The major complications of an ongoing Zika virus outbreak in the Americas and Asia are congenital defects caused by the virus’s ability to cross the placenta and infect the fetal brain. The ability to generate molecular tools to analyze viral isolates from the current outbreak is essential for furthering our understanding of how these viruses cause congenital defects. The majority of existing viral isolates and infectious cDNA clones generated from them have undergone various numbers of passages in cell culture and/or suckling mice, which is likely to result in the accumulation of adaptive mutations that may affect viral properties. The approach described herein allows rapid generation of new, fully functional Zika virus isolates directly from deep sequencing data from virus-infected tissues without the need for prior virus passaging and for the generation and propagation of full-length cDNA clones. The approach should be applicable to other medically important flaviviruses and perhaps other positive-strand RNA viruses.
KEYWORDS: infectious DNA, Zika virus, flavivirus, mouse model
ABSTRACT
Zika virus (ZIKV) has recently emerged and is the etiological agent of congenital Zika syndrome (CZS), a spectrum of congenital abnormalities arising from neural tissue infections in utero. Herein, we describe the de novo generation of a new ZIKV isolate, ZIKVNatal, using a modified circular polymerase extension reaction protocol and sequence data obtained from a ZIKV-infected fetus with microcephaly. ZIKVNatal thus has no laboratory passage history and is unequivocally associated with CZS. ZIKVNatal could be used to establish a fetal brain infection model in IFNAR−/− mice (including intrauterine growth restriction) without causing symptomatic infections in dams. ZIKVNatal was also able to be transmitted by Aedes aegypti mosquitoes. ZIKVNatal thus retains key aspects of circulating pathogenic ZIKVs and illustrates a novel methodology for obtaining an authentic functional viral isolate by using data from deep sequencing of infected tissues.
IMPORTANCE The major complications of an ongoing Zika virus outbreak in the Americas and Asia are congenital defects caused by the virus’s ability to cross the placenta and infect the fetal brain. The ability to generate molecular tools to analyze viral isolates from the current outbreak is essential for furthering our understanding of how these viruses cause congenital defects. The majority of existing viral isolates and infectious cDNA clones generated from them have undergone various numbers of passages in cell culture and/or suckling mice, which is likely to result in the accumulation of adaptive mutations that may affect viral properties. The approach described herein allows rapid generation of new, fully functional Zika virus isolates directly from deep sequencing data from virus-infected tissues without the need for prior virus passaging and for the generation and propagation of full-length cDNA clones. The approach should be applicable to other medically important flaviviruses and perhaps other positive-strand RNA viruses.
INTRODUCTION
Zika virus (ZIKV) is a mosquito-borne flavivirus that has recently reemerged, with transmission now reported in >70 countries and territories (1). In 2016, the World Health Organization declared the ZIKV pandemic a public health emergency of international concern (2). In humans, an estimated 80% of primary infections with ZIKV appear to be asymptomatic (3), with the majority of symptomatic cases showing mild disease (including rash, conjunctivitis, arthralgia, myalgia, and fever), although infection can occasionally lead to Guillain-Barré syndrome (1, 3, 4). The primary concern is infection of pregnant mothers, which can lead to the virus infecting (and thereby damaging) the fetal brain, resulting in a range of congenital birth defects now recognized as congenital Zika syndrome (CZS) (1, 3). In a recent study of symptomatic mothers, abnormal clinical or brain imaging findings were seen in 55% of the infants born to mothers infected in the first trimester, 52% of the infants born to mothers infected in the second trimester, and 29% of the infants born to mothers infected in the third trimester (5). In that study, microcephaly was evident in 3.4% of the neonates examined, with microcephaly usually associated with fetal growth restriction. There is also emerging evidence that children who were infected in utero but showed no overt congenital defects at birth manifest a range of disabilities over time (1, 5, 6).
One of the first cases of ZIKV-associated severe microcephaly was identified in 2015 in an aborted fetus from a mother who was infected with ZIKV during the first trimester of pregnancy while living in the Natal region of Brazil (7). Viral RNA, viral proteins, and viral particles were detected in fetal brain tissues (7), and a complete genome sequence of the virus was obtained by next-generation sequencing of brain tissue (GenBank accession number KU527068). This ZIKVNatal sequence therefore represents a ZIKV sequence unequivocally associated with a human case of microcephaly and is free of any potential adaptive mutations arising from serial passage, for instance, in cells in vitro or in suckling mice.
Flavivirus infectious cDNA clones have facilitated numerous scientific discoveries over the years; however, the well-documented instability of plasmids harboring full-length flavivirus cDNA sequences during propagation in bacteria remains an issue. Recently, three groups have independently reported that the toxicity of ZIKV sequences has hindered the stable propagation in bacteria of plasmid DNAs harboring full-length ZIKV cDNA (8–10), with insertion of introns required to resolve the issue (9, 10).
We have recently developed and optimized a protocol to generate de novo infectious flaviviruses that does not involve the generation of DNA plasmids encoding the complete viral genome or in vitro RNA transcription. The method, based on the circular polymerase extension cloning protocols described previously (11), significantly simplifies and accelerates the process of de novo virus generation (12, 13). The protocol involves the generation of a circular DNA (with high-fidelity polymerase) that encompasses the entire viral cDNA sequence in vitro. A cytomegalovirus (CMV) promoter sits directly upstream of the first nucleotide of viral cDNA, and following the transfection of circular polymerase extension reaction (CPER) products into mammalian cells, viral RNA is synthesized and infectious virus is recovered. The protocol has been successfully used to generate wild-type (WT) and mutant West Nile viruses (13), chimeric viruses consisting of different West Nile virus strains (12), and chimeric viruses consisting of West Nile virus and Brazilian Rocio virus (14).
Herein, we describe the application of a modified CPER protocol to generate an infectious ZIKVNatal isolate de novo from a published sequence. We describe the behavior of this new isolate in vitro, in mouse models, and in Aedes aegypti mosquitoes.
RESULTS
Recovery of infectious ZIKVNatal from the published sequence.
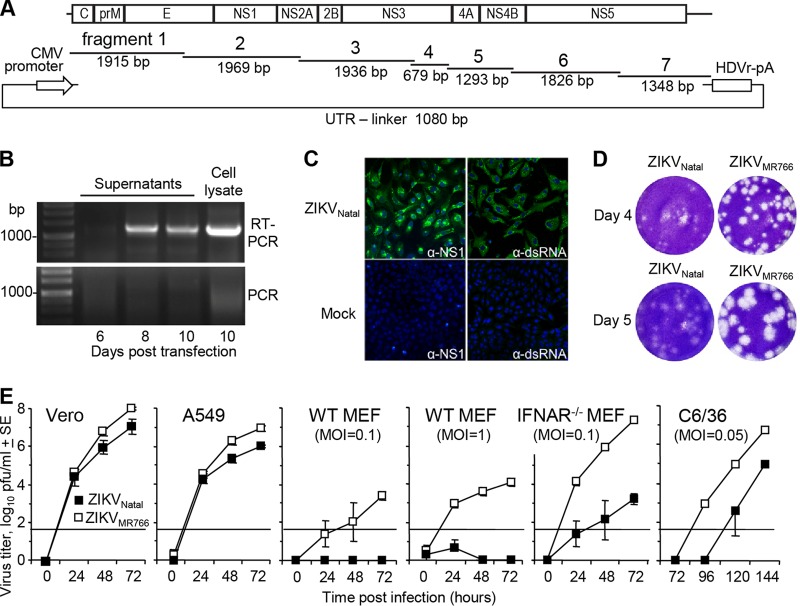
The sequence of ZIKVNatal (derived by deep sequencing of an infected fetal brain [7]) was obtained from GenBank (accession number KU527068). Seven overlapping double-stranded DNA (dsDNA) fragments covering the entire viral sequence (Fig. 1A) were cloned into pUC19 vectors. An eighth pUC19 plasmid contained the untranslated region (UTR) linker, which comprised the minimal CMV promoter and the first 22 nucleotides (nt) of the viral sequence at one end and the last 22 nt of the viral sequence, the hepatitis delta virus ribozyme (HDVr) site, and a polyadenylation (pA) signal at the other (Fig. 1A). The CMV promoter initiates viral RNA transcription, and the HDVr sequence ensures authentic formation of the 3′ UTR. These plasmids were then used to generate cDNA fragments by PCR, with the resulting eight dsDNA fragments mixed in equimolar amounts and subjected to 12 cycles of CPER with Q5 DNA polymerase. The CPER products were then transfected into Vero cells, and on days 6, 8, and 10 after transfection, the tissue culture supernatants were harvested. The presence of viral RNA in the supernatants was demonstrated by reverse transcription (RT)-PCR of isolated RNA with fragment 5-specific primers, with the minus RT control showing no bands (Fig. 1B). Further confirmation of virus recovery was obtained by immunofluorescent-antibody staining of Vero cells with anti-NS1 and anti-dsRNA antibodies 3 days after infection with the day 10 supernatant (Fig. 1C).
FIG 1 .
De novo generation of infectious ZIKVNatal by CPER. (A) Schematic representation of CPER assembly with seven synthetic DNA fragments covering the entire genome of ZIKVNatal with ~22-nt overlapping ends. The fragments were mixed with the UTR linker, which contained the CMV promoter followed by the first 22 nt of ZIKVNatal and, at the other end, the last 22 nt of ZIKVNatal, an HDVr site, and a poly(A) tail (pA). After CPER, the products were transfected into Vero cells. (B) On the posttransfection days indicated, RNA was isolated from Vero cell supernatants and a cell lysate and subjected to RT-PCR with fragment 5-specific primers (RT-PCR). To demonstrate that the bands were not due to contaminating DNA, a control (RT minus) PCR was performed in parallel (PCR). (C) Vero cells were infected with ZIKVNatal (culture supernatant from day 10 posttransfection), and mock-infected cells were used as controls. At 3 days postinfection, Vero cells were analyzed by immunofluorescent-antibody staining with anti-NS1 (4G4) and anti-dsRNA (3G1) antibodies (green) and DAPI counterstain (blue). (D) Plaque morphology following infection of Vero cells with ZIKVNatal and ZIKVMR766. Vero cells were fixed and stained with crystal violet on the postinfection days indicated. (E) Growth kinetics of ZIKVNatal and ZIKVMR766 in the cell lines indicated. Infection was performed at an MOI of 0.1, unless otherwise indicated. Virus titers in the culture supernatants were determined by plaque assays on Vero cells. The data and standard errors (SE) shown are from six independent experiments with Vero cells, A549 cells, and WT MEFs (MOI of 1) and three independent experiments with WT MEFs (MOI of 0.1), IFNAR−/− MEFs, and C6/36 cells. The horizontal line represents the limit of detection (50 PFU/ml).
The viral titer in the day 10 supernatant was 5 × 105 PFU/ml, as determined by plaque assay on Vero cells. This virus (passage 0) was amplified in C6/36 cells in two independent expansions to produce passage 1 virus stocks that were used in subsequent experiments, i.e., (i) infection at a multiplicity of infection (MOI) of 0.001, reaching 7.6 log10 50% cell culture infective doses (CCID50) by day 5, and (ii) infection at an MOI of 0.003, reaching 4.5 × 107 PFU/ml on day 6 and 1.25 × 108 PFU/ml on day 8. In a separate experiment, a different DNA polymerase, PrimeSTAR GXL, was used to generate circular DNA by CPER. Transfection of this DNA resulted in the recovery of higher virus titers by day 10 after transfection (passage 0, 2.6 × 107 PFU/ml). These results illustrate that the modified CPER protocol can efficiently produce high titers of ZIKV either directly after transfection or after a single passage.
Sequencing of ZIKVNatal.
RNA from virus recovered from day 10 supernatants (Fig. 1B) was used to generate amplicons by RT-PCR, which were then subjected to deep sequencing with the Illumina MiSeq platform. A total of 451,583 reads were mapped to 100% of the genome, with a mean read coverage of 5,734. The sequencing quality scores were 83.4% at Q20 and 76.2% at Q30. The only changes from the consensus published sequence (accession no. KU527068) were polymorphisms at nt 4328 at the start of the NS2B gene (C to T; a read coverage, 9,170, 83% T, 14% C); nt 4574 at the end of the NS2B gene (A to G; read coverage, 10,322; 98% G); and nt 5900 in the NS3 gene (C to T; read coverage, 3,960; 47% C, 51% T). None of these nucleotide changes introduced amino acid changes. The remaining sequence matched the published sequence.
In vitro growth properties of recovered ZIKVNatal compared with those of the prototype African MR766 isolate.
The plaque morphology of ZIKVNatal was compared with that of the prototype mouse-adapted African isolate ZIKVMR766 in Vero cells. The plaques generated by ZIKVNatal were generally smaller and less distinct than those produced by ZIKVMR766 (Fig. 1D).
Analysis of growth kinetics revealed that ZIKVNatal replicated less efficiently than ZIKVMR766 in Vero, A549, and C6/36 cells and in IFNAR−/− mouse embryonic fibroblasts (MEFs) infected at an MOI of 0.1 (determined on the basis of viral titers in Vero cells) (Fig. 1E). ZIKVNatal was unable to replicate efficiently in WT MEFs infected at an MOI of 0.1 or 1 on the basis of viral titers determined in Vero cells (Fig. 1E, WT MEFs). In contrast, ZIKVMR766 did replicate in these cells, perhaps reflecting the fact that ZIKVMR766 has been mouse adapted, with more than 100 serial passages in mouse brains (15, 16). ZIKVMR766 also grew substantially better in IFNAR−/− MEFs than ZIKVNatal did (Fig. 1E, IFNAR−/− MEFs).
ZIKVNatal infection of WT, IFNAR−/−, and IRF7−/− mice.
ZIKV infection of WT mice generally does not produce detectable viremia (17). We similarly were unable to detect viremia in WT (C57BL/6J) mice after subcutaneous (s.c.) infection with ZIKVMR766 or ZIKVNatal by CCID50 assays. Even intravaginal infection (18) with a dose of 6 log10 CCID50 of ZIKVNatal failed to produce detectable viremia (data not shown).
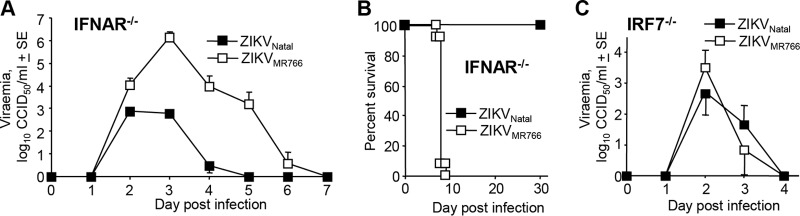
A number of mouse ZIKV models have used IFNAR−/− mice (17), and ZIKVNatal (103 CCID50 s.c.) produced a 4- to 5-day viremia in IFNAR−/− mice that was ~1 to 4 logs lower than that produced by infection with the same dose of ZIKVMR766 (Fig. 2A). Increasing the s.c. ZIKVNatal infection dose to 6 log10 CCID50 increased the peak viremia by 2 to 3 logs (see Fig. S1A in the supplemental material).
FIG 2 .
ZIKVNatal infection of IFNAR−/− and IRF7−/− mice. (A) Viremia in IFNAR−/− mice (female, 8 to 12 weeks old) after infection with ZIKVNatal or ZIKVMR766 (s.c., 3 log10 CCID50), determined by CCID50 assays (n = 5 for ZIKVNatal, n = 11 for ZIKVMR766). Limit of detection, 2 log10 CCID50. (B) Survival of IFNAR−/− mice (female, 8 to 12 weeks old) after s.c. infection with ZIKVNatal or ZIKVMR766. The dose of ZIKVMR766 was 103 CCID50 (n = 13). A range of viral doses of ZIKVNatal were tested, i.e., 103 CCID50 (n = 8), 104 CCID50 (n = 5), 105 CCID50 (n = 5), and 106 CCID50 (n = 6), with no mice requiring euthanasia. (C) Viremia in IRF7−/− mice (female, 8 to 12 weeks old) after infection with ZIKVNatal or ZIKVMR766 (s.c., 3 log10 CCID50) (n = 3 to 5). SE, standard error.
(A). ZIKVNatal infection (s.c., 6 log10 CCID50) of IFNAR−/− and IRF7−/− mice (>7 weeks old). No clinical symptoms were observed. n = 5 per group (B). Survival of IFNAR−/− mice after ZIKVNatal infection of young mice and i.p. inoculation (n = 3 for 4-week-old mice; n = 5 for the remaining groups). Mice were euthanized when hind leg weakness reached an ethically defined level of severity. Groups with deaths also showed 1 to 2 days of mild hunching (C). Infection of male IFNAR−/− mice (8 to 12 weeks old) with 5 log10 CCID50 of ZIKVNatal s.c. (n = 3). Testes were tested on day 7, and the titer is indicated by the red data point. Download FIG S1, TIF file, 0.5 MB (549.7KB, tif) .
Copyright © 2017 Setoh et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
ZIKVMR766 infection of IFNAR−/− mice was always highly symptomatic, and all mice reached ethically defined endpoints requiring euthanasia by days 7 to 9 (Fig. 2B), consistent with previous reports (17, 19). In contrast, s.c. ZIKVNatal infection of female IFNAR−/− mice (>8 weeks of age) with a range of viral inoculation doses (3 to 6 log10 CCID50) resulted in a 100% survival rate, with no animals displaying any symptoms (Fig. 2B). Nevertheless, ZIKVNatal could produce symptomatic infections in IFNAR−/− mice requiring euthanasia in 30 to 40% of the animals when (i) 4-week-old IFNAR−/− mice were infected s.c. with 3 log10 CCID50 (consistent with age-dependent susceptibility in flavivirus models [20]) or when (ii) 8- to 12-week-old mice were infected intraperitoneally with 5 log10 CCID50 (see Fig. S1B).
ZIKVNatal and ZIKVMR766 infections of IRF7−/− mice were also examined, with both viruses producing detectable viremia (Fig. 2C). Although mild hunching was observed for 1 to 2 days after ZIKVMR766 infection, no IRF7−/− mice required euthanasia after s.c. infection with either virus at 3 log10 CCID50. Infection of IRF7−/− mice with increasing s.c. doses of ZIKVNatal ranging from 3 (Fig. 3C) to 6 log10 CCID50 brought forward but did not significantly increase the peak viremia, and the mice did not show any symptoms (see Fig. S1A).
FIG 3 .
Pregnancy outcomes of ZIKVNatal-infected IFNAR−/− dams. (A) Examples of E17.5/E18.5 fetuses from IFNAR−/− × IFNAR−/− mating after s.c. infection of dams (>8 weeks of age) at E6 (or E12.5 [*]) with the ZIKVNatal doses indicated. Fetuses with clear signs of IUGR are shown facing left. *, Severely deformed fetus and placenta. Severely deformed fetal/placental masses are also shown in the right two images in row 10^5 and all of the images in row 10^6. (B) The percentages of fetuses that appeared normal, showed IUGR, and were severally deformed are shown. Data are from three pregnancies per group (mean of 7.78 ± 1.55 [standard deviation] fetuses per pregnancy). White bars indicate that one (33%) or two (66%) of the dams aborted. (C) Placenta and head virus titers of fetuses after large- and small-dose ZIKVNatal infection of dams at E6 or E12.5. *, Fetus with IUGR shown at the top right of panel A; **, severally deformed fetal/placental masses. ND, not detected.
ZIKVNatal fetal infection of IFNAR−/− dams.
Highly symptomatic infection of dams complicates the ability to establish an ethically acceptable fetal ZIKV infection model. As s.c. ZIKVNatal infection of IFNAR−/− mice >8 weeks old was asymptomatic, these conditions were used to infect pregnant dams at embryonic day 6.5 (E6.5; early pregnancy) and E12.5 (mid pregnancy), nominally the first and second trimesters (21), with a range of ZIKVNatal doses. Dams and fetuses were euthanized at E17.5/E18.5, and (i) fetuses were photographed (Fig. 3A), (ii) the pregnancy outcomes were quantitated (Fig. 3B), and (iii) the viral tissue titers in the placentas and heads of selected fetuses were determined by CCID50 assays (Fig. 3C). Intrauterine growth restriction (IUGR) (described previously in mice [18, 22] and humans [5]) was observed at doses of 3 to 4 log10 CCID50 and infection at E6.5 (Fig. 3A and B, fetuses facing left). With larger doses (4 to 6 log10 CCID50), severely deformed fetal/placental masses were evident (Fig. 3A), with such outcomes and abortions (reported previously [21]) increasing with the inoculation dose (Fig. 3B).
Most placentas contained high titers of virus after inoculation with large or small doses of ZIKANatal at either E6.5 or E12.5 (Fig. 3C). As might be expected, the head of a fetus with IUGR and the deformed fetal/placental masses contained infectious ZIKV (Fig. 3C). The heads of some outwardly normal fetuses were also infected, with titers 2 to 3 logs lower than the corresponding placental titers (Fig. 3C). At the 3 log10 CCID50 dose, many head titers were below the limit of detection, which was not the case at 6 log10 CCID50 (Fig. 3C).
Infection of IRF7−/− dams at E6.5 with a s.c. dose of 4 or 6 log10 CCID50 of ZIKVNatal did not result in detectable replicating virus in the placentas or the fetal heads (n = 3 for each dose). Male IFNAR−/− mice could also be infected, with the testes becoming infected (see Fig. S1C), consistent with previous reports (19).
ZIKVNatal infection of A. aegypti mosquitoes.
The main vector species in the Brazilian outbreak of ZIKV was A. aegypti (23–25). To determine whether ZIKVNatal retains the ability to be transmitted by A. aegypti, standard artificial membrane feeding (with blood containing ZIKVNatal) was undertaken and mosquito infection was assessed by CCID50 assays after 14 days. Half of the successfully blood-fed mosquitoes contained detectable replicating virus, with whole-body titers ranging from 4.5 to 7.5 log10/CCID50/ml (one mosquito was macerated in 1 ml of medium) (Fig. 4A). Saliva was also collected from individual mosquitoes in a separate group of 20 (fed as described above and also tested 14 days postfeeding) by allowing the mosquitoes to salivate into capillary tubes containing a sucrose solution. Salivary expectorant from three mosquitoes (15%) contained detectable ZIKV, supporting the view that ZIKVNatal retains the ability to be transmitted by A. aegypti. As mosquitoes expectorate an estimated 0.11 to 24 nl of saliva (26), the 2.6-log10 CCID50/ml titer (Fig. 3B) represents an estimated titer in saliva of 5.2 to 7.6 log10 CCID50/ml.
FIG 4 .
A. aegypti infection with ZIKVNatal. (A) A. aegypti mosquitoes were artificially fed blood meals containing ZIKVNatal, and after 14 days, the mosquitoes were homogenized in 1 ml of medium and viral titers were determined by CCID50 assay. The limit of detection was 2 log10 CCID50/ml of medium. (B) Same as for panel A, except that saliva was collected from a separate group of mosquitoes by allowing them to salivate into capillary tubes containing 10 μl of a sucrose-FBS solution. Titers were determined by CCID50 assay and represent the titers per milliliter of sucrose-FBS solution. The limit of detection was 2.6 log10 CCID50/ml. (C) Same as for panel A, except that mosquitoes were examined by fluorescence immunohistochemistry with 4G4, a flavivirus-specific antibody. M, midgut; H, head; S, salivary gland.
Immunofluorescent-antibody staining of whole mosquitoes (fed as described above, 14 days postfeeding) with 4G4 (pan-flavivirus anti-NS1 antibody) showed abundant staining of midgut cells, and in some mosquitoes, clear staining of the salivary glands was also evident (Fig. 3C). Taken together, these experiments argue that ZIKVNatal remains mosquito transmission competent.
DISCUSSION
Herein, we describe the de novo generation of ZIKVNatal by a modified CPER protocol, with the resulting virus sequence identical (except for three synonymous nucleotide changes) to the published sequence obtained by next-generation sequencing of infected fetal brain tissue (7). ZIKVNatal therefore represents the first ZIKV isolate without any passage history in cells and/or mice. In addition, as no virus isolate was obtained from the original brain tissues (7), this study also illustrates a novel pathway for obtaining a new virus isolate (next-generation sequencing of infected tissues, followed by de novo virus generation via CPER). Furthermore, ZIKVNatal was able to infect fetal brains and cause IUGR in IFNAR−/− fetuses and appears to retain the ability to be transmitted by A. aegypti mosquitoes. The CPER protocol thus provides a rapid method for obtaining an infectious flavivirus isolate from publically available sequence data without the need for international transport of infectious material.
A similar PCR-based protocol to generate African and Asian ZIKV isolates, as well as chimeric viruses, was recently reported that uses three DNA fragments covering the genome overlapping by ~70 to 80 nt (27). However, no joining of these fragments into circular DNA was performed. This may explain the relatively inefficient virus recovery observed in the study of Atieh et al., which required two passages to obtain viral titers of ~4 log10 CCID50/ml for the Asian isolate and ~5 log10 CCID50/ml for the African isolate. In contrast, the titers of passage 0 ZIKVNatal generated in two different CPER and transfection experiments in our study were 5 × 105 and 2.6 × 107 PFU/ml, respectively, and after one passage of the former, the virus reached a titer of 1.25 × 108 PFU/ml or 7.6 log10 CCID50/ml. The difference in the titers of viruses obtained in two different CPER assembly and transfection experiments is likely due to the differences in the processivity of the Q5 and PrimeSTAR GXL DNA polymerases, as well as the different cycling conditions used (more cycles and longer extension times for PrimeSTAR GXL DNA polymerase produced a higher virus titer), rather than the potential presence of mutations that could affect the infectivity of CPER-generated DNAs. Notably, CPER does not amplify DNA as conventional PCR would; it only doubles the amount of DNA by extending the gaps between annealed regions of fragments for each strand. Once polymerase reaches the next annealed region, it stops. Thus, unlike PCR, CPER is not an amplification process and therefore will not accumulate mutations as PCR could. In addition, both polymerases are high fidelity; hence, the introduction of different nucleotide changes in different independent experiments that could result in different infectivities of each CPER-generated DNA is unlikely.
A range of mouse models to study fetal infections by ZIKV have been reported (17). A range of strategies have been used to overcome the poor replication of ZIKV in WT mice, including (i) injection with anti-Ifnar1 monoclonal antibody (28, 29), (ii) infection via the intraperitoneal route later in pregnancy (E13.5) (30) or infection via the intravaginal route (18) (although fetal brain infection was only detectable by immunofluorescent-antibody staining), (iii) the use of SJL mice and very high doses of virus (22), (vi) direct intrauterine inoculation (31), and (v) infection of neonates (32). However, ZIKV does replicate in type I interferon response-deficient mice, with the use of IFNAR−/− mice widely adopted (17). Infection of IFNAR−/− mice with many ZIKV isolates is often highly symptomatic and lethal (17), although some recent isolates are showing reduced lethality in these mice (33, 34). A model using IFNAR−/− females mated with WT males to produce IFNAR+/− fetuses has been described, although maternal illness and demise and resorption of the majority of fetuses were noted (29). As ZIKVNatal infection of IFNAR−/− mice (>8 weeks old) via the s.c. route was asymptomatic and nonlethal, this system allowed infection of pregnant dams with minimal ethical concerns. Fetal head infections and the occurrence of IUGR in ZIKVNatal-infected IFNAR−/− fetuses illustrate that this model recapitulates key elements of CZS.
In ZIKVNatal-infected IFNAR−/− fetuses, when heads were infected, the corresponding placentas always had higher viral titers. In IFNAR−/− mice, a range of placental cells appear to be infected (29). In humans, infection is largely restricted to Hofbauer cells, with placental trophoblasts thought to be protected from ZIKV infection by gamma interferon (35, 36), an activity that would clearly be absent in IFNAR−/− mice. How, exactly, the virus traverses the placenta in humans remains unclear (3, 35–38). Although transplacental infection with other members of the flavivirus genus has only rarely been reported (3), perhaps of note, a virus in the pestivirus genus (family Flaviviridae), bovine viral diarrhea virus, can infect placental trophoblasts, leading to transplacental infection and congenital abnormalities (39, 40).
Our results support the view that ZIKVNatal retains the ability to be transmitted by A. aegypti mosquitoes, with the difference in the percentage of mosquitoes infected and the percentage with infectious saliva broadly consistent with the reported rates for a variety of ZIKV isolates in these mosquitoes (23, 25). Previous studies have shown that Australian A. aegypti can transmit the ZIKAMR766 isolate (23), and our results now extend these findings to include a Brazilian ZIKV isolate associated with CZS. Australia has seen 128 imported ZIKV cases, 85 confirmed and the rest suspected (http://health.gov.au/internet/main/publishing.nsf/Content/ohp-vectorborne-overseas-acquired.htm; accessed 3 November 2017).
In summary, we have generated a new ZIKV isolate, ZIKVNatal, from sequence data and characterized the in vitro growth properties of the virus against those of the prototype, ZIKVMR766. Additionally, we have shown that it can recapitulate key aspects of CZS in a mouse model and can be transmitted by A. aegypti mosquitoes. ZIKVNatal should thus find utility in future laboratory studies on ZIKV and provide a system for testing new interventions against ZIKV.
MATERIALS AND METHODS
Generation of ZIKVNatal by CPER.
Seven dsDNA fragments covering the entire viral sequence (accession number KU527068) (Fig. 1A) were purchased from Integrated DNA Technologies, Inc. (Baulkham Hills, NSW, Australia), as Gblocks, and each was cloned into a separate pUC19 vector. For fragment 7, the last nucleotide (T) from the Gblocks gene fragment was omitted, as all flaviviruses end with TCT at the 3′ UTR, rather than the published TCTT sequence. These plasmids were grown in DH5α, the inserted sequences were verified, and the plasmids were then used to generate viral cDNA fragments by PCR (Fig. 1A). An additional PCR fragment (UTR linker) was generated from plasmid pUC19 containing the minimal CMV promoter, the first and the last 22 nt of the ZIKVNatal sequence, the HDVr, and a pA signal (Fig. 1A). PCR fragments were generated with high-fidelity Q5 DNA polymerase and primer pairs that have complementary ends with a 24- to 30-nucleotide overlap (see Table S1). The resulting eight DNA fragments were then mixed in equimolar amounts (0.1 pmol each) and subjected to CPER with Q5 DNA polymerase (an initial 3 min of incubation at 98°C; 2 cycles of 30 s at 98°C, 30 s at 55°C, and 6 min at 72°C; and 10 cycles of 30 s at 98°C, 30 s at 55°C, and 72°C for 8 min) to generate circular DNA (Fig. 1A). In a separate CPER protocol, PrimeSTAR GXL DNA polymerase (TaKaRa) was used with the same eight PCR fragments (0.1 pmol each) but different cycling conditions (an initial 2 min of denaturation at 98°C; 20 cycles of 10 s at 98°C, 15 s at 55°C, and 12 min at 68°C; and a final extension for 12 min at 68°C.
Primers used for PCR amplification of fragments used in CPER. Download TABLE S1, DOCX file, 0.01 MB (14.1KB, docx) .
Copyright © 2017 Setoh et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
The CPER products were then transfected directly (without additional purification) into Vero cells with Lipofectamine LTX Plus transfection reagent (Life Technologies, Inc.), in accordance with the manufacturer’s instructions. At 6, 8, and 10 days after transfection, the culture supernatants were harvested. Day 10 supernatant (from the Q5 polymerase-generated CPER transfection) was expanded once in C6/36 cells (to produce passage 1 virus), aliquoted, and stored at −80°C prior to use in further experiments.
RT-PCR for detection of viral RNA.
The cell culture supernatants collected at various time points posttransfection were subjected to RQ1 DNase (Promega) treatment to digest any CPER DNA left over from transfection; this was followed by viral RNA isolation with the NucleoSpin RNA Virus kit (Macherey-Nagel) and RT-PCR amplification with the SuperScript III One-Step RT-PCR System and platinum Taq DNA polymerase (Life Technologies, Inc.) with primers specific for the amplification of fragment 5 (30 min at 55°C; 2 min at 94°C; 35 cycles of 15 s at 94°C, 30 s at 55°C, and 2 min at 68°C; and a final extension for 5 min at 68°C). Total cell RNA was also isolated from transfected cells at day 10 after transfection and subjected to RT-PCR with the same primers and under the same cycling conditions. PCR amplification of viral RNA isolated from transfected cell supernatant and of total RNA from transfected cells was performed without RT with Q5 DNA polymerase (NEB) (3 min at 98°C; 35 cycles of 15 s at 98°C, 30 s at 55°C, and 2 min at 72°C; and a final extension for 5 min at 72°C) to confirm the lack of CPER DNA contamination.
Immunofluorescent-antibody staining.
Vero cells were infected with the 10-day transfection supernatant, fixed with 100% acetone 3 days after infection, and stained with 4G4 (anti-NS1) (41) and 3G1 (anti-dsRNA) (42) antibodies and an Alexa Fluor 488-labeled goat anti-mouse IgG secondary antibody (Life Technologies, Inc.). Coverslips were mounted with ProLong Gold Antifade Mountant with 4',6-diamidino-2-phenylindole (DAPI; Life Technologies, Inc.), and immunofluorescence microscopy was performed with a Zeiss LSM710 confocal scanning microscope.
Deep sequencing of viral RNA.
Viral RNA was isolated with the NucleoSpin RNA Virus kit (see above). RT-PCR amplicons (fragments 1, 2, 3, 4 plus 5, 6, and 7, Fig. 1A) for deep sequencing were generated from viral RNA isolated from 10-day culture fluid with the SuperScript III One-Step RT-PCR System and Platinum Taq DNA Polymerase (Life Technologies, Inc.). These represent the same pairs of primers that were used to generate PCR fragments for CPER, except that fragments 4 and 5 were combined into one amplicon. Libraries were prepared with the Nextera XT DNA Sample Preparation kit (Illumina Inc., San Diego, CA). The prepared library was sequenced on an Illumina NextSeq500 platform 2 × 150 bp PE run with V2 chemistry. Reads were mapped to the ZIKVNatal genome (accession no. KU527068) with Bowtie 2 (1.1.2).
Infection of cell lines for virus growth kinetics.
C6/36 cells (ATCC CRL-1660), Vero cells (CCL-81), A549 cells (ATCC CCL-185), WT MEFs, and IFNAR−/− MEFs (43) were infected with passage 1 of a C6/36-derived stock of ZIKVNatal or a C6/36-derived stock of ZIKVMR766 at the MOIs indicated, and 200 μl of culture supernatant was collected from each sample well at the postinfection times indicated. Six independent experiments were conducted with each cell line, with the exception of WT MEFs (MOI of 0.1), IFNAR−/− MEFs, and C6/36 cells (n = 3). ZIKVNatal titers were determined by standard plaque assay on Vero cells. Briefly, Vero cells were seeded into six-well plates and infected with 10-fold serial dilutions of virus samples for 1 h at 37°C, after which 2 ml of 0.75% low-melting-point agarose in Dulbecco’s modified Eagle’s medium supplemented with 5% fetal bovine serum was overlaid onto the cells and allowed to solidify before incubation at 37°C in 5% CO2. At 5 days postinfection, the cells were fixed with 4% formaldehyde for 30 min at room temperature. The agar overlay was then removed, and the fixed cells were stained with 0.2% crystal violet solution and the plaques were counted.
Ethics statement.
All mouse work was conducted in accordance with the Australian Code for the Care and Use of Animals for Scientific Purposes, as defined by the National Health and Medical Research Council of Australia. Animal experiments were approved by the QIMR Berghofer Medical Research Institute animal ethics committee.
Mice and ZIKV infection.
C57BL/6J mice were purchased from the Animal Resources Centre (Canning Vale, WA, Australia). IRF7−/− mice were generated by T. Taniguchi (University of Tokyo) and provided by M. S. Diamond (Washington University School of Medicine, St. Louis, MO). IFNAR−/− mice were provided by P. Hertzog (Monash University, Melbourne, VIC, Australia). The latter mice were on a C57BL/6J background (44, 45).
ZIKVMR766 (ATCC VR-84) stocks were prepared in low-passage-number C6/36 cells, aliquoted, and stored at −80°C (titer, 8 log10 CCID50/ml). Mice were infected s.c. at the base of the tail or i.p. in 50 μl of medium with the ZIKV doses indicated. A scorecard system was used to evaluate animal wellbeing and included scores for posture, mobility, swelling, fur ruffling, hind leg weakness, and injection site reactions. Mice were euthanized with CO2 when ethically defined endpoints were reached.
ZIKV CCID50 assays.
ZIKV CCID50 assays for viremia and tissue titers were performed as previously described (46, 47), with minor modifications. Briefly, serum or supernatants from tissues or mosquitoes (bead macerated in medium) were collected and titrated in duplicate in 10-fold serial dilutions on low-passage-number C6/36 cells (ATCC CRL-1660). After 5 days, 50-μl volumes of supernatants were individually transferred onto parallel plates (i.e., A1 to A1, A2 to A2 … H12 to H12) containing low-passage-number Vero E6 cells (ATCC CRL-1586). After another 5 days, the plates were stained with crystal violet to visualize cytopathic effects. The titers were calculated by the method of Reed and Muench (50).
A. aegypti infection.
An A. aegypti colony was established (at the insectory facilities of the QIMR Berghofer Medical Research Institute) from eggs of Wolbachia-free adult females collected at Innisfail, Australia, in April 2016. Mosquitoes were reared as previously described (48). Adult 3- to 4-day-old A. aegypti mosquitoes were fed blood meals containing a 1:1 mixture of ZIKVNatal (7.6 log10 CCID50/ml) and defibrinated sheep blood by membrane feeder as previously described (47). Fed mosquitoes were collected and kept for 14 days at 28°C and 75% humidity as previously described (47). The mosquitoes were then anesthetized with CO2 and ice and homogenized (1 mosquito/ml of medium) as previously described (47), and after centrifugation, ZIKV titers in supernatants were determined by CCID50 assays as described above. Fluorescence immunohistochemistry of whole mosquitoes was also done as previously described (47), with pan-flavivirus, anti-NS1 protein monoclonal antibody 4G4 (41). Saliva was also collected from individual mosquitoes in a separate group of 20 at 14 days postfeeding by allowing the mosquitoes to salivate into capillary tubes containing 10 μl of a solution containing 10% sucrose and 10% fetal bovine serum (FBS) for 20 min as previously described (49), and titers were determined by CCID50 assays.
ACKNOWLEDGMENTS
This work was primarily funded by a seeding grant from the Australian Infectious Diseases Research Centre.
We thank Nicola Angel (Australian Centre for Ecogenomics, University of Queensland) for help in deep sequencing of viral RNA. We thank Roy Hall for the 4G4 and anti-dsRNA antibodies, Julian Druce (VIDRL) and Cameron Simmons (Doherty Institute) for providing MR766 virus, I. Anraku for his assistance with management of the biosafety level 3 facility at QIMR B, C. Winterford and his team at Histology Services (QIMR B) for their assistance with mosquito staining, N. Beebe (University of Queensland/CSIRO) for mosquito collections, and O. Ong (QIMR B) and F. Frentiu (Queensland University of Technology, Brisbane) for assistance with mosquito processing.
Footnotes
This paper was submitted via the mSphereDirect™ pathway.
Contributor Information
W. Paul Duprex, Boston University School of Medicine.
Matthew Evans, Icahn School of Medicine at Mount Sinai.
Ted Pierson, National Institute of Allergy and Infectious Diseases.
REFERENCES
- 1.Kapogiannis BG, Chakhtoura N, Hazra R, Spong CY. 2017. Bridging knowledge gaps to understand how Zika virus exposure and infection affect child development. JAMA Pediatr 171:478–485. doi: 10.1001/jamapediatrics.2017.0002. [DOI] [PubMed] [Google Scholar]
- 2.Falcao MB, Cimerman S, Luz KG, Chebabo A, Brigido HA, Lobo IM, Timerman A, Angerami RN, da Cunha CA, Bacha HA, Alves JR, Barbosa AN, Teixeira RF, Weissmann L, Oliveira PR, Cyrillo MA, Bandeira AC. 2016. Management of infection by the Zika virus. Ann Clin Microbiol Antimicrob 15:57. doi: 10.1186/s12941-016-0172-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ritter JM, Martines RB, Zaki SR. 2017. Zika virus: pathology from the pandemic. Arch Pathol Lab Med 141:49–59. doi: 10.5858/arpa.2016-0397-SA. [DOI] [PubMed] [Google Scholar]
- 4.Weaver SC. 2017. Emergence of epidemic Zika virus transmission and congenital Zika syndrome: are recently evolved traits to blame? mBio 8:e02063-16. doi: 10.1128/mBio.02063-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brasil P, Pereira JP Jr., Moreira ME, Ribeiro Nogueira RM, Damasceno L, Wakimoto M, Rabello RS, Valderramos SG, Halai UA, Salles TS, Zin AA, Horovitz D, Daltro P, Boechat M, Raja Gabaglia C, Carvalho de Sequeira P, Pilotto JH, Medialdea-Carrera R, Cotrim da Cunha D, Abreu de Carvalho LM, Pone M, Machado Siqueira A, Calvet GA, Rodrigues Baião AE, Neves ES, Nassar de Carvalho PR, Hasue RH, Marschik PB, Einspieler C, Janzen C, Cherry JD, Bispo de Filippis AM, Nielsen-Saines K. 2016. Zika virus infection in pregnant women in Rio de Janeiro. N Engl J Med 375:2321–2334. doi: 10.1056/NEJMoa1602412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Miranda-Filho Dde B, Martelli CM, Ximenes RA, Araujo TV, Rocha MA, Ramos RC, Dhalia R, Franca RF, Marques Junior ET, Rodrigues LC. 2016. Initial description of the presumed congenital Zika syndrome. Am J Public Health 106:598–600. doi: 10.2105/AJPH.2016.303115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mlakar J, Korva M, Tul N, Popović M, Poljšak-Prijatelj M, Mraz J, Kolenc M, Resman Rus K, Vesnaver Vipotnik T, Fabjan Vodušek V, Vizjak A, Pižem J, Petrovec M, Avšič Županc T. 2016. Zika virus associated with microcephaly. N Engl J Med 374:951–958. doi: 10.1056/NEJMoa1600651. [DOI] [PubMed] [Google Scholar]
- 8.Shan C, Xie X, Muruato AE, Rossi SL, Roundy CM, Azar SR, Yang Y, Tesh RB, Bourne N, Barrett AD, Vasilakis N, Weaver SC, Shi PY. 2016. An infectious cDNA clone of Zika virus to study viral virulence, mosquito transmission, and antiviral inhibitors. Cell Host Microbe 19:891–900. doi: 10.1016/j.chom.2016.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tsetsarkin KA, Kenney H, Chen R, Liu G, Manukyan H, Whitehead SS, Laassri M, Chumakov K, Pletnev AG. 2016. A full-length infectious cDNA clone of Zika virus from the 2015 epidemic in Brazil as a genetic platform for studies of virus-host interactions and vaccine development. mBio 7:e01114-16. doi: 10.1128/mBio.01114-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schwarz MC, Sourisseau M, Espino MM, Gray ES, Chambers MT, Tortorella D, Evans MJ. 2016. Rescue of the 1947 Zika virus prototype strain with a cytomegalovirus promoter-driven cDNA clone. mSphere 1:e00246-16. doi: 10.1128/mSphere.00246-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Quan J, Tian J. 2009. Circular polymerase extension cloning of complex gene libraries and pathways. PLoS One 4:e6441. doi: 10.1371/journal.pone.0006441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Setoh YX, Prow NA, Rawle DJ, Tan CS, Edmonds JH, Hall RA, Khromykh AA. 2015. Systematic analysis of viral genes responsible for differential virulence between American and Australian West Nile virus strains. J Gen Virol 96:1297–1308. doi: 10.1099/vir.0.000069. [DOI] [PubMed] [Google Scholar]
- 13.Edmonds J, van Grinsven E, Prow N, Bosco-Lauth A, Brault AC, Bowen RA, Hall RA, Khromykh AA. 2013. A novel bacterium-free method for generation of flavivirus infectious DNA by circular polymerase extension reaction allows accurate recapitulation of viral heterogeneity. J Virol 87:2367–2372. doi: 10.1128/JVI.03162-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Amarilla AA, Setoh YX, Periasamy P, Peng NY, Pali G, Figueiredo LT, Khromykh AA, Aquino VH. 2017. Chimeric viruses between Rocio and West Nile: the role for Rocio prM-E proteins in virulence and inhibition of interferon-α/β signaling. Sci Rep 7:44642. doi: 10.1038/srep44642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Haddow AD, Schuh AJ, Yasuda CY, Kasper MR, Heang V, Huy R, Guzman H, Tesh RB, Weaver SC. 2012. Genetic characterization of Zika virus strains: geographic expansion of the Asian lineage. PLoS Negl Trop Dis 6:e1477. doi: 10.1371/journal.pntd.0001477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dick GW, Kitchen SF, Haddow AJ. 1952. Zika virus. I. Isolations and serological specificity. Trans R Soc Trop Med Hyg 46:509–520. doi: 10.1016/0035-9203(52)90042-4. [DOI] [PubMed] [Google Scholar]
- 17.Morrison TE, Diamond MS. 2017. Animal models of Zika virus infection, pathogenesis, and immunity. J Virol 91:e00009-17. doi: 10.1128/jvi.00009-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yockey LJ, Varela L, Rakib T, Khoury-Hanold W, Fink SL, Stutz B, Szigeti-Buck K, Van den Pol A, Lindenbach BD, Horvath TL, Iwasaki A. 2016. Vaginal exposure to Zika virus during pregnancy leads to fetal brain infection. Cell 166:1247–1256.e4. doi: 10.1016/j.cell.2016.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lazear HM, Govero J, Smith AM, Platt DJ, Fernandez E, Miner JJ, Diamond MS. 2016. A mouse model of Zika virus pathogenesis. Cell Host Microbe 19:720–730. doi: 10.1016/j.chom.2016.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Clark DC, Brault AC, Hunsperger E. 2012. The contribution of rodent models to the pathological assessment of flaviviral infections of the central nervous system. Arch Virol 157:1423–1440. doi: 10.1007/s00705-012-1337-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mysorekar IU, Diamond MS. 2016. Modeling Zika virus infection in pregnancy. N Engl J Med 375:481–484. doi: 10.1056/NEJMcibr1605445. [DOI] [PubMed] [Google Scholar]
- 22.Cugola FR, Fernandes IR, Russo FB, Freitas BC, Dias JL, Guimarães KP, Benazzato C, Almeida N, Pignatari GC, Romero S, Polonio CM, Cunha I, Freitas CL, Brandão WN, Rossato C, Andrade DG, Faria Dde P, Garcez AT, Buchpigel CA, Braconi CT, Mendes E, Sall AA, Zanotto PM, Peron JP, Muotri AR, Beltrão-Braga PC. 2016. The Brazilian Zika virus strain causes birth defects in experimental models. Nature 534:267–271. doi: 10.1038/nature18296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hall-Mendelin S, Pyke AT, Moore PR, Mackay IM, McMahon JL, Ritchie SA, Taylor CT, Moore FA, van den Hurk AF. 2016. Assessment of local mosquito species incriminates Aedes aegypti as the potential vector of Zika virus in Australia. PLoS Negl Trop Dis 10:e0004959. doi: 10.1371/journal.pntd.0004959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Costa-da-Silva AL, Ioshino RS, Araújo HR, Kojin BB, Zanotto PM, Oliveira DB, Melo SR, Durigon EL, Capurro ML. 2017. Laboratory strains of Aedes aegypti are competent to Brazilian Zika virus. PLoS One 12:e0171951. doi: 10.1371/journal.pone.0171951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chouin-Carneiro T, Vega-Rua A, Vazeille M, Yebakima A, Girod R, Goindin D, Dupont-Rouzeyrol M, Lourenço-de-Oliveira R, Failloux AB. 2016. Differential susceptibilities of Aedes aegypti and Aedes albopictus from the Americas to Zika Virus. PLoS Negl Trop Dis 10:e0004543. doi: 10.1371/journal.pntd.0004543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ye YH, Carrasco AM, Frentiu FD, Chenoweth SF, Beebe NW, van den Hurk AF, Simmons CP, O’Neill SL, McGraw EA. 2015. Wolbachia reduces the transmission potential of dengue-infected Aedes aegypti. PLoS Negl Trop Dis 9:e0003894. doi: 10.1371/journal.pntd.0003894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Atieh T, Baronti C, de Lamballerie X, Nougairède A. 2016. Simple reverse genetics systems for Asian and African Zika viruses. Sci Rep 6:39384. doi: 10.1038/srep39384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sapparapu G, Fernandez E, Kose N, Bin C, Fox JM, Bombardi RG, Zhao H, Nelson CA, Bryan AL, Barnes T, Davidson E, Mysorekar IU, Fremont DH, Doranz BJ, Diamond MS, Crowe JE. 2016. Neutralizing human antibodies prevent Zika virus replication and fetal disease in mice. Nature 540:443–447. doi: 10.1038/nature20564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Miner JJ, Cao B, Govero J, Smith AM, Fernandez E, Cabrera OH, Garber C, Noll M, Klein RS, Noguchi KK, Mysorekar IU, Diamond MS. 2016. Zika virus infection during pregnancy in mice causes placental damage and fetal demise. Cell 165:1081–1091. doi: 10.1016/j.cell.2016.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wu KY, Zuo GL, Li XF, Ye Q, Deng YQ, Huang XY, Cao WC, Qin CF, Luo ZG. 2016. Vertical transmission of Zika virus targeting the radial glial cells affects cortex development of offspring mice. Cell Res 26:645–654. doi: 10.1038/cr.2016.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vermillion MS, Lei J, Shabi Y, Baxter VK, Crilly NP, McLane M, Griffin DE, Pekosz A, Klein SL, Burd I. 2017. Intrauterine Zika virus infection of pregnant immunocompetent mice models transplacental transmission and adverse perinatal outcomes. Nat Commun 8:14575. doi: 10.1038/ncomms14575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Manangeeswaran M, Ireland DD, Verthelyi D. 2016. Zika (PRVABC59) infection is associated with T cell infiltration and neurodegeneration in CNS of immunocompetent neonatal C57BL/6 mice. PLoS Pathog 12:e1006004. doi: 10.1371/journal.ppat.1006004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tripathi S, Balasubramaniam VR, Brown JA, Mena I, Grant A, Bardina SV, Maringer K, Schwarz MC, Maestre AM, Sourisseau M, Albrecht RA, Krammer F, Evans MJ, Fernandez-Sesma A, Lim JK, García-Sastre A. 2017. A novel Zika virus mouse model reveals strain specific differences in virus pathogenesis and host inflammatory immune responses. PLoS Pathog 13:e1006258. doi: 10.1371/journal.ppat.1006258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Widman DG, Young E, Yount BL, Plante KS, Gallichotte EN, Carbaugh DL, Peck KM, Plante J, Swanstrom J, Heise MT, Lazear HM, Baric RS. 2017. A reverse genetics platform that spans the Zika virus family tree. mBio 8:e02014-16. doi: 10.1128/mBio.02014-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Simoni MK, Jurado KA, Abrahams VM, Fikrig E, Guller S. 2017. Zika virus infection of Hofbauer cells. Am J Reprod Immunol 77:e12613. doi: 10.1111/aji.12613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bayer A, Lennemann NJ, Ouyang Y, Bramley JC, Morosky S, Marques ET Jr., Cherry S, Sadovsky Y, Coyne CB. 2016. Type III interferons produced by human placental trophoblasts confer protection against Zika virus infection. Cell Host Microbe 19:705–712. doi: 10.1016/j.chom.2016.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang ZW, Li ZL, Yuan S. 2016. The role of secretory autophagy in Zika virus transfer through the placental barrier. Front Cell Infect Microbiol 6:206. doi: 10.3389/fcimb.2016.00206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Noronha Ld, Zanluca C, Azevedo ML, Luz KG, Santos CN. 2016. Zika virus damages the human placental barrier and presents marked fetal neurotropism. Mem Inst Oswaldo Cruz 111:287–293. doi: 10.1590/0074-02760160085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hansen TR, Smirnova NP, Webb BT, Bielefeldt-Ohmann H, Sacco RE, Van Campen H. 2015. Innate and adaptive immune responses to in utero infection with bovine viral diarrhea virus. Anim Health Res Rev 16:15–26. doi: 10.1017/S1466252315000122. [DOI] [PubMed] [Google Scholar]
- 40.Fredriksen B, Press CM, Løken T, Odegaard SA. 1999. Distribution of viral antigen in uterus, placenta and foetus of cattle persistently infected with bovine virus diarrhoea virus. Vet Microbiol 64:109–122. doi: 10.1016/S0378-1135(98)00263-6. [DOI] [PubMed] [Google Scholar]
- 41.Prow NA, Setoh YX, Biron RM, Sester DP, Kim KS, Hobson-Peters J, Hall RA, Bielefeldt-Ohmann H. 2014. The West Nile virus-like flavivirus Koutango is highly virulent in mice due to delayed viral clearance and the induction of a poor neutralizing antibody response. J Virol 88:9947–9962. doi: 10.1128/JVI.01304-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.O’Brien CA, Hobson-Peters J, Yam AWY, Colmant AMG, McLean BJ, Prow NA, Watterson D, Hall-Mendelin S, Warrilow D, Ng ML, Khromykh AA, Hall RA. 2015. Viral RNA intermediates as targets for detection and discovery of novel and emerging mosquito-borne viruses. PLoS Negl Trop Dis 9:e0003629. doi: 10.1371/journal.pntd.0003629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Melian EB, Edmonds JH, Nagasaki TK, Hinzman E, Floden N, Khromykh AA. 2013. West Nile virus NS2A protein facilitates virus-induced apoptosis independently of interferon response. J Gen Virol 94:308–313. doi: 10.1099/vir.0.047076-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Swann JB, Hayakawa Y, Zerafa N, Sheehan KC, Scott B, Schreiber RD, Hertzog P, Smyth MJ. 2007. Type I IFN contributes to NK cell homeostasis, activation, and antitumor function. J Immunol 178:7540–7549. doi: 10.4049/jimmunol.178.12.7540. [DOI] [PubMed] [Google Scholar]
- 45.Rudd PA, Wilson J, Gardner J, Larcher T, Babarit C, Le TT, Anraku I, Kumagai Y, Loo YM, Gale M Jr., Akira S, Khromykh AA, Suhrbier A. 2012. Interferon response factors 3 and 7 protect against Chikungunya virus hemorrhagic fever and shock. J Virol 86:9888–9898. doi: 10.1128/JVI.00956-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gardner J, Anraku I, Le TT, Larcher T, Major L, Roques P, Schroder WA, Higgs S, Suhrbier A. 2010. Chikungunya virus arthritis in adult wild-type mice. J Virol 84:8021–8032. doi: 10.1128/JVI.02603-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hugo LE, Prow NA, Tang B, Devine G, Suhrbier A. 2016. Chikungunya virus transmission between Aedes albopictus and laboratory mice. Parasit Vectors 9:555. doi: 10.1186/s13071-016-1838-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sikulu-Lord MT, Maia MF, Milali MP, Henry M, Mkandawile G, Kho EA, Wirtz RA, Hugo LE, Dowell FE, Devine GJ. 2016. Rapid and non-destructive detection and identification of two strains of Wolbachia in Aedes aegypti by near-infrared spectroscopy. PLoS Negl Trop Dis 10:e0004759. doi: 10.1371/journal.pntd.0004759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Poole-Smith BK, Hemme RR, Delorey M, Felix G, Gonzalez AL, Amador M, Hunsperger EA, Barrera R. 2015. Comparison of vector competence of Aedes mediovittatus and Aedes aegypti for dengue virus: implications for dengue control in the Caribbean. PLoS Negl Trop Dis 9:e0003462. doi: 10.1371/journal.pntd.0003462. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A). ZIKVNatal infection (s.c., 6 log10 CCID50) of IFNAR−/− and IRF7−/− mice (>7 weeks old). No clinical symptoms were observed. n = 5 per group (B). Survival of IFNAR−/− mice after ZIKVNatal infection of young mice and i.p. inoculation (n = 3 for 4-week-old mice; n = 5 for the remaining groups). Mice were euthanized when hind leg weakness reached an ethically defined level of severity. Groups with deaths also showed 1 to 2 days of mild hunching (C). Infection of male IFNAR−/− mice (8 to 12 weeks old) with 5 log10 CCID50 of ZIKVNatal s.c. (n = 3). Testes were tested on day 7, and the titer is indicated by the red data point. Download FIG S1, TIF file, 0.5 MB (549.7KB, tif) .
Copyright © 2017 Setoh et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Primers used for PCR amplification of fragments used in CPER. Download TABLE S1, DOCX file, 0.01 MB (14.1KB, docx) .
Copyright © 2017 Setoh et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.