: http://dx.doi.org/10.1161/CIRCEP.116.004161. [DOI] [PMC free article] [PubMed]
- 83.Rooryck C, Kyndt F, Bozon D, Roux-Buisson N, Sacher F, Probst V, et al. New Family With Catecholaminergic Polymorphic Ventricular Tachycardia Linked to the Triadin Gene. J Cardiovasc Electrophysiol. 2015;26:1146–1150. doi: 10.1111/jce.12763. [DOI] [PubMed] [Google Scholar]
- 84.Di Pasquale E, Lodola F, Miragoli M, Denegri M, Avelino-Cruz JE, Buonocore M, et al. CaMKII inhibition rectifies arrhythmic phenotype in a patient-specific model of catecholaminergic polymorphic ventricular tachycardia. [Last accessed 11/1/2016];Cell Death Dis. 2013 4:e843. doi: 10.1038/cddis.2013.369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Kujala K, Paavola J, Lahti A, Larsson K, Pekkanen-Mattila M, Viitasalo M, et al. Cell model of catecholaminergic polymorphic ventricular tachycardia reveals early and delayed afterdepolarizations. [Last accessed 11/1/2016];PLoS One. 2012 7:e44660. doi: 10.1371/journal.pone.0044660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.MacLennan DH, Chen SRW. Store overload-induced Ca2+ release as a triggering mechanism for CPVT and MH episodes caused by mutations in RYR and CASQ genes. J Physiol. 2009;587:3113–3115. doi: 10.1113/jphysiol.2009.172155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Liu N, Ruan Y, Denegri M, Bachetti T, Li Y, Colombi B, et al. Calmodulin kinase II inhibition prevents arrhythmias in RyR2(R4496C+/−) mice with catecholaminergic polymorphic ventricular tachycardia. J Mol Cell Cardiol. 2011;50:214–222. doi: 10.1016/j.yjmcc.2010.10.001. [DOI] [PubMed] [Google Scholar]
- 88.Gomes J, Finlay M, Ahmed AK, Ciaccio EJ, Asimaki A, Saffitz JE, et al. Electrophysiological abnormalities precede overt structural changes in arrhythmogenic right ventricular cardiomyopathy due to mutations in desmoplakin-A combined murine and human study. Eur Heart J. 2012;33:1942–1953. doi: 10.1093/eurheartj/ehr472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Kim C, Wong J, Wen J, Wang S, Wang C, Spiering S, et al. Studying arrhythmogenic right ventricular dysplasia with patient-specific iPSCs. Nature. 2013;494:105–110. doi: 10.1038/nature11799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Ma D, Wei H, Lu J, Ho S, Zhang G, Sun X, et al. Generation of patient-specific induced pluripotent stem cell-derived cardiomyocytes as a cellular model of arrhythmogenic right ventricular cardiomyopathy. Eur Heart J. 2013;34:1122–1133. doi: 10.1093/eurheartj/ehs226. [DOI] [PubMed] [Google Scholar]
- 91.Caspi O, Huber I, Gepstein A, Arbel G, Maizels L, Boulos M, et al. Modeling of arrhythmogenic right ventricular cardiomyopathy with human induced pluripotent stem cells. Circ Cardiovasc Genet. 2013;6:557–568. doi: 10.1161/CIRCGENETICS.113.000188. [DOI] [PubMed] [Google Scholar]
- 92.Wen J-Y, Wei C-Y, Shah K, Wong J, Wang C, Chen H-SV. Maturation-Based Model of Arrhythmogenic Right Ventricular Dysplasia Using Patient-Specific Induced Pluripotent Stem Cells. Circ J. 2015;79:1402–1408. doi: 10.1253/circj.CJ-15-0363. [DOI] [PubMed] [Google Scholar]
- 93.Nademanee K, Veerakul G, Nimmannit S, Chaowakul V, Bhuripanyo K, Likittanasombat K, et al. Arrhythmogenic marker for the sudden unexplained death syndrome in Thai men. Circulation. 1997;96:2595–2600. doi: 10.1161/01.cir.96.8.2595. [DOI] [PubMed] [Google Scholar]
- 94.Vatta M, Dumaine R, Varghese G, Richard TA, Shimizu W, Aihara N, et al. Genetic and biophysical basis of sudden unexplained nocturnal death syndrome (SUNDS), a disease allelic to Brugada syndrome. Hum Mol Genet. 2002;11:337–345. doi: 10.1093/hmg/11.3.337. [DOI] [PubMed] [Google Scholar]
- 95.Mizusawa Y, Wilde AAM. Brugada Syndrome. Circ Arrhythm Electrophysiol. 2012;5:606–616. doi: 10.1161/CIRCEP.111.964577. [DOI] [PubMed] [Google Scholar]
- 96.Wilde AAM, Antzelevitch C, Borggrefe M, Brugada J, Brugada R, Brugada P, et al. Proposed Diagnostic Criteria for the Brugada Syndrome. Circulation. 2002;106:2514–2519. doi: 10.1161/01.cir.0000034169.45752.4a. [DOI] [PubMed] [Google Scholar]
- 97.Bezzina C, Veldkamp MW, van den Berg MP, Postma AV, Rook MB, Viersma J-W, et al. A Single Na+ Channel Mutation Causing Both Long-QT and Brugada Syndromes. Circ Res. 1999;85:1206–1213. doi: 10.1161/01.res.85.12.1206. [DOI] [PubMed] [Google Scholar]
- 98.Remme CA, Verkerk AO, Wilde AAM, Veldkamp MW, de Bakker JMT, Bezzina CR. Diversity in cardiac sodium channel disease phenotype in transgenic mice carrying a single SCN5A mutation. Neth Heart J. 2007;15:235–238. doi: 10.1007/BF03085988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Veerman CC, Mengarelli I, Guan K, Stauske M, Barc J, Tan HL, et al. hiPSC-derived cardiomyocytes from Brugada Syndrome patients without identified mutations do not exhibit clear cellular electrophysiological abnormalities. Sci Rep. 2016;6:30967. doi: 10.1038/srep30967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Strong A, Musunuru K. Genome editing in cardiovascular diseases. Nat Rev Cardiol. 2016 doi: 10.1038/nrcardio.2016.139. Internet. Available from: http://dx.doi.org/10.1038/nrcardio.2016.139. [DOI] [PubMed]
- 101.Lee MP, Hu RJ, Johnson LA, Feinberg AP. Human KVLQT1 gene shows tissue-specific imprinting and encompasses Beckwith-Wiedemann syndrome chromosomal rearrangements. Nat Genet. 1997;15:181–185. doi: 10.1038/ng0297-181. [DOI] [PubMed] [Google Scholar]
- 102.Saenen JB, Vrints CJ. Molecular aspects of the congenital and acquired Long QT Syndrome: clinical implications. J Mol Cell Cardiol. 2008;44:633–646. doi: 10.1016/j.yjmcc.2008.01.006. [DOI] [PubMed] [Google Scholar]
- 103.Saenen JB, Paulussen ADC, Jongbloed RJ, Marcelis CL, Gilissen RAHJ, Aerssens J, et al. A single hERG mutation underlying a spectrum of acquired and congenital long QT syndrome phenotypes. J Mol Cell Cardiol. 2007;43:63–72. doi: 10.1016/j.yjmcc.2007.04.012. [DOI] [PubMed] [Google Scholar]
- 104.Zhang M, D’Aniello C, Verkerk AO, Wrobel E, Frank S, Ward-van Oostwaard D, et al. Recessive cardiac phenotypes in induced pluripotent stem cell models of Jervell and Lange-Nielsen syndrome: disease mechanisms and pharmacological rescue. [Last accessed 11/1/2016];Proc Natl Acad Sci U S A. 2014 111:E5383–92. doi: 10.1073/pnas.1419553111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Bellin M, Casini S, Davis RP, D’Aniello C, Haas J, Ward-van Oostwaard D, et al. Isogenic human pluripotent stem cell pairs reveal the role of a KCNH2 mutation in long-QT syndrome. EMBO J. 2013;32:3161–3175. doi: 10.1038/emboj.2013.240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Sala L, Yu Z, Ward-van Oostwaard D, van Veldhoven JP, Moretti A, Laugwitz K-L, et al. A new hERG allosteric modulator rescues genetic and drug-induced long-QT syndrome phenotypes in cardiomyocytes from isogenic pairs of patient induced pluripotent stem cells. EMBO Mol Med. 2016;8:1065–1081. doi: 10.15252/emmm.201606260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Lieve KV, Wilde AA, van der Werf C. The Role of Flecainide in the Management of Catecholaminergic Polymorphic Ventricular Tachycardia. Arrhythm Electrophysiol Rev. 2016;5:45–49. doi: 10.15420/aer.2016.3.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Qu Z, Weiss JN. Mechanisms of ventricular arrhythmias: from molecular fluctuations to electrical turbulence. Annu Rev Physiol. 2015;77:29–55. doi: 10.1146/annurev-physiol-021014-071622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Tischer D, Weiner OD. Illuminating cell signalling with optogenetic tools. Nat Rev Mol Cell Biol. 2014;15:551–558. doi: 10.1038/nrm3837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Molitoris JM, Paliwal S, Sekar RB, Blake R, Park J, Trayanova NA, et al. Precisely parameterized experimental and computational models of tissue organization. Integr Biol. 2016;8:230–242. doi: 10.1039/c5ib00270b. [DOI] [PMC free article] [PubMed] [Google Scholar]