Abstract
Coxiella burnetii is a Gram-negative intracellular pathogen and is the causative agent of the zoonotic disease Q fever. To cause disease, C. burnetii requires a functional type IVB secretion system (T4BSS) to transfer effector proteins required for the establishment and maintenance of a membrane-bound parasitophorous vacuole (PV) and further modulation of host cell process. However, it is not clear how the T4BSS interacts with the PV membrane since neither a secretion pilus nor an extracellular pore forming apparatus has not been described. To address this, we used the acidified citrate cysteine medium (ACCM) along with cell culture infection and immunological techniques to identify the cellular and extracellular localization of T4BSS components. Interestingly, we found that DotA and IcmX were secreted/released in a T4BSS-dependent manner into the ACCM. Analysis of C. burnetii-infected cell lines revealed that DotA colocalized with the host cell marker CD63 (LAMP3) at the PV membrane. In the absence of bacterial protein synthesis, DotA also became depleted from the PV membrane. These data are the first to identify the release/secretion of C. burnetii T4BSS components during axenic growth and the interaction of a T4BSS component with the PV membrane during infection of host cells.
Keywords: Q fever; type 4B secretion system; DotA, Coxiella; intracellular pathogenesis; ACCM
The T4BSS component protein DotA of Coxiella burnetii is released during infection of host cells and localizes to the membrane of the parasitophorous vacuole in a T4BSS-dependent manner.
INTRODUCTION
Coxiella burnetii is a Gram-negative intracellular pathogen and is the causative agent of acute Q fever as well as chronic diseases. Transmission of C. burnetii is predominantly through the aerosol route and one to ten organisms is sufficient to cause disease (Marrie 2004). Post inhalation, C. burnetii targets alveolar macrophages to establish an intravacuolar replicative niche termed the parasitophorous vacuole (PV), which has many of the attributes of a mature phagolysosome (Voth and Heinzen 2007). During the ∼6-day infectious cycle, C. burnetii alternates between the environmentally stable, metabolically quiescent small cell variant (SCV) and the replicative large cell variant (LCV) (Heinzen, Hackstadt and Samuel 1999). This cycle is defined by a morphogenesis of the SCV into the LCV by 6–8 h post infection (hpi) and then at 6 days the LCV asynchronously converts back to the environmentally stable SCV form (Coleman et al. 2004). After cellular uptake and entering the endocytic pathway, infectious C. burnetii delay the trafficking of the early PV for ∼2 hpi (Howe and Mallavia 2000). During this time, the PV gains the host markers early endosome antigen 1, Rab5 and LC3 (Beron et al. 2002; Romano et al. 2007). As the PV matures the early markers dissociate and Rab7 (Beron et al. 2002), lysosome-associated membrane protein (LAMP) 1, LAMP 2 and LAMP 3 (CD63) (Voth and Heinzen 2007), vacuolar H+ ATPase (Heinzen et al. 1996), and flotillin 1 and 2 (Howe and Heinzen 2006) decorate the PV membrane. The development of the PV is essential for the replication and subsequent pathogenesis of C. burnetii and is mediated by the type 4B secretion system (T4BSS) (Howe et al. 2003b; Beare et al. 2011).
From genomic sequence analysis, it was determined that C. burnetii possesses homologs to 23 of the 26 Legionella pneumophila T4BSS components (Seshadri et al. 2003). Legionella pneumophila utilizes the T4BSS as an essential virulence factor for the translocation of effector proteins into the cytoplasm of the host cell (Segal and Shuman 1997; Bardill, Miller and Vogel 2005). Legionella pneumophila has been used as a surrogate to identify T4BSS-dependent secretion of several putative C. burnetii effector proteins (Pan et al. 2008; Chen et al. 2010). This indicates a degree of functional similarity between the T4BSSs of each organism, and is further supported by the ability of C. burnetii dotB, icmS, icmT and icmW genes to complement the corresponding L. pneumophila mutants although complementation was not observed for dotL, dotM, dotN, dotO, icmQ and icmX-deficient mutants (Zamboni et al. 2003; Zusman, Yerushalmi and Segal 2003). The ability to grow C. burnetii axenically and generate site-specific mutants has shown that the cellular pathogenesis of C. burnetii is dependent on a functional T4BSS (Omsland and Heinzen 2011; Beare et al. 2012). Deletion of C. burnetii dotA and dotB (Beare et al. 2012) and transposon mutagenesis of dotE, dotF, dotG, dotL1, dotH, dotK, dotM, dotN, icmD, icmV and icmX have been shown to prevent the development of a replicative PV during infection of cells (Beare et al. 2011; Carey et al. 2011; Martinez et al. 2014). These findings demonstrate the crucial role that the C. burnetii T4BSS plays in infection and replication within host cells. However, unlike the model T4ASS of Agrobacterium tumefaciens, a pilus for the C. burnetii and L. pneumophila T4BSS has not been demonstrated (Vincent and Vogel 2008) despite the core structure of the L. pneumophila T4BSS being similar in architecture and likely secretion mechanism as the T4ASS (Kubori et al. 2014; Ghosal et al. 2017). This lack of a secretion pilus in the T4BSS further confounds how C. burnetii and L. pneumophila, which are contained in a membrane-bound vacuole during infection, engages the PV membrane for the transfer of effector proteins into the host cell cytoplasm.
Interestingly, it was observed that during growth in axenic media L. pneumophila secretes, or releases, DotA and IcmX in a T4BSS-dependent manner (Matthews and Roy 2000; Nagai and Roy 2001). Although predicted to be a polytopic inner membrane protein, DotA from L. pneumophila was found to form hollow ring structures when harvested from the growth medium and observed by transmission electron microscopy (TEM). It was hypothesized that this structure might interact with the host or vacuolar membrane to mediate the transfer of effector molecules into the host cytoplasm during infection, but direct evidence of DotA interacting with the vacuolar membrane during infection was not achievable (Nagai and Roy 2001). In addition, IcmX from L. pneumophila is also released into the axenic media during growth although it is primarily localized to the periplasmic space (Matthews and Roy 2000) and requires a functional type II secretion system for subsequent T4BSS-dependent secretion (DebRoy et al. 2006). The significance and function of IcmX in the periplasm or away from the cell and the release of DotA are unclear, yet both proteins are essential for L. pneumophila pathogenesis.
Here, we sought to identify C. burnetii T4BSS components with secretion signals that are potentially secreted or released from the bacteria during growth in the axenic acidified citrate cysteine medium (ACCM) and subsequently to characterize their localization in the host cell during infection. Our hypothesis is that T4BSS components found secreted in the ACCM will localize extracellularly in an infectious setting. Previously, we have shown that the T4BSS localizes to the pole(s) of C. burnetii and that the bacterium can often be observed in direct contact with the PV (Morgan, Luedtke and Shaw 2010). This suggests that an intimate interaction of the T4BSS with the PV membrane occurs, but it is not clear how effector molecules are translocated without clear evidence of a secretion pilus. In this study, we demonstrate that C. burnetii DotA and IcmX are secreted, or released, into the ACCM during C. burnetii growth by a mechanism that is dependent on an intact T4BSS. During infection, we have identified DotA colocalizing with the PV membrane.
MATERIALS AND METHODS
Bacterial cultivation and storage
Avirulent Coxiella burnetii Nine Mile phase II RSA 439 (NMII) and virulent C. burnetii Nine Mile phase I RSA 493 (NMI) were cultivated and the SCV form purified as previously described (Coleman et al. 2004; Morgan, Luedtke and Shaw 2010). All work with NMI was performed in a CDC approved biosafety level-3 (BSL-3) with proper personal protection. NMII and NMI were stored at –80°C in SPG buffer (0.7 M sucrose, 3.7 mM KH2PO4, 6.0 mM K2HPO4, 0.15 M KCl, 5.0 mM glutamic acid, pH 7.4). In addition, C. burnetii NMII and C. burnetii NMII ΔdotA, ΔdotB and ΔicmD mutants (kindly provided by Dr Robert Heinzen) (Beare et al. 2011, 2012) were propagated using ACCM and stored at –80°C as previously described (Omsland et al. 2009).
Cell culture and infection
Vero, HeLa and RK13 (rabbit kidney) cell lines were propagated in antibiotic free RPMI 1640 medium supplemented with 5% FBS and incubated in a humidified chamber at 37°C with 5% CO2. Vero and HeLa cells were infected with NMII using centrifugation at 600 g for 15 min at 25°C (termed spinoculation) (Wike et al. 1972). After spinoculation, unphagocytosed bacteria were removed using three washes of pre-warmed RPMI 1640 medium. NMI infection of RK13 cells was accomplished without spinoculation by inoculating the cell culture with NMI and incubating for 2 h at 37°C, then washing cells with pre-warmed RPMI 1640 three times to remove extracellular bacteria. Infected cells were cultured at 37°C, 5% CO2 atmosphere in a humidified chamber for the desired length of hpi with time zero being immediately post washing.
Antibody production
Full-length C. burnetii icmX, dotH, dotN, com1 and nucleotides corresponding to amino acids 177–298 of DotA were amplified by PCR for cloning into the pET200/D-TOPO vector as described by the manufacturer (Invitrogen, Carlsbad, CA, USA). Expression and isolation of recombinant protein as well as subsequent polyclonal antibody production was performed as described (Morgan, Luedtke and Shaw 2010). Protein A cross-linked beads (Pierce, Rockford, IL Rockford, IL, USA) were used to enrich IgG from the serum, and the antibody was absorbed against Escherichia coli BL21 to eliminate non-specific reactivity (Morgan, Luedtke and Shaw 2010). Rabbit antibodies against C. burnetii IcmD were kindly provided by Dr Robert Heinzen (Beare et al. 2011). Rabbit polyclonal antibodies against C. burnetii DotB and DotC were raised using the synthetic peptides KSQRYPNEPSRFEPK, TLTRRELSNAETSDL and HYEIRPNRNERFRYR and LNSLPPGSGQINNIR, LNLADDDTIRTADKT and TAQSALQPDSSHWNP, respectively. Polyclonal antibodies against whole, heat killed sucrose gradient purified C. burnetii NMII were produced in guinea pigs using a custom antibody production service (Rockland Immunochemicals, Gilbertsville, PA, USA). The specificity of each antibody against the T4BSS homologs IcmX, DotB, DotC, DotH, DotN and the major outer membrane protein Com1 was determined by immunoblot against the respective recombinant protein and compared to NMII lysate. Recombinant protein was subsequently produced for DotB and DotC in full length to analyze the specificity of the antibodies against the synthetic peptides. The specificity of antibody against DotA was addressed by comparing NMII and NMII ΔdotA lysates by immunoblot and by absorbing the antibody with recombinant DotA (rDotA). In addition, the DotA and DotH antibodies were utilized as previously described (Beare et al. 2014)
Reverse transcriptase-PCR
Reverse transcriptase-PCR (RT-PCR) was performed on total RNA harvested from NMII cultures grown in ACCM for 4 and 7 days post inoculation as previously described (Morgan et al. 2010). Briefly, nucleic acids were isolated using Tri-Reagent® (Ambion, Carlsbad, CA, USA), and DNA was removed using RQ1 DNase (Promega, Madison, WI, USA). Primer sets for RT-PCR of dotH (forward 5΄-ATTGGGGCCAGTATCATTCC-3΄, reverse 5΄-ATGGAGTGTGCGGATTTGAT-3΄), icmT (forward 5΄-ATGAAATCTCTCGATGAGG-3΄, reverse 5΄-GTTATCCCACCATGCTATGG-3΄), icmV (forward 5΄-ATGATTCTTTTGGAGTCTTCC-3΄, reverse 5΄-TTGTTTGGACCCCTTAAAGGTG-3΄) and icmW (forward 5΄-ATGCCAGATCTGTCGC-3΄, reverse 5΄-TAAACCACCTTCCTCAAGAG-3΄) were internal of the open reading frames. To confirm samples contained no DNA, SuperScript® III RT (ThermoFisher, Carlsbad, CA, USA) was not included in paired reactions. The ability of primer sets to amplify DNA was validated using C. burnetii genomic DNA.
Immunoblot analysis
NMII cultures were grown in ACCM for 96 and 168 hpi and were harvested by centrifugation at 13 000 g for 15 min at 4°C. The supernatant (media) was carefully removed and Halt protease inhibitor cocktail (Pierce, Rockford, IL, USA) was added prior to storage at –80°C. The resulting NMII pellet was washed with sterile PBS pH 7.4, pelleted again as above and resuspended in protein running buffer containing Halt protease inhibitor cocktail (Pierce) and stored at –80°C. To analyze the ACCM for C. burnetii secreted/released proteins, the medium was concentrated using I-Con 9kDa spin concentrators (Pierce). Halt protease inhibitor cocktail (Pierce) was added to samples and stored at –80°C. Total protein was not boiled prior to being separated using 12% SDS-PAGE to prevent the insolubility or aggregation of T4BSS membrane proteins. After SDS-PAGE, total proteins were immobilized on a nitrocellulose membrane (Whatman, Dassel, Germany). Immunoblot analysis was carried out with overnight incubation at 4°C with the respective primary antibody all at 1:3000 and followed by HRP-conjugated secondary antibody at 1:20 000 for 1 h. The labeled proteins were detected using the Pico Western Chemiluminescence Kit (Pierce) following the manufacturer's directions. Digital images were captured using a FluorChem HD2 Imaging System (Alpha Innotech Corporation, Leandro, CA). All micrographs were processed equally and analyzed using ImageJ version 1.46h (Rasband 1997–2012).
Immunoflurocescent microscopy
Uninfected cell lines were seeded on 12-mm glass coverslips in 24-well tissue culture plates at 105 cells/well. Cells were allowed at least 16 h to adhere to the coverslip before infecting. Infections were carried out as described above. At various hpi, cells were fixed for 5 min using 100% ice-cold methanol. In some cases, the protein synthesis of C. burnetii was inhibited using chloramphenicol (10 μg/mL) beginning at 72 hpi. Antigen was stained using the polyclonal antibodies developed against the DotA177-298 epitope (1:500), IcmX (1:250) and whole C. burnetii (1:2500) as previously described here. Monoclonal antibody against the host protein CD63 was purchased commercially (BD Biosciences, San Diego, CA). Host cell lysate, for the respective cell line, was added at a 1:100 dilution to the diluted primary antibodies and allowed to incubate at 25°C for 1 h before applying to the fixed cells. The primary antibodies were detected using antibody conjugated with either Alexa Fluor® 488, 555 or 633 all at a 1:2000 dilution and previously absorbed as described for the primary antibodies (Molecular Probes®, Carlsbad, CA). Total nucleic acid was stained with 4΄,6-diamidino-2-phenylindole (DAPI). Images were captured using a Nikon Eclipse TE 2000 SE microscope with a Nikon DS FI1 camera or a Leica DMIRB2 SP2 laser scanning confocal microscope. All images were processed equally using ImageJ version 1.46h (Rasband 1997–2012), and the Coloc 2 plugin was used to determine areas of protein colocalization (Schindelin et al. 2012).
Immunoelectron microscopy
Vero cells were seeded on sterilized ACLAR film (Ted Pella, Redding, CA) in six-well tissue culture plates and allowed to adhere at least 16 h prior to infection. The cells were infected using spinoculation as described above. At 96 hpi, all cells were fixed using a periodate-lysine-paraformaldehyde (PLP) fixative as previously described, with slight modifications (McLean and Nakane 1974; Brown and Farquhar 1989; Bannantine, Rockey and Hackstadt 1998). Briefly, media was removed from cells and 0.01M periodate, 0.075M lysine, 0.075M sodium phosphate, 1% paraformaldehyde and 0.05% glutaraldehyde were applied for 3 h at room temperature and then washed with PBS. For immunoelectron microscopy, the cells on ACLAR were infiltrated overnight in the cryoprotectant 2.3M sucrose/20% polyvinyl pyrrolidone in PIPES/MgCl2 at 4°C. To permeabilize cells for antibody labeling, cells on the ACLAR were plunge frozen in liquid nitrogen and subsequently thawed in PBS at room temperature. Samples were probed with the rabbit anti-DotA (1:500) followed by FluoroNanogold anti-rabbit Fab (1:250) (Nanoprobes, Yaphank, NY, USA) and silver enhancement for 2 to 5 min (HQ silver enhancement kit, Nanoprobes). Samples were then processed for standard resin embedding. Prelabeling experiments were conducted in parallel with controls omitting the primary antibody. These controls were consistently negative at the concentration of Nanoprobes-conjugated secondary.
RESULTS
The Coxiella burnetii T4BSS is expressed at the mRNA and protein levels during growth in ACCM
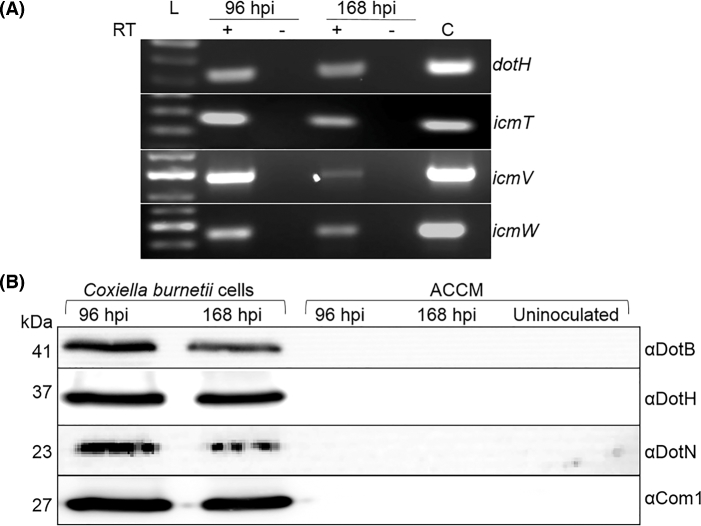
With little existing analysis of C. burnetii T4BSS gene expression during growth in ACCM, we sought to further confirm/define the expression of select homologs, which were previously confirmed using tissue culture infection (Morgan et al. 2010), in the axenic medium. Coxiella burnetii was grown to and harvested at 96 and 168 h post inoculation, which represents the mid log and stationary growth phases, respectively. These time points were selected to represent the metabolically active LCVs at 96 h post inoculation and the mixed SCV/LCV population in the 168 h post inoculation (Omsland et al. 2009), in which T4BSS transcripts and protein is most likely to be at a detectable level. Using total RNA harvested from ACCM cultures grown for 96 and 168 h post inoculation, non-quantitative RT-PCR analysis revealed that the C. burnetii T4BSS is transcribed during axenic growth (Fig. 1A). Transcripts of dotH, icmT, icmV and icmW are amplified in the plus-RT lanes and absent from the minus–RT controls (Fig. 1A). The detection of C. burnetii T4BSS proteins during ACCM growth was performed by immunoblot analysis of whole C. burnetii harvested at 96 and 168 h post inoculation. Analysis of antibody specificity showed that each of the antibodies respectively detected IcmX, DotA, DotB, DotC, DotH, DotN and Com1 at the expected molecular weight and were specific to the corresponding T4BSS protein in the NMII lysate when compared to the recombinant protein (data not shown). Using antibodies against C. burnetii T4BSS homologs shown by Vincent et al. (2006) to reside in the cytoplasm (DotB), the inner membrane (DotN) and the outer membrane (DotH), all of these proteins were found to be expressed at both time points during ACCM growth. Com1, which was previously found to be equally present for SCVs and LCVs during axenic growth, was used to normalize the loading volume between C. burnetii lysates (Omsland et al. 2008). In addition, the ACCM did not contain detectable amounts of DotB, DotH, DotN and Com1, which represents each of the cellular fractions (Fig. 1B). These results demonstrate that C. burnetii T4BSS proteins are expressed during bacterial growth in ACCM, and the absence of DotB, DotH, DotN and Com1 in the ACCM from 96 and 168 h post inoculation suggests that during growth in ACCM and preparation of the samples that the cellular structure of the bacteria is not compromised. (Fig. 1B).
Figure 1.
The C. burnetii T4BSS is expressed during growth in ACCM. (A) Total C. burnetii RNA was harvested from ACCM cultures at 96 and 168 hpi. Transcripts for dotH, icmT, icmV and icmW are detectable only when reverse transcriptase (RT) is added to the 96 and 168 hpi RT-PCR reactions. (C) Represents a chromosomal DNA PCR-positive control. (B) Total C. burnetii protein was harvested from the 96 and 168 hpi cultures. Equal volumes of the uninoculated and spent media (ACCM) from the respective cultures were retained and concentrated to equal volumes. The loading volumes of the 96 and 168 hpi cell samples were normalized using the membrane protein Com1. Predicted molecular weights (kDa) are indicated on the left. Immunoblot was used to detect the predicted cytoplasmic protein DotB, the predicted inner membrane protein DotN and the predicted outer membrane protein DotH. These proteins were detectable in all of the C. burnetii lysates and could not be detected in the ACCM.
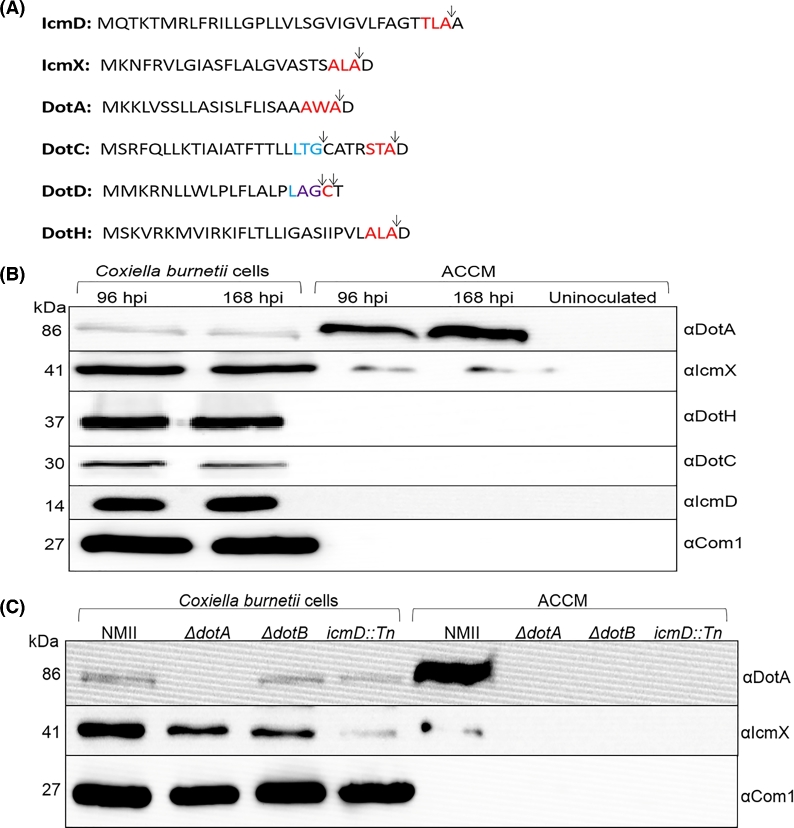
Coxiella burnetii T4BSS homologs contain secretion sequences and DotA and IcmX are secreted/released into the ACCM during growth in a T4BSS-dependent manner
With the observation of C. burnetii T4BSS transcripts and proteins being synthesized during growth in ACCM, the conditioned growth medium at 96 and 168 h post inoculation was investigated for the secretion or release of T4BSS proteins. We hypothesized that C. burnetii T4BSS components with an N-terminal secretion signal sequence could be secreted into the ACCM. In silico analysis (using SignalP 4.0) of the 23 C. burnetii T4BSS proteins revealed 6 proteins with predicted secretion sequences (Petersen et al. 2011). Coxiella burnetii T4BSS proteins with a predicted secretion signal sequence for type I signal peptidase cleavage included DotA, DotC, DotD, DotH, IcmD and IcmX while DotC and DotD had an additional secretion signal sequence for type II signal peptidase, which is associated with lipoproteins (Fig. 2A). Immunoblot analysis of these proteins, with the exception of DotD (no existing antibody), shows that IcmX and DotA are released into the ACCM during growth while DotC, DotH and IcmD could not be detected in the ACCM but were found associated with the C. burnetii lysates (Fig. 2B). To determine whether the secretion/release of C. burnetii IcmX and DotA requires a functional T4BSS, C. burnetii mutants deficient for T4B secretion were grown in ACCM and the medium was probed for IcmX and DotA. IcmX and DotA from secretion competent C. burnetii were consistently found in the ACCM, while in ΔdotA, ΔdotB and icmD::Tn C. burnetii strains (Beare et al. 2011, 2012) neither IcmX nor DotA could be detected in the media (Fig. 2C). These results demonstrate that the secretion/release of these proteins is dependent on a functional C. burnetii T4BSS.
Figure 2.
Prediction of T4BSS secretion signal sequences, detection of T4BSS components in the ACCM during growth of C. burnetii and dependence of a functional T4BSS secretion system for the release of DotA and IcmX. (A) SignalP 4.0 was used to identify N-terminal secretion signals amongst the 23 T4BSS proteins (Petersen et al. 2011). IcmD, IcmX, DotA, DotC, DotD and DotH contained a secretion signal for type I peptidase (red text). In addition, DotC and DotD also contained a secretion signal for type II peptidase (blue text). In DotD, the type I and type II signals overlapped (purple text). Arrows designate predicted cleavage location. (B)Coxiella burnetii grown in ACCM were harvested at 96 and 168 hpi and the ACCM from these cultures was retained. T4BSS components with predicted secretion signals were independently probed for by immunoblot using antibodies against DotA, IcmX, DotH, DotC and IcmD. All T4BSS components with a predicted secretion signal are detectable in the C. burnetii lysate and only DotA and IcmX are released into the ACCM. The amount of total C. burnetii protein loaded was normalized using Com1. Equal starting volumes of an uninoculated ACCM (negative control) and the 96 and 168 hpi ACCM samples were concentrated to an equal volume and equal volumes loaded. Predicted molecular weights (kDa) are indicated on the left. (C) ACCM cultures of C. burnetii NMII and the C. burnetii NMII T4BSS ΔdotA, ΔdotB and icmD::Tn mutants were grown to 168 h post inoculation and the ACCM from the respective cultures retained. Coxiella burnetii lysates were normalized to Com1 and equal volumes of the conditioned ACCM were concentrated to an equivalent volume. Proteins were detected by immunoblot using antibodies against DotA and IcmX. Predicted molecular weights (kDa) are indicated on the left. DotA and IcmX were not detectable in the ACCM from mutant strains and are only found in ACCM from the C. burnetii NMII culture and the C. burnetii cell lysates.
Coxiella burnetii DotA localizes to the host cell PV membrane and cytoplasmic vesicles during infection
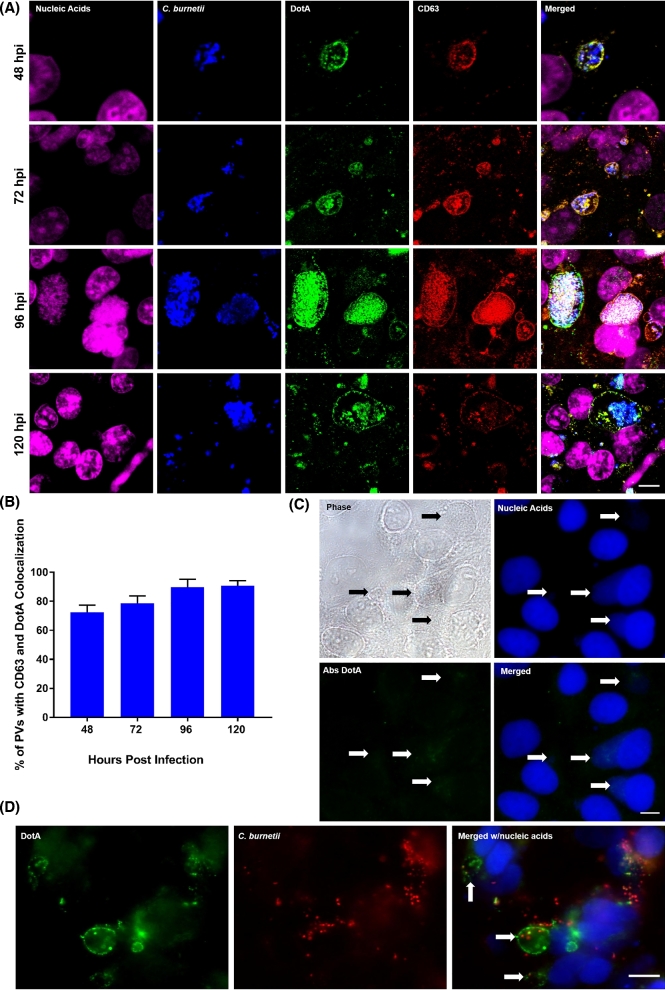
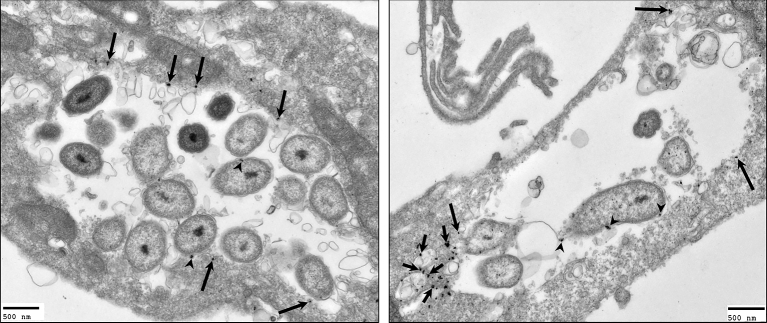
With the finding that C. burnetii IcmX and DotA are released into the conditioned ACCM, we sought to determine whether the release of IcmX and DotA occurred during C. burnetii infection of host cells. Using polyclonal antibodies against C. burnetii, IcmX and DotA, and monoclonal antibody against the established host cell PV marker, CD63, multiplex IFAs were performed on NMII-infected HeLa cells over the course of a 120-h infection. In infected cells, IcmX was not found to localize to areas away from the bacteria (data not shown). However, in the infected cells, the fluorescent signal indicating the presence of DotA localized to the PV membrane and colocalized with CD63 as early as 48 hpi and remained through 120 hpi (Fig. 3A). This colocalization was observed in ∼72%, 79%, 90% and 91% of infected cells for times 48, 72, 96 and 120 hpi, respectively (Fig. 3B). In addition, the specificity of the antibodies against DotA was observed by absorbing the reactive antibodies against rDotA. The rDotA effectively neutralized the staining of DotA and no fluorescence was detectable at the PV membrane or on the C. burnetii (Fig. 3C). To determine whether DotA secretion/release occurs during infection with virulent C. burnetii, RK13 cells were infected with NMI and IFA analysis was performed. In agreement with the C. burnetii NMII observations, DotA staining localized to the PV membrane of the infected RK13 cells (Fig. 3D). In addition, the use of RK13 cells infected with NMI demonstrates that the localization of DotA to the PV membrane is not a cell line-specific observation. To further confirm our IFA observations and gain a more detailed visualization of DotA localization in an infected host cell, IEM was utilized. Using IEM to observe DotA localization in Vero cells infected for 96 h with NMII, it was found that DotA localizes to the PV membrane and to host cell cytoplasmic structures, which are devoid of C. burnetii, that resemble multivesciular bodies (MVBs) (Fig. 4).
Figure 3.
Temporal analysis of DotA localization during infection of host cells, specificity of DotA antibody and DotA staining using virulent C. burnetii. (A) Representative fluorescence micrographs of HeLa cells infected with C. burnetii over the course of infection. Infections were initiated with C. burnetii SCVs and fixed at 48, 72, 96 and 120 hpi. Fixed cells were labeled for total nucleic acids (pseudo-colored pink), C. burnetii (pseudo-colored blue), DotA (green) and CD63 (red). DotA is detectable by 48 hpi and in the merged images remains colocalized with CD63 through the course of infection. Scale bar equals 10 μm. (B) The average number of infected cells with PVs that were positive for CD63 and DotA colocalization enumerated from 100 infected cells from three separate replicates at 48, 72, 96, and 120 hpi. The percentage of PVs with CD63 and DotA colocalization remains consistent through the course of infection. (C) Representative phase contrast and fluorescent micrograph of infected Vero cells. Vero cells were fixed at 96 hpi and probed with antibodies against DotA that were previously absorbed with rDotA (Abs DotA). Arrows indicate the PV within the infected cells. No DotA staining is observed in the infected cells. Scale bar equals 10 μm. (D) Representative fluorescence micrographs of RK13 cells infected with C. burnetii NMI. RK13 cells were fixed at 72 hpi and labeled for DotA (green), C. burnetii (red) and total nucleic acids (blue). DotA staining appears at the PV membrane (arrows) as observed with the avirulent C. burnetii NMII-infected HeLa and Vero cell lines. Scale bar equals 10 μm.
Figure 4.
Localization of DotA using immunoelectron microscopy. Infected Vero cells were fixed using a modified version of the PLP method at 96 hpi for immunoelectron microscopy. Fixed cells were probed with antibodies against DotA and counterstained with gold-conjugated secondary antibodies. The gold particles (arrows) were observed localizing to the PV membrane (long arrows), multivesicular-like bodies (short arrows) and C. burnetii (arrow heads). Scale bars equal 500 nm.
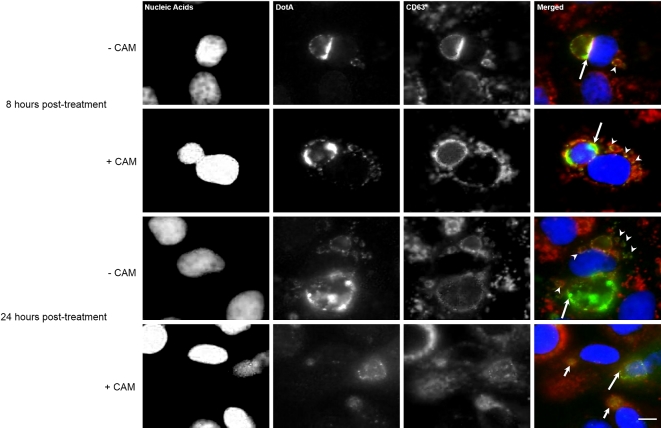
Chloramphenicol treatment of Coxiella burnetii-infected cells depletes DotA from the PV membrane and cytoplasmic vacuoles
It is known that C. burnetii protein synthesis is required for the fusogenicity of the PV with host cell vesicles, but vesicle trafficking or recycling of membrane originating from the PV has not been observed (Howe et al. 2003b). However, it is presumed to occur due to the decrease in PV size when infected cells are treated with antimicrobial agents (Howe et al. 2003b; Mahapatra, Ayoubi and Shaw 2010). Using DotA as a marker for PV-derived membrane, NMII-infected cells at 72 hpi were mock treated or treated with a bacteriostatic concentration (10 μg/mL) of chloramphenicol. By 8 h post treatment, DotA signal in both cytoplasmic vesicles and the PV was still observable regardless of treatment (Fig. 5); however, by 24 h post treatment with chloramphenicol, DotA no longer appeared to be associated with CD63-positive structures in the host cell cytoplasm and the PV membrane had reduced DotA staining (Fig. 5). Interestingly, DotA was found in the lumen of the collapsed PVs. The CD63 within the cytoplasm of the host cell returned to pre-infection distribution, resembling what has been previously reported for uninfected cells (Larson and Heinzen 2017) (Fig. 5).
Figure 5.
DotA–host cell membrane association is dependent on bacterial protein synthesis. Representative monochrome and respective merged fluorescence micrographs of Vero cells infected with C. burnetii and either mock treated (–CAM) or treated with 10 μg/mL chloramphenicol (+CAM) at 72 hpi. Top group: Infected cells were fixed 8 h after the mock treatment and chloramphenicol treatment. Nucleic acids (blue), DotA (green) and CD63 (red) were fluorescently labeled. At 8 h post treatment (hpt), DotA is visible in the PV membrane (long arrows) and cytoplasmic vesicles (arrowheads) for the both the mock and chloramphenicol-treated cells. Bottom group: Infected cells were fixed 24 h after the mock treatment and chloramphenicol treatment. Nucleic acids (blue), DotA (green) and CD63 (red) were fluorescently labeled. By 24 hpi, the +CAM cells had DotA associated only with PVs that are considered spacious (long arrow) and absent from collapsed PVs (short arrows) and also is no longer observed within the host cell cytoplasm while in the –CAM cells had DotA present in the PV (long arrow) and the cytoplasmic vesicles (arrowheads). Scale bars equal 10 μm.
DISCUSSION
The T4BSS has been shown to be an essential virulence mechanism for Legionella pneumophila and Coxiella burnetii pathogenesis (Andrews, Vogel and Isberg 1998; Beare et al. 2011; Carey et al. 2011). However, the details of assembly, functionality and structure of the T4BSS are limited when compared to what is known for T4ASSs. The knowledge gap between the two systems is primarily due to (i) a lack of significant homology between the two systems, (ii) the T4BSS is composed of twice as many protein components and (iii) the difficulties associated with culturing intracellular bacteria when compared to the genetically malleable model systems historically available for T4ASS studies (Christie et al. 2005; Nagai and Kubori 2011). The analysis of the L. pneumophila T4BSS has revealed clues to the functionality of some of the T4BSS proteins and their localization within the bacterial cell. Vincent et al. (2006) was the first to begin to detail the subcellular localization of the L. pneumophila T4BSS components and provided predictions of protein interactions thought to be involved in the formation of subcomplexes. A model outlining the predicted assembly and localization of the subcomplexes and proteins was drawn from these studies. In Vincent's model, a pilus structure was not predicted and has not been identified, leaving a question as to how effector proteins are translocated through the bacterial membranes, and across the host membrane to reach the host cell cytoplasm. To examine this question, we sought to utilize axenic cultivation of C. burnetii as a tool to analyze the extracellular localization of C. burnetii T4BSS components.
The development of an axenic growth medium for the propagation of C. burnetii has created an opportunity to understand and gain insight into the physiology and pathogenesis of this unique bacterium (Omsland et al. 2009, 2011). However, our understanding of the expression of the T4BSS components at the transcriptional and translational level during growth in ACCM is limited. Our detection of transcripts for T4BSS genes and proteins suggests that the C. burnetii T4BSS is produced during growth in ACCM. However, the functionality of the C. burnetii T4BSS during growth in ACCM remains unclear as the detection of known T4BSS effector proteins has not been reported during growth in ACCM (Carey et al. 2011) while one effector protein, which the authors suspect to be a false positive, was found in ACCM2 (Stead et al. 2013). The lack of detectable effector proteins further suggests that a host cell or environmental signal is required for the maturation and competence of the secretion system as it is in other bacterial virulence secretion systems (van der Goot et al. 2004; Epler et al. 2009; Jimenez-Soto et al. 2009). Moreover, the identification of PmrA, a part of a two-component regulatory system, as a modulator of T4BSS and effector protein expression in L. pneumophila and C. burnetii further suggests that an environmental signal(s) are involved in the development of the T4BSS (Beare et al. 2014). Indeed, multiple approximate 20 nm in length ‘needle-like structures’ have been observed by TEM that appear to extend from a C. burnetii cell towards what appears to be the PV membrane (Larson et al. 2016).
Our observation of DotA and IcmX being secreted by a T4BSS-dependent mechanism agrees with observations of this process occurring in liquid cultures of L. pneumophila (Matthews and Roy 2000; Nagai and Roy 2001). However, the functional significance of DotA secretion by L. pneumophila was debated by Vincent and Vogel (2008) who claimed that the secretion of a polytopic membrane protein from a bacterial secretion system has no precedent and that it is unclear how DotA could function as both an inner membrane protein and a T4B secretion substrate. Interestingly, our in silico detection of a conventional type I signal peptidase signal suggests that DotA could be targeted for transport across the inner membrane, and our observation of DotA being secreted into ACCM by a T4BSS-dependent mechanism agrees with observations of this process occurring in liquid cultures of L. pneumophila (Nagai and Roy 2001). With the detection of DotA in the ACCM, which was absent of detectable amounts of protein from the cytoplasm (DotB and DotN), inner membrane (DotN and IcmD), and outer membrane (DotC, DotH and Com1) (Figs 1 and 2), in combination with the observation of DotA associating with CD63-positive membrane supports the mobility of DotA beyond the inner membrane and is likely not an artifact from growth in axenic media or from sample preparation. In addition, it is unlikely that DotA is inadvertently released into the ACCM via outer membrane vesicles (OMVs) since DotA was not detected in ACCM2 by a highly sensitive method such as mass spectrometry despite the EM observation of OMVs at the C. burnetii cell surface during ACCM2 growth. (Stead et al. 2013). Moreover, OMVs were not found to be associated with the release of DotA in L. pneumophila (Nagai and Roy 2001) and the release of DotA has been used as an indicator of T4BSS function (Kuroda et al. 2015).
Additional support for the secretion of DotA in an infectious setting, by Nagai and Roy (2001), was absent, but they speculated that DotA could serve as a pore in the L. pneumophila-containing vacuole membrane for the translocation of effector proteins into the host cytoplasm. With the drastic difference in infectious cycle rates between L. pneumophila and C. burnetii, it is possible that cells infected with L. pneumophila would not have detectable levels of DotA in the vacuolar membrane prior to host cell lysis as we were not able to clearly detect DotA in the PV membrane until 48 hpi (Alli et al. 2000). In addition, the cellular fixation method used by Nagai and Roy could have masked the anti-DotA immunoreactive epitopes (Kakuk et al. 2006). We found that using a standard paraformaldehyde with detergent fixation method (Morgan, Luedtke and Shaw 2010) did not provide a distinct fluorescent signal, which suggests an inability of the anti-DotA antibodies to find the corresponding eptiopes (data not shown). It is reasonable to believe that the use of methanol to fix the infected cells for IFA would extract lipids from the PV membrane and MVBs and denature proteins, which would expose the immunoreactive epitopes (DiDonato and Brasaemle 2003). Moreover, for TEM, the best DotA detection was done using the alternative PLP fixation method, which can be used to cross-link proteins bound in membrane (Brown and Farquhar 1989) and was also found to provide the best results for staining the Chlamydia psittaci IncC protein that associates with the inclusion membrane (Bannantine, Rockey and Hackstadt 1998). IcmX was also found to be secreted within tissue culture cells, but the detection of the protein in a localized area of the cell could not be elucidated (Matthews and Roy 2000). We were also unable to detect IcmX that was not localized to C. burnetii during infection. This could be due to a proteolysis of IcmX in the PV or insufficient localized antigen to produce a detectable signal. Further research is required to understand the importance of the secretion of IcmX in relation to T4BSS functionality and pathogenesis.
These data provide evidence that a component of the T4BSS interacts at the PV membrane during infection. The observation of DotA colocalizing with CD63 at the PV membrane suggests that DotA engages the membrane and maybe capable of membrane insertion. However, at this time it is not clear if the interaction of DotA with the PV membrane is dependent on C. burnetii associating directly with the PV membrane. Indeed, some T4SSs are dependent on contact with a host cell for the structural alteration of the secretion complex (Hayes, Aoki and Low 2010). With the previous visualization of DotA as a hollow ring structure (Nagai and Roy 2001), it could be hypothesized that DotA and IcmX form a T4BSS translocon to allow the transfer of effector proteins across the PV membrane.
The host cell process of trafficking endocytic and autophagic vesicles is important for degrading foreign particles as well as damaged or unnecessary cellular components for recycling within the cell. Some intracellular parasites have evolved mechanisms to alter vesicle trafficking as a way to form a replicative niche and/or acquire essential nutrients (Hackstadt 2000; Wang, Weiss and Orlofsky 2009). During infection, the C. burnetii PV is highly fusogenic with autophagic and endocytic vesicles; however, the trafficking of PV membrane away from the PV has not been described (Howe et al. 2003a; Romano et al. 2007). Our observation of DotA colocalization with CD63-positive vesicles, which do not contain C. burnetii, in the host cytoplasm by 72 hpi suggests that membrane and/or components from the PV lumen are entering a host cell membrane recycling pathway as transferrin receptor and clathrin associate with the PV (Latomanski et al. 2016; Larson and Heinzen 2017). Moreover, this could also explain the previous observation of increased CD63-positive vesicles in the host cell cytoplasm during infection (Larson and Heinzen 2017). However, it is not clear which host protein(s) or C. burnetii protein(s) are involved in this process, but it is likely that this process is C. burnetii driven as after the 24-h treatment with a bacteriostatic concentration of chloramphenicol, the infected cells had no detectable DotA colocalization with CD63-positive vesicles in the cytoplasm, and agrees with previous studies showing C. burnetii alters the normal host cell vesicle trafficking, recycling, and fusion for PV development and maintenance (Larson et al. 2013, 2015; Latomanski et al. 2016; Larson and Heinzen 2017).
The T4BSS-dependent development and maintenance of the PV is essential for C. burnetii pathogenesis. Our use of ACCM and tissue culture to characterize the localization of the C. burnetii T4BSS has provided data that demonstrates an array of C. burnetii T4BSS genes are expressed at the mRNA and protein level during growth in ACCM. The analysis of C. burnetii T4BSS homologs containing a predicted secretion signal determined that C. burnetii IcmX and DotA are released into the ACCM in a manner that is dependent on a functional T4BSS. Here we showed C. burnetii T4BSS proteins being released during growth in liquid media as well as during infection of host cells. Moreover, we demonstrate that a T4BSS protein colocalizes with the host cell/PV membrane during infection. In addition, we show that DotA can be found associated with MVBs/vesicles that are devoid of C. burnetii and located away from the PV. This finding may allow DotA to be used as a marker of PV-derived membrane for trafficking/recycling studies in infected host cells and could provide a better understanding of how C. burnetii interacts with the host cell from within the membrane-bound PV.
Acknowledgments
We wish to thank Dr Anders Omsland, Rocky Mountain Laboratories, Dr Dan Voth, University of Arkansas College of Medicine, for technical expertise and advice on the in vitro growth of C. burnetii, and Dr Wandy Beatty Washington University in St. Louis School of Medicine, for expertise and processing of samples for immunoelectron microscopy.
FUNDING
This research was supported by National Institutes of Health grant A1072710 (EIS).
Conflict of interest. None declared.
REFERENCES
- Alli OA, Gao LY, Pedersen LL et al. Temporal pore formation-mediated egress from macrophages and alveolar epithelial cells by Legionella pneumophila. Infect Immun 2000;68:6431–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews HL, Vogel JP, Isberg RR. Identification of linked Legionella pneumophila genes essential for intracellular growth and evasion of the endocytic pathway. Infect Immun 1998;66:950–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bannantine JP, Rockey DD, Hackstadt T. Tandem genes of Chlamydia psittaci that encode proteins localized to the inclusion membrane. Mol Microbiol 1998;28:1017–26. [DOI] [PubMed] [Google Scholar]
- Bardill JP, Miller JL, Vogel JP. IcmS-dependent translocation of SdeA into macrophages by the Legionella pneumophila type IV secretion system. Mol Microbiol 2005;56:90–103. [DOI] [PubMed] [Google Scholar]
- Beare PA, Gilk SD, Larson CL et al. Dot/Icm type IVB secretion system requirements for Coxiella burnetii growth in human macrophages. MBio 2011;2:e00175–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beare PA, Larson CL, Gilk SD et al. Two systems for targeted gene deletion in Coxiella burnetii. Appl Environ Microb 2012;78:4580–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beare PA, Sandoz KM, Larson CL et al. Essential role for the response regulator PmrA in Coxiella burnetii type 4B secretion and colonization of mammalian host cells. J Bacteriol 2014, DOI: 10.1128/JB.01532-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beron W, Gutierrez MG, Rabinovitch M et al. Coxiella burnetii localizes in a Rab7-labeled compartment with autophagic characteristics. Infect Immun 2002;70:5816–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown WJ, Farquhar MG. Immunoperoxidase methods for the localization of antigens in cultured cells and tissue sections by electron microscopy. Methods Cell Biol 1989;31:553–69. [DOI] [PubMed] [Google Scholar]
- Carey KL, Newton HJ, Lührmann A et al. The Coxiella burnetii Dot/Icm system delivers a unique repertoire of type IV effectors into host cells and is required for intracellular replication. PLoS Pathog 2011;7:e1002056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C, Banga S, Mertens K et al. Large-scale identification and translocation of type IV secretion substrates by Coxiella burnetii. P Natl Acad Sci USA 2010;107:21755–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christie PJ, Atmakuri K, Krishnamoorthy V et al. Biogenesis, architecture, and function of bacterial type IV secretion systems. Annu Rev Microbiol 2005;59:451–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coleman SA, Fischer ER, Howe D et al. Temporal analysis of Coxiella burnetii morphological differentiation. J Bacteriol 2004;186:7344–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DebRoy S, Dao J, Soderberg M et al. Legionella pneumophila type II secretome reveals unique exoproteins and a chitinase that promotes bacterial persistence in the lung. P Natl Acad Sci USA 2006;103:19146–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiDonato D, Brasaemle DL. Fixation methods for the study of lipid droplets by immunofluorescence microscopy. J Histochem Cytochem 2003;51:773–80. [DOI] [PubMed] [Google Scholar]
- Epler CR, Dickenson NE, Olive AJ et al. Liposomes recruit IpaC to the Shigella flexneri type III secretion apparatus needle as a final step in secretion induction. Infect Immun 2009;77:2754–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosal D, Chang YW, Jeong KC et al. In situ structure of the Legionella Dot/Icm type IV secretion system by electron cryotomography. EMBO Rep 2017, DOI: 10.15252/embr.201643598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hackstadt T. Redirection of host vesicle trafficking pathways by intracellular parasites. Traffic 2000;1:93–9. [DOI] [PubMed] [Google Scholar]
- Hayes CS, Aoki SK, Low DA. Bacterial contact-dependent delivery systems. Annu Rev Genet 2010;44:71–90. [DOI] [PubMed] [Google Scholar]
- Heinzen RA, Hackstadt T, Samuel JE. Developmental biology of Coxiella burnettii. Trends Microbiol 1999;7:149–54. [DOI] [PubMed] [Google Scholar]
- Heinzen RA, Scidmore MA, Rockey DD et al. Differential interaction with endocytic and exocytic pathways distinguish parasitophorous vacuoles of Coxiella burnetii and Chlamydia trachomatis. Infect Immun 1996;64:796–809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howe D, Heinzen RA. Coxiella burnetii inhabits a cholesterol-rich vacuole and influences cellular cholesterol metabolism. Cell Microbiol 2006;8:496–507. [DOI] [PubMed] [Google Scholar]
- Howe D, Mallavia LP. Coxiella burnetii exhibits morphological change and delays phagolysosomal fusion after internalization by J774A.1 cells. Infect Immun 2000;68:3815–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howe D, Melnicakova J, Barak I et al. Fusogenicity of the Coxiella burnetii parasitophorous vacuole. Ann N Y Acad Sci 2003a;990:556–62. [DOI] [PubMed] [Google Scholar]
- Howe D, Melnicakova J, Barak I et al. Maturation of the Coxiella burnetii parasitophorous vacuole requires bacterial protein synthesis but not replication. Cell Microbiol 2003b;5:469–80. [DOI] [PubMed] [Google Scholar]
- Jimenez-Soto LF, Kutter S, Sewald X et al. Helicobacter pylori type IV secretion apparatus exploits beta1 integrin in a novel RGD-independent manner. PLoS Pathog 2009;5:e1000684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kakuk A, Friedlander E, Vereb G Jr et al. Nucleolar localization of phosphatidylinositol 4-kinase PI4K230 in various mammalian cells. Cytometry A 2006;69:1174–83. [DOI] [PubMed] [Google Scholar]
- Kubori T, Koike M, Bui XT et al. Native structure of a type IV secretion system core complex essential for Legionella pathogenesis. P Natl Acad Sci USA 2014;111:11804–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuroda T, Kubori T, Thanh Bui X et al. Molecular and structural analysis of Legionella DotI gives insights into an inner membrane complex essential for type IV secretion. Sci Rep 2015;5:10912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larson CL, Beare PA, Howe D et al. Coxiella burnetii effector protein subverts clathrin-mediated vesicular trafficking for pathogen vacuole biogenesis. P Natl Acad Sci USA 2013;110:E4770–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larson CL, Beare PA, Voth DE et al. Coxiella burnetii effector proteins that localize to the parasitophorous vacuole membrane promote intracellular replication. Infect Immun 2015;83:661–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larson CL, Heinzen RA. High-content imaging reveals expansion of the endosomal compartment during Coxiella burnetii parasitophorous vaculole maturation. Front Cell Infect Microbiol 2017, DOI: 10.3389/fcimb.2017.00048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larson CL, Martinez E, Beare PA et al. Right on Q: genetics begin to unravel Coxiella burnetii host cell interactions. Future Microbiol 2016;11:919–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latomanski EA, Newton P, Khoo CA et al. The effector Cig57 Hijacks FCHO-Mediated vesicular trafficking to facilitate intracellular replication of Coxiella burnetii. PLoS Pathog 2016;12:e1006101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLean IW, Nakane PK. Periodate-lysine-paraformaldehyde fixative. A new fixation for immunoelectron microscopy. J Histochem Cytochem 1974;22:1077–83. [DOI] [PubMed] [Google Scholar]
- Mahapatra S, Ayoubi P, Shaw EI. Coxiella burnetii Nine Mile II proteins modulate gene expression of monocytic host cells during infection. BMC Microbiol 2010;10:244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marrie TJ. Q fever pneumonia. Curr Opin Infect Dis 2004;17:137–42. [DOI] [PubMed] [Google Scholar]
- Martinez E, Cantet F, Fava L et al. Identification of OmpA, a Coxiella burnetii protein involved in host cell invasion, by multi-phenotypic high-content screening. PLoS Pathog 2014;10:e1004013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthews M, Roy CR. Identification and subcellular localization of the Legionella pneumophila IcmX protein: a factor essential for establishment of a replicative organelle in eukaryotic host cells. Infect Immun 2000;68:3971–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan JK, Luedtke BE, Shaw EI. Polar localization of the Coxiella burnetii type IVB secretion system. FEMS Microbiol Lett 2010;305:177–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan JK, Luedtke BE, Thompson HA et al. Coxiella burnetii type IVB secretion system region I genes are expressed early during the infection of host cells. FEMS Microbiol Lett 2010;311:61–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagai H, Kubori T. Type IVB secretion systems of Legionella and other Gram-negative bacteria. Front Microbiol 2011;2:136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagai H, Roy CR. The DotA protein from Legionella pneumophila is secreted by a novel process that requires the Dot/Icm transporter. Embo J 2001;20:5962–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omsland A, Beare PA, Hill J et al. Isolation from animal tissue and genetic transformation of Coxiella burnetii are facilitated by an improved axenic growth medium. Appl Environ Microb 2011;77:3720–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omsland A, Cockrell DC, Fischer ER et al. Sustained axenic metabolic activity by the obligate intracellular bacterium Coxiella burnetii. J Bacteriol 2008;190:3203–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omsland A, Cockrell DC, Howe D et al. Host cell-free growth of the Q fever bacterium Coxiella burnetii. P Natl Acad Sci USA 2009;106:4430–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omsland A, Heinzen RA. Life on the outside: the rescue of Coxiella burnetii from its host cell. Annu Rev Microbiol 2011;65:111–28. [DOI] [PubMed] [Google Scholar]
- Pan X, Luhrmann A, Satoh A et al. Ankyrin repeat proteins comprise a diverse family of bacterial type IV effectors. Science 2008;320:1651–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen TN, Brunak S, von Heijne G et al. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat Methods 2011;8:785–6. [DOI] [PubMed] [Google Scholar]
- Rasband WS. ImageJ, 1.46h edn Bethesda, MD: U.S. National Institutes of Health, 1997–2012. [Google Scholar]
- Romano PS, Gutierrez MG, Beron W et al. The autophagic pathway is actively modulated by phase II Coxiella burnetii to efficiently replicate in the host cell. Cell Microbiol 2007;9:891–909. [DOI] [PubMed] [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E et al. Fiji: an open-source platform for biological-image analysis. Nat Methods 2012;9:676–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segal G, Shuman HA. Characterization of a new region required for macrophage killing by Legionella pneumophila. Infect Immun 1997;65:5057–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seshadri R, Paulsen IT, Eisen JA et al. Complete genome sequence of the Q-fever pathogen Coxiella burnetii. P Natl Acad Sci USA 2003;100:5455–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stead CM, Omsland A, Beare PA et al. Sec-mediated secretion by Coxiella burnetii. BMC Microbiol 2013;13:222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Goot FG, Tran van Nhieu G, Allaoui A et al. Rafts can trigger contact-mediated secretion of bacterial effectors via a lipid-based mechanism. J Biol Chem 2004;279:47792–8. [DOI] [PubMed] [Google Scholar]
- Vincent CD, Friedman JR, Jeong KC et al. Identification of the core transmembrane complex of the Legionella Dot/Icm type IV secretion system. Mol Microbiol 2006;62:1278–91. [DOI] [PubMed] [Google Scholar]
- Vincent CD, Vogel JP. The Dot/Icm type IVB secretion system of Legionella. In: Klaus H, Swanson M (eds.) Legionella: Molecular Microbiology, Poole, UK: Horizon Scientific Press, 2008, 165–79. [Google Scholar]
- Voth DE, Heinzen RA. Lounging in a lysosome: the intracellular lifestyle of Coxiella burnetii. Cell Microbiol 2007;9:829–40. [DOI] [PubMed] [Google Scholar]
- Wang Y, Weiss LM, Orlofsky A. Host cell autophagy is induced by Toxoplasma gondii and contributes to parasite growth. J Biol Chem 2009;284:1694–701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wike DA, Tallent G, Peacock MG et al. Studies of the rickettsial plaque assay technique. Infect Immun 1972;5:715–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamboni DS, McGrath S, Rabinovitch M et al. Coxiella burnetii express type IV secretion system proteins that function similarly to components of the Legionella pneumophila Dot/Icm system. Mol Microbiol 2003;49:965–76. [DOI] [PubMed] [Google Scholar]
- Zusman T, Yerushalmi G, Segal G. Functional similarities between the icm/dot pathogenesis systems of Coxiella burnetii and Legionella pneumophila. Infect Immun 2003;71:3714–23. [DOI] [PMC free article] [PubMed] [Google Scholar]