Abstract
BMI1 is a component of the PRC1 complex that is overexpressed in breast and other cancers, and promotes self-renewal of cancer stem-like cells. The oncogenic mucin 1 (MUC1) C-terminal (MUC1-C) subunit is similarly overexpressed in human carcinoma cells and has been linked to their self-renewal. There is no known relationship between MUC1-C and BMI1 in cancer. The present studies demonstrate that MUC1-C drives BMI1 transcription by a MYC-dependent mechanism in breast and other cancer cells. In addition, we show that MUC1-C blocks miR-200c-mediated downregulation of BMI1 expression. The functional significance of this MUC1-C→BMI1 pathway is supported by the demonstration that targeting MUC1-C suppresses BMI1-induced ubiquitylation of H2A and thereby derepresses homeobox HOXC5 and HOXC13 gene expression. Notably, our results further show that MUC1-C binds directly to BMI1 and promotes occupancy of BMI1 on the CDKN2A promoter. In concert with BMI1-induced repression of the p16INK4a tumor suppressor, we found that targeting MUC1-C is associated with induction of p16INK4a expression. In support of these results, analysis of three gene expresssion datasets demonstrated highly significant correlations between MUC1-C and BMI1 in breast cancers. These findings uncover a previously unrecognized role for MUC1-C in driving BMI1 expression and in directly interacting with this stem cell factor, linking MUC1-C with function of the PRC1 in epigenetic gene silencing.
Keywords: MUC1-C, BMI1, PRC1, H2A, HOXC5, HOXC13, p16INK4a tumor suppressor
Introduction
The B cell-specific Moloney murine leukemia virus integration site 1 (BMI1) protein is a component of the polycomb repressive complex 1 (PRC1) (1; 2). BMI1 binds to the catalytic RING2 subunit to form an E3 ubiquitin ligase that catalyzes the mono-ubiquitination of histone H2A and thereby promotes gene silencing (3–6). BMI1 contributes to the self-renewal of normal stem cells, at least in part, by repressing the CDNK2A locus, which encodes the p16INK4a and p14ARF tumor suppressors (1; 2). BMI1 is overexpressed in breast and other carcinomas and is associated with poor outcomes (7; 8; 2). In addition, BMI1-induced suppression of p16INK4a expression has been attributed to involvement of BMI1 in promoting the self-renewal and tumorigenic potential of cancer stem-like cells (CSCs) (1; 9; 2; 10). BMI1 is also critical for the self-renewal of leukemic stem cells (11; 12). In concert with a role in stemness, BMI1 has been linked to (i) stabilization of SNAIL, (ii) downregulation of the PTEN tumor suppressor, (iii) induction of the epithelial-mesenchymal transition (EMT), and (iv) chemoresistance (13; 14; 2). Additionally, BMI1-mediated ubiquitylation of H2A and gH2AX facilitates the DNA damage response (DDR) and repair of double-stranded DNA breaks (DSBs) (15). Those findings and the inhibitory effects of BMI1 on DSB-induced CHK1 and CHK2 checkpoint activation have supported the notion that BMI1 promotes genomic instability and transformation (16–18; 15). BMI1 has thus emerged as an attractive target for the treatment of cancer; however, there are presently no clinically available BMI1 inhibitors (10). Other strategies, such as downregulation of BMI1 translation by miR-200c (19), have therefore been explored as approaches for targeting BMI1 in cancer cells.
Mucin 1 (MUC1) is a heterodimeric protein that is aberrantly overexpressed in breast and diverse other carcinomas (20; 21). The transmembrane MUC1 C-terminal (MUC1-C) subunit induces transformation in part by interacting with receptor tyrosine kinases at the cell membrane and promoting their activation and downstream signaling pathways (21–23). In addition, MUC1-C is imported into the nucleus, where it associates with β-catenin/TCF4 and drives activation of the WNT pathway CCND1 and MYC genes (24–26). MUC1-C also activates the inflammatory TAK1→IKK→NF-κB p65 pathway, binds directly to NF-κB p65 and promotes the induction of NF-κB target genes (27–29). In this way, MUC1-C/NF-κB p65 complexes activate transcription of the ZEB1 gene, which encodes an EMT-inducing transcription factor (30). In turn, ZEB1 suppresses miR-200c and activates the EMT program (30). MUC1-C also promotes EMT by activating the LIN28B→Let-7 pathway (31). Other studies have demonstrated that MUC1-C is necessary for the CSC phenotype as evidenced by the demonstration that targeting MUC1-C inhibits self-renewal capacity and tumorigenicity (32; 33; 23). The findings that MUC1-C is of importance for EMT and stemness invoked the possibility that MUC1-C may also be involved in the epigenetic regulatory mechanisms that control these programs (34). Indeed, subsequent work has shown that MUC1-C induces the DNMT1 and DNMT3b genes encoding DNA methyltransferases and thereby regulates global and gene-specific DNA methylation patterns (35; 36). Interestingly, MUC1-C-induced DNMT1 and DNMT3b expression is conferred by an NF-κB p65-dependent mechanism, linking MUC1-C to the inflammatory TAK1→IKK→NF-κB and the epigenetic regulation of EMT and stemness (35). These findings have also supported the notion that MUC1-C may control other epigenetic regulatory mechanisms, such as modifications of chromatin-associated histones, to achieve additional changes in gene expression.
The present studies demonstrate that targeting MUC1-C in diverse carcinoma cells is associated with downregulation of BMI1, RING1 and RING2 expression, indicating that MUC1-C induces the major components of the PRC1 complex. We have focused on MUC1-C-mediated regulation of BMI1 and demonstrate that MUC1-C (i) activates BMI1 transcription by a MYC-dependent mechanism, and (ii) blocks miR-200c-mediated downregulation of BMI1 expression. In concert with these results, we show that targeting MUC1-C decreases ubiquitylation of H2A and derepresses homeobox (HOX) genes. We also show that MUC1-C interacts with BMI1 on the CDKN2A promoter and contributes to repression of the p16INK4a tumor suppressor.
Results
Silencing MUC1-C downregulates expression of the PRC1 complex
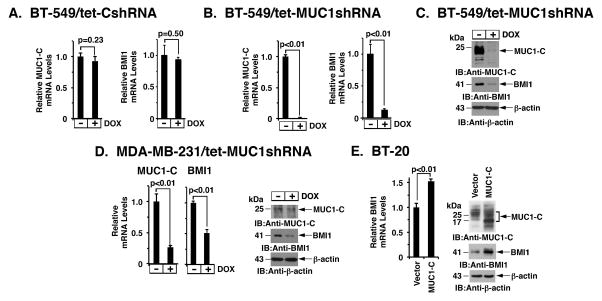
The PRC1 complex includes BMI1, RING1 and RING2. To assess the potential role of MUC1-C in regulating this complex, we established BT-549 breast cancer cells that stably express a tetracycline-inducible tet-on control shRNA (tet-CshRNA) or MUC1 shRNA (tet-MUC1shRNA). Treatment of BT-549/tet-CshRNA cells with doxycycline (DOX) had no significant effect on MUC1-C or BMI1 mRNA levels (Fig. 1A, left and right). In contrast, DOX treatment of BT-549/tet-MUC1shRNA cells resulted in suppression of both MUC1-C and BMI1 expression (Fig. 1B, left and right). In concert with these results, BMI1 protein was also downregulated in response to MUC1-C silencing (Fig. 1C). Intriguingly, targeting MUC1-C was also associated with suppression of RING1 and RING2 mRNAs (Supplemental Fig. S1A), indicating that MUC1-C induces expression of multiple members of the PRC1 complex. BMI1 is upregulated in diverse cancers and confers a poor prognosis (7; 8; 2). Accordingly, we have focused our studies here on the regulation of BMI1. In support of the results obtained with BT-549 cells, DOX treatment of MDA-MB-231/tet-MUC1shRNA, but not MDA-MB-231/tet-CshRNA, cells was similarly associated with downregulation of BMI1 mRNA (Fig. 1D, left; Supplemental Fig. S1B) and protein (Fig. 1D, right). Moreover, similar results were obtained with BT-20 (Supplemental Fig. S2A) and MDA-MB-468 (Supplemental Fig. S2B) TNBC cells. To extend this line of investigation, we found that targeting MUC1-C in KRAS mutant A549 (Supplemental Fig. S2C) and H460 (Supplemental Fig. S2D) lung cancer cells also results in suppression of BMI1 expression, indicating that this response is not restricted to breast cancer. To further investigate the relationship between MUC1-C and BMI1, we stably overexpressed MUC1-C in BT-20 cells. Upregulation of MUC1-C was associated with increases in BMI1 mRNA and protein levels (Fig. 1E, left and right), providing further support for the notion that MUC1-C induces BMI1 expression.
Figure 1. Silencing MUC1-C downregulates BMI1 expression.
A–C. BT-549 cells were stably transduced to express a tetracycline-inducible control shRNA (tet-CshRNA) (A) or a MUC1 shRNA (tet-MUC1shRNA) (B). Cells treated with 200 ng/ml DOX for 4 d were analyzed for MUC1 and BMI1 mRNA levels by qRT-PCR. The results (mean±SD) are expressed as relative mRNA levels compared to that obtained for control DOX-untreated cells (assigned a value of 1). Cell lysates treated with 200 ng/ml DOX for 7 d were immunoblotted with the indicated antibodies (C). D. MDA-MB-231/tet-MUC1shRNA cells treated with 200 ng/ml DOX for 4 d were analyzed for MUC1 and BMI1 mRNA levels by qRT-PCR (left). Cell lysates treated with 200 ng/ml DOX for 7 d were immunoblotted with the indicated antibodies (right). E. BT-20 cells stably expressing a control or MUC1-C vector were analyzed for BMI1 mRNA levels by qRT-PCR. The results (mean±SD) are expressed as relative BMI1 mRNA levels compared to that obtained for vector cells (assigned a value of 1) (left). Lysates were immunoblotted with the indicated antibodies (right).
Targeting the MUC1-C CQC motif downregulates BMI1
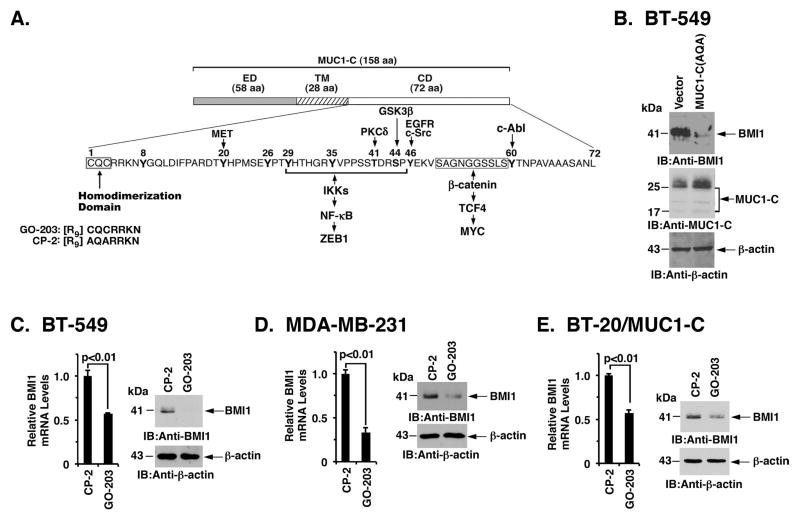
The MUC1-C cytoplasmic domain includes a CQC motif, which is essential for the formation of MUC1-C homodimers and thereby the MUC1-C oncogenic function (Fig. 2A). As a result, enforced expression of MUC1-C(AQA) in which the CQC motif has been mutated to AQA blocks MUC1-C homodimerization and functions as a dominant-negative for transformation (37). Consistent with a role for MUC1-C in driving BMI1, expression of MUC1-C(AQA) in BT-549 cells was associated with downregulation of BMI1 (Fig. 2B). Studies were also performed with the MUC1-C inhibitor GO-203 (Fig. 2A), which binds to the MUC1-C CQC motif and blocks homodimerization (38; 39). Treatment of BT-549 cells with GO-203, but not the control peptide CP-2, was associated with downregulation of BMI1 mRNA and protein (Fig. 2C, left and right). Similar results were obtained in MDA-MB-231 (Fig. 2D, left and right) and BT-20/MUC1-C (Fig. 2E, left and right) cells, confirming that targeting MUC1-C suppresses BMI1 expression.
Figure 2. Targeting the MUC1-C cytoplasmic domain downregulates BMI1 expression.
A. Schema of the MUC1-C subunit with the 58 aa extracellular domain (ED), the 28 aa transmembrane domain (TM), and the sequence of the 72 aa cytoplasmic domain (CD). The MUC1-C cytoplasmic domain contains a CQC motif that is necessary and sufficient for MUC1-C homodimerization and oncogenic function. GO-203 is a cell-penetrating peptide that binds the CQC motif and blocks MUC1-C homodimerization. Highlighted are MUC1-C-induced pathways that confer the activation of ZEB1 and MYC. B. BT-549 cells were transfected with a control or MUC1-C(AQA) vector in which the CQC motif had been mutated to AQA. Lysates were immunoblotted with the indicated antibodies. C–E. BT-549 (C), MDA-MB-231 (D), and BT-20/MUC1-C (E) cells treated with 5 μM CP-2 or 5 μM GO-203 for 12 h were analyzed for BMI1 mRNA levels by qRT-PCR. The results (mean±SD) are expressed as relative BMI1 mRNA levels compared to that obtained for CP-2 (assigned a value of 1) (left). Cell lysates treated with 5 μM CP-2 or 5 μM GO-203 for 48 h were immunoblotted with the indicated antibodies (right).
MUC1-C drives BMI1 transcription by a MYC-dependent mechanism
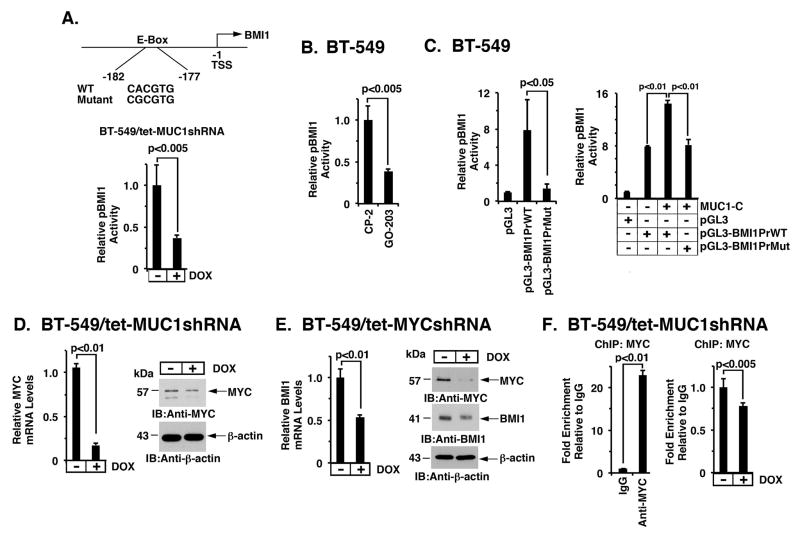
The upregulation of BMI1 in cancer cells has been attributed, at least in part, to MYC-induced activation of the BMI1 promoter, which contains a consensus E-Box (CACGTG) for MYC binding (Fig. 3A, upper panel) (40). In studies of BT-549/tet-MUC1shRNA cells, DOX treatment was associated with downregulation of a BMI1 promoter-reporter (pGL3-BMI1PrWT; Fig. 3A, lower panel). Moreover, treatment of BT-549 cells with GO-203 was associated with downregulation of the BMI1 promoter-reporter (Fig. 3B), indicating that MUC1-C activates BMI1 transcription. Studies in BT-549 cells further demonstrated that activation of the pBMI1 promoter is abrogated by mutation of the E-Box (pGL3-BMI1PrMut; Fig. 3C, left). In addition, cotransfection of MUC1-C and pGL3-BMI1PrWT or pGL3-BMI1PrMut further demonstrated that MUC1-C activates the BMI1 promoter-reporter by a MYC-mediated mechanism (Fig. 3C, right). Notably, MUC1-C drives MYC expression in NSCLC (25), multiple myeloma (26) and breast cancer (41) cells by activation of the WNT/β-catenin pathway (Fig. 2A). In this way, targeting MUC1-C in BT-549 cells resulted in the downregulation of MYC mRNA and protein (Fig. 3D, left and right). Moreover, targeting MYC in BT-549 cells decreased BMI1 expression (Fig. 3E, left and right), supporting the notion that the MUC1-C→MYC pathway induces BMI1 expression. In further support of such a mechanism, silencing MUC1-C was associated with decreased occupancy of MYC on the BMI1 promoter (Fig. 3F, left and right).
Figure 3. MUC1-C activates BMI1 transcription by a MYC-dependent mechanism.
A. Schema of the BMI1 promoter region with positioning of the putative MYC binding site at −177 to −182 bp upstream of the transcription start site. The BMI1 promoter-luciferase (Luc) pGL3-BMI1PrWT vector includes the wild-type BMI1 promoter and pGL3-BMI1PrMut contains a mutation in the E-Box sequences (CACGTG has been mutated to CGCGTG)(upper panel). BT-549/tet-MUC1shRNA cells cultured with or without DOX for 5 d were transfected with the pGL3-Basic Luc or pGL3-BMI1PrWT reporter for 48 h and then analyzed for luciferase activity. The results (mean±SD of 3 determinations) are expressed as the relative luciferase activity compared to that obtained with pGL3-Basic Luc (assigned a value of 1)(lower panel). B. BT-549 cells were transfected with the pGL3-Basic Luc or pGL3-BMI1PrWT reporter for 6 h and then treated with 5 μM CP-2 or GO-203 for an additional 42 h. The results (mean±SD of 3 determinations) are expressed as the relative luciferase activity compared to that obtained with CP-2-treated cells (assigned a value of 1). C. BT-549 cells were transfected with the pGL3-Basic Luc, pGL3-BMI1PrWT or pGL3-BMI1PrMut reporter for 48 h and then analyzed for luciferase activity (left). BT-549 cells were transfected with (i) a control vector or one expressing MUC1-C, and (ii) the pGL3-Basic Luc, pGL3-BMI1PrWT or pGL3-BMI1PrMut reporter for 72 h and then analyzed for luciferase activity (right). The results (mean±SD of 3 determinations) are expressed as the relative luciferase activity compared to that obtained with pGL3-Basic Luc (assigned a value of 1). D. BT-549/tet-MUC1shRNA cells were treated with or without DOX for 5 d. MYC mRNA levels were determined by qRT-PCR. The results (mean±SD) are expressed as relative MYC mRNA levels compared to that obtained for control DOX-untreated cells (assigned a value of 1) (left). Lysates were immunoblotted with the indicated antibodies (right). E. BT-549/tet-MYCshRNA cells cultured with or without DOX for 12 h were analyzed for BMI1 mRNA levels by qRT-PCR. The results (mean±SD) are expressed as relative BMI1 mRNA levels compared to that obtained for control DOX-untreated cells (assigned a value of 1) (left). Cell lysates treated with DOX for 48 h were immunoblotted with the indicated antibodies (right). F. Soluble chromatin from BT-549/tet-MUC1shRNA cells was precipitated with anti-MYC or a control IgG (left). The final DNA samples were amplified by qPCR with primers for the BMI1 promoter. The results (mean±SD of three determinations) are expressed as the relative fold enrichment compared with that obtained with the IgG control (assigned a value of 1). Soluble chromatin from 549/tet-MUC1shRNA cells cultured with or without DOX for 5 d was precipitated with anti-MYC or a control IgG. The final DNA samples were amplified by qPCR. The results (mean±SEM of three determinations) are expressed as the relative fold enrichment compared to that obtained for control DOX-untreated cells (assigned a value of 1) (right).
MUC1-C regulates BMI1 expression by an NF-κB-mediated mechanism
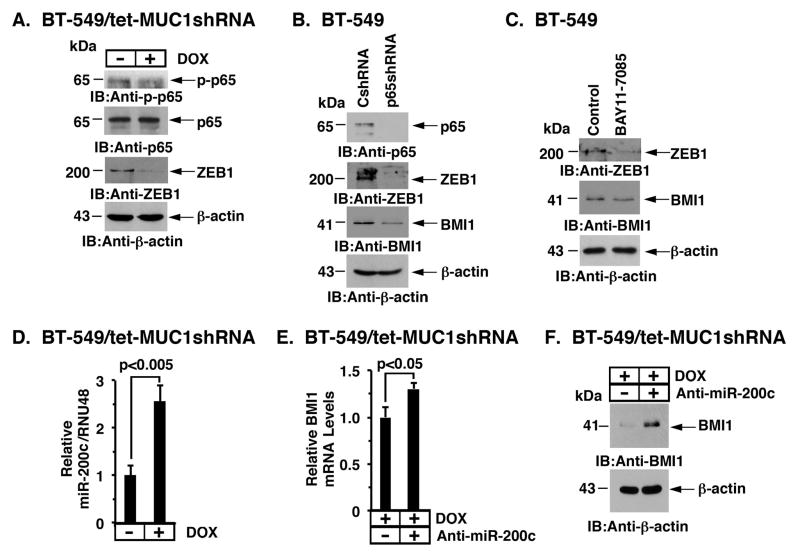
In addition to driving MYC (Fig. 2A), MUC1-C also activates NF-κB p65 (27–29) and thereby induces expression of the ZEB1 transcriptional repressor (30). In concert with this MUC1-C→NF-κB p65→ZEB1 pathway, we found that DOX-induced silencing of MUC1-C in BT-549/tet-MUC1shRNA cells is associated with decreases in phospho-NF-κB p65 (p-p65) and ZEB1 (Fig. 4A). Similar results were obtained in experiments with MDA-MB-231 cells (Supplemental Fig. S3A). The importance of NF-κB p65 in induction of ZEB1 expression was further supported by the demonstration that silencing NF-κB p65 is associated with decreases in ZEB1; however, surprisingly, we also found that NF-κB p65 functions upstream to BMI1 (Fig. 4B). Inhibition of NF-κB activity with BAY-11-7085 also suppressed ZEB1 and BMI1 expression (Fig. 4C and Supplemental Fig. S3B). MUC1-C/ZEB1 complexes bind to highly conserved Z-boxes in the miR-200c promoter and thereby suppresses miR-200c expression (30). In this context, targeting MUC1-C resulted in induction of miR-200c expression (Fig. 4D and Supplemental Fig. S3C). Consistent with the reported role of miR-200c in suppressing BMI1 mRNA levels (42), targeting miR-200c was associated with upregulation of BMI1 mRNA (Fig. 4E) and protein (Fig. 4F). Our findings thus demonstrate that MUC1-C induces BMI1 expression as a result of (i) transcriptional activation by a MYC-mediated mechanism, and (ii) post-transcriptional upregulation by the suppression of miR-200c.
Figure 4. MUC1-C blocks miR-200c-mediated downregulation of BMI1 expression.
A. BT-549/tet-MUC1shRNA cells were treated with or without DOX for 4 d. Lysates were immunoblotted with the indicated antibodies. B. BT-549 cells were transduced with lentiviral vectors to stably express a control shRNA (CshRNA) or a NF-κB p65 shRNA. Lysates were immunoblotted with the indicated antibodies. C. BT-549 cells were treated with control DMSO vehicle or BAY-11-7085 for 16 h. Lysates were immunoblotted with the indicated antibodies. D. BT-549/tet-MUC1shRNA cells were treated with or without DOX for 4 d. The cells were analyzed for miR-200c levels by qRT-PCR. The results (mean±SD) are expressed as relative miR-200c/RNU48 levels compared to that obtained for control DOX-untreated cells (assigned a value of 1). E and F. BT-549/tet-MUC1shRNA cells cultured with DOX for 7 d were transfected with 12.5 nM anti-miR-200c or a negative control oligonucleotide for 4 d. The cells were then analyzed for BMI1 mRNA levels by qRT-PCR. The results (mean±SD) are expressed as relative BMI1 mRNA levels compared to that obtained for control anti-miR-200c-untreated cells (assigned a value of 1) (E). Lysates were immunoblotted with the indicated antibodies (F).
Targeting MUC1-C decreases ubiquitylation of H2A and activates the HOX gene expression
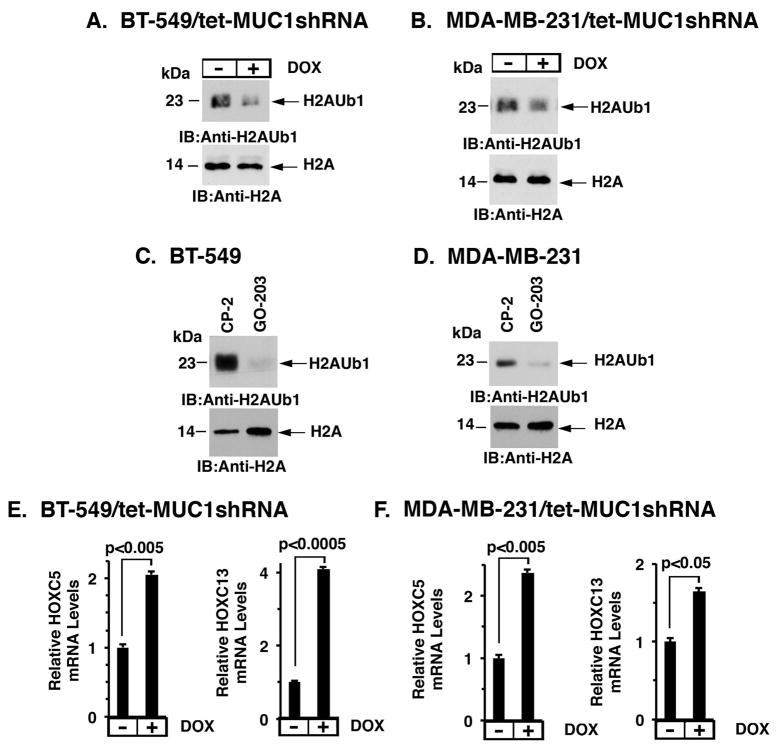
Studies in mouse models and human HeLa cells have demonstrated the Bmi1/BMI1 is of importance for H2A ubiquitylation and homeobox (HOX) gene silencing (5; 43). Accordingly, we first investigated whether targeting MUC1-C affects ubiquitylation of H2A (H2AK119Ub1). Indeed, DOX-induced MUC1-C silencing in BT-549/tet-MUC1shRNA cells was associated with decreases in H2AK119Ub1 levels (Fig. 5A). Similar results were obtained in DOX-treated MDA-MB-231/tet-MUC1shRNA cells (Fig. 5B). Moreover, inhibition of MUC1-C with GO-203, but not the control CP-2, resulted in marked downregulation of H2AK119Ub1 in BT-549 (Fig. 5C) and MDA-MB-231 (Fig. 5D) cells. H2AK119Ub1 localizes to the 5′ regulatory region of the HOXC5 gene and represses HOXC5 transcription (43). In concert with involvement of MUC1-C in driving BMI1 and H2AK119Ub1, targeting MUC1-C in BT-549/tet-MUC1shRNA (Fig. 5E, left) and MDA-MB-231/tet-MUC1shRNA (Fig. 5F, left) cells significantly induced HOXC5 mRNA levels. Loss of H2AK119Ub1 has also been linked to upregulation of HOXC13 gene expression (5). In this respect, we also found that targeting MUC1-C is associated with increases in HOXC13 mRNA levels (Figs. 5E, right and 5F, right). Studies in BT-20 cells further demonstrated that overexpression of MUC1-C results in downregulation of HOXC5 and HOXC13 mRNA levels (Supplemental Fig. S4, left and right), confirming that MUC1-C represses expression of these HOX genes.
Figure 5. Targeting MUC1-C decreases ubiquitylation of H2A and derepresses HOX gene expression.
A and B. BT-549/tet-MUC1shRNA (A) and MDA-MB-231/tet-MUC1shRNA (B) cells were treated with or without DOX for 5 d. Lysates were immunoblotted with the indicated antibodies. C and D. BT-549 (C) and MDA-MB-231 (D) cells were treated with 5 μM CP-2 or 5 μM GO-203 for 48 h. Lysates were immunoblotted with the indicated antibodies. E and F. BT-549/tet-MUC1shRNA (E) and MDA-MB-231/tet-MUC1shRNA (F) cells were treated with or without DOX for 7 d. HOXC5 (left) and HOXC13 (right) mRNA levels were determined by qRT-PCR. The results (mean±SD) are expressed as relative mRNA levels compared to that obtained for control DOX-untreated cells (assigned a value of 1).
MUC1-C binds directly to BMI1 and promotes BMI1-mediated repression of CDKN2A
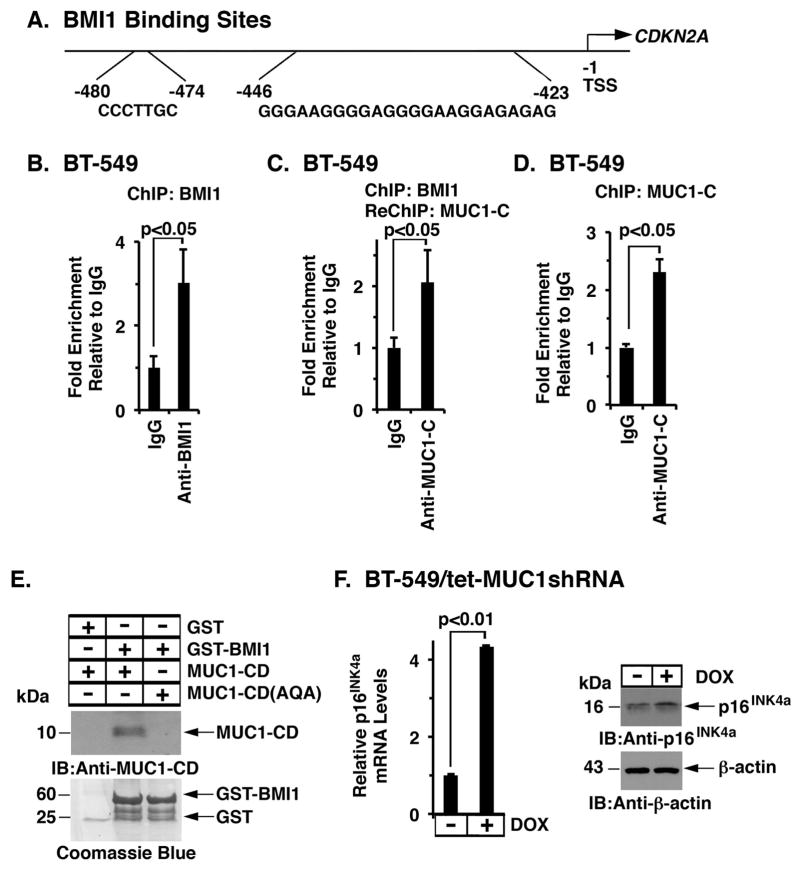
BMI1 silences the INK4a/ARF (CDKN2A) tumor suppressor locus, which encodes the p16INK4a protein inhibitor of CDK4 and CDK6 (44–46), by binding directly to the BMI1-response element (BRE) in CDKN2A promoter (47) (Fig. 6A). By extension, ChIP analysis of BT-549 cell chromatin demonstrated occupancy of BMI1 on the CDKN2A promoter (Fig. 6B). Moreover and surprisingly, re-ChIP studies further demonstrated occupancy of BMI1 with MUC1-C (Fig. 6C). In concert with this result, a primary ChIP with anti-MUC1-C confirmed localization of MUC1-C on the CDKN2A promoter (Fig. 6D). To determine if BMI1 associates with MUC1-C, in vitro studies were performed with GST-tagged BMI1 and purified MUC1-C cytoplasmic domain (MUC1-CD). The results showed direct binding of these proteins (Fig. 6E). Mutation of the MUC1-CD CQC motif to AQA abrogated the binding (Fig. 6E), indicating that the interaction between BMI1 and MUC1-C is conferred by the Cys residues. Notably, targeting MUC1-C in BT-549/tet-MUC1shRNA cells resulted in induction of p16INK4a mRNA and protein (Fig. 6F, left and right). Similar results were obtained in DOX-treated MDA-MB-468/tet-MUC1shRNA cells (Supplemental Fig. S5A). Additionally, stable overexpression of MUC1-C in BT-20 cells was associated with downregulation of p16INK4a mRNA and protein (Supplemental Fig. S5B). BMI1 functions as a repressor of the CDKN2A and CDH1 genes (2; 13). Therefore, to extend this line of investigation, we silenced BMI1 in the BT-20/MUC1-C overexpressing cells and found upregulation of p16INK4a expression (Supplemental Fig. S5C), supporting the premise that MUC1-C→BMI1 signaling represses expression of the p16INK4a tumor suppressor and well-established marker of senescence (48). Silencing BMI1 in the BT-20/MUC1-C cells was also associated with induction of E-cadherin (Supplemental Fig. S5C), which is downregulated in association with the CSC phenotype of EMT, migration and invasion (49).
Figure 6. MUC1-C/BMI1 complexes occupy the CDKN2A promoter.
A. Schema of the CDKN2A promoter with positioning of the BMI1-response element (BRE) at −423 to −446 and −474 to −480 bp upstream to the transcription start site. B. Soluble chromatin from BT−549 cells was precipitated with anti-BMI1 or a control IgG. C. In the re-ChIP analysis, BMI1 precipitates were released and re-immunoprecipitated with anti-MUC1-C and a control IgG. D. Soluble chromatin from BT-549 cells was precipitated with anti-MUC1-C or a control IgG. The final DNA samples were amplified by qPCR with primers for the CDKN2A promoter. The results (mean±SD of three determinations) are expressed as the relative fold enrichment compared with that obtained with the IgG control (assigned a value of 1). E. GST and GST-BMI1 were incubated with either purified MUC1-CD or MUC1-CD(AQA). The adsorbates were immunoblotted with anti-MUC1-C. Input of the GST proteins was assessed by Coomassie blue staining. F. BT-549/tet-MUC1shRNA cells were treated with or without DOX for 7 d. p16INK4a mRNA levels were determined by qRT-PCR. The results (mean±SD) are expressed as relative p16INK4a mRNA levels compared to that obtained for control DOX-untreated cells (assigned a value of 1) (left). Cell lysates cultured with or without DOX for 12 d were immunoblotted with the indicated antibodies (right).
MUC1-C correlates with BMI1 expression in breast cancers
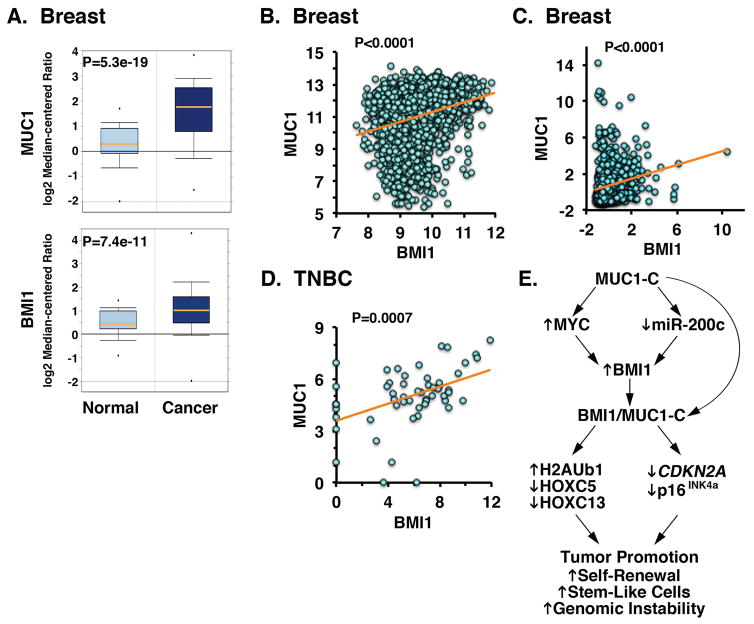
Based on the above findings, we analyzed microarray datasets to determine whether MUC1 expression is associated with that of BMI1 in breast tumors. MUC1 expression was significantly elevated in primary breast cancer samples (n=389) as compared to that in the normal breast tissues (n=61) (Fig. 7A, upper panel). Additionally and like MUC1, levels of BMI1 expression were significantly increased in breast cancer samples (Fig. 7A, lower panel). To investigate the correlation between MUC1 and BMI1, we performed bioinformatics analysis on publicly available datasets from cBioPortal and Gene Expression Omnibus (GEO). Analysis of breast tumors (n=1980) demonstrated that MUC1 and BMI1 expression correlate positively (Fig. 7B). Similar findings were obtained in the TCGA breast cancer dataset (n=528), in which MUC1 expression positively correlated with BMI1 levels (Fig. 7C). We further investigated the nature of this relationship in TNBCs and found a strong positive correlation between MUC1 and BMI1 (Fig. 7D). In concert with the findings that MUC1-C drives RING1 and RING2 (Supplemental Fig. S1), we also found significant correlations between expression of MUC1 and these members of the PRC1 complex (Supplemental Figures S6A and S6B).
Figure 7. Correlation between MUC1 and BMI1 in breast cancer samples.
A. Microarray data from Oncomine database are expressed as box plots (25th–75th percentiles) for MUC1 (upper panel) and BMI1 (lower panel) expression in normal breast tissues (n=61) and breast cancer samples (n=389). B–D. MUC1 and BMI1 gene expression data from METABRIC (B; n=1980), TCGA (C; n=528), and GSE41970 (D; n=54) datasets was assessed for correlation using the Pearson’s correlation coefficient, where p<0.05 was considered as statistically significant. E. Schema depicting the proposed effects of MUC1-C on BMI1 expression and function. MUC1-C drives BMI1 expression by (i) inducing MYC and in turn MYC-mediated activation of the BMI1 gene, and (ii) downregulating miR-200c and thereby inhibiting the suppressive effects of miR-200c on BMI1 mRNA. As a result, MUC1-C promotes the ubiquitylation of H2A and repression of HOXC5 and HOXC13. MUC1-C also interacts directly with BMI1 and contributes to repression of the CDKN2A promoter and suppression of p16INK4a expression. These findings and the demonstration that MUC1 and BMI1 significantly correlate in breast tumors supports the notion that MUC1-C is, at least in part, responsible for the upregulation of BMI1 in human cancers, which has been linked to tumor promotion by increasing self-renewal capacity, cancer stem-like cells and genomic instability.
Discussion
Overexpression of MUC1-C in cancer cells promotes self-renewal capacity, tumorigenicity and stemness (50; 32; 33; 23; 31). Additionally, MUC1-C has been linked to induction of EMT (30). BMI1 has also been characterized as a cancer stem-like cell factor that drives EMT and invasion (1; 13; 51; 2; 10). However, there has been no known association between MUC1-C and BMI1. The present studies demonstrate that targeting MUC1-C in cancer cells results in suppression of BMI1 expression, invoking the possibility that MUC1-C participates in activation of the BMI1 gene. Indeed, our results demonstrate that MUC1-C activates BMI1 transcription. Little is known about regulation of the BMI1 gene in cancer cells; however, BMI1 has been linked to repression of the Dickkopf (DKK) family of WNT inhibitors and thereby activation of the WNT/β-catenin pathway and an autoinductive loop involving MYC (52). In contrast to the DKK mechanism, MUC1-C binds directly to and stabilizes β-catenin, and thereby activates WNT/β-catenin signaling (53; 54; 24). MUC1-C thus forms complexes with β-catenin and TCF4 on the promoters of WNT target genes, such as CCND1 and MYC, and activates their transcription (24–26). In this way, MUC1-C drives MYC expression in NSCLC and multiple myeloma cells (25; 26). The present work demonstrates that MUC1-C similarly upregulates MYC in breast cancer cells and, as found in neuroblastomas (40), activates the BMI1 gene. We also found that MUC1-C blocks miR-200c-mediated downregulation BMI1 expression (19). In this context, MUC1-C drives an inflammatory TAK1→IKK→NF-κB autoinductive pathway that integrates activation of ZEB1 with suppression of miR-200c and induction of EMT (27; 28; 30; 29). These findings thus supported a model in which MUC1-C drives BMI1 expression by transcriptional and post-transcriptional mechanisms.
PRC1 includes BMI1, RING1 and RING2, and possesses H2A-K119 ubiquitin E3 ligase activity (3). RING2 is the catalytic subunit, whereas BMI1 is necessary for activity by maintaining integrity of the complex (5) and directly stimulates RING2 function (43). Interestingly, we found that, in addition to BMI1, targeting MUC1-C is associated with downregulation of RING1 and RING2 expression, indicating that MUC1-C induces multiple PRC1 members. Moreover, we found that expression of MUC1 in breast cancers significantly correlates with that of BMI1, RING1 and RING2, supporting our in vitro findings. The present work focused on how MUC1-C drives BMI1 expression; nonetheless, subsequent studies will be needed to define how MUC1-C directs RING1 and RING2 expression in cancer cells. In concert with the function of PRC1 as an H2A ubiquitin E3 ligase, targeting MUC1-C with silencing or the GO-203 inhibitor resulted in marked downregulation of H2AK119Ub1 levels. Ubiquitylation of H2A, which is found in 5–15% of total cellular H2A, has been associated with gene repression (55; 3). Specifically, PRC1-mediated ubiquitylation of H2A represses HOX genes, which encode transcriptional regulators involved in spatial animal development (56). By extension, we found that targeting MUC1-C with downregulation of H2A ubiquitylation was associated with depression of HOXC5 (43) and HOXC13 (5). Genome-wide mapping has identified groups of genes with H2AK119Ub1 enrichment that are targets of PRC1 silencing (57). These genes maintain embryonic stem cell identity, indicating that H2A ubiquitylation is of importance for stem cell self-renewal (57). PRC1-mediated H2A ubiquitylation may also contribute to transformation by regulating repair of double-stranded DNA breaks (DSBs) (15) and inhibiting of DSB-induced CHK1 and CHK2 checkpoint activation (16–18; 15). Therefore and in addition to HOX gene repression, a role for MUC1-C in driving BMI1-mediated H2A ubiquitylation could also contribute to self-renewal and genomic instability of cancer cells.
Evidence is accumulating that MUC1-C is of importance for the epigenetic regulation of gene expression in cancer. MUC1-C induces expression of DNMT1 and DNMT3b in breast and other cancer cells (35). MUC1-C occupies the DNMT1 and DNMT3b promoters in complexes with NF-κB p65 and activates their transcription (35). In this way, MUC1-C regulates global and gene promoter-specific DNA methylation patterns in cancer cells (35; 36). The present work extends the involvement of MUC1-C in epigenetic regulation by demonstrating that MUC1-C binds directly to BMI1 and occupies the CDKN2A promoter in complexes with BMI1, indicating that MUC1-C may function as a component of PRC1. Moreover, the finding that MUC1-C contributes to BMI1 occupancy on the CDKN2A promoter could be function of the direct binding to BMI1 and/or the demonstration that MUC1-C promotes expression of BMI1 and other PRC1 members. Of significance is the demonstration that targeting MUC1-C induces expression of the p16INK4a tumor suppressor, a finding in concert with derepression of the CDKN2A promoter. Somewhat paradoxically and in contrast to p16INK4a, targeting MUC1-C had little effect on activation the ARF locus, suggesting that MUC1-C occupancy on the CDKN2A promoter may differentially repress the p16INK4a and ARF genes. Our studies on MUC1-C-induced DNMT expression linked changes in DNA methylation patterns to the repression of CDH1 and other TSGs (35; 36). Epigenetic silencing of TSGs has been proposed to be an early driving event in oncogenesis (58). Therefore, the effects of MUC1-C on the epigenetic functions of DNMTs and BMI1 may be selective at least in part for TSG silencing. Given the above, PRC1 complexes are recruited to chromatin at sites modified by PRC2, which catalyzes the trimethylation of K27 on histone H3 (H3K27me3). Accordingly, it will be of interest to determine whether MUC1-C is involved in the regulation of PRC2 activity.
Finally, with regard to potential clinical impact of the present findings, a Phase I trial of GO-203 has been completed in patients with advanced solid tumors. In addition, based on the marked synergy between GO-203 and decitabine in AML (36), a Phase I/II trial of this combination is underway for patients with relapsed/refractory AML. We have also formulated GO-203 in novel polymeric nanoparticles for sustained delivery of this agent to target the cancer epigenome in the clinic (59).
Materials and Methods
Cell culture
Human BT-549, A549/KRAS(G12S), and H460/KRAS(Q61H) cells were grown in RPMI1640 medium (ATCC, Manassas, VA, USA). MDA-MB-231 and MDA-MB-468 cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM) (Corning, Manassas, VA, USA). BT-20 cells were cultured in Eagle’s Minimum Essential Medium (EMEM) (ATCC). Media were supplanted with 10% heat-inactivated fetal bovine serum (HI-FBS), 100 U/ml penicillin and 100 μg/ml streptomycin. These cells were selected for study based on their comparatively high and low levels of MUC1-C expression (Supplemental Fig. S7). Cells stably expressing MUC1-C were generated as described (60). Cells were treated with the MUC1-C inhibitor GO-203 or the control CP-2 peptide (32). Authentication of cells was performed by short tandem repeat (STR) analysis. Cell were monitored for mycoplasma contamination using the MycoAlert® Mycoplasma Detection Kit (Lonza, Rockland, MA, USA).
Tetracycline-inducible MUC1 and MYC silencing
MUC1shRNA#1 (MISSION shRNA; Sigma, TRCN0000122938), MUC1shRNA#2 (MISSION shRNA; Sigma, TRCN0000122937), MYCshRNA (MISSION shRNA; Sigma, TRCN0000039642) or a control scrambled CshRNA (Sigma) was inserted into the pLKO-tet-puro vector (Addgene, Cambridge, MA, USA; Plasmid #21915). The viral vectors were produced in HEK293T cells as previously described (60; 61). BT-549, A549, H460, BT-20 and MDA-MB-468 cells expressing tet-MUC1shRNA#1, MDA-MB-231 cells expressing tet-MUC1shRNA#2 and cells expressing tet-MYCshRNA or tet-CshRNA were selected for growth in 1–3 μg/ml puromycin. Cells were treated with doxycycline (DOX; Sigma, St. Louis, MO, USA).
Transient cell transfections
Cells were cultured to 60% to 80% confluence and transiently transfected with (i) anti-miR-200c or a negative control oligonucleotide (Ambion, Carlsbad, CA, USA) and (ii) a BMI1 siRNA or control siRNA (Cell Signaling Technology, Danvers, MA, USA) using Lipofectamine RNAiMax (Invitrogen, Carlsbad, CA, USA) in accordance with the protocol provided by the manufacturer.
RNA extraction and real-time quantitative reverse-transcription PCR (qRT-PCR)
Total RNA was isolated using with Trizol reagent (Invitrogen) following the manufacturer’s protocol. Complementary DNA was synthesized from 2.0 μg total RNA using the High Capacity cDNA Reverse Transcription Kit (Applied Biosystems, Foster City, CA, USA) as described (62). The Power SYBR Green PCR Master Mix (Applied Biosystems) was used with 1 μl of diluted cDNA for each sample. The samples were amplified using the 7300 Realtime PCR System (Applied Biosystems). Primers used for RT–PCR analysis are listed in Supplemental Table S1.
Real-time qRT-PCR for miRNA expression
The reverse transcription reaction was performed with the TaqMan MicroRNA RT Kit (Applied Biosystems). Real-time quantitative PCR was performed with TaqMan MicroRNA Assays (Applied Biosystems) and the 7300 Realtime PCR System (Applied Biosystems). The miR-200c signal was normalized relative to that of the endogenous control, RNU48. Data were analyzed according to the comparative Ct method.
BMI1 promoter luciferase reporter assays
pGL-BMI1PrWT and pGL-BMI1PrMut reporter vectors were kindly provided by Dr. Goberdhan P. Dimri (52). pGL3-BMI1PrWT contains the +45 to -233 region of the BMI1 promoter and untranslated region of BMI1 mRNA. pGL3-BMI1PrMut contains a mutation in the MYC binding sequences (CACGTG mutated to CGCGTG). Cells growing in 24-well plates were transfected with pGL-BMI1PrWT or pGL-BMI1PrMut reporter vector and SV-40-Renilla-Luc in the presence of Lipofectamine™ 3000 Reagent (Invitrogen). At 48 h after transfection, cell extracts were prepared with passive lysis buffer using the Luciferase® Assay System (Promega, Madison, WI, USA). Luminescence was measured with the Dual-Luciferase® Reporter Assay System (Promega) according to the manufacturer’s instructions.
Chromatin immunoprecipitation (ChIP) assay
Soluble chromatin was precipitated with anti-MUC1-C (NeoMarkers, Fremont, CA, USA), anti-MYC (Abcam, Cambridge, MA, USA), anti-BMI1 (Cell Signaling Technology) or a control non-immune IgG (Santa Cruz Biotechnology, Dallas, TX, USA). For re-ChIP analysis, anti-BMI1 complexes from the primary ChIP were eluted and re-immunoprecipitated with anti-MUC1-C (NeoMarkers). For real-time ChIP qPCR, the SYBR green system was used with the ABI Prism 7300 sequence detector (Applied Biosystems). Data are reported as relative fold enrichment (35). Primers used for ChIP qPCR of the BMI1 and CDKN2A promoter are listed in the Supplementary Table S2.
Immunoblot analysis
Western blot analysis was performed as described previously (60). Whole cells were lysed in NP-40 buffer, containing phosphatase Inhibitor and protease inhibitor cocktail. Immunoblotting was performed with anti-MUC1-C (NeoMarkers), anti-BMI1, ZEB1, anti-phospho-p65(Ser-536), anti-Ubiquityl-Histone H2A (Lys119), anti-H2A (Cell Signaling Technology), anti-MYC, anti-CDKN2A/p16INK4a (Abcam), anti-E-cadherin, anti-NF-κB p65 (Santa Cruz Biotechnology), and anti-β-actin (Sigma).
In vitro direct binding assays
GST, GST-MUC1-CD, and GST-MUC1-CD(AQA) were prepared as described (28). GST-tagged BMI1 was generated by PCR amplification of the pT3-EF1a-BMI1 plasmid (Addgene) and subcloning into the pGEX-5X-1 expression vector (GE Healthcare, Pittsburg, PA, USA). Purified GST-MUC1-CD and GST-MUC1-CD(AQA) were cleaved with thrombin to remove the GST moiety (30). For bindings assays, purified proteins were incubated for 2 h at room temperature. Adsorbates to glutathione-conjugated beads were analyzed by immunoblotting.
Statistical analysis
Each experiment was repeated at least three times. Data are expressed as mean±SD. The unpaired Student’s t-test was used to examine differences between means of two groups. A p-value <0.05 was considered a statistically significant difference.
Bioinformatics analysis
Clinical data of breast cancer patients was obtained from cBioPortal METABRIC and TCGA datasets (63). The correlations between MUC1 and BMI1 were assessed using Pearson’s correlation coefficient. Additionally, dataset of TNBC patients was downloaded from Gene Expression Omnibus (GEO) under the accession number of GSE41970. The data was log2 transformed and the correlation between MUC1 and BMI1 was assessed using Pearson’s correlation coefficient (26; 29).
Supplementary Material
Acknowledgments
This work was supported by Grants from the National Cancer Institute of the National Institutes of Health under award numbers CA97098 and CA166480 and by the Lung Cancer Research Foundation.
The authors thank Dr. Goberdhan P. Dimri, Department of Biochemistry and Molecular Medicine, School of Medicine and Health Sciences, The George Washington University, Washington, DC for kindly providing the pGL-BMI1PrWT and pGL-BMI1PrMut reporter vectors.
Abbreviations
- BMI1
B cell-specific Moloney murine leukemia virus integration site 1
- PcG
polycomb group proteins
- PRC1
polycomb repressive complex 1
- PRC2
polycomb repressive complex 2
- CSCs
cancer stem-like cells
- EMT
epithelial-mesenchymal transition
- DDR
DNA damage response
- DSB
double-stranded DNA break
- MUC1
mucin 1
- MUC1-C
MUC1 C-terminal subunit
- DNMT
DNA methyltransferase
- TSG
tumor suppressor gene
- DOX
doxycycline
- TNBC
triple-negative breast cancer
Footnotes
Conflict of Interest
The authors declare competing financial interests: D.K. holds equity in Genus Oncology and is a consultant to the company. The other authors disclosed no potential conflicts of interest.
References
- 1.Park IK, Morrison SJ, Clarke MF. Bmi1, stem cells, and senescence regulation. J Clin Invest. 2004;113:175–179. doi: 10.1172/JCI20800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Siddique HR, Saleem M. Role of BMI1, a stem cell factor, in cancer recurrence and chemoresistance: preclinical and clinical evidences. Stem Cells. 2012;30:372–8. doi: 10.1002/stem.1035. [DOI] [PubMed] [Google Scholar]
- 3.Wang H, Wang L, Erdjument-Bromage H, Vidal M, Tempst P, Jones RS, et al. Role of histone H2A ubiquitination in Polycomb silencing. Nature. 2004;431:873–8. doi: 10.1038/nature02985. [DOI] [PubMed] [Google Scholar]
- 4.de Napoles M, Mermoud JE, Wakao R, Tang YA, Endoh M, Appanah R, et al. Polycomb group proteins Ring1A/B link ubiquitylation of histone H2A to heritable gene silencing and X inactivation. Dev Cell. 2004;7:663–76. doi: 10.1016/j.devcel.2004.10.005. [DOI] [PubMed] [Google Scholar]
- 5.Cao R, Tsukada Y, Zhang Y. Role of Bmi-1 and Ring1A in H2A ubiquitylation and Hox gene silencing. Mol Cell. 2005;20:845–54. doi: 10.1016/j.molcel.2005.12.002. [DOI] [PubMed] [Google Scholar]
- 6.Ohtsubo M, Yasunaga S, Ohno Y, Tsumura M, Okada S, Ishikawa N, et al. Polycomb-group complex 1 acts as an E3 ubiquitin ligase for Geminin to sustain hematopoietic stem cell activity. Proc Natl Acad Sci U S A. 2008;105:10396–401. doi: 10.1073/pnas.0800672105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang Y, Zhe H, Ding Z, Gao P, Zhang N, Li G. Cancer stem cell marker Bmi-1 expression is associated with basal-like phenotype and poor survival in breast cancer. World J Surg. 2012;36:1189–94. doi: 10.1007/s00268-012-1514-3. [DOI] [PubMed] [Google Scholar]
- 8.Crea F, Paolicchi E, Marquez VE, Danesi R. Polycomb genes and cancer: time for clinical application? Crit Rev Oncol Hematol. 2012;83:184–93. doi: 10.1016/j.critrevonc.2011.10.007. [DOI] [PubMed] [Google Scholar]
- 9.Richly H, Aloia L, Di Croce L. Roles of the Polycomb group proteins in stem cells and cancer. Cell Death Dis. 2011;2:e204. doi: 10.1038/cddis.2011.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kreso A, van Galen P, Pedley NM, Lima-Fernandes E, Frelin C, Davis T, et al. Self-renewal as a therapeutic target in human colorectal cancer. Nat Med. 2014;20:29–36. doi: 10.1038/nm.3418. [DOI] [PubMed] [Google Scholar]
- 11.Lessard J, Sauvageau G. Bmi-1 determines the proliferative capacity of normal and leukaemic stem cells. Nature. 2003;423:255–60. doi: 10.1038/nature01572. [DOI] [PubMed] [Google Scholar]
- 12.Rizo A, Olthof S, Han L, Vellenga E, de Haan G, Schuringa JJ. Repression of BMI1 in normal and leukemic human CD34(+) cells impairs self-renewal and induces apoptosis. Blood. 2009;114:1498–505. doi: 10.1182/blood-2009-03-209734. [DOI] [PubMed] [Google Scholar]
- 13.Song LB, Li J, Liao WT, Feng Y, Yu CP, Hu LJ, et al. The polycomb group protein Bmi-1 represses the tumor suppressor PTEN and induces epithelial-mesenchymal transition in human nasopharyngeal epithelial cells. J Clin Invest. 2009;119:3626–36. doi: 10.1172/JCI39374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yang MH, Hsu DS, Wang HW, Wang HJ, Lan HY, Yang WH, et al. Bmi1 is essential in Twist1-induced epithelial-mesenchymal transition. Nat Cell Biol. 2010;12:982–92. doi: 10.1038/ncb2099. [DOI] [PubMed] [Google Scholar]
- 15.Lin X, Ojo D, Wei F, Wong N, Gu Y, Tang D. A Novel Aspect of Tumorigenesis-BMI1 Functions in Regulating DNA Damage Response. Biomolecules. 2015;5:3396–415. doi: 10.3390/biom5043396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu J, Cao L, Chen J, Song S, Lee IH, Quijano C, et al. Bmi1 regulates mitochondrial function and the DNA damage response pathway. Nature. 2009;459:387–92. doi: 10.1038/nature08040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ginjala V, Nacerddine K, Kulkarni A, Oza J, Hill SJ, Yao M, et al. BMI1 is recruited to DNA breaks and contributes to DNA damage-induced H2A ubiquitination and repair. Mol Cell Biol. 2011;31:1972–82. doi: 10.1128/MCB.00981-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nacerddine K, Beaudry JB, Ginjala V, Westerman B, Mattiroli F, Song JY, et al. Akt-mediated phosphorylation of Bmi1 modulates its oncogenic potential, E3 ligase activity, and DNA damage repair activity in mouse prostate cancer. J Clin Invest. 2012;122:1920–32. doi: 10.1172/JCI57477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dimri M, Kang M, Dimri GP. A miR-200c/141-BMI1 autoregulatory loop regulates oncogenic activity of BMI1 in cancer cells. Oncotarget. 2016 doi: 10.18632/oncotarget.8811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kufe D. Mucins in cancer: function, prognosis and therapy. Nature Reviews Cancer. 2009;9:874–885. doi: 10.1038/nrc2761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kufe D. MUC1-C oncoprotein as a target in breast cancer: activation of signaling pathways and therapeutic approaches. Oncogene. 2013;32:1073–1081. doi: 10.1038/onc.2012.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Raina D, Uchida Y, Kharbanda A, Rajabi H, Panchamoorthy G, Jin C, et al. Targeting the MUC1-C oncoprotein downregulates HER2 activation and abrogates trastuzumab resistance in breast cancer cells. Oncogene. 2014;33:3422–3431. doi: 10.1038/onc.2013.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kharbanda A, Rajabi H, Jin C, Tchaicha J, Kikuchi E, Wong K, et al. Targeting the oncogenic MUC1-C protein inhibits mutant EGFR-mediated signaling and survival in non-small cell lung cancer cells. Clin Cancer Res. 2014;20:5423–5434. doi: 10.1158/1078-0432.CCR-13-3168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rajabi H, Ahmad R, Jin C, Kosugi M, Alam M, Joshi M, et al. MUC1-C oncoprotein induces TCF7L2 transcription factor activation and promotes cyclin D1 expression in human breast cancer cells. J Biol Chem. 2012;287:10703–10713. doi: 10.1074/jbc.M111.323311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bouillez A, Rajabi H, Pitroda S, Jin C, Alam M, Kharbanda A, et al. Inhibition of MUC1-C suppresses MYC expression and attenuates malignant growth in KRAS mutant lung adenocarcinomas. Cancer Res. 2016;76:1538–1548. doi: 10.1158/0008-5472.CAN-15-1804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tagde A, Rajabi H, Bouillez A, Alam M, Gali R, Bailey S, et al. MUC1-C drives MYC in multiple myeloma. Blood. 2016;127:2587–2597. doi: 10.1182/blood-2015-07-659151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ahmad R, Raina D, Trivedi V, Ren J, Rajabi H, Kharbanda S, et al. MUC1 oncoprotein activates the IκB kinase β complex and constitutive NF-κB signaling. Nat Cell Biol. 2007;9:1419–1427. doi: 10.1038/ncb1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ahmad R, Raina D, Joshi MD, Kawano T, Kharbanda S, Kufe D. MUC1-C oncoprotein functions as a direct activator of the NF-κB p65 transcription factor. Cancer Res. 2009;69:7013–7021. doi: 10.1158/0008-5472.CAN-09-0523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Takahashi H, Jin C, Rajabi H, Pitroda S, Alam M, Ahmad R, et al. MUC1-C activates the TAK1 inflammatory pathway in colon cancer. Oncogene. 2015;34:5187–5197. doi: 10.1038/onc.2014.442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rajabi H, Alam M, Takahashi H, Kharbanda A, Guha M, Ahmad R, et al. MUC1-C oncoprotein activates the ZEB1/miR-200c regulatory loop and epithelial-mesenchymal transition. Oncogene. 2014;33:1680–1689. doi: 10.1038/onc.2013.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Alam M, Ahmad R, Rajabi H, Kufe D. MUC1-C induces the LIN28B→LET-7→HMGA2 axis and self-renewal in NSCLC cells. Mol Cancer Res. 2015;13:449–460. doi: 10.1158/1541-7786.MCR-14-0363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Alam M, Rajabi H, Ahmad R, Jin C, Kufe D. Targeting the MUC1-C oncoprotein inhibits self-renewal capacity of breast cancer cells. Oncotarget. 2014;5:2622–2634. doi: 10.18632/oncotarget.1848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kharbanda A, Rajabi H, Jin C, Alam M, Wong K, Kufe D. MUC1-C confers EMT and KRAS independence in mutant KRAS lung cancer cells. Oncotarget. 2014;5:8893–8905. doi: 10.18632/oncotarget.2360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tam WL, Weinberg RA. The epigenetics of epithelial-mesenchymal plasticity in cancer. Nat Med. 2013;19:1438–1449. doi: 10.1038/nm.3336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rajabi H, Tagde A, Alam M, Bouillez A, Pitroda S, Suzuki Y, et al. DNA methylation by DNMT1 and DNMT3b methyltransferases is driven by the MUC1-C oncoprotein in human carcinoma cells. Oncogene. 2016 doi: 10.1038/onc.2016.180. [Epub ahead of print May 23] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tagde A, Rajabi H, Stroopinsky D, Gali R, Alam M, Bouillez A, et al. MUC1-C induces DNA methyltransferase 1 and represses tumor suppressor genes in acute myeloid leukemia. Oncotarget. 2016 doi: 10.18632/oncotarget.9777. [Epub ahead of print Jun 1] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kufe D. Functional targeting of the MUC1 oncogene in human cancers. Cancer Biol Ther. 2009;8:1201–1207. doi: 10.4161/cbt.8.13.8844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Raina D, Ahmad R, Rajabi H, Panchamoorthy G, Kharbanda S, Kufe D. Targeting cysteine-mediated dimerization of the MUC1-C oncoprotein in human cancer cells. Int J Oncol. 2012;40:1643–1649. doi: 10.3892/ijo.2011.1308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Raina D, Agarwal P, Lee J, Bharti A, McKnight C, Sharma P, et al. Characterization of the MUC1-C cytoplasmic domain as a cancer target. PLoS One. 2015;10:e0135156. doi: 10.1371/journal.pone.0135156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Huang R, Cheung NK, Vider J, Cheung IY, Gerald WL, Tickoo SK, et al. MYCN and MYC regulate tumor proliferation and tumorigenesis directly through BMI1 in human neuroblastomas. FASEB J. 2011;25:4138–49. doi: 10.1096/fj.11-185033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Alam M, Bouillez A, Tagde A, Ahmad R, Rajabi H, Maeda T, et al. MUC1-C Represses the Crumbs Complex Polarity Factor CRB3 and Downregulates the Hippo Pathway. Mole Cancer Res. 2016 doi: 10.1158/1541-7786.MCR-16-0233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shimono Y, Zabala M, Cho RW, Lobo N, Dalerba P, Qian D, et al. Downregulation of miRNA-200c links breast cancer stem cells with normal stem cells. Cell. 2009;138:592–603. doi: 10.1016/j.cell.2009.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wei J, Zhai L, Xu J, Wang H. Role of Bmi1 in H2A ubiquitylation and Hox gene silencing. J Biol Chem. 2006;281:22537–44. doi: 10.1074/jbc.M600826200. [DOI] [PubMed] [Google Scholar]
- 44.Jacobs JJ, Scheijen B, Voncken JW, Kieboom K, Berns A, van Lohuizen M. Bmi-1 collaborates with c-Myc in tumorigenesis by inhibiting c-Myc-induced apoptosis via INK4a/ARF. Genes Dev. 1999;13:2678–90. doi: 10.1101/gad.13.20.2678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jacobs JJ, Kieboom K, Marino S, DePinho RA, van Lohuizen M. The oncogene and Polycomb-group gene bmi-1 regulates cell proliferation and senescence through the ink4a locus. Nature. 1999;397:164–168. doi: 10.1038/16476. [DOI] [PubMed] [Google Scholar]
- 46.Bracken AP, Kleine-Kohlbrecher D, Dietrich N, Pasini D, Gargiulo G, Beekman C, et al. The Polycomb group proteins bind throughout the INK4A-ARF locus and are disassociated in senescent cells. Genes Dev. 2007;21:525–30. doi: 10.1101/gad.415507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Meng S, Luo M, Sun H, Yu X, Shen M, Zhang Q, et al. Identification and characterization of Bmi-1-responding element within the human p16 promoter. J Biol Chem. 2010;285:33219–29. doi: 10.1074/jbc.M110.133686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Campisi J. Senescent cells, tumor suppression, and organismal aging: good citizens, bad neighbors. Cell. 2005;120:513–22. doi: 10.1016/j.cell.2005.02.003. [DOI] [PubMed] [Google Scholar]
- 49.Polyak K, Weinberg RA. Transitions between epithelial and mesenchymal states: acquisition of malignant and stem cell traits. Nat Rev Cancer. 2009;9:265–273. doi: 10.1038/nrc2620. [DOI] [PubMed] [Google Scholar]
- 50.Alam M, Ahmad R, Rajabi H, Kharbanda A, Kufe D. MUC1-C oncoprotein activates ERK→C/EBPβ-mediated induction of aldehyde dehydrogenase activity in breast cancer cells. J Biol Chem. 2013;288:30829–30903. doi: 10.1074/jbc.M113.477158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Valk-Lingbeek ME, Bruggeman SW, van Lohuizen M. Stem cells and cancer; the polycomb connection. Cell. 2004;118:409–18. doi: 10.1016/j.cell.2004.08.005. [DOI] [PubMed] [Google Scholar]
- 52.Cho JH, Dimri M, Dimri GP. A positive feedback loop regulates the expression of polycomb group protein BMI1 via WNT signaling pathway. J Biol Chem. 2013;288:3406–18. doi: 10.1074/jbc.M112.422931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yamamoto M, Bharti A, Li Y, Kufe D. Interaction of the DF3/MUC1 breast carcinoma-associated antigen and β-catenin in cell adhesion. J Biol Chem. 1997;272:12492–12494. doi: 10.1074/jbc.272.19.12492. [DOI] [PubMed] [Google Scholar]
- 54.Huang L, Chen D, Liu D, Yin L, Kharbanda S, Kufe D. MUC1 oncoprotein blocks GSK3β-mediated phosphorylation and degradation of β-catenin. Cancer Res. 2005;65:10413–10422. doi: 10.1158/0008-5472.CAN-05-2474. [DOI] [PubMed] [Google Scholar]
- 55.Fleming A, Osley MA. Silence of the rings. Cell. 2004;119:449–51. doi: 10.1016/j.cell.2004.11.001. [DOI] [PubMed] [Google Scholar]
- 56.Mallo M, Alonso CR. The regulation of Hox gene expression during animal development. Development. 2013;140:3951–63. doi: 10.1242/dev.068346. [DOI] [PubMed] [Google Scholar]
- 57.Endoh M, Endo TA, Endoh T, Isono K, Sharif J, Ohara O, et al. Histone H2A mono-ubiquitination is a crucial step to mediate PRC1-dependent repression of developmental genes to maintain ES cell identity. PLoS Genet. 2012;8:e1002774. doi: 10.1371/journal.pgen.1002774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kazanets A, Shorstova T, Hilmi K, Marques M, Witcher M. Epigenetic silencing of tumor suppressor genes: Paradigms, puzzles, and potential. Biochim Biophys Acta. 2016;1865:275–88. doi: 10.1016/j.bbcan.2016.04.001. [DOI] [PubMed] [Google Scholar]
- 59.Hasegawa M, Sinha RK, Kumar M, Alam M, Yin L, Raina D, et al. Intracellular targeting of the oncogenic MUC1-C protein with a novel GO-203 nanoparticle formulation. Clin Cancer Res. 2015;21:2338–47. doi: 10.1158/1078-0432.CCR-14-3000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hiraki M, Suzuki Y, Alam M, Hinohara K, Hasegawa M, Jin C, et al. MUC1-C stabilizes MCL-1 in the oxidative stress response of triple-negative breast cancer cells to BCL-2 inhibitors. Sci Rep. 2016:6. doi: 10.1038/srep26643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hasegawa M, Takahashi H, Rajabi H, Alam M, Suzuki Y, Yin L, et al. Functional interactions of the cystine/glutamate antiporter, CD44v and MUC1-C oncoprotein in triple-negative breast cancer cells. Oncotarget. 2016;7:11756–69. doi: 10.18632/oncotarget.7598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hiraki M, Nishimura J, Takahashi H, Wu X, Takahashi Y, Miyo M, et al. Concurrent targeting of KRAS and AKT by MiR-4689 is a novel treatment against mutant KRAS colorectal cancer. Mol Ther Nucleic Acids. 2015;4:e231. doi: 10.1038/mtna.2015.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6:pl1. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.