Abstract
Oxidative stress can induce cell dysfunction and lead to a broad range of degenerative alterations, including carcinogenesis, aging, and other oxidative stress-related conditions. To avoid undergoing carcinogenesis in response to oxidative stress, cells trigger a succession of checkpoint responses, including premature senescence and apoptosis. Increasing evidence indicates that H2O2, an important cause of oxidative stress, functions as an important physiological regulator of intracellular signaling pathways that participate in regulation of cell premature senescence and apoptosis. However, the precise mechanisms underlying this process remain to be studied extensively. In this study, we describe the importance of Pim-1 kinase in this checkpoint response to oxidative stress. Pim-1 binds to and phosphorylates the transcription factor high mobility group box transcription factor 1 (HBP1), activating it. H2O2 enhances the interaction between Pim-1 and HBP1 and promotes HBP1 accumulation. In turn, HBP1 rapidly and selectively up-regulates Pim-1 expression in H2O2-stimulated cells, thereby creating a Pim-1-HBP1 positive feedback loop that regulates H2O2-induced premature senescence and apoptosis. Furthermore, the Pim-1-HBP1 positive feedback loop exerts its effect by regulating the senescence markers DNMT1 and p16 and the apoptosis marker Bax. The Pim-1-HBP1 axis thus constitutes a novel checkpoint pathway critical for the inhibition of tumorigenesis.
Keywords: apoptosis, cellular senescence, gene transcription, hydrogen peroxide, phosphorylation, HBP1, Pim-1, positive feedback loop
Introduction
Oxidative stress has long been recognized as the major mechanism by which cells are damaged. It affects a variety of physiological processes, including cell proliferation, differentiation, invasion, apoptosis, senescence, and survival (1–4). The role of reactive oxygen species in cellular damage and death is well documented with effects on a broad range of degenerative alterations, including carcinogenesis, aging, and other oxidative stress-related conditions. H2O2 is widely used as an activator for establishing cellular models of oxidative stress-induced apoptosis and premature senescence (5, 6). The effects of H2O2 are concentration-dependent and vary among different cell types and densities (7, 8). H2O2 controls cell signaling and stimulates proliferation at low levels, whereas in high concentrations it initiates premature senescence and apoptosis. Although it has historically been regarded as purely harmful, recently a growing body of evidence suggests that H2O2 also functions as an important physiological regulator of intracellular signaling pathways (9, 10). However, the mechanisms underlying the physiological functions of H2O2 have not been extensively studied.
Pim-1 is a proto-oncogene encoding a serine/threonine protein kinase that regulates cell proliferation and growth. In response to various types of stress, Pim-1 is hyperactivated to modulate the expression of numerous target genes (11–13). Pim-1 contributes to cancer development by phosphorylating multiple target substrates that are related to cell proliferation and antiapoptotic effects. Traditionally, the proto-oncogene Pim-1 has been thought to mainly promote tumorigenesis. Overexpression of Pim-1 promotes cell proliferation and inhibits apoptosis, thus leading to tumor formation in mice (12, 14, 15). However, recent studies suggest that other, unconventional activities of Pim-1 can lead to it acting as a tumor suppressor. A number of studies indicate that Pim-1 overexpression elicits senescence rather than enhancing growth in normal fibroblasts and cancer cells (16–19). However, the molecular mechanisms driving this regulation are still poorly understood.
High mobility group (HMG)2 box-containing protein 1 (HBP1) is homologous to the sequence-specific HMG family of transcriptional factors (20–22). Previous evidence suggests that HBP1 is a tumor suppressor. HBP1 maps to chromosome 7q31.1, a region that has been reported to be frequently deleted in numerous cancer types (23–28), and has been shown to be required for oncogene-mediated premature senescence, which is a feature lost in malignant transformation (29). Thus, HBP1 has many features that are consistent with it being a regulator of tumorigenesis. As a transcription factor, HBP1 has dual and complex transcriptional functions. HBP1 directly represses transcription via a high-affinity element on target genes, including p47phox, N-MYC, MIF, DNMT1, and EZH2 (22, 29, 31, 32). Additionally, HBP1 can transcriptionally activate genes such as those encoding p16, p21, myeloperoxidase (MPO), and histone H1 (33–37). The dual transcriptional repression and activation activities of HBP1 on different genes are dictated by differences in the DNA elements bound, differential acetylation, and differential promoter DNA and histone methylation. Given its regulatory effect on important cell cycle regulators, it is not surprising that overexpression of HBP1 induces cell cycle arrest and premature senescence in numerous types of cells and in organs (29, 32, 33).
Protein phosphorylation is a key posttranslational modification that regulates protein stability, activity, and localization. Indeed, over 70% of cellular proteins are regulated by phosphorylation (38–41). In this study, we investigated the effect of Pim-1 kinase on phosphorylation of the transcription factor HBP1 and found an intricate posttranslational modification mechanism involved in regulating premature cellular senescence and apoptosis. A previous publication has shown that p38, a kinase activated by H2O2, phosphorylates HBP1 and increases its stability (42). However, the pathway by which HBP1 regulates H2O2-activated premature cellular senescence and apoptosis is not well understood. Here, we found that Pim-1 interacts physically with HBP1 to promote HBP1 phosphorylation and enhance HBP1 protein stability and transcriptional activity. The interaction of Pim-1 and HBP1 is enhanced in H2O2-induced prematurely senescent and apoptotic cells and is followed by HBP1 activation. Furthermore, the Pim-1 gene itself is a target for positive transcriptional regulation by HBP1. Thus, the Pim-1-HBP1 axis constitutes a novel positive feedback pathway that causes premature senescence and apoptosis and is critical for maintenance of proper cellular metabolism following oxidative stress.
Results
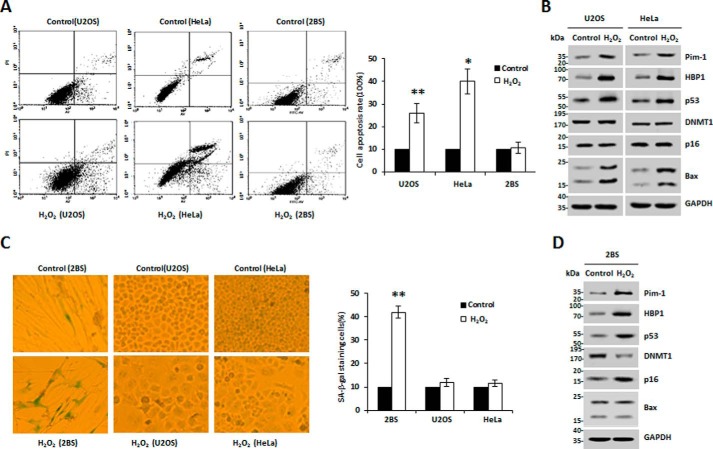
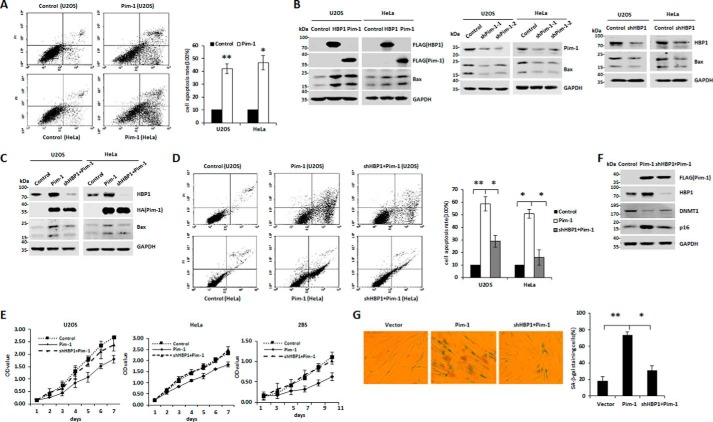
Both Pim-1 and HBP1 expression are up-regulated in H2O2-induced prematurely senescent and apoptotic cells
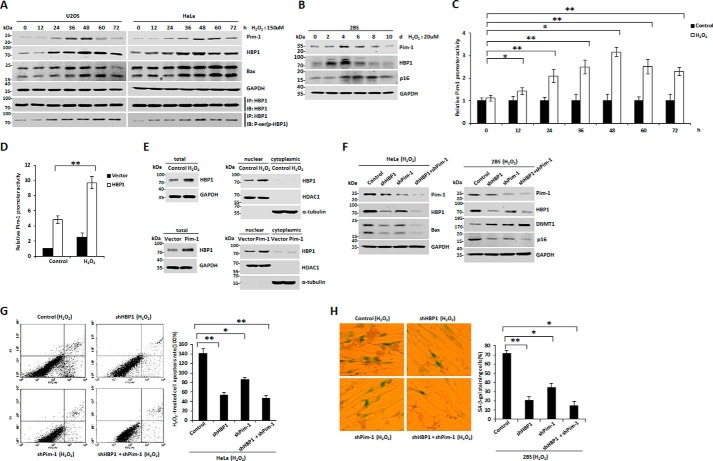
Mao and co-workers (16) and others (43) previously reported that Pim-1 levels were increased in H2O2-induced prematurely senescent or apoptotic cells. Our data show that HBP1 expression was up-regulated in response to H2O2-induced oxidative stress, suggesting a syntropic relationship. Intriguingly, we observed a statistically significant positive correlation between Pim-1 and HBP1 expression in cervical cancer and pancreatic cancer based on data from public databases (supplemental Fig. S1, A and B), suggesting that the positive correlation between Pim-1 and HBP1 expression may be involved in some specific carcinogenesis. Thus, we hypothesized that HBP1 is a target of Pim-1. We planned to examine the relative expression of Pim-1 and HBP1 in response to H2O2-induced oxidative stress in three different cell lines: 2BS (human diploid fibroblasts), HeLa (cervical cancer), and U2OS (osteosarcoma). First, we tested a complete profile of senescence and apoptosis markers for all three cell lines. The apoptosis markers (cell apoptosis rate and Bax protein expression) were tested by FACS and Western blotting, respectively. The results show that the apoptosis markers remained almost unchanged in H2O2-induced 2BS cells but increased in H2O2-induced HeLa or U2OS cells (Fig. 1, A, B, and D). Meanwhile, the senescence markers (senescence-associated (SA) β-gal staining of cells and p16 and DNMT1 protein expression) were tested by SA-β-gal staining and Western blotting, respectively. The results show that the senescence markers also remained unchanged in H2O2-induced HeLa or U2OS cells but significantly changed in H2O2-induced 2BS cells (Fig. 1, B–D). We also measured p53 status of the cell lines with Western blotting. As shown in Fig. 1, B and D, the levels of p53 protein increased in the three cell lines treated with H2O2. Altogether, the data indicate that H2O2 induces apoptosis in U2OS or HeLa cells and premature senescence in 2BS cells. We speculate that H2O2 activates p53 and p16 signal pathways in normal cells; thus the synergistic roles of the two signal pathways cause senescence in normal cells, whereas H2O2 only activates the p53 signal pathway, thereby causing apoptosis in tumor cells. Additionally, Pim-1 and HBP1 protein levels increased in the three cell lines in response to H2O2 treatment (Fig. 1, B and D). Our data indicate that expression of both Pim-1 and HBP1 is up-regulated in H2O2-induced prematurely senescent and apoptotic cells.
Figure 1.
Both Pim-1 and HBP1 expression are up-regulated in H2O2-induced premature senescent or apoptotic cells. A, U2OS and HeLa cells were treated with or without 150 μm H2O2 for 48 h, and 2BS cells were treated with or without 20 μm H2O2 for 1 week. Then the cells were stained with Annexin V-FITC and PI, and cell apoptosis was analyzed by FACS. Error bars represent S.D. *, p < 0.05; **, p < 0.01. B, U2OS and HeLa cells were treated as indicated in A, and the expression of the endogenous Pim-1, HBP1, p53, DNMT1, p16, and Bax was measured by Western blotting. C, U2OS, HeLa, and 2BS cells were treated as indicated in A, and then cells were stained for SA-β-gal. Error bars represent S.D. *, p < 0.05; **, p < 0.01. D, 2BS cells were treated as indicated in A, and the protein levels of Pim-1, HBP1, p53, DNMT1, p16, and Bax are shown. GAPDH was used as loading control.
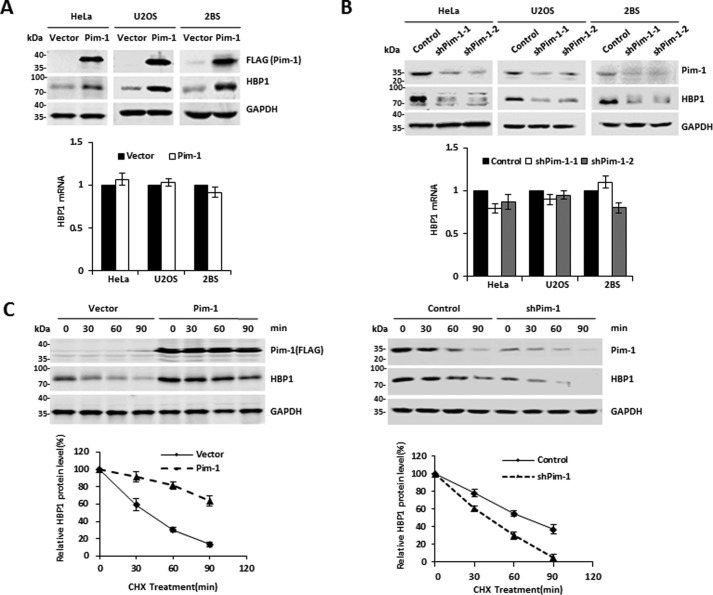
Pim-1 kinase promotes HBP1 expression by enhancing the stability of HBP1 protein
Next, we investigated whether Pim-1 kinase has a causative role in regulating HBP1. Exogenous Pim-1 increased HBP1 protein levels but had no effect on HBP1 mRNA expression (Fig. 2A). Moreover, Pim-1 knockdown decreased HBP1 protein levels but had no effect on HBP1 mRNA (Fig. 2B). Thus, Pim-1 regulated HBP1 expression at the posttranscriptional level. We next investigated whether the Pim-1-induced increase in HBP1 protein levels by was due to enhanced protein stability. Overexpression of Pim-1 caused a dramatic increase in HBP1 stability, whereas Pim-1 knockdown by shRNA reduced HBP1 stability (Fig. 2C). These results indicate that Pim-1 kinase promotes HBP1 expression by enhancing the stability of HBP1 protein.
Figure 2.
The Pim-1 kinase regulates HBP1 expression. A, exogenous Pim-1 regulates HBP1 expression. The protein levels of Pim-1 and HBP1 were measured by Western blotting in HeLa, U2OS, and 2BS cells transfected with either pITA-FLAG-Pim-1 or pITA (as a control) (top). The mRNA levels of HBP1 were measured by real-time PCR (bottom). B, endogenous Pim-1 regulates HBP1 expression. The protein levels of Pim-1 and HBP1 were measured by Western blotting in HeLa, U2OS, and 2BS cells transfected with pLL3.7-shPim-1-1, pLL3.7-shPim-1-2, or pLL3.7 (as a control) (top). The mRNA levels of HBP1 were measured by real-time PCR (bottom). C, Pim-1 prolongs the half-life of HBP1. HeLa cells were stably transfected with pITA, pITA-FLAG-Pim-1, pLL3.7, or pLL3.7-shPim-1 through lentiviral infection, respectively. Cells were incubated with the protein translation inhibitor CHX for 0, 30, 60, or 90 min before harvest. Pim-1 and HBP1 protein levels were detected by Western blotting. GAPDH was used as the loading control (top). Quantification of HBP1 protein levels was determined using TotalLab software and normalized to GAPDH, and densitometry was plotted for the mean ± S.D. (error bars) of three independent experiments (bottom). Error bars represent S.D.
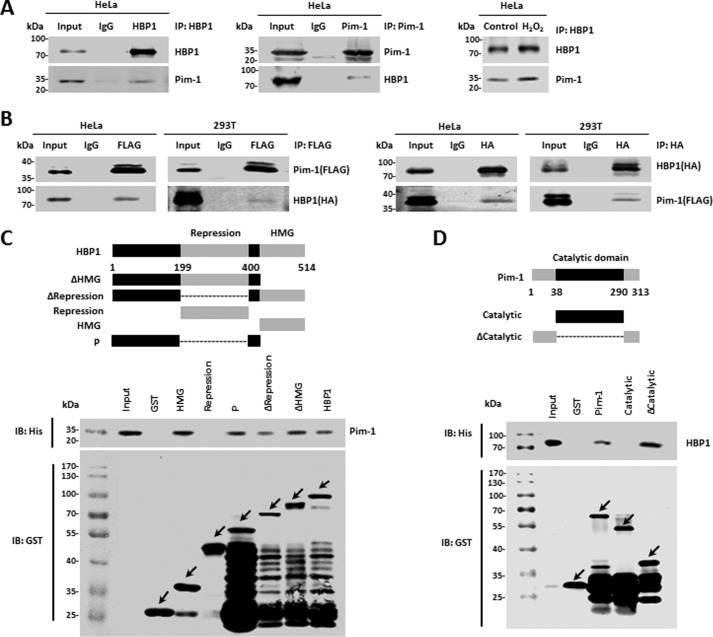
Pim-1 interacts with HBP1, and H2O2 enhances this interaction
We next sought to elucidate the potential mechanism by which Pim-1 enhances HBP1 protein stability. We hypothesized that there may be physical interaction between Pim-1 and HBP1 proteins and that HBP1 may be a target of Pim-1. First, we conducted co-immunoprecipitation with HeLa cells. Endogenous Pim-1 interacted with endogenous HBP1, and H2O2 enhanced this interaction (Fig. 3A). Next, we cotransfected FLAG-tagged Pim-1 and HA-tagged HBP1 into HeLa and HEK293T cells. Similarly to the endogenous proteins, exogenous Pim-1 interacted with exogenous HBP1 in HeLa and HEK293T cells (Fig. 3B).
Figure 3.
Pim-1 interacts with HBP1 in vivo and in vitro. A and B, Pim-1 interacts with HBP1 in vivo, and H2O2 enhances the interaction. HeLa cells treated with or without H2O2 were lysed with IP lysis buffer and then subjected to immunoprecipitation with anti-HBP1 or anti-Pim-1 antibodies followed by Western blotting with anti-HBP1 or anti-Pim-1 antibodies (A). HeLa or HEK293T cells were cotransfected with FLAG-Pim-1 and HA-HBP1. An IP assay was carried out using anti-FLAG/HA antibody followed by Western blotting with anti-HA or anti-FLAG antibodies. The same samples were immunoblotted (IB) against FLAG or HA to determine immunoprecipitation efficiency (B). C and D, Pim-1 interacts with HBP1 in vitro. Shown is a schematic representation of N-terminal GST-tagged full-length HBP1 along with its various deletion mutants (C, top) and Pim-1 along with its various deletion mutants (D, top). P domain, the regions outside of the HMG box binding and the repression domains. GST pulldown assay was carried out to determine the domain of HBP1 essential for its interaction with Pim-1 (C, bottom) and the domains of Pim-1 essential for its interaction with HBP1 (D, bottom). GST pulldown efficiency was evaluated by Western blotting with anti-GST antibody. Arrows represent the proteins of GST-tagged HBP1, Pim-1, or their deletion mutants.
To determine whether the interaction between Pim-1 and HBP1 is direct, we next performed a GST pulldown assay. GST-HBP1, but not GST alone, pulled down Pim-1 in vitro. A reciprocal GST pulldown assay further demonstrated an interaction between Pim-1 and HBP1 in vitro. In summary, Pim-1 directly interacts with HBP1 in vivo and in vitro. To further determine the specific domain required for this interaction, we constructed a set of HBP1 and Pim-1 deletion mutants in the pGEX-4T-1 vector and used these in GST pulldown assays. These assays indicated that the repression domain of HBP1 (designated as Repression) did not interact with Pim-1, whereas HBP1 lacking repression domain (designated as ΔRepression) interacted with Pim-1. The weaker binding of ΔRepression to Pim-1 compared with the other mutants might be due to the conformational changes caused by repression domain deletion (Fig. 3C). A subsequent reciprocal GST pulldown assay showed that the catalytic domain of Pim-1 did not bind HBP1, whereas a Pim-1 mutant lacking the catalytic domain (ΔCatalytic) bound HBP1 (Fig. 3D). These results suggest that the interaction between HBP1 and Pim-1 is mediated by diverse domains of the two proteins but not by the repression domain of HBP1 or the catalytic domain of Pim-1.
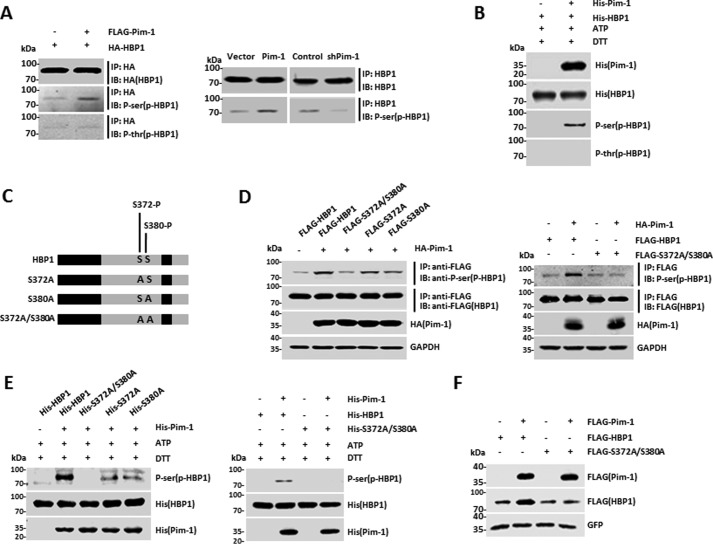
HBP1 is phosphorylated at Ser-372/Ser-380 by Pim-1
To determine whether HBP1 is phosphorylated by Pim-1, HEK293T cells were cotransfected with HBP1 expression vector with or without Pim-1 expression vector. Subsequently, anti-phosphoserine and anti-phosphothreonine antibodies were used to detect phospho-HBP1 in the Pim-1-expressing cells. Immunoprecipitation (IP) with anti-HA antibody followed by probing with anti-phosphoserine or anti-phosphothreonine antibodies confirmed the presence of exogenous phospho-HBP1 in the Pim-1-expressing cells and demonstrated that the HBP1 phosphorylation occurred specifically at serine (Fig. 4A, left). Then endogenous HBP1 phosphorylation in HeLa cells was confirmed (Fig. 4A, right). We also investigated HBP1 phosphorylation in vitro. Purified His-tagged HBP1 protein was incubated with or without His-Pim-1 in the presence of adenosine triphosphate (ATP). The reaction products were separated by SDS-PAGE and immunoblotted with anti-His, anti-phosphoserine, or anti-phosphothreonine antibodies. The result further demonstrated that HBP1 was phosphorylated by Pim-1 at serine in vitro (Fig. 4B).
Figure 4.
HBP1 is phosphorylated at Ser-372/Ser-380 by Pim-1. A, HBP1 is phosphorylated by Pim-1 in vivo. HEK293T cells were cotransfected HA-HBP1 with or without FLAG-Pim-1. An IP assay was carried out using anti-HA antibody followed by Western blotting with anti-HA, -phosphoserine, or -phosphothreonine antibodies. HeLa cells were stably transfected with pITA, pITA-FLAG-Pim-1, pLL3.7, or pLL3.7-shPim-1 through lentiviral infection, respectively. An IP assay was carried out using anti-HBP1 antibody followed by Western blotting with anti-HBP1 or anti-phosphoserine antibodies. B, HBP1 is phosphorylated by Pim-1 in vitro. Purified His-tagged HBP1 protein was incubated with or without His-Pim-1 in the presence of ATP. The reaction products were separated by SDS-PAGE and immunoblotted (IB) with the anti-His, -phosphoserine, or -phosphothreonine antibodies. C, schematic representation of wild-type HBP1 along with its single or double point mutants. D, HBP1 is phosphorylated at Ser-372 and Ser-380 by Pim-1 in vivo. HEK293T cells were cotransfected wild-type HBP1 or its single or double point mutants with or without HA-Pim-1. An IP assay was carried out using anti-FLAG, and the phosphorylation of wild-type and mutant HBP1 was assessed by Western blotting using antibodies against FLAG and phosphoserine (HBP1). E, HBP1 is phosphorylated at Ser-372 and Ser-380 by Pim-1 in vitro. Purified His-tagged wild-type HBP1 or its mutants were incubated with or without His-Pim-1 in the presence of ATP. The reaction products were separated by SDS-PAGE and immunoblotted with the anti-His or -phosphoserine (HBP1) antibodies. F, the increase of HBP1 protein levels induced by Pim-1 depend on Ser-372/Ser-380 phosphorylation. The protein levels of HBP1 and Pim-1 were measured by Western blotting in HEK293T cells transfected with FLAG-tagged HBP1, double point mutant S372A/S380A, or GFP with or without Pim-1 transfection. Levels of GFP protein are shown as equal transfection efficiency.
We next performed mass spectrometry to identify the exact amino acids that were phosphorylated by Pim-1. Ser-372 and Ser-380 of HBP1 were phosphorylated in the presence of Pim-1 (supplemental Fig. S2, A and B). To confirm this result, we designed three phosphorylation-deficient mutants: S372A, S380A, and S372A/S380A (Fig. 4C). Wild-type HBP1 was phosphorylated by Pim-1, consistent with the previous data. The HBP1 single-residue mutants (S372A and S380A) were partially phosphorylated by Pim-1, whereas for the double-residue mutant, S372A/S380A, the phospho-HBP1 level was close to the control level, suggesting that Pim-1-mediated HBP1 phosphorylation had been abolished (Fig. 4D). Furthermore, an in vitro kinase assay confirmed that HBP1 is phosphorylated at Ser-372/Ser-380 by Pim-1 (Fig. 4E). Because Pim-1 increases HBP1 protein levels, we next explored whether HBP1 phosphorylation at Ser-372/Ser-380 is required for Pim-1 induction of HBP1. Pim-1 increased wild-type HBP1 protein levels but had no effect on levels of the double-residue mutant S372A/S380A, suggesting that Pim-1 increases HBP1 protein levels by phosphorylating HBP1 at Ser-372/Ser-380 (Fig. 4F).
HBP1 participates in Pim-1-induced apoptosis and premature senescence
Our data suggest that HBP1 protein is a target of Pim-1 kinase and can be phosphorylated at Ser-372/Ser-380 to enhance its stability. We next investigated whether HBP1 participates in Pim-1-induced apoptosis or premature senescence in tumor cells or normal cells and the potential role of HBP1 phosphorylation by Pim-1 in this process. FACS analysis demonstrated that Pim-1 induced apoptosis in U2OS and HeLa cells (Fig. 5A). Pim-1 or HBP1 overexpression increased levels of the apoptosis-associated protein Bax, whereas Pim-1 or HBP1 knockdown decreased Bax protein levels (Fig. 5B). Furthermore, HBP1 knockdown attenuated induction of Bax by Pim-1 (Fig. 5C), diminished Pim-1-induced apoptosis (Fig. 5D), and abolished Pim-1-induced growth arrest (Fig. 5E) in U2OS and HeLa cells. These results indicate that HBP1 participates in Pim-1-induced apoptosis in tumor cells. Additionally, Pim-1 overexpression increased levels of the senescence-associated protein p16 and decreased levels of the DNA methyltransferase DNMT1, which is known to negatively correlate with senescence (32) (Fig. 5F). Conversely, HBP1 knockdown attenuated the effect of Pim-1 on the expression of these two genes. HBP1 knockdown also attenuated Pim-1-induced senescence as indicated by SA-β-gal staining (Fig. 5G) and growth arrest in 2BS cells (Fig. 5E), suggesting that HBP1 also participates in Pim-1-induced premature senescence in normal cells.
Figure 5.
HBP1 participates in Pim-1-induced apoptosis and premature senescence. A, Pim-1 induces apoptosis in tumor cells. U2OS and HeLa cells were transfected with pcDNA3-HA-Pim-1 or pcDNA3 (as a control), and then cell apoptosis was analyzed by FACS after 48 h (left). Results of cell apoptosis rate were representative of three independent experiments (right). Error bars represent S.D. *, p < 0.05; **, p < 0.01. B, both HBP1 and Pim-1 increase Bax expression. The protein levels of Pim-1, HBP1, or Bax were measured by Western blotting in HeLa or U2OS cells transfected with pcDNA3 (as a control), FLAG-HBP1, or FLAG-Pim-1 individually (left). The protein levels of Pim-1 and Bax were measured by Western blotting in HeLa or U2OS cells transfected with pLL3.7-shPim-1-1, pLL3.7-shPim-2, or pLL3.7 (as a control) individually (middle). The protein levels of Pim-1 and Bax were measured by Western blotting in HeLa or U2OS cells transfected with pLL3.7-shHBP1 or pLL3.7 (as a control) individually (right). C and D, Pim-1 induces Bax expression and apoptosis in an HBP1-dependent manner. The protein levels of Pim-1, HBP1, and Bax were measured by Western blotting, and cell apoptosis rates were measured by FACS in HeLa or U2OS cells transfected with vector, HA-Pim-1, or HA-Pim-1 + shHBP1. Error bars represent S.D. *, p < 0.05; **, p < 0.01. E, Pim-1 induces cell growth arrest in an HBP1-dependent manner. Cell proliferation rates were measured by MTT assay in HeLa, U2OS, or 2BS cells transfected with vector, Pim-1, or Pim-1 + shHBP1. F and G, HBP1 knockdown attenuates Pim-1-induced premature senescence. 2BS cells were transfected with vector, Pim-1, or shHBP1 + Pim-1. The protein levels of Pim-1, HBP1, DNMT1, and p16 were determined by Western blotting (F), and cell senescence rates were measured by SA-β-gal staining (G). Error bars represent S.D. *, p < 0.05; **, p < 0.01.
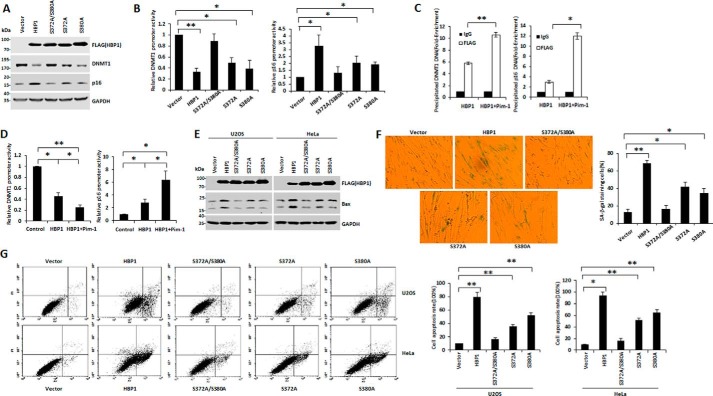
We next investigated the role of HBP1 phosphorylation by Pim-1 in HBP1-induced apoptosis and premature senescence. Expression of wild-type HBP1 decreased DNMT1 protein levels but increased p16 protein levels, whereas expression of the S372A/S380A mutant reduced this HBP1-mediated decrease in DNMT1 and increase in p16 to nearly control levels. Expression of the two single-residue mutants (S372A and S380A) resulted in partial HBP1-mediated reduction in DNMT1 levels and an increase in p16 levels (Fig. 6A). To explore the mechanisms underlying this effect, we conducted a reporter gene assay. Expression of wild-type HBP1 repressed the DNMT1 promoter and activated the p16 promoter, but the repression of the DNMT1 gene and activation of p16 gene were defective in cells expressing S372A/S380A. Cells expressing the single-residue mutants S372A and S380A partially retained the effects of HBP1 on the DNMT1 and p16 promoters (Fig. 6B). Furthermore, Pim-1 enhanced binding of HBP1 to the DNMT1 and p16 promoters (Fig. 6C) and thus promoted the transcriptional regulation effect of HBP1 on these promoters (Fig. 6D). These data suggest that phosphorylation of HBP1 at Ser-372/Ser-380 by Pim-1 is required for HBP1-mediated DNMT1 repression and p16 activation.
Figure 6.
The phosphorylation at Ser-372/Ser-380 by Pim-1 is indispensable for HBP1-induced premature senescence and apoptosis. A, phosphorylation at Ser-372/Ser-380 is required for the regulation of DNMT1 and p16 by HBP1. The protein levels of HBP1, DNMT1, and p16 were measured by Western blotting in 2BS cells transfected with vector, HBP1, S372A/S380A, S372A, and S380A individually. B, phosphorylation at Ser-372/Ser-380 is required for the transactivation of DNMT1 and p16 by HBP1. Relative activities of HBP1 and associated mutants on the native DNMT1 or p16 promoter were determined by luciferase reporter gene assay. HEK293T cells were cotransfected with 0.1 μg of the indicated reporters and 0.8 μg of HBP1 or mutant expression plasmids. Error bars represent S.D. *, p < 0.05; **, p < 0.01. C, Pim-1 enhances the binding of HBP1 to DNMT1 or p16 promoter. HEK293T cells were transfected with HBP1 or HBP1 + Pim-1. A ChIP assay was used to test the binding of exogenous HBP1 to the endogenous DNMT1 or p16 promoter. Error bars represent S.D. *, p < 0.05; **, p < 0.01. D, Pim-1 enhances the transactivation of HBP1 on DNMT1 or p16 promoter. HEK293T cells were cotransfected with the indicated reporters and HBP1 or HBP1 + Pim-1 expression plasmids. Error bars represent S.D. *, p < 0.05; **, p < 0.01. E, the phosphorylation at Ser-372/Ser-380 is required for the induction of Bax by HBP1. U2OS and HeLa cells were transfected with vector, HBP1, S372A/S380A, S372A, or S380A individually. After 48-h transfection, Bax protein expression was analyzed by Western blotting. F, phosphorylation at Ser-372/Ser-380 contributes to HBP1-induced SA-β-gal staining. 2BS cells (PD20) were stably transfected with vector, HBP1, S372A/S380A, S372A, or S380A through lentiviral infection, respectively, and then cells were stained for SA-β-gal. Error bars represent S.D. *, p < 0.05; **, p < 0.01. G, phosphorylation at Ser-372/Ser-380 contributes to HBP1-induced apoptosis. U2OS and HeLa cells were transfected with vector, HBP1, S372A/S380A, S372A, or S380A individually. After 48-h transfection, cell apoptosis rates were measured by FACS. Error bars represent S.D. *, p < 0.05; **, p < 0.01.
We next investigated the role of HBP1 phosphorylation in induction of apoptosis and premature senescence. The two single-residue mutants S372A and S380A were partially defective for HBP1-induced Bax expression and apoptosis, whereas the double-residue mutant S372A/S380A exhibited total abolition of HBP1-induced Bax expression and apoptosis (Fig. 6, E and G). Furthermore, the percentage of SA-β-gal-positive cells in S372A/S380A-expressing cells was not increased compared with control cells, whereas this percentage was slightly increased in S372A- and S380A-expressing cells, indicating that mutation of the two sites (Ser-372 and Ser-380) abolished induction of premature senescence by HBP1 (Fig. 6F). Thus, phosphorylation at Ser-372/Ser-380 is necessary for transactivation of HBP1, thereby allowing it to participate in Pim-1-induced premature senescence and apoptosis.
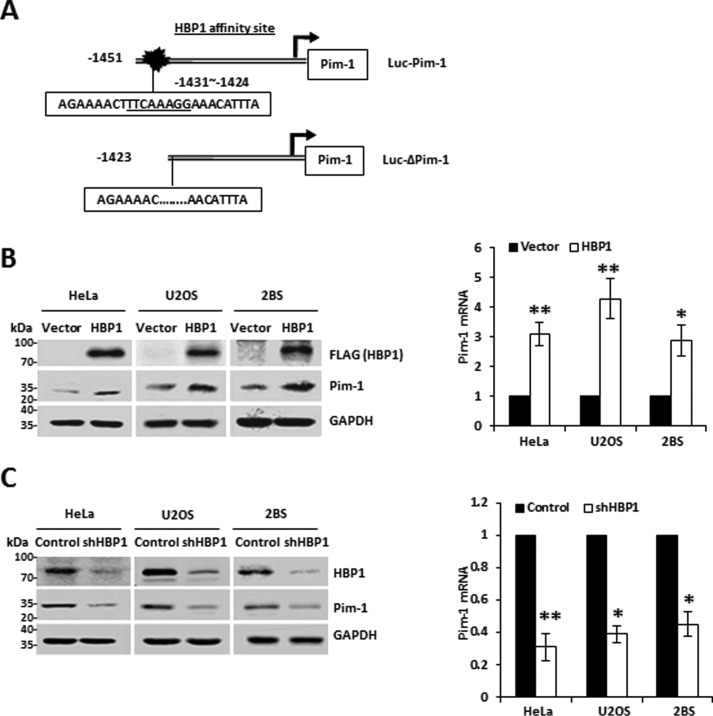
HBP1 enhances Pim-1 expression by binding a high-affinity site in the Pim-1 promoter
Intriguingly, we identified a high-affinity HBP1-binding site in the Pim-1 promoter by bioinformatics (Fig. 7A). Thus, we hypothesized that HBP1 is also a regulator of the Pim-1 gene. We found that exogenous HBP1 expression elevated Pim-1 protein and mRNA levels in all three cell lines tested (Fig. 7B). Moreover, HBP1 knockdown reduced Pim-1 protein and mRNA levels (Fig. 7C). Thus, we conclude that HBP1 regulates the Pim-1 gene.
Figure 7.
HBP1 also regulates Pim-1 expression. A, schematic diagram of the Pim-1 promoter. Shown is the HBP1 affinity site within the Pim-1 promoter at positions bp −1423 to −1451 from the transcriptional start site (Luc-Pim-1; top). Luc-ΔPim-1 is a mutant Pim-1 promoter with a deletion in the HBP1 affinity site (bottom). B, exogenous HBP1 regulates Pim-1 expression. The protein levels of HBP1 and Pim-1 were measured by immunoblotting in HeLa, U2OS, or 2BS cells transfected with vector (as control) or HBP1 expression plasmid (left). The mRNA levels of HBP1 and Pim-1 were measured by real-time PCR in HeLa, U2OS, or 2BS cells transfected with vector (as control) or HBP1 expression plasmid (right). Error bars represent S.D. *, p < 0.05; **, p < 0.01. C, endogenous HBP1 regulates Pim-1 expression. The protein levels of HBP1 and Pim-1 were measured by immunoblotting in HeLa, U2OS, or 2BS cells transfected with pLL3.7 (as a control) or pLL3.7-shHBP1 (left). The mRNA levels of HBP1 and Pim-1 were measured by real-time PCR in HeLa, U2OS, or 2BS cells transfected with pLL3.7 (as a control) or pLL3.7-shHBP1(right). Error bars represent S.D. *, p < 0.05; **, p < 0.01.
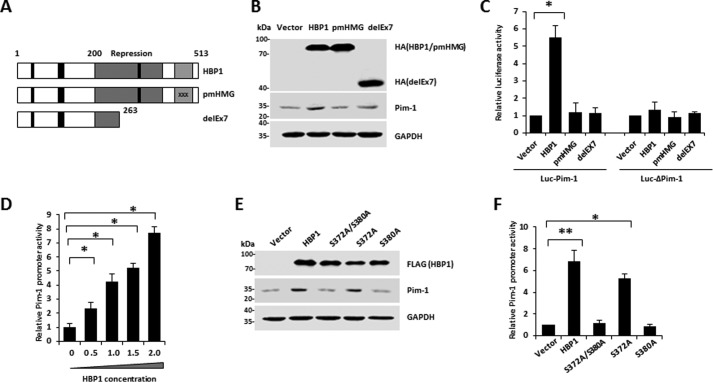
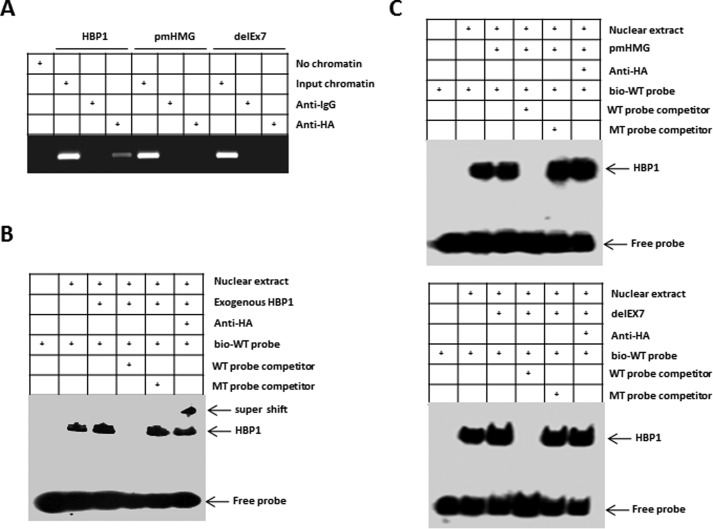
We next explored whether HBP1 transcriptionally activates the Pim-1 promoter through sequence-specific DNA binding. HBP1 is a member of the sequence-specific HMG box family of transcriptional factors with a central repression domain (amino acids 191–400) and a C-terminal HMG box DNA-binding domain (amino acids 431–509) as well as a retinoblastoma- and p38 MAPK-binding regulatory region (42, 44, 45). To examine the domains required for DNA binding, we used two mutants, pmHMG and delEx7 (Fig. 8A). pmHMG carries a triple-residue mutation in the HMG box DNA-binding domain that abolishes DNA binding. The delEx7 mutant was isolated in our previous breast cancer study and represents a naturally occurring mutant of HBP1 that is associated with invasive breast cancer (44). An exon-skipping deletion in delEx7 creates a premature termination signal, resulting in deletion of the DNA-binding domain and much of the repression domain. Wild-type HBP1 overexpression increased Pim-1 protein levels, but expression of the DNA-binding-defective mutants pmHMG and delEx7 had no effect on Pim-1 protein levels relative to levels in control cells (Fig. 8B). We designed two Pim-1 promoter-luciferase reporters with a native Pim-1 segment (Luc-Pim-1) or with a Pim-1 segment with a deletion that abolishes the HBP1 affinity site (Luc-ΔPim-1) (Fig. 7A). Wild-type HBP1 expression activated the Pim-1 promoter; in contrast, it had no effect on the Pim-1 promoter that lacked the high-affinity site. As expected, the HBP1 mutants defective in sequence-specific DNA binding (pmHMG and delEx7) had no effect on either the native or mutant Pim-1 promoters (Fig. 8C). These results indicate that the HBP1 DNA-binding domain and integrity of the high-affinity HBP1 promoter element are indispensable for Pim-1 gene activation by HBP1.
Figure 8.
HBP1 enhances Pim-1 promoter activity in a DNA-binding-dependent manner. A, schematic diagram of wild-type HBP1 and associated mutants. B, expression of exogenous HBP1 increases Pim-1 protein level. HEK293T cells were transfected with HA-HBP1, HA-pmHBP1, or delEx7. Western blotting was performed on protein lysates using anti-HA, Pim-1, or GAPDH antibodies. C, relative activities of HBP1 and associated mutants on the native Pim-1 promoter (Luc-Pim-1) or the mutant Pim-1 promoter (Luc-ΔPim-1). HEK293T cells were cotransfected with 0.1 μg of the indicated reporters and 0.8 μg of HBP1 or mutant expression plasmids. Error bars represent S.D. *, p < 0.05; **, p < 0.01. D, HBP1 enhances Pim-1 promoter activity in a dose-dependent manner. HEK293T cells were cotransfected with 0.1 μg of Luc-Pim-1 and different doses (0–2.0 μg) of HBP1. Error bars represent S.D. *, p < 0.05; **, p < 0.01. E, the phosphorylation of HBP1 at Ser-380 is indispensable for its induction of Pim-1 protein. The protein levels of HBP1 and its associated mutants or Pim-1 were measured by Western blotting in HeLa cells transfected with vector, HBP1, or its associated mutants (S372A/S380A, S372A, or S380A) individually. F, the phosphorylation of HBP1 at Ser-380 is indispensable for its enhancement of Pim-1 promoter. HEK293T cells were cotransfected with 0.1 μg of Pim-1 reporter and 0.8 μg of HBP1 or associated mutant expression plasmids. Error bars represent S.D. *, p < 0.05; **, p < 0.01.
We next investigated the effect of HBP1 and the phosphorylation mutants on Pim-1 gene activation. With the addition of exogenous HBP1, the relative activity of the native Pim-1 promoter increased significantly in a dose-dependent manner (Fig. 8D). Expression of wild-type HBP1 and the S372A mutant enhanced Pim-1 promoter activity and protein expression, whereas the S380A and S372A/S380A mutants exhibited defective activation of the Pim-1 gene and protein expression (Fig. 8, E and F). These data indicate that phosphorylation of HBP1 at Ser-380 is indispensable for its induction of the Pim-1 gene.
We next investigated whether HBP1 binds to the Pim-1 gene promoter. A chromatin immunoprecipitation assay indicated that wild-type HA-HBP1 bound to the endogenous Pim-1 promoter near the high-affinity site (Fig. 9A). HA-pmHMG, which is defective in DNA binding, and delEx7, which lacks the DNA-binding domain, did not bind the endogenous Pim-1 promoter. Furthermore, an electrophoretic mobility shift assay showed that endogenous HBP1 could bind to a specific oligonucleotide probe containing the affinity site and biotin label (TAGAAAACTTTCAAAGGAAACATTTA-biotin), and transfection of wild-type HBP1 resulted in increased HBP1 binding (Fig. 9B). The mutant HBP1 defective for DNA binding, pmHMG, did not bind to the probe. Another mutant, delEx7, which lacks the HMG box DNA-binding domain, also did not bind to the probe (Fig. 9C). The HBP1 electrophoretic mobility shift assay signal was specific as determined by competition with a 100-fold excess of unlabeled probe (wild-type probe) but not with a mutant probe bearing point mutations in the high-affinity site (Fig. 9, B and C). Moreover, the stronger bands in the lanes for the transfected HA-HBP1 were supershifted with anti-HA antibody, confirming that HBP1 binds to the high-affinity site probe specifically (Fig. 9B). Together, these results indicate that HBP1 specifically binds to and activates the endogenous Pim-1 promoter, resulting in the observed increase in Pim-1 expression.
Figure 9.
HBP1 occupies its affinity site in the Pim-1 promoter. A, HBP1 binding to the endogenous Pim-1 promoter requires the HMG domain. ChIPs were used to test the binding of exogenous HBP1 to the endogenous Pim-1 gene. HEK293T cells were transfected with HA-HBP1, HA-pmHMG, or HA-delEx7. The region from position bp −1423 to position bp −1451 contains the HBP1 element and was analyzed by specific PCR. Anti-HA antibody or control IgG was used in the indicated lanes. B and C, EMSAs were performed by using a biotin-labeled WT probe (bio-WT; containing the HBP1 affinity site). 10-μg amounts of nuclear extracts from HEK293T cells expressing HA-HBP1 (B), HA-pmHMG, or HA-delEx7 (C) were used. The probe (TAGAAAACTTTCAAAGGAAACATTTA) and the mutant probe (TAGAAAACTTTACCCTGAAACATTTA) were used as unlabeled competitors at a 100-fold excess. The presence of specific complexes, including supershifted HA-HBP1 in the complexes, is indicated (B).
A positive feedback loop between Pim-1 and HBP1 contributes to H2O2-induced premature senescence and apoptosis
We next examined the role of the Pim-1-HBP1 positive feedback loop in H2O2-induced premature senescence and apoptosis. Pim-1 and HBP1 protein levels initially increased during H2O2 treatment but then declined later in the treatment period (Fig. 10, A and B). Two protein markers, Bax and p16, were increased for the entire duration of H2O2 treatment. To confirm that the feedback between Pim-1 and HBP1 really happens in the context of H2O2 damage, we performed immunoprecipitation assay to test the kinetics of HBP1 phosphorylation in response to H2O2 induction. As shown in Fig. 10A, the level of phospho-HBP1 increased with H2O2 treatment in HeLa and U2OS cells, and phosphorylation of HBP1 was initially activated at 12 or 24 h in HeLa or U2OS cells treated with H2O2, coincident with the increased expression pattern of HBP1 protein. Additionally, we also performed a luciferase assay to test whether H2O2 affects HBP1 transactivation on Pim-1 promoter. As shown in Fig. 10C, H2O2 enhanced transactivation of endogenous HBP1 on Pim-1 promoter. Furthermore, H2O2 also enhanced transactivation of exogenous HBP1 on Pim-1 promoter (Fig. 10D). To explore the mechanism of H2O2 enhancement of transactivation of HBP1 on Pim-1 promoter, HeLa cells were treated with H2O2, and then nuclear and cytoplasmic HBP1 protein levels were determined by Western blotting (Fig. 10E, top). The data indicate that H2O2 treatment causes nuclear accumulation of HBP1, thus enhancing its transactivation on Pim-1 promoter. In turn, Pim-1 overexpression also caused nuclear accumulation of HBP1 (Fig. 10E, bottom), thereby further enhancing HBP1 transcription activity. Our data indicate that the positive feedback loop between HBP1 and Pim-1 occurs upon H2O2 exposure. These results also suggest that Pim-1 and HBP1 proteins act as key regulators to prevent propagation of damaged cells during H2O2 exposure initially but that complex cell metabolic processes and activation of other signaling pathways may occur and influence the activity of the Pim-1-HBP1 positive feedback loop upon longer H2O2 exposure. However, p16 and Bax protein levels remain elevated throughout the exposure period and thus eventually induce premature senescence and apoptosis. To further explore how the Pim-1-HBP1 positive loop affects H2O2-induced apoptosis and premature senescence, we transfected HeLa and 2BS cell lines with shHBP1, shPim-1, shHBP1 + shPim-1, and control vector. All of these cell lines were treated with H2O2. HBP1 or Pim-1 knockdown by shRNA partly attenuated H2O2 induction of Bax, DNMT1, and p16, and double knockdown of HBP1 and Pim-1 enhanced this attenuation (Fig. 10F).
Figure 10.
A positive feedback loop between Pim-1 and HBP1 contributes to premature senescence and apoptosis induced by H2O2. A, the protein levels of Pim-1, HBP1, and phosphoserine (phospho-HBP1 (p-HBP1)) increase with time progression in tumor cells with initial H2O2 treatment. U2OS and HeLa cells were treated with 150 μm H2O2 for different lengths of time (0, 12, 24, 36, 48, 60, and 72 h). The protein levels of Pim-1, HBP1, Bax, and phosphoserine (phospho-HBP1 (p-HBP1)) were determined by Western blotting. B, the protein levels of Pim-1 and HBP1 increase with time progression in normal fibroblasts with initial H2O2 treatment. 2BS cells were treated with 20 μm H2O2 for different lengths of time (0, 2, 4, 6, 8, and 10 days). The protein levels of Pim-1, HBP1, p16, and GAPDH (as loading control) were determined by Western blotting. C, H2O2 enhanced transactivation of endogenous HBP1 on Pim-1 promoter. HeLa cells transfected with Luc-Pim-1 were treated with H2O2 for different lengths of time (0, 12, 24, 36, 48, 60, and 72 h). The luciferase activity measurements of the group treated with H2O2 were normalized to luciferase activity of the group without H2O2. Error bars represent S.D. *, p < 0.05; **, p < 0.01. D, H2O2 enhanced transactivation of exogenous HBP1 on Pim-1 promoter. HeLa cells transfected with Luc-Pim-1 were treated with H2O2 with or without HBP1. Error bars represent S.D. *, p < 0.05; **, p < 0.01. E, HeLa cells were treated with H2O2 or transfected with Pim-1 expression plasmid, and then nuclear and cytoplasmic HBP1 protein levels were determined by Western blotting. F, the positive feedback loop between Pim-1 and HBP1 regulates the expression of senescence markers (DNMT1 and p16) and apoptosis marker (Bax). HeLa and 2BS cells were stably transfected with vector, shHBP1, shPim-1, or shHBP1 + shPim-1. After 4 weeks of selection, HeLa cells were treated with 150 μm H2O2 for 48 h, and 2BS cells were treated with 20 μm H2O2 for 1 week. Then the cells were harvested for Western blotting using anti-Pim-1, anti-HBP1, anti-Bax, anti-DNMT1, anti-p16, or anti-GAPDH antibodies (as loading control). G and H, the positive feedback loop between Pim-1 and HBP1 promotes premature senescence and apoptosis in response to H2O2 induction. HeLa or 2BS cells were treated as in F, HeLa cells were stained with Annexin V-FITC and PI, and cell apoptosis was analyzed by FACS (G). 2BS cells were stained for SA-β-gal (H, left), and the percentage of cells positive for SA-β-gal in 2BS cells was determined in three independent experiments (H, right). Error bars represent S.D. *, p < 0.05; **, p < 0.01.
In accordance with these results, HBP1 or Pim-1 knockdown partly attenuated H2O2 induction of apoptosis and premature senescence, and double knockdown of HBP1 and Pim-1 enhanced this attenuation (Fig. 10, G and H). The results indicate that the positive feedback loop between Pim-1 and HBP1 promotes premature senescence and apoptosis in response to H2O2 exposure by regulating the senescence markers DNMT1 and p16 and the apoptosis marker Bax.
Discussion
H2O2, like other oxidants, has historically been considered a damaging entity that causes oxidative stress-related diseases such as cardiovascular disease, pulmonary fibrosis, and cancer. The rationale for this relationship is that H2O2 oxidizes various biomolecules, resulting in their loss of function and causing damage that ultimately compromises cellular function. However, the precise nature by which H2O2 functions as a cellular signaling agent has remained unclear.
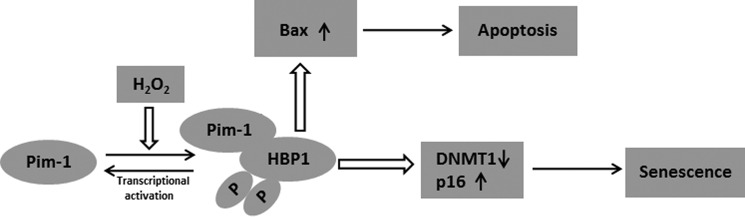
Here, we show that H2O2 promoted apoptosis and premature senescence through a Pim-1-HBP1 signaling pathway. We have identified both Pim-1 and HBP1 as important factors in regulating H2O2-induced premature senescence and apoptosis. Our work is consistent with a model in which H2O2 induces cell growth arrest by activation of a Pim-1-HBP1 positive feedback loop (see model in Fig. 11). More specifically, we suggest that, in response to H2O2 induction, Pim-1 kinase binds to and phosphorylates HBP1 (Figs. 3 and 4), which leads to rapid initial HBP1 activation. The activated HBP1 binds preferentially to the Pim-1 promoter and triggers prompt, selective induction of Pim-1 gene expression in cells treated with H2O2 (Figs. 7–9). This further elevates Pim-1 protein levels, which in turn further drives HBP1 activation. Eventually, expression of HBP1 target genes such as p16, DNMT1, and Bax is induced, which leads to premature senescence and apoptosis. Our data implicate Pim-1 kinase activity in HBP1 induction and transactivation of Pim-1 by HBP1 following oxidative stress. The hypothesis that Pim-1 and HBP1 are components of a common checkpoint pathway in response to oxidative stress is also supported by the finding that the loss of either protein attenuates H2O2-induced premature senescence and apoptosis (Fig. 10, F–H).
Figure 11.
Model for the proposed role of Pim-1 and HBP1 in the response to H2O2-induced premature senescence and apoptosis. In response to H2O2 induction, Pim-1 can positively regulate HBP1 by binding and phosphorylating at the position of Ser-372/Ser-380. This leads to HBP1 activation, which selectively turns on Pim-1 transcription, resulting in up-regulation of the Pim-1 level and further stabilization and induction of HBP1. The potent HBP1 activation leads to regulation of many other HBP1 target genes, including p16, DNMT1, and Bax, thus inducing premature senescence and apoptosis.
We also found that, in cells undergoing oxidative damage, HBP1 activation is initiated first, well before growth arrest occurs. Further HBP1 activation builds up gradually thereafter through escalation of the output of the Pim-1-HBP1 positive feedback loop (Fig. 10, A and B). Unexpectedly, the amplified action of the Pim-1-HBP1 positive feedback loop is abolished on longer exposure to H2O2 (over 60 h in HeLa or U2OS cells or 6 days in 2BS cells). These results indicate that the positive feedback loop between Pim-1-HBP1 only occurs during initial H2O2 exposure and contributes to premature senescence and apoptosis. We suggest that complex cell metabolic processes and activation of other signaling pathways may interfere with activity of the Pim-1-HBP1 positive feedback loop upon longer H2O2 exposure. However, p16 and Bax protein levels remain elevated and thus eventually induce premature senescence and apoptosis. This suggests that H2O2-exposed cells that undergo growth arrest can avoid tumorigenesis.
Proliferative arrest is a major characteristic of senescent and apoptotic cells. Increasing evidence indicates that cellular senescence and apoptosis act as protective mechanisms against tumorigenesis (46, 47). Pim-1 is a serine/threonine kinase that regulates multiple cellular functions, including the cell cycle, cell survival, and drug resistance. Aberrant elevation of Pim-1 kinase is associated with various types of cancer (12, 48, 49). Unexpectedly, in this study, we found that expression of the oncogene Pim-1 is induced in response to oxidative stress and that loss of Pim-1 impairs the cellular senescence or apoptosis response in normal fibroblast cells and tumor cells, suggesting that Pim-1 may have tumor-suppressive properties. Some studies have reported that a high Pim-1 level correlates with good prognosis in prostate adenocarcinoma, pancreatic ductal carcinoma, and non-small-cell lung cancer (50–53). This would be in keeping with Pim-1 being part of the cellular senescence or apoptosis mechanism. Based on the data that Pim-1 acts as a key regulator to induce senescence and apoptosis and thereby prevents propagation of damaged cells during H2O2 exposure, we speculate that inhibition of Pim-1 expression alone probably induces tumorigenesis in normal cells and enhances malignancy in tumor cells in response to oxidative stress. Thus, caution must be exercised when considering Pim-1 inhibitors as potential therapeutic interventions for cancer (16).
Our current study demonstrated that Pim-1 phosphorylates HBP1 protein on Ser-372 and Ser-380 to enhance its stability and that Ser-380 phosphorylation of HBP1 is indispensable for its activation of the Pim-1 promoter. We previously reported that p38 MAPK phosphorylates HBP1 on Ser-401 to contribute to Ras-induced premature senescence (42). Our data support a molecular framework in which HBP1 contributes to cell cycle inhibition by acting as a novel target of specific protein kinases. In the present work, we provide evidence that phosphorylation of HBP1 by Pim-1 enhances Pim-1 transcription to form a positive feedback loop and enhances the expression of target genes, including p16, DNMT1, and Bax, to induce cell cycle inhibition in response to oxidative stress. Our work underscores the importance of a Pim-1-HBP1 positive feedback loop for regulating cellular senescence and apoptosis in response to oxidative stress, which is a G1 arrest checkpoint control, especially during the early period of the stress. Hence, tumors may conceivably bypass optimal G1 arrest checkpoint control and accrue malignancy by disabling either Pim-1 or HBP1.
Experimental procedures
Cell culture, transfection, and lentivirus gene expression
2BS, HEK293T, HeLa, and U2OS cells were cultured in DMEM containing 10% fetal bovine serum. H2O2 was purchased from Sigma. The lentivirus vector pLL3.7-shHBP1 expresses shRNA that targets HBP1 mRNA (5′-ACTGTGAGTGCCACTTCTC-3′ and 5′-CACATGGAGCTTGATGACC-3′, and the lentivirus vector pLL3.7-shPim-1 expresses shRNA that targets Pim-1 mRNA (5′-TATTCCTTTCGAGCATG AC-3′ and 5′-GTTTCGTCCTGATCCTGGA-3′).
Real-time PCR
Total RNA was isolated using the RNAsimple Total RNA kit (Tiangen). The cDNA was synthesized using ReverAid First Strand cDNA Synthesis kit (Thermo Scientific) and then analyzed by real-time PCR analysis with Maxima SYBR Green qPCR Master Mix (Thermo Scientific). The DNA sequences of the human HBP1 primers are 5′-TGAAGGCTGTGATAATGAGGAAGAT-3′ and 5′-CATAGAAAGGGTGGTCCAGCTTA-3′. These primers result in a product of 191 bp. The DNA sequences of the human Pim-1 primers are 5′-CGAGCATGACGAAGAGATCAT-3′ and 5′-TCGAAGGTTGGCCTATCTGA-3′. These primers result in a 119-bp product. The human GAPDH primers are 5′-CCATGGAGAAGGCTGGGG-3′ and 5′-CAAAGTTGTCATGGATGACC-3′ with a 195-bp product. GAPDH is applied as an internal control for normalizing the real-time PCR results. Results are representative of three independent experiments, and values are the mean ± S.D. (error bars). *, p < 0.05; **, p < 0.01.
Western blotting and antibodies
The experiment was performed as described previously (30). The whole-cell lysates for Western blot analysis were prepared in radioimmune precipitation assay buffer (Thermo Scientific) containing protease inhibitor mixture (Sigma). Cytoplasmic and nuclear extracts were obtained using nuclear and cytoplasmic protein extraction reagents (Beyotime) containing protease inhibitor mixture (Sigma), and then protein concentrations were measured by the BCA Protein Assay kit (Pierce). 20–60 μg of protein was separated by SDS-PAGE and transferred to nitrocellulose membranes (Pall Corp.). The primary antibodies used for immunoblot analysis were against FLAG (F1804, Sigma), HA (MMS-101P, Covance), GST (IT003M, M&C Gene Technology), His (66005-1-Ig, Proteintech), Pim-1 (sc-13513, Santa Cruz Biotechnology), Bax (sc-7480, Santa Cruz Biotechnology), DNMT1 (sc-10222, Santa Cruz Biotechnology), p53 (sc-126, Santa Cruz Biotechnology), phosphoserine (sc-81514, Santa Cruz Biotechnology), phosphothreonine (9381S, Cell Signaling Technology), HBP1 (11746-1-AP, Proteintech), p16 (10883-1-AP, Proteintech), HDAC1 (AH379, Beyotime), α-tubulin (RM2007, Rayantibody), GFP (CW8006, CWBiotech), and GAPDH (KM9002, Sungene). The secondary antibodies IRDye 800-conjugated anti-mouse IgG antibody (610-132-121) and DyLight 800-conjugated affinity-purified anti-rabbit IgG (611-145-002) were purchased from Rockland.
GST pulldown assay
The control GST and GST-tagged proteins were expressed in Escherichia coli strain BL21 (DE3). Bacterial lysates were prepared in ice-cold binding buffer (PBS) by sonication and incubated with glutathione-Sepharose beads (GE Healthcare) overnight at 4 °C with rocking. After the incubation, His-tagged proteins were added to each tube for 4 h at 4 °C. The beads were washed with binding buffer three times and eluted with elution buffer (50 mm Tris-HCl, pH 8.0) containing 10 mm reduced glutathione. The elution was separated by SDS-PAGE, and the interactions were analyzed by Western blotting with the specified antibody.
In vitro kinase assay and identification of HBP1 phosphorylation sites by mass spectrometry
For the in vitro kinase assays, 2 μg of His-Pim-1 and 8 μg of His-HBP1 were incubated in kinase buffer (Cell Signaling Technology) for 30 min at 30 °C in the presence of 200 μm ATP. Then SDS-PAGE loading buffer was added to stop the reaction. Phosphorylation of HBP1 was analyzed by Western blotting with anti-phosphoserine or anti-phosphothreonine antibodies. To identify HBP1 phosphorylation sites, the reaction products were resolved by SDS-PAGE, and gels were stained with Coomassie Blue. The protein bands were retrieved and analyzed by mass spectrometry.
Protein half-life assay
HeLa cells were transfected with plasmids as indicated in individual experiments. 48 h after transfection, 100 μg/ml cycloheximide (CHX) was added to the dishes, and the CHX treatment was terminated at 0-, 30-, 60-, and 90-min time points as indicated. Whole-cell lysates were prepared, and 60 μg of total protein from each sample was analyzed by Western blotting with anti-HBP1 antibody. Quantification of HBP1 protein was determined using TotalLab software and normalized to GAPDH.
Electrophoretic mobility shift assay (EMSA)
The EMSA was performed as described previously (34). The DNA sequences of the probes used were as follows: Pim-1-WT, 5′-TAGAAAACTTTCAAAGGAAACATTTA-3′ and 5′-TAAATGTTTCCTTTGAAAGTTTTCTA-3′; Pim-1-MT, 5′-TAGAAAACTTTACCCTGAAACATTTA-3′ and 5′-TAAATGTTTCAGGGTAAAGTTTTCTA-3′.
Chromatin immunoprecipitation (ChIP)
ChIP assays were performed as described previously (34). For the Pim-1 promoter, the PCR primer sequences were 5′-ATAAAAGTTGTGGAGGATCTGGG-3′ and 5′-AATTGAATTGTCAGTGGCCTAAGTA-3′. For the DNMT1 promoter, the PCR primer sequences were 5′-AGATGGAGGTTGGATTGGA-3′ and 5′-AGAGGCGATACCCTGTGC-3′ (32). For the p16 promoter, the PCR primer sequences were 5′-CCTTCCAATGACTCCCTC-3′ and 5′-AACCTTCCTAACTGCCAAA-3′ (33).
Reporter gene assay
Cells were transfected with plasmids as indicated in individual experiments. Cell lysates were prepared with the Dual-Luciferase Reporter Assay kit (Promega) according to the manufacturer's instructions at 24- 48 h posttransfection. The firefly luciferase activity measurements were normalized to Renilla luciferase activity for the same sample. The luciferase assay was performed on three biological replicates, and each replicate was measured at least three times. The luciferase activities were expressed as the means ± S.D. (error bars) from four experiments. *, p < 0.05; **, p < 0.01.
3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay
HeLa, U2OS, and 2BS cells were stably transfected with plasmids as indicated in individual experiments. After puromycin (0.4 μg/ml) and/or G418 (300 μg/ml) selection, cells were seeded into 96-well plates at a density of 2000 cells/well. After culturing for 1, 2, 3, 4, 5, 6, 7, 9, or 11 days, 15 μl of MTT solution (5 mg/ml) was added to each well followed by further incubation at 37 °C for 4 h. Medium was removed, and 200 μl of DMSO was added to each well to dissolve the formazan crystals. The absorbance at 490 nm was read using a microplate reader. The MTT assay was performed on three biological replicates, and each replicate was measured at least three times.
SA-β-gal staining
The experiment was performed using a Senescence β-Galactosidase Staining kit (Beyotime) following the manufacturer's instructions. Cells were washed once in PBS, fixed for 15 min at room temperature in 3% formaldehyde, and washed three times with PBS again. Then cells were incubated overnight at 37 °C with freshly prepared SA-β-gal stain solution. At least 300 cells were counted in randomly chosen fields, and the percentage of cells positive for SA-β-Gal was determined in three independent experiments and expressed as mean ± S.D. (error bars). *, p < 0.05; **, p < 0.01.
Apoptosis assay
The apoptosis assay was conducted using the Annexin V-FITC/propidium iodide (PI) Dual Staining kit (BestBio). Briefly, cells were resuspended in 400 μl of 1× binding buffer and incubated with Annexin V-FITC for 15 min and then with PI for another 5 min at 4 °C in the dark. After staining, cells were analyzed using a flow cytometer (BD Biosciences). The apoptosis assay was measured at least three times with independent samples. Results of cell apoptosis rate were representative of three independent experiments and were expressed as mean ± S.D. (error bars). *, p < 0.05, **, p < 0.01.
Statistical analysis
The data are expressed as the mean ± S.D. (error bars) from three independent experiments as indicated in the figure legends. The statistical analysis was done by using Student's t test, and p < 0.01 or 0.05 was considered significant. *, p < 0.05, **, p < 0.01.
Author contributions
S. W. performed and analyzed the experiments and contributed to the manuscript writing. Z. C. and J. X. provided technical assistance and contributed to the preparation of the figures. H. L., W. J., and Y. C. contributed to the GST pulldown assay. G. L. contributed to the manuscript writing. X. Z. coordinated the study and wrote the paper. All authors reviewed the results and approved the final version of the manuscript.
Supplementary Material
Acknowledgment
We thank Dr. Zebin Mao for providing expression vector of Pim-1.
This work was supported by National Natural Science Foundation of China Grants 81672717, 31471292, and 81572705 and Beijing Natural Science Foundation Grant 5162013. The authors declare that they have no conflicts of interest with the contents of this article.
This article contains supplemental Figs. S1 and S2.
- HMG
- high mobility group
- HBP1
- HMG box transcription factor 1
- SA
- senescence-associated
- IP
- immunoprecipitation
- Luc
- luciferase
- CHX
- cycloheximide
- MTT
- 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide
- PI
- propidium iodide.
References
- 1. Schieber M., and Chandel N. S. (2014) ROS function in redox signaling and oxidative stress. Curr. Biol. 24, R453–462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Chance B., Sies H., and Boveris A. (1979) Hydroperoxide metabolism in mammalian organs. Physiol. Rev. 59, 527–605 [DOI] [PubMed] [Google Scholar]
- 3. Finkel T. (2011) Signal transduction by reactive oxygen species. J. Cell Biol. 194, 7–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Finkel T., and Holbrook N. J. (2000) Oxidants, oxidative stress and the biology of ageing. Nature 408, 239–247 [DOI] [PubMed] [Google Scholar]
- 5. Lombard D. B., Chua K. F., Mostoslavsky R., Franco S., Gostissa M., and Alt F. W. (2005) DNA repair, genome stability, and aging. Cell 120, 497–512 [DOI] [PubMed] [Google Scholar]
- 6. Lee K. Y., Koh S. H., Noh M. Y., Park K. W., Lee Y. J., and Kim S. H. (2007) Glycogen synthase kinase-3beta activity plays very important roles in determining the fate of oxidative stress-inflicted neuronal cells. Brain Res. 1129, 89–99 [DOI] [PubMed] [Google Scholar]
- 7. Valencia A., and Morán J. (2004) Reactive oxygen species induce different cell death mechanisms in cultured neurons. Free Radic. Biol. Med. 36, 1112–1125 [DOI] [PubMed] [Google Scholar]
- 8. Holmström K. M., and Finkel T. (2014) Cellular mechanisms and physiological consequences of redox-dependent signalling. Nat. Rev. Mol. Cell Biol. 15, 411–421 [DOI] [PubMed] [Google Scholar]
- 9. Nogueira V., Park Y., Chen C. C., Xu P. Z., Chen M. L., Tonic I., Unterman T., and Hay N. (2008) Akt determines replicative senescence and oxidative or oncogenic premature senescence and sensitizes cells to oxidative apoptosis. Cancer Cell 14, 458–470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Woo H. A., Yim S. H., Shin D. H., Kang D., Yu D. Y., and Rhee S. G. (2010) Inactivation of peroxiredoxin I by phosphorylation allows localized H2O2 accumulation for cell signaling. Cell 140, 517–528 [DOI] [PubMed] [Google Scholar]
- 11. Li Z., Lin F., Zhuo C., Deng G., Chen Z., Yin S., Gao Z., Piccioni M., Tsun A., Cai S., Zheng S. G., Zhang Y., and Li B. (2014) PIM1 kinase phosphorylates the human transcription factor FOXP3 at serine 422 to negatively regulate its activity under inflammation. J. Biol. Chem. 289, 26872–26881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Morishita D., Katayama R., Sekimizu K., Tsuruo T., and Fujita N. (2008) Pim kinases promote cell cycle progression by phosphorylating and down-regulating p27Kip1 at the transcriptional and posttranscriptional levels. Cancer Res. 68, 5076–5085 [DOI] [PubMed] [Google Scholar]
- 13. Yang J., Wang J., Chen K., Guo G., Xi R., Rothman P. B., Whitten D., Zhang L., Huang S., and Chen J. L. (2013) eIF4B phosphorylation by pim kinase plays a critical role in cellular transformation by Abl oncogenes. Cancer Res. 73, 4898–4908 [DOI] [PubMed] [Google Scholar]
- 14. Lilly M., Sandholm J., Cooper J. J., Koskinen P. J., and Kraft A. (1999) The PIM-1 serine kinase prolongs survival and inhibits apoptosis-related mitochondrial dysfunction in part through a bcl-2-dependent pathway. Oncogene 18, 4022–4031 [DOI] [PubMed] [Google Scholar]
- 15. Wang Z., Bhattacharya N., Weaver M., Petersen K., Meyer M., Gapter L., and Magnuson N. S. (2001) Pim-1: a serine/threonine kinase with a role in cell survival, proliferation, differentiation and tumorigenesis. J. Vet. Sci. 2, 167–179 [PubMed] [Google Scholar]
- 16. Jin B., Wang Y., Wu C. L., Liu K. Y., Chen H., and Mao Z. B. (2014) PIM-1 modulates cellular senescence and links IL-6 signaling to heterochromatin formation. Aging Cell 13, 879–889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hogan C., Hutchison C., Marcar L., Milne D., Saville M., Goodlad J., Kernohan N., and Meek D. (2008) Elevated levels of oncogenic protein kinase Pim-1 induce the p53 pathway in cultured cells and correlate with increased Mdm2 in mantle cell lymphoma. J. Biol. Chem. 283, 18012–18023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Zemskova M. Y., Song J. H., Cen B., Cerda-Infante J., Montecinos V. P., and Kraft A. S. (2015) Regulation of prostate stromal fibroblasts by the PIM1 protein kinase. Cell. Signal. 27, 135–146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Zemskova M., Lilly M. B., Lin Y. W., Song J. H., and Kraft A. S. (2010) p53-dependent induction of prostate cancer cell senescence by the PIM1 protein kinase. Mol. Cancer Res. 8, 1126–1141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Yee A. S., Shih H. H., and Tevosian S. G. (1998) New perspectives on retinoblastoma family functions in differentiation. Front. Biosci. 3, D532–D547 [DOI] [PubMed] [Google Scholar]
- 21. Zhuma T., Tyrrell R., Sekkali B., Skavdis G., Saveliev A., Tolaini M., Roderick K., Norton T., Smerdon S., Sedgwick S., Festenstein R., and Kioussis D. (1999) Human HMG box transcription factor HBP1: a role in hCD2 LCR function. EMBO J. 18, 6396–6406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Berasi S. P., Xiu M., Yee A. S., and Paulson K. E. (2004) HBP1 repression of the p47phox gene: cell cycle regulation via the NADPH oxidase. Mol. Cell. Biol. 24, 3011–3024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Driouch K., Briffod M., Bièche I., Champème M. H., and Lidereau R. (1998) Location of several putative genes possibly involved in human breast cancer progression. Cancer Res. 58, 2081–2086 [PubMed] [Google Scholar]
- 24. Koike M., Tasaka T., Spira S., Tsuruoka N., and Koeffler H. P. (1999) Allelotyping of acute myelogenous leukemia: loss of heterozygosity at 7q31.1 (D7S486) and q33–34 (D7S498, D7S505). Leuk. Res. 23, 307–310 [DOI] [PubMed] [Google Scholar]
- 25. Liang H., Fairman J., Claxton D. F., Nowell P. C., Green E. D., and Nagarajan L. (1998) Molecular anatomy of chromosome 7q deletions in myeloid neoplasms: evidence for multiple critical loci. Proc. Natl. Acad. Sci. U.S.A. 95, 3781–3785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Zenklusen J. C., Thompson J. C., Klein-Szanto A. J., and Conti C. J. (1995) Frequent loss of heterozygosity in human primary squamous cell and colon carcinomas at 7q31.1: evidence for a broad range tumor suppressor gene. Cancer Res. 55, 1347–1350 [PubMed] [Google Scholar]
- 27. Zenklusen J. C., Thompson J. C., Troncoso P., Kagan J., and Conti C. J. (1994) Loss of heterozygosity in human primary prostate carcinomas: a possible tumor suppressor gene at 7q31.1. Cancer Res. 54, 6370–6373 [PubMed] [Google Scholar]
- 28. Zenklusen J. C., Weitzel J. N., Ball H. G., and Conti C. J. (1995) Allelic loss at 7q31.1 in human primary ovarian carcinomas suggests the existence of a tumor suppressor gene. Oncogene 11, 359–363 [PubMed] [Google Scholar]
- 29. Zhang X., Kim J., Ruthazer R., McDevitt M. A., Wazer D. E., Paulson K. E., and Yee A. S. (2006) The HBP1 transcriptional repressor participates in RAS-induced premature senescence. Mol. Cell. Biol. 26, 8252–8266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Wang W., Chen Y., Wang S., Hu N., Cao Z., Wang W., Tong T., and Zhang X. (2014) PIASxα ligase enhances SUMO1 modification of PTEN protein as a SUMO E3 ligase. J. Biol. Chem. 289, 3217–3230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Chen Y. C., Zhang X. W., Niu X. H., Xin D. Q., Zhao W. P., Na Y. Q., and Mao Z. B. (2010) Macrophage migration factor is a direct target of HBP1-mediated transcriptional repression that is overexpressed in prostate cancer. Oncogene 29, 3067–3078 [DOI] [PubMed] [Google Scholar]
- 32. Pan K., Chen Y., Roth M., Wang W., Wang S., Yee A. S., and Zhang X. (2013) HBP1-mediated transcriptional regulation of DNA methyltransferase 1 and its impact on cell senescence. Mol. Cell. Biol. 33, 887–903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Li H., Wang W., Liu X., Paulson K. E., Yee A. S., and Zhang X. (2010) Transcriptional factor HBP1 targets P16 (INK4A), upregulating its expression and consequently is involved in Ras-induced premature senescence. Oncogene 29, 5083–5094 [DOI] [PubMed] [Google Scholar]
- 34. Chen Y., Pan K., Wang P., Cao Z., Wang W., Wang S., Hu N., Xue J., Li H., Jiang W., Li G., and Zhang X. (2016) HBP1-mediated regulation of p21 protein through the Mdm2/p53 and TCF4/EZH2 pathways and its impact on cell senescence and tumorigenesis. J. Biol. Chem. 291, 12688–12705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Gartel A. L., Goufman E., Tevosian S. G., Shih H., Yee A. S., and Tyner A. L. (1998) Activation and repression of p21 (WAF1/CIP1) transcription by RB binding proteins. Oncogene 17, 3463–3469 [DOI] [PubMed] [Google Scholar]
- 36. Yao C. J., Works K., Romagnoli P. A., and Austin G. E. (2005) Effects of overexpression of HBP1 upon growth and differentiation of leukemic myeloid cells. Leukemia 19, 1958–1968 [DOI] [PubMed] [Google Scholar]
- 37. Lemercier C., Duncliffe K., Boibessot I., Zhang H., Verdel A., Angelov D., and Khochbin S. (2000) Involvement of retinoblastoma protein and HBP1 in histone H1(0) gene expression. Mol. Cell. Biol. 20, 6627–6637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Dephoure N., Zhou C., Villén J., Beausoleil S. A., Bakalarski C. E., Elledge S. J., and Gygi S. P. (2008) A quantitative atlas of mitotic phosphorylation. Proc. Natl. Acad. Sci. U.S.A. 105, 10762–10767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Macek B., Mann M., and Olsen J. V. (2009) Global and sitespecific quantitative phosphoproteomics: principles and applications. Annu. Rev. Pharmacol. Toxicol. 49, 199–221 [DOI] [PubMed] [Google Scholar]
- 40. Olsen J. V., Blagoev B., Gnad F., Macek B., Kumar C., Mortensen P., and Mann M. (2006) Global, in vivo, and site-specific phosphorylation dynamics in signaling networks. Cell 127, 635–648 [DOI] [PubMed] [Google Scholar]
- 41. Olsen J. V., Vermeulen M., Santamaria A., Kumar C., Miller M. L., Jensen L. J., Gnad F., Cox J., Jensen T. S., Nigg E. A., Brunak S., and Mann M. (2010) Quantitative phosphoproteomics reveals widespread full phosphorylation site occupancy during mitosis. Sci. Signal. 3, ra3. [DOI] [PubMed] [Google Scholar]
- 42. Xiu M., Kim J., Sampson E., Huang C. Y., Davis R. J., Paulson K. E., and Yee A. S. (2003) The transcriptional repressor HBP1 is a target of the p38 mitogen-activated protein kinase pathway in cell cycle regulation. Mol. Cell. Biol. 23, 8890–8901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Samse K., Emathinger J., Hariharan N., Quijada P., Ilves K., Völkers M., Ormachea L., De La Torre A., Orogo A. M., Alvarez R., Din S., Mohsin S., Monsanto M., Fischer K. M., Dembitsky W. P., et al. (2015) Functional effect of Pim1 depends upon intracellular localization in human cardiac progenitor cells. J. Biol. Chem. 290, 13935–13947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Paulson K. E., Rieger-Christ K., McDevitt M. A., Kuperwasser C., Kim J., Unanue V. E., Zhang X., Hu M., Ruthazer R., Berasi S. P., Huang C. Y., Giri D., Kaufman S., Dugan J. M., Blum J., et al. (2007) Alterations of the HBP1 transcriptional repressor are associated with invasive breast cancer. Cancer Res. 67, 6136–6145 [DOI] [PubMed] [Google Scholar]
- 45. Swanson K. A., Knoepfler P. S., Huang K., Kang R. S., Cowley S. M., Laherty C. D., Eisenman R. N., and Radhakrishnan I. (2004) HBP1 and Mad1 repressors bind the Sin3 corepressor PAH2 domain with opposite helical orientations. Nat. Struct. Mol. Biol. 11, 738–746 [DOI] [PubMed] [Google Scholar]
- 46. Lanigan F., Geraghty J. G., and Bracken A. P. (2011) Transcriptional regulation of cellular senescence. Oncogene 30, 2901–2911 [DOI] [PubMed] [Google Scholar]
- 47. Yin X., Zhang Y., Su J., Hou Y., Wang L., Ye X., Zhao Z., Zhou X., Li Y., and Wang Z. (2016) Rottlerin exerts its anti-tumor activity through inhibition of Skp2 in breast cancer cells. Oncotarget 7, 66512–66524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Mochizuki T., Kitanaka C., Noguchi K., Muramatsu T., Asai A., and Kuchino Y. (1999) Physical and functional interactions between Pim-1 kinase and Cdc25A phosphatase. Im-plications for the Pim-1-mediated activation of the c-Myc signaling pathway. J. Biol. Chem. 274, 18659–18666 [DOI] [PubMed] [Google Scholar]
- 49. Nawijn M. C., Alendar A., and Berns A. (2011) For better or for worse: the role of Pim oncogenes in tumorigenesis. Nat. Rev. Cancer 11, 23–34 [DOI] [PubMed] [Google Scholar]
- 50. Dhanasekaran S. M., Barrette T. R., Ghosh D., Shah R., Varambally S., Kurachi K., Pienta K. J., Rubin M. A., and Chinnaiyan A. M. (2001) Delineation of prognostic biomarkers in prostate cancer. Nature 412, 822–826 [DOI] [PubMed] [Google Scholar]
- 51. Reiser-Erkan C., Erkan M., Pan Z., Bekasi S., Giese N. A., Streit S., Michalski C. W., Friess H., and Kleeff J. (2008) Hypoxia-inducible proto-oncogene Pim-1 is a prognostic marker in pancreatic ductal adenocarcinoma. Cancer Biol. Ther. 7, 1352–1359 [DOI] [PubMed] [Google Scholar]
- 52. Warnecke-Eberz U., Bollschweiler E., Drebber U., Pohl A., Baldus S. E., Hoelscher A. H., and Metzger R. (2008) Frequent down-regulation of pim-1 mRNA expression in non-small cell lung cancer is associated with lymph node metastases. Oncol. Rep. 20, 619–624 [PubMed] [Google Scholar]
- 53. Zhang Y., Parsanejad M., Huang E., Qu D., Aleyasin H., Rousseaux M. W., Gonzalez Y. R., Cregan S. P., Slack R. S., and Park D. S. (2010) Pim-1 kinase as activator of the cell cycle pathway in neuronal death induced by DNA damage. J. Neurochem. 112, 497–510 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.