Abstract
Lipid peroxidation is an endogenous source of aldehydes that gives rise to covalent modification of proteins in various pathophysiological states. In this study, a strategy for the comprehensive detection and comparison of adducts was applied to find a biomarker for lipid peroxidation-modified proteins in vivo. This adductome approach utilized liquid chromatography with electrospray ionization tandem mass spectrometry (LC-ESI-MS/MS) methods designed to detect the specific product ions from positively ionized adducts in a selected reaction monitoring mode. Using this procedure, we comprehensively analyzed lysine and histidine adducts generated in the in vitro oxidized low-density lipoproteins (LDL) and observed a prominent increase in several adducts, including a major lysine adduct. Based on the high resolution ESI-MS of the adduct and on the LC-ESI-MS/MS analysis of the synthetic adduct candidates, the major lysine adduct detected in the oxidized LDL was identified as Nϵ-(8-carboxyoctanyl)lysine (COL). Strikingly, a significantly higher amount of COL was detected in the sera from atherosclerosis-prone mice and from patients with hyperlipidemia compared with the controls. These data not only offer structural insights into protein modification by lipid peroxidation products but also provide a platform for the discovery of biomarkers for human diseases.
Keywords: biomarker, lipid oxidation, lipoprotein, mass spectrometry (MS), protein chemical modification, aldehyde
Introduction
Lipid peroxidation in tissues represents a degradation process, which is the consequence of the production and the propagation of free radical reactions primarily involving membrane polyunsaturated fatty acids (PUFAs), and has been implicated in the pathogenesis of various diseases, including atherosclerosis, cancer, rheumatoid arthritis, and diabetes, as well as aging (1). Lipid peroxidation leads to the generation of a broad array of different products with diverse and potent biological activities. Among them are a variety of different aldehydes. The primary products of the lipid peroxidation reaction, lipid hydroperoxides, can undergo carbon-carbon bond cleavage via alkoxyl radicals in the presence of transition metals giving rise to the formation of short-chain unesterified aldehydes of 3–9 carbons in length, and a second class of aldehydes still esterified to the parent lipid (2). These reactive aldehydes are considered important mediators of cellular injury due to their ability to covalently modify biomolecules, which can disrupt essential cellular functions and cause mutations (2). Indeed, the covalent adduction of aldehydes to apolipoprotein B in low-density lipoproteins (LDL) has been strongly implicated in the mechanism by which LDL is converted into an atherogenic form that is taken up by macrophages, leading to the formation of foam cells.
Based on many reports concerning the chemical and immunochemical detection of lipid peroxidation-derived aldehyde adducts in human diseases, there is no doubt that the steady-state levels of lipid peroxidation products increase under pathophysiological conditions associated with oxidative stress. Considerable progress has recently been made toward understanding the mechanisms of action of lipid peroxidation products. However, there are intrinsic difficulties associated with measuring reactive molecules. Like other reactive species, such as reactive oxygen species, many lipid peroxidation products can readily react with biomolecules to form conjugates, whereas only a few studies have addressed the concentration of the aldehyde adducts in vitro and in vivo. This may be partly due to the fact that many adducts generated in the modified proteins are unstable under the strong acid conditions of conventional acid hydrolysis. In addition, because of technical constraints, the covalent modification of proteins by aldehydes has been individually studied, and comprehensive analysis of protein modification has rarely been performed.
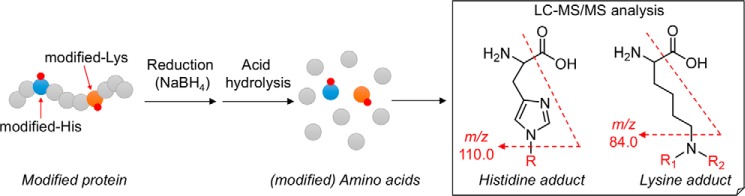
To simultaneously detect a variety of known and unknown adducts in protein samples, we adapted a mass spectrometry-based method for the comprehensive analysis of adducts (“adductome”). We expected that, although adductome analysis is semiquantitative, it would help to provide a comprehensive picture of the adducts. Our strategy for the adductome analysis is illustrated in Fig. 1. This method is based on the fact that aldehydes form adducts with specific amino acid residues possessing carbonyl groups or a Schiff base, which upon reaction with NaBH4 can be converted into their reduced derivatives. It was anticipated that these derivatives after borohydride reduction were resistant to the process of acid hydrolysis. In this study, based on the adductome analysis of oxidatively modified LDL, we successfully detected a number of histidine and lysine adducts, including a major lysine adduct, by high-performance-liquid chromatography with on-line electrospray ionization-tandem mass spectrometry (LC-ESI-MS/MS).2 Moreover, we established a stable isotope dilution-based absolute quantification method for the major lysine adduct and determined it in the sera from atherosclerosis-prone mice and from patients with hyperlipidemia. These data not only offer structural insights into protein modification by lipid peroxidation products, but also provide a platform for the discovery of biomarkers for human diseases.
Figure 1.
Strategy of histidine and lysine adductome analysis.
Results
Strategy for adductome analysis of aldehyde-modified proteins
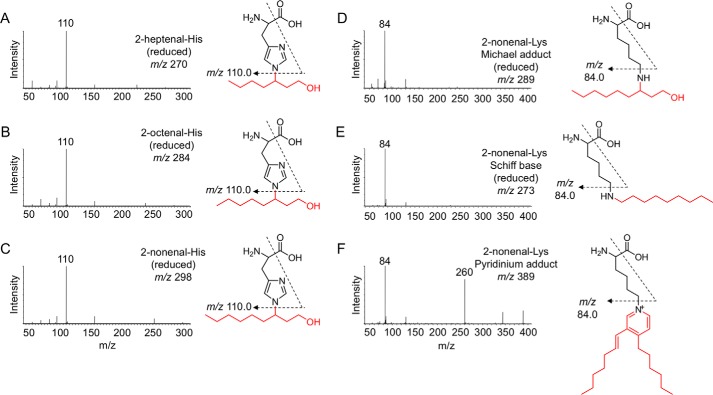
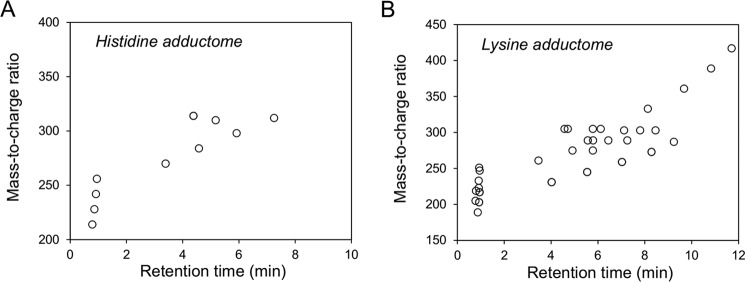
The histidine and lysine residues in proteins are two representative targets of covalent modification by lipid peroxidation-derived aldehydes (3). Therefore, we focused on adducts originating from these amino acids and attempted to identify a common fragment ion from the MS/MS product-ion analysis of the authentic adducts. When we analyzed the product ions of three authentic 2-alkenal-histidine adducts (2-hexenal-histidine, 2-octenal-histidine, and 2-nonenal-histidine Michael addition-type adducts), a common fragment ion at m/z 110, corresponding to a histidine immonium ion, was detected (Figs. 1 and 2, A–C). Similarly, the authentic lysine adducts (2-nonenal-lysine Michael addition-type, 2-nonenal-lysine Schiff base-type, and 2-nonenal-lysine pyridinium-type adducts) gave a common fragment ion at m/z 84, corresponding to the loss of NH3 from the lysine immonium ion (Figs. 1 and 2, D–F). It was expected that these characteristic common fragment ions might allow the comprehensive analysis of histidine and lysine adducts using LC-ESI-MS/MS. To test the validity of this procedure, native and modified BSA with the lipid peroxidation-derived aldehydes, such as 2-alkenals, 4-hydroxy-2-alkenals, and 4-hydroperoxy-2-nonenal, were treated with NaBH4 to stabilize the adducts, hydrolyzed under the conditions of conventional acidic hydrolysis, and then subjected to the LC-ESI-MS/MS analyses. As shown in Fig. 3, the MS data could be visualized as a two-dimensional image, in which the x axis represents the LC retention time, and the y axis represents the mass-to-charge ratio (m/z) for the individual detected adducts.
Figure 2.
Collision-induced dissociation of the [M + H]+ of histidine and lysine adducts at the collision energy of 25 V and proposed structures of individual ions. A, reduced 2-heptenal-His (m/z 270). B, reduced 2-octenal-His (m/z 284). C, reduced 2-nonenal-His (m/z 298). D, reduced 2-nonenal-Lys Michael adduct (m/z 289). E, reduced 2-nonenal-Lys Schiff base (m/z 273). F, 2-nonenal-Lys pyridinium adduct (m/z 389).
Figure 3.
Adductome maps of known histidine adducts (A) and lysine adducts (B). The specific product ions from positively ionized histidine and lysine adducts were analyzed by LC-ESI-MS/MS in the SRM mode transmitting the [M + H]+ >110 (for histidine adduct, A) or [M + H]+ >84 (for lysine adduct, B) transition.
Adductome analysis of oxidized LDL
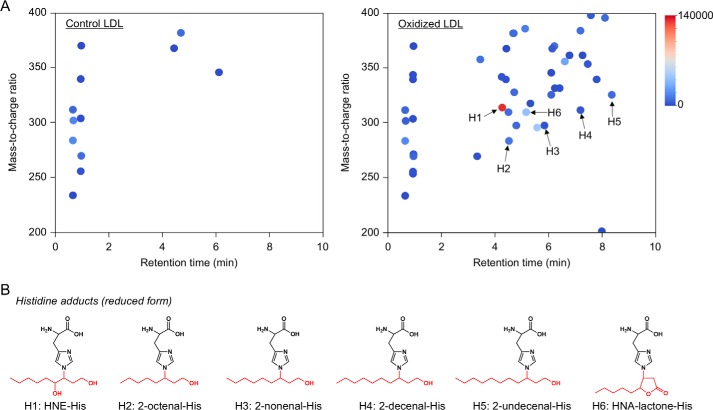
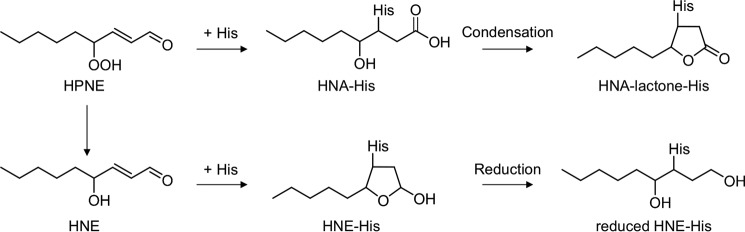
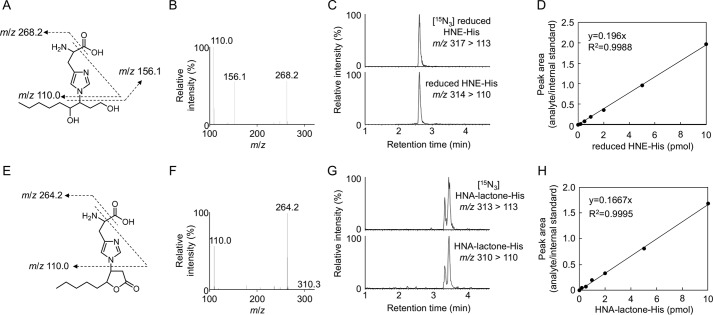
An important part of the pathogenesis of atherosclerosis has been implicated by the oxidative modification of low-density lipoproteins (LDL) (4, 5). The modification of LDL involves the peroxidation of PUFAs included in the LDL particle along with the appearance of lipid peroxidation products, such as aldehydes (2, 6). Hence, we applied the adductome analysis to the native and Cu2+-oxidized LDL to simultaneously detect a variety of histidine and lysine adducts. Fig. 4A shows the adductome maps of the putative histidine adducts generated in the native and oxidized LDL. The adductome maps are shown with a color gradient encoding the relative abundance from blue (low) to red (high). Product identification was tentatively made by comparison of the retention time and the mass-to-charge ratio of each adduct detected in the aldehyde-modified proteins (Fig. 3). The most abundant product H1 was suggested to be identical to the reduced form of the 4-hydroxy-2-nonenal (HNE)-histidine Michael adduct (Fig. 4B). In addition, products H2, H3, H4, and H5 were putatively identified as the reduced form of 2-octenal-histidine, 2-nonenal-histidine, 2-decanal-histidine, and 2-undecanal-histidine Michael addition-type adducts, respectively (Fig. 4B). The adductome maps suggested that these 2-alkenal-histidine adducts might be relatively minor products. It was also suggested that product H6 was one of the major histidine adducts with an M + H+ at 310, representing an increase of m/z 155. This change in the molecular weight was previously observed in an aldehyde-oxidized product, Nϵ-4-hydroxynonanoic acid-lysine, originating from the reaction of lysine with 4-hydroperoxy-2-nonenal (HPNE), the 4-hydroperoxy analog of HNE (7). Indeed, the hydroxynonanoic acid-histidine adduct was detected as one of the major products in the reaction of histidine with HPNE (data not shown). Thus, it was revealed that histidine residues could be converted to the HNE- and HPNE-derived adducts during oxidative modification of the LDL (Fig. 5).
Figure 4.
Adductome analysis of histidine adducts detected in the oxidized LDL. A, adductome maps of histidine adducts detected in the control (left) or Cu2+-oxidized LDL (right). The adductome maps are shown with a color gradient encoding the relative abundance from blue (low) to red (high). The known histidine adducts are indicated by arrows labeled H1–H6. B, chemical structure of histidine adducts (H1–H6) detected in the Cu2+-oxidized LDL.
Figure 5.
Proposed mechanism for the formation of HNE-His and HNA-lactone-His adducts.
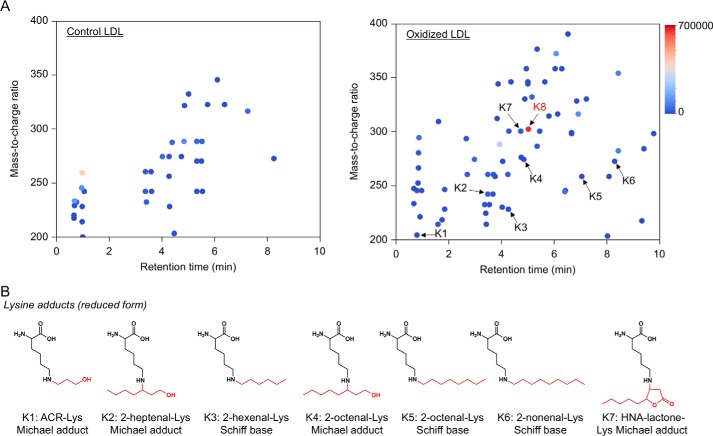
In contrast, the profile of lysine adducts generated in the oxidized LDL was much more complex than that of the histidine adducts (Fig. 6). In contrast to the histidine adducts, the 4-hydroxy-2-alkenal-derived lysine adducts were barely detected in the oxidized LDL. At least six minor products were putatively identified as the 2-alkenal-lysine adducts, such as the acrolein-lysine (K1), 2-heptenal-lysine (K2), 2-hexenal-lysine (K3), 2-octenal-lysine (K4 and K5), and 2-noxenal-lysine (K6) adducts. In addition, an HPNE-derived Nϵ-4-hydroxynonanoic acid-lysine adduct (HNA-lactone-Lys) was also detected as the minor product (K7). In addition to these minor products, we detected K8 as one of the most abundant lysine adducts generated in the oxidized LDL. However, the product showed no identity to any of the putative adducts detected in the modified proteins with authentic aldehydes (Fig. 3B). Therefore, we sought to identify the adduct K8.
Figure 6.
Adductome analysis of lysine adducts detected in the oxidized LDL. A, adductome maps of lysine adducts detected in the control (left) or Cu2+-oxidized LDL (right). The adductome maps are shown with a color gradient encoding the relative abundance from blue (low) to red (high). The known lysine adducts are indicated by arrows labeled K1–K7. The adduct K8 indicates a putative adduct that was detected at a high level. B, chemical structure of lysine adducts (K1–K7) detected in the Cu2+-oxidized LDL.
Identification of a major lysine adduct
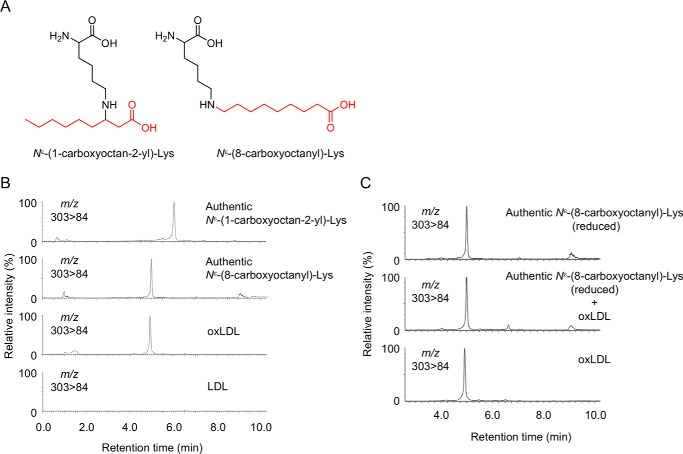
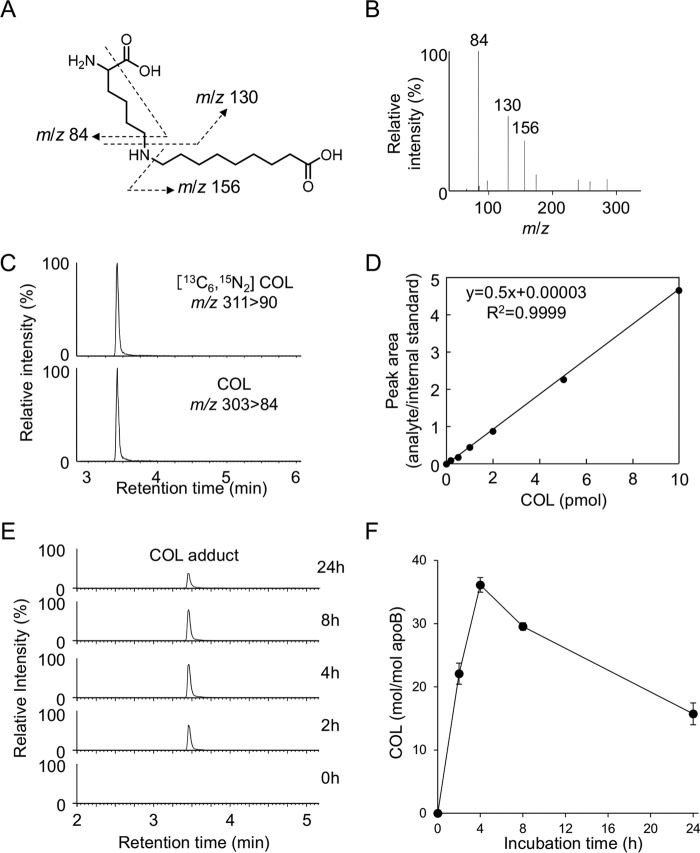
The LC-MS of the most abundant lysine adduct K8 in the positive ion mode showed a molecular ion at m/z of 303 ([M + H]+), corresponding to a 157-Da increase in the mass value of the unmodified lysine. The high resolution ESI-MS showed a molecular ion peak at m/z 303.22783, [M + H]+, corresponding to the molecular formula of C15H31N2O4. These data suggest that the adducted moiety may have the molecular formula of C9H17O2. Because this moiety was likely to be originated from lipid peroxidation, the structure of the adduct was speculated to be either Nϵ-(1-carboxyoctan-2-yl)lysine, a Michael addition adduct of lysine with 2-nonenoic acid, or Nϵ-(8-carboxyoctanyl)lysine, a Schiff base adduct of lysine with 9-oxononanoic acid (Fig. 7A). To confirm the structure, both adducts were chemically prepared and analyzed by LC-ESI-MS/MS. The data revealed that K8 was indistinguishable from Nϵ-(8-carboxyoctanyl)lysine (hereinafter referred to as “COL”) (Fig. 7, B and C).
Figure 7.
Identification of COL adduct as a major lysine adduct in oxidized LDL. A, chemical structure of Nϵ-(1-carboxyoctan-2-yl)-lysine (left) and COL adduct (right). B, LC-ESI-MS/MS analysis of authentic COL adduct. C, co-injection experiment on the LC-ESI-MS/MS of synthetic COL adduct with the acid-hydrolyzed sample from oxidized LDL.
Identification of a lipid-derived aldehyde responsible for the formation of COL
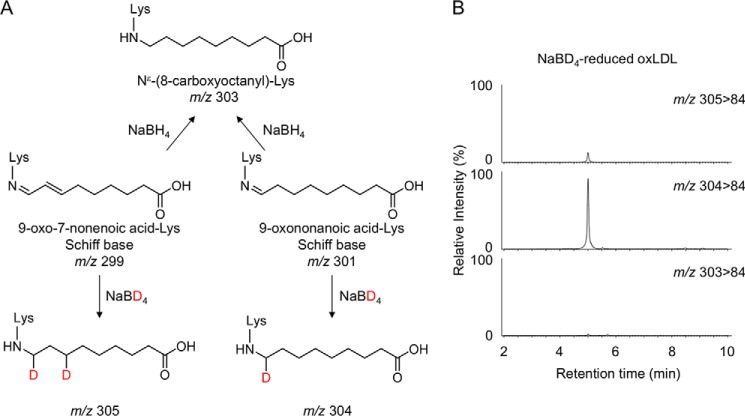
We next sought to identify the lipid-derived aldehyde responsible for the formation of COL in the oxidized LDL. The presence of a carboxyl group suggested that it might have originated from the reaction of lysine with aldehydes still esterified to the parent molecules of lipid esters, namely core aldehydes. Hence, we attempted to identify an oxidized fatty acid moiety in the core aldehydes. The most likely candidates might be the 9-oxo-7-nonenoic and 9-oxononanoic acids. Upon reaction with lysine, they form distinct Schiff base adducts but could be converted to the same product (COL) via reduction with NaBH4 (Fig. 8A). Hence, to determine which adducts are actually formed in the oxidized LDL, we performed the deuterium incorporation experiments using sodium borodeuteride (NaBD4). It was anticipated that reduction with NaBD4 allowed us to discriminate between the 9-oxononanoic acid-lysine and 9-oxo-7-nonenoic acid-lysine adducts; upon the NaBD4 reduction of the 9-oxo-7-nonenoic acid-lysine adduct (m/z 299), the molecular mass of COL increased by 6 Da (m/z 305), whereas the reduction of the 9-oxononanoic acid-lysine adduct (m/z 301) resulted in an increase of 3 Da (m/z 304) (Fig. 8A). This approach using NaBD4 was then applied to the oxidized LDL. The oxidized LDL was reduced with NaBD4, hydrolyzed, and analyzed by LC-ESI-MS/MS for the detection of COL. As shown in Fig. 8B, both deuterided COLs were detected; however, a monodeuterided product was detected much more prominently than the dideuterided product. These data strongly suggest that COL generated in the oxidized LDL mainly originated from the 9-oxononanoic acid-lysine adduct.
Figure 8.
Deuterium incorporation experiments using NaBD4. A, reduction of 9-oxo-7-nonenoic acid-Lys and 9-oxononanoic acid-Lys Schiff base adducts by NaBD4. B, LC-ESI-MS/MS analysis of the NaBD4-reduced oxidized LDL. The oxidized LDL was analyzed by LC-ESI-MS/MS in the SRM mode (upper, 305 > 84; middle, 304 > 84; lower, 303 > 84) following NaBD4 reduction and acid hydrolysis.
Determination of COL in the oxidized LDL
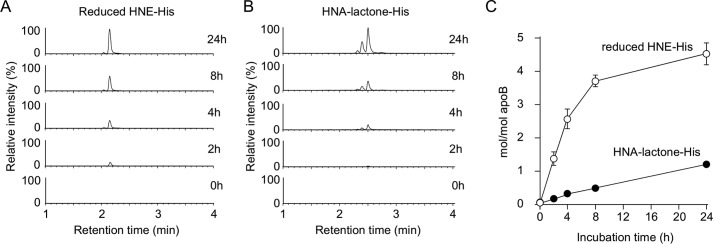
We then sought to determine whether COL could be a biomarker for the lipid peroxidation modification of proteins in vivo. To this end, we established a highly sensitive and specific method for the measurement of COL using LC-ESI-MS/MS coupled with a stable isotope dilution method. The collision-induced dissociation of COL produced relevant daughter ions at m/z 156, 130, and 84 (Fig. 9, A and B). The amount of COL was quantified by the ratio of the peak area of the target products and of the stable isotope-labeled internal standard (Fig. 9, C and D). We then attempted to detect COL in the Cu2+-oxidized LDL using LC-ESI-MS/MS. When the isolated human plasma LDL (1 mg/ml) was incubated at 37 °C with Cu2+ (5 μm), the formation of COL steadily increased up to 4 h and thereafter decreased (Fig. 9, E and F). Strikingly, the maximum yield of the adduct was 36 mol/mol LDL. This exceptionally high yield was in good agreement with the observation that COL was detected as one of the most abundant lysine adducts in the adductome analysis (Fig. 6). In contrast, the yields of the most abundant histidine adducts, i.e. the HNE-histidine and HPNE-histidine (HNA-lactone-His) adducts, were less than 5 mol/mol LDL (Figs. 10 and 11). COL was also detected in the minimally oxidized LDL with isolated lipoxygenase (2.40 ± 0.10 mol/mol LDL).
Figure 9.
Quantification of COL adduct in Cu2+-oxidized LDL. A and B, collision-induced dissociation of the [M + H]+ of COL adduct at the collision energy of 25 V. A, proposed structures of individual ions. C, LC-ESI-MS/MS analysis of [13C6,15N3]COL adduct. Upper, SRM for [13C6,15N3]COL adduct (m/z 311 > 90); lower, SRM for COL adduct (m/z 303 84). D, calibration curves for COL adduct. E and F, time-dependent formation of COL adduct in the Cu2+-oxidized LDL. The LDLs were analyzed by LC-ESI-MS/MS in the SRM mode following NaBH4 reduction and acid hydrolysis.
Figure 10.
Collision-induced dissociation of the [M + H]+ of reduced HNE-histidine and HNA-lactone-histidine adducts at the collision energy of 25 V. A–D, reduced HNE-His adducts (m/z 314). E–H, HNA-lactone-His adduct (m/z 310). Proposed structures of individual ions (A and E). C, LC-ESI-MS/MS analysis of 15N3-reduced HNE-His adduct. Upper, SRM for 15N3-reduced HNE-His (m/z 317 > 113); lower, SRM for reduced HNE-His (m/z 314 > 110). D, calibration curves for reduced HNE-His adduct. G, LC-ESI-MS/MS analysis of [15N3]HNA-lactone-His adduct. Upper, SRM for [15N3]HNA-lactone-His (m/z 313 > 113); lower, SRM for reduced HNA-lactone-His (m/z 310 > 110). H, calibration curves for HNA-lactone-His adduct.
Figure 11.
Quantification of histidine adducts in Cu2+-oxidized LDL. Time-dependent formation of reduced HNE-His (A and C) and HNA-lactone-His adducts (B and C) in the Cu2+-oxidized LDL. The LDLs were analyzed by LC-ESI-MS/MS in the SRM mode following NaBH4 reduction and acid hydrolysis.
Determination of COL in the sera from spontaneously hyperlipidemic mice and hyperlipidemia patients
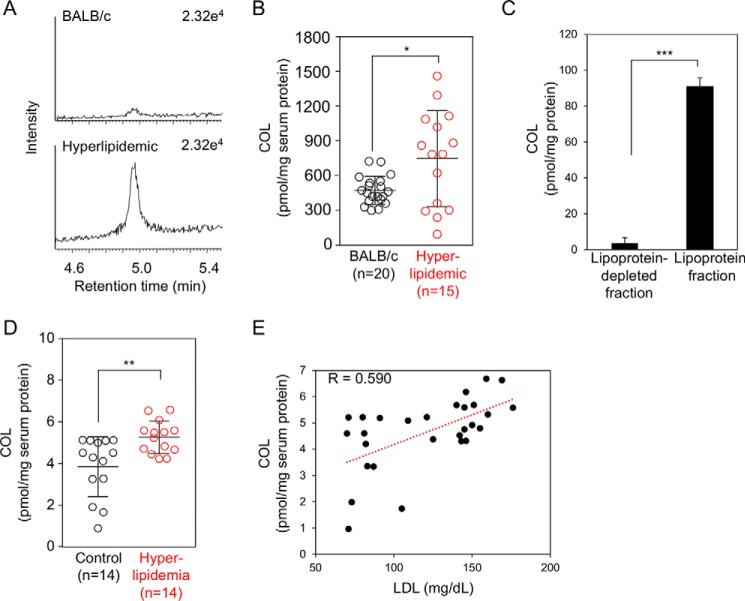
To evaluate the clinical utility of COL as a biomarker for human diseases, we determined the adduct in the sera from apoE-deficient C.KOR/StmSlc-Apoesh mice, spontaneously hyperlipidemic mice with a genetic background of BALB/c. We confirmed that the total cholesterol and triglyceride levels were significantly increased in the hyperlipidemic mice compared with that in the wild-type controls, although there were no significant differences in the glucose level and body weight (Table 1). The sera from the control BALB/c and hyperlipidemic mice were analyzed for COL using a stable isotope dilution-based LC-ESI-MS/MS technique. As shown in Fig. 12, A and B, consistent with the importance of the oxidation in the hyperlipidemic mice, the COL levels in the sera of hyperlipidemic mice were significantly higher than those of the normal mice. The average amount of COL in the control and hyperlipidemic mice was about 473 and 748 pmol/mg serum protein, respectively (Fig. 12B). In addition, we separated lipoprotein fractions from pooled fresh sera of hyperlipidemic mice and analyzed COL by LC-ESI-MS/MS. The amount of COL in the lipoprotein fractions was about 90.1 pmol/mg protein, whereas the adduct in lipoprotein-depleted fractions was only about 3.6 pmol/mg protein (Fig. 12C).
Table 1.
Fasting serum biochemistry in BALB/c and spontaneously hyperlipidemic mice
Data are shown as mean ± S.E. *, p < 0.05; ***, p < 0.005.
| BALB/c (n = 20) | Hyperlipidemic mice (n = 15) | |
|---|---|---|
| COL (pmol/mg protein) | 473.2 ± 27.3 | 748.3 ± 107.9* |
| Total cholesterol (mg/dl) | 90.8 ± 2.4 | 1018 ± 24*** |
| Triglyceride (mg/dl) | 36.3 ± 1.9 | 161.2 ± 9.9*** |
| Glucose (mg/dl) | 30.4 ± 3.1 | 27.4 ± 2.2 |
| Body weight (g) | 23.1 ± 0.3 | 23.5 ± 0.3 |
Figure 12.
Quantification of COL adduct in vivo. A and B, formation of COL adduct in the sera from control and spontaneously hyperlipidemic mice. *, p < 0.05. C, formation of COL adduct in lipoproteins factions from spontaneously hyperlipidemic mice. ***, p < 0.005. D, formation of COL adduct in the sera from normal subjects and patients with hyperlipidemia. **, p < 0.01. E, correlation between the levels of COL and those of LDL.
We then measured COL in the sera from patients with hyperlipidemia. Hyperlipidemia was defined as the total cholesterol level >220 mg/dl, an LDL level >140 mg/dl, and triglycerides >150 mg/dl (Table 2). Fourteen hyperlipidemia patients meeting these specific criteria and 14 healthy individuals were analyzed to measure the COL. The COL was detected in both the normal and hyperlipidemia individuals, but significantly higher amounts were present in the hyperlipidemia group (normal (n = 14), 3.94 ± 0.39 pmol/mg protein; hyperlipidemia (n = 14), 5.36 ± 0.21 pmol/mg protein (mean ± S.E.), p < 0.01 by Student's unpaired t test) (Fig. 12D). In addition, a significant but moderate correlation between the levels of COL and those of LDL was observed (Fig. 12E). This finding is consistent with the data that the exceptionally high yield of COL was detected as one of the most abundant lysine adducts in the adductome analysis of the in vitro-oxidized LDL. These clinical data suggest that COL could be a candidate biomarker for hyperlipidemia-related diseases, such as atherosclerosis.
Table 2.
Serum biochemistry in control and hyperlipidemia patients
Data are shown as mean ± S.E. **, p < 0.01; ***, p < 0.005.
| Control (n = 14) | Hyperlipidemia (n = 14) | |
|---|---|---|
| Female/male | 6/8 | 5/9 |
| COL(pmol/mg protein) | 3.94 ± 0.39 | 5.36 ± 0.21** |
| Age | 67.3 ± 2.7 | 66.1 ± 2.2 |
| LDL (mg/dl) | 89.2 ± 5.0 | 151.9 ± 2.9*** |
| Triglyceride (mg/dl) | 95.1 ± 7.7 | 168.6 ± 12.7*** |
| Total cholesterol (mg/dl) | 167.4 ± 6.99 | 246.6 ± 6.4*** |
| Glucose (mg/dl) | 90.9 ± 2.6 | 168.9 ± 13.8*** |
Discussion
In this study, to discover new biomarkers for the systematic detection and monitoring of lipid peroxidation-specific products, we adapted a mass spectrometry-based adductome approach. The major advantage of this approach was to facilitate the visualization of putative adduct patterns and their relative levels of incidence, differentiating between various modes of modification and assessing the contributions of a given mode to a particular disease state. By comparing adduct spots across parallel samples, this approach may lead to the identification of unique adduct spots capable of distinguishing different samples, a feature that has the potential use for biomarker discovery. The adductome approach was first applied by Matsuda and co-workers (8) for a survey of DNA adducts with lipid peroxidation products. They designed a strategy based on the principle that DNA adducts are prone to lose 2-deoxyribose from positively ionized 2-deoxynucleoside adducts during the fragmentation process, i.e. fragment ion peaks showing a loss of the 2-deoxyribose moiety from a precursor ion in the MS spectrum were presumed to be derived from the DNA adducts. Using this procedure, they performed a comprehensive analysis of the DNA adducts generated in human tissues (9–11).
Based on the fact that the generation of covalently modified proteins with lipid peroxidation products is associated with a number of pathological conditions (12), we expected that the adductome approach might also be useful for the discovery of new biomarkers for the lipid peroxidation modification of proteins. Taking advantage of the fact that the authentic histidine and lysine adducts gave the specific fragment ions that were observed at m/z 110 and 84, respectively, we analyzed modified proteins with the lipid peroxidation-derived aldehydes, such as 2-alkenals, 4-hydroxy-2-alkenals, and 4-hydroperoxy-2-nonenal, and we demonstrated that the data obtained from analyzing the complex adduct mixtures by LC-ESI-MS/MS could be visualized as a two-dimensional plot (Fig. 3). The identification of these fragment ions and the visualization of the adduct patterns were crucial because they allowed the comprehensive and comparative analysis of histidine and lysine adducts for the analysis of more complex biological samples.
The oxidative modification of LDL in the artery wall has been implicated as one of the major physiologically relevant mechanisms for the pathogenesis of atherosclerosis. There is widespread evidence supporting the role for lipid peroxidation in the molecular mechanism of the formation of the oxidized LDL as a pathogenic factor. Many of the lipid peroxidation products exhibit a high reactivity with proteins, generating a variety of inter- and intramolecular covalent adducts. Significant fractions of the lysine and histidine residues are modified during the Cu2+-catalyzed oxidation of the LDL (13). The PUFA-derived reactive aldehydes are thought to be responsible for such modifications by formation of adducts to the imidazole groups of the histidine residues and the ϵ-amino groups of the lysine residues. However, a large fraction of lysine residues modified during the LDL oxidation remains unquantified and even unidentified. Because of the extensively recognized biological effects of the oxidized LDL, identification and quantification of the adducts, especially the lysine adducts, are essential for clarifying the role of the oxidized LDL in the pathogenesis of atherosclerosis and other diseases. Hence, to find a biomarker for assessing the lipid peroxidation modification of protein in vivo, we analyzed the oxidized LDL by the adductome approach and identified 14 putative adducts (6 histidine and 8 lysine adducts) as biomarker candidates (Figs. 4 and 6). Although the identities of many other formed products remain to be determined, the adductome analysis of the in vitro oxidized LDL enabled the identification of the majority of the major histidine and lysine adducts.
The histidine adductome of the oxidized LDL revealed that HNE is mainly responsible for modification of the histidine residues in the LDL apoB (Fig. 4). This result is consistent with the previous finding that the yield of the HNE-histidine adducts generated in the oxidized LDL was about 6 mol/mol LDL, accounting for nearly 70% of the histidine residues lost (14). The adductome analysis of the oxidized LDL also allowed detection of a histidine adduct with a five-membered lactone ring (H6). The origin was speculated to be an HPNE adduct, 4-hydroxynonenoic acid-histidine. HPNE indeed reacted by a Michael addition mechanism with the imidazole ring of a histidine analog to generate the 4-hydroxynonenoic acid-histidine adduct, which was further converted to the lactone ring-containing product after acid hydrolysis (Fig. 5). Other adducts identified by the adductome included the reduced form of the 2-alkenal-histidine adducts, such as 2-octenal-histidine, 2-nonenal-histidine, 2-decanal-histidine, and 2-undecanal-histidine Michael addition-type adducts (Fig. 4B).
Based on the lysine adductome analysis of the oxidized LDL, we putatively identified COL as a potential biomarker for the lipid peroxidation modification of proteins. Strikingly, the maximum yield of COL was 36 mol/mol LDL, which is probably the most abundant among the adducts that had been detected in the in vitro oxidized LDL. In addition, COL was also detected in the lipoxygenase-catalyzed minimally oxidized LDL, suggesting that COL could be formed under a biologically relevant lipid oxidation state. Using the deuterium incorporation experiments (Fig. 8), we identified 9-oxononanoic acid as a source of COL. It has been reported that this aldehyde can be formed by the Hock cleavage reaction during the peroxidation of linoleic acid (15), the most abundant fatty acid at the sn-2 position of the membrane phospholipids (16). The presence of a carboxyl group suggested that the origin of COL might be lysine adducts with phospholipid and/or cholesteryl ester core aldehydes. Several previous studies have indeed demonstrated the covalent binding of oxidized phospholipids to lysine residues of the LDL apoB (17, 18). Itabe et al. (19) reported that oxidized phospholipids form complexes with lysine residues on proteins due to the presence of 9-oxononanoylphosphatidylcholine. Gillotte et al. (20) proved by measuring the phosphorus incorporated into the LDL apoB that the majority of adducts generated in the oxidized LDL was attributed to oxidized phospholipids. In addition to the adduction of oxidized phospholipids to apoB, the formation of the lysine Schiff-base adduct with oxidized cholesteryl esters during the LDL oxidation was also confirmed by LC-MS analysis (21) and by an immunochemical procedure (22).
Hyperlipidemia is a major risk factor for atherosclerotic cardiovascular disease (23). It is caused by impaired lipid metabolism and is marked by elevation of the serum total cholesterol, triglycerides, low-density lipoprotein cholesterol, and relative reduction of high-density lipoprotein cholesterol (24). Lipid-lowering drugs, such as statins, fibrates, and nicotinic acid, are commonly used for the treatment of hyperlipidemia. Using LC-ESI-MS/MS coupled with a stable isotope dilution method, a significantly higher amount of COL was detected in the sera from the atherosclerosis-prone, spontaneously hyperlipidemic mice. In addition, significantly higher amounts of COL were present in the patients with hyperlipidemia. Interestingly, the COL level in the sera from the mice was 100-fold higher than that of the human samples (Fig. 12). Although we have no data to explain the difference, this might be associated with the differences in the composition and/or metabolism of lipoproteins between mouse and human.
A correlation between the COL and LDL (Fig. 12) suggests that COL may arise from the peroxidation of lipoproteins, such as LDL. This speculation is also supported by our following observations: (i) the exceptionally high yield of COL was detected as one of the most abundant lysine adducts in the adductome analysis of the in vitro-oxidized LDL (Fig. 6), and (ii) COL was mainly detected in the lipoprotein fractions from sera of hyperlipidemic mice (Fig. 12). The oxidative modification of LDL-associated lipids is directly involved in the initiation of the atherosclerotic process, and the LDL quality directly influences the cardiovascular risk (25). Thus, COL may serve as a useful biochemical index of lipoprotein peroxidation and/or LDL quality in vivo. COL measurements may also facilitate a variety of investigations into the pathophysiology of both atherosclerosis- and age-related complications. These would include clinical studies aimed at elucidating the benefit of strict LDL control in preventing hyperlipidemia and atherosclerosis, as well as experimental investigations of the role of the lipid peroxidation modification of proteins in the pathogenesis of atherosclerosis. We also attempted to detect COL in the athero-prone legions from hyperlipidemic mice. Although COL was detectable in both control and athero-prone legions, a significant difference between control and athero-prone legions was not observed (data not shown). The COL levels in the tissues were extremely low (∼10 pmol/mg protein or less) in comparison with those detected in the sera from hyperlipidemic mice (∼700 pmol/mg protein) (Fig. 12). Although details remain unclear, these data suggest the presence of a specific production mechanism of COL in the circulating system and the utility and importance of COL as a serum biomarker for hyperlipidemia.
We hypothesized that if the lipid content is higher, more adducts are likely to be formed even when there is a “normal level” of lipid peroxidation. To confirm this, we normalized the amounts of COL to the triglyceride content. However, the normalization showed a significant decline of the COL levels in the hyperlipidemic mice compared with the control mice (data not shown). Similar results were also obtained in the hyperlipidemic patients (data not shown). These results suggest that although hyperlipidemia results in increased accumulation of lipids and thereby elevated plasma lipid peroxidation product levels, which in turn are responsible for the increase in COL, the individual lipids in the hyperlipidemic mice are less prone to lipid peroxidation. It may be associated with the fact that fatty acid composition can be changed in both plasma and liver samples in hyperlipidemia mice. In fact, the increase in the percentages of myristic acid (C14:0), palmitoleic acid (C16:1), and oleic acid (C18:1) has been reported in hyperlipidemia (26).
To examine the effect of high-fat diet on the COL adduct formation, we attempted to analyze the COL level in sera from a high-fat diet-fed hyperlipidemic mice. Contrary to expectations, the COL adduct was significantly reduced in the high-fat diet-fed hyperlipidemic mice compared with control diet-fed hyperlipidemic mice.3 These results may be ascribed to the high amounts of saturated and monounsaturated fatty acids (∼89%) and relatively low amounts of polyunsaturated fatty acids, including linoleic acid, a precursor of COL, in the high-fat diet. This can be the reason why the COL level was significantly reduced in the sera from the high-fat diet-fed hyperlipidemic mice. Although these results are very interesting, we would like to investigate more about this phenomenon in our future study.
In summary, to assess the totality of the covalent modification of histidine and lysine residues in protein, we developed a novel method for the comprehensive analysis of histidine and lysine modification based on LC-ESI-MS/MS. The adductome analysis of the oxidized LDL in this study allowed the identification of the most abundant lysine adduct, COL. In addition, a significantly higher amount of COL was detected in the sera from the atherosclerosis-prone mice and from the patients with hyperlipidemia, compared with the controls. These data not only offer structural insights into protein modification by lipid peroxidation products but also provide a platform for the discovery of biomarkers for human diseases.
Experimental procedures
Materials
HPNE was prepared by the autoxidation of (3Z)-nonenal as described previously (7). All of the other reagents used in the study were of analytical grade and obtained from commercial sources.
Preparation aldehyde-modified protein
Human serum albumin or BSA (1.0 mg/ml) was incubated with 1.0 mm each aldehyde in PBS at 37 °C for 24 h under atmospheric oxygen.
In vitro peroxidation of LDL
LDL (1.019–1.063 g/ml) was prepared from the plasma of healthy humans by sequential ultracentrifugation and then extensively dialyzed against PBS (pH 7.4) containing 100 μm EDTA at 4 °C. The LDL used for the oxidative modification by Cu2+ was dialyzed against a 1000-fold volume of PBS at 4 °C. It was sterilized with a Mille-GV filter (Millipore) after dialysis. The protein concentration of LDL was measured using the bicinchoninic acid protein assay reagent (Thermo Fisher Scientific). The oxidation of LDL (1 mg of protein/ml) by 5 μm Cu2+ was carried out at 37 °C under air in PBS (pH 7.4). Enzymatically modified LDL was prepared using soybean lipoxygenase (Sigma). LDL (1 mg of protein/ml) was incubated with or without lipoxygenase (1000 units/ml) for 8 h at 37 °C in PBS (pH 7.4) containing EDTA (1 mm).
Sample reduction and hydrolysis
The protein samples were reduced with 100 mm NaBH4 at room temperature for 3 h and then treated with an equal volume of 20% trichloroacetic acid on ice for 1 h. After centrifugation at 4000 × g for 30 min at 4 °C, the proteins were hydrolyzed in vacuo with 2 ml of 6 n HCl for 24 h at 110 °C.
Protein adductome analysis using LC-ESI-MS/MS
Acid-hydrolyzed protein used for the adductome analysis was redissolved in ethanol and then subjected to the protein adductome analysis using a TQD triple stage quadrupole mass spectrometer (Waters) equipped with an ACQUITY ultra-performance LC system (Waters). The sample injection volumes of 10 μl each were separated on a Develosil HB-C30-UG 3-μm column (100 × 2.0 mm) (Nomura Chemical) at the flow rate of 0.3 ml/min. A discontinuous gradient was used by solvent A (H2O containing 0.1% formic acid) with solvent B (methanol) as follows: 1% B at 0 min, 1% B at 1 min, 99% B at 15 min, and 99% B at 20 min. Selected reaction monitoring (SRM) was performed in the positive ion mode using nitrogen as the nebulizing gas. The experimental conditions were as follows: ion source temperature, 120 °C; desolvation temperature, 350 °C; cone voltage, 25 V; collision energy, 25 eV; desolvation gas flow rate, 700 liters/h; cone gas flow rate, 50 liters/h; collision gas, argon. The strategy was designed to detect the product ion (m/z 84.0, for lysine adducts; m/z 110.0, for histidine adducts) from positively ionized lysine and histidine adducts by monitoring the samples transmitting their [M + H]+ >84.0 (for lysine adducts) and [M + H]+ >110.0 (for His adducts) transitions.
Intra- and inter-assay variation of adductome analysis
To confirm the accuracy and reproducibility of adductome analysis, the intra-assay precision was determined in five repeats within one LC-MS/MS run using the same sample from oxidized human LDL. Inter-assay variation was investigated in three independent experimental runs performed on 3 days. The average variations in inter-assay experiments for known His adducts (four adducts) and Lys adducts (seven adducts) were 9.0 and 9.6%, respectively. The average variations in intra-assay experiments for known His adducts (four adducts) and Lys adducts (seven adducts) were 3.3 and 7.8%, respectively.
Quantification of HNE-histidine and HNA-lactone-histidine adducts by LC-ESI-MS/MS
Mass spectrometric analyses were performed using an ACQUITY TQD system (Waters) equipped with an ESI probe and interfaced with a UPLC system (Waters). The sample injection volumes of 10 μl each were separated on a Waters BEH C18 1.7-μm column (100 × 2.1 mm) at a flow rate of 0.3 ml/min. A discontinuous gradient was used by solvent A (H2O containing 0.1% formic acid) with solvent B (acetonitrile containing 0.1% formic acid) as follows: 5% B at 0 min, 95% B at 6 min, and 95% B at 7 min. Mass spectrometric analyses were performed on line using ESI-MS/MS in the positive ion mode with the SRM mode (cone potential 30 eV/collision energy 30 eV for HNE-histidine; cone potential 35 eV/collision energy 30 eV for HNA-lactone-His). The monitored SRM transitions were as follows: [15N3]HNE-histidine, m/z 317.0 > 110.0, and HNE-histidine, m/z 314.0 > 110.0; [15N3]HNA-lactone-histidine, m/z 313.0 > 113.0, and HNA-lactone-histidine, m/z 310.0 > 110.0. The amounts of the adducts were quantified by the ratio of the peak area of the target adducts and of the stable isotope. QuanLynx software (Waters) was used to create the standard curves and to calculate the adduct concentrations.
Preparation of COL
The 9-oxononanoic acid was prepared using the Grubbs catalyst second generation (Sigma). To a solution of 7-octenoic acid (2.5 mmol, 1 eq) and acrolein (7.4 mmol, 3 eq) in dry oxygen-free CH2Cl2 (12.5 ml) was added the Grubbs catalyst second generation (21.2 mg, 0.01 eq). After stirring for 3 h at room temperature, the solvent was evaporated, and the residue was purified by column chromatography (hexane/ethyl acetate, 50:50) to give the 9-oxononanoic acid. Nα-Acetyl-l-lysine (10 mm) was incubated with oxononanoic acid (10 mm) in PBS at 37 °C for 24 h. After incubation, the resulting reaction solution was reduced with 100 mm NaBH4 at room temperature for 3 h and then hydrolyzed with HCl for 24 h at 110 °C. After acid hydrolysis, the COL was purified by reverse-phase HPLC.
Preparation of Nϵ-(1-carboxyoctan-2-yl)-lysine
The Nϵ-(1-carboxyoctan-2-yl)-lysine adduct was prepared by the oxidation of the trans-2-nonenal-lysine Michael adduct as described previously (27). Briefly, trans-2-nonenal-treated HSA was treated with NaClO2 for oxidation of the aldehyde group to the carboxylic acid.
Quantification of COL adduct by LC-ESI-MS/MS
The sample injection volumes of 10 μl each were separated on a Develosil HB-C30-UG 3-μm column (100 × 2.0 mm) (Nomura Chemical) at the flow rate of 0.3 ml/min. A discontinuous gradient was used by solvent A (H2O containing 0.1% formic acid) with solvent B (methanol) as follows: 1% B at 0 min, 99% B at 6 min. Mass spectrometric analyses were performed on line using ESI-MS/MS in the positive ion mode with the SRM mode (cone potential 40 eV/collision energy 30 eV). The monitored SRM transitions were as follows: [13C6,15N2]COL, m/z 339.0 > 90.0; COL, m/z 331.0 > 84.0. The amounts of the adducts were quantified by the ratio of the peak area of the target adducts and of the stable isotope. QuanLynx software (Waters) was used to create the standard curves and to calculate the adduct concentrations. The intra-assay precision was determined in five repeats within one LC-MS/MS run using the hydrolyzed sample from serum of a single hyperlipidemic mouse. The variations in inter-assay experiments for the COL adduct was 5.3%.
Animal experiments
Male BALB/c and male spontaneously hyperlipidemic mice (C.KOR/StmSlc-Apoeshl), a line of apoE-deficient spontaneously hyperlipidemic mice with a genetic background of BALB/c, were purchased from Japan SLC (Hamamatsu, Japan). The mice were housed in a temperature-controlled pathogen-free room with light from 7:00 to 19:00 h (daytime) and had free access to standard food and water. All procedures were approved by the Animal Experiment Committee in the Graduate School of Bioagricultural Sciences, Nagoya University. Twenty BALB/c and 20 spontaneously hyperlipidemic mice were used as serum donors at 10 weeks of age. Among them, five of the hyperlipidemic mice, which were housed in the same cage, were excluded because of significant loss of body weight. Blood was collected from the tail vein and allowed to stand for 2 h at room temperature to coagulate the blood, after which the sera were collected by centrifugation at 3500 rpm for 10 min and stored at −80 °C until used.
The mouse sera (10–30 μl, 1 mg of protein/sample) were reduced with 100 mm NaBH4 at room temperature for 3 h and then treated with an equal volume of 20% trichloroacetic acid on ice for 1 h. After centrifugation at 4000 × g for 30 min at 4 °C, the proteins mixed with the internal standard (1 nmol) were hydrolyzed in vacuo with 2 ml of 6 n HCl for 24 h at 110 °C. The resulting hydrolysates were dissolved with 400 μl of ethanol and analyzed by LC-MS/MS.
Plasma/serum glucose, triglyceride, low density lipoprotein, and total cholesterol were examined by the Nagoya University Clinical Laboratory. The lipoprotein fraction from sera of hyperlipidemic mice was precipitated with dextran sulfate and calcium chloride. The 10% dextran sulfate solution (4 μl) and 1 m CaCl2 (100 μl) were added in 100 μl of pooled fresh sera from four hyperlipidemic mice (male, 9 weeks old). After incubation for 10 min at room temperature with mixing, the samples were centrifuged at 10,000 × g for 10 min at 4 °C. The resulting precipitate was used as a lipoprotein fraction.
Human serum samples
Serum samples were obtained from 14 healthy individuals and 14 patients with hyperlipidemia who underwent diagnostic evaluation at the Nagoya University Hospital (Nagoya, Japan). This study was approved by the Ethical Committee of the Nagoya University School of Medicine. Patients were considered to have this lipid disorder based on clinical and biochemical criteria as follows: high total cholesterol (>220 mg/dl), triglyceride (>150 mg/dl), and LDL (>140 mg/dl) levels at the baseline.
The human frozen sera (10–30 μl, 1 mg of protein/sample) were thawed and reduced with 100 mm NaBH4 at room temperature for 3 h and then treated with an equal volume of 20% trichloroacetic acid on ice for 1 h. After centrifugation at 4000 × g for 30 min at 4 °C, the proteins mixed with the internal standard (1 nmol) were hydrolyzed in vacuo with 2 ml of 6 N HCl for 24 h at 110 °C. The resulting hydrolysates were dissolved with 400 μl of ethanol and analyzed by LC-MS/MS.
Author contributions
K. U. designed research; T. S., K. S., K. H., F. N., and R. K. performed research; R. K., and T. M. contributed human samples; T. S., R. K., T. M., and K. U. analyzed data; T. S. and K. U. wrote the paper.
Acknowledgments
We thank Yuki Hondoh and Emi Inuzuka for their excellent editorial and technical support.
This work was supported in part by Grant-in-aid for Scientific Research (A) 26252018 (to K. U.), Grant-in-aid for Scientific Research on Innovative Areas “Oxygen Biology: a new criterion for integrated understanding of life” 26111011 (to K. U.), and the Japan Science and Technology Agency PRESTO program (JPMJPR1334 to T. S.) of the Ministry of Education, Sciences, Sports, Technology (MEXT), Japan. The authors declare that they have no conflicts of interest with the contents of this article.
T. Shibata, K. Shimizu, and K. Uchida, unpublished observations.
- LC-ESI-MS/MS
- high performance liquid chromatography with on-line electrospray ionization tandem mass spectrometry
- COL
- Nϵ-(8-carboxyoctanyl)lysine
- HNA
- 4-hydroxynonanoic acid
- HNE
- 4-hydroxy-2-nonenal
- HPNE
- 4-hydroperoxy-2-nonenal
- SRM
- selected reaction monitoring.
References
- 1. Gutteridge J. M., and Halliwell B. (1990) The measurement and mechanism of lipid peroxidation in biological systems. Trends Biochem. Sci. 15, 129–135 [DOI] [PubMed] [Google Scholar]
- 2. Esterbauer H., Schaur R. J., and Zollner H. (1991) Chemistry and biochemistry of 4-hydroxynonenal, malonaldehyde and related aldehydes. Free Radic. Biol. Med. 11, 81–128 [DOI] [PubMed] [Google Scholar]
- 3. Uchida K. (2003) Histidine and lysine as targets of oxidative modification. Amino Acids 25, 249–257 [DOI] [PubMed] [Google Scholar]
- 4. Steinberg D., Parthasarathy S., Carew T. E., Khoo J. C., and Witztum J. L. (1989) Beyond cholesterol. Modifications of low-density lipoprotein that increase its atherogenicity. N. Engl. J. Med. 320, 915–924 [DOI] [PubMed] [Google Scholar]
- 5. Steinberg D. (1995) Role of oxidized LDL and antioxidants in atherosclerosis. Adv. Exp. Med. Biol. 369, 39–48 [DOI] [PubMed] [Google Scholar]
- 6. Jürgens G., Lang J., and Esterbauer H. (1986) Modification of human low-density lipoprotein by the lipid peroxidation product 4-hydroxynonenal. Biochim. Biophys. Acta 875, 103–114 [DOI] [PubMed] [Google Scholar]
- 7. Shimozu Y., Hirano K., Shibata T., Shibata N., and Uchida K. (2011) 4-Hydroperoxy-2-nonenal is not just an intermediate but a reactive molecule that covalently modifies proteins to generate unique intramolecular oxidation products. J. Biol. Chem. 286, 29313–29324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kanaly R. A., Hanaoka T., Sugimura H., Toda H., Matsui S., and Matsuda T. (2006) Development of the adductome approach to detect DNA damage in humans. Antioxid. Redox Signal. 8, 993–1001 [DOI] [PubMed] [Google Scholar]
- 9. Kanaly R. A., Matsui S., Hanaoka T., and Matsuda T. (2007) Application of the adductome approach to assess intertissue DNA damage variations in human lung and esophagus. Mutat. Res. 625, 83–93 [DOI] [PubMed] [Google Scholar]
- 10. Chou P.-H., Kageyama S., Matsuda S., Kanemoto K., Sasada Y., Oka M., Shinmura K., Mori H., Kawai K., Kasai H., Sugimura H., and Matsuda T. (2010) Detection of lipid peroxidation-induced DNA adducts caused by 4-oxo-2(E)-nonenal and 4-oxo-2(E)-hexenal in human autopsy tissues. Chem. Res. Toxicol. 23, 1442–1448 [DOI] [PubMed] [Google Scholar]
- 11. Matsuda T., Tao H., Goto M., Yamada H., Suzuki M., Wu Y., Xiao N., He Q., Guo W., Cai Z., Kurabe N., Ishino K., Matsushima Y., Shinmura K., Konno H., et al. (2013) Lipid peroxidation-induced DNA adducts in human gastric mucosa. Carcinogenesis 34, 121–127 [DOI] [PubMed] [Google Scholar]
- 12. Stadtman E. R., and Levine R. L. (2000) Protein oxidation. Ann. N.Y. Acad. Sci. 899, 191–208 [DOI] [PubMed] [Google Scholar]
- 13. Uchida K., Toyokuni S., Nishikawa K., Kawakishi S., Oda H., Hiai H., and Stadtman E. R. (1994) Michael addition-type 4-hydroxy-2-nonenal adducts in modified low-density lipoproteins: markers for atherosclerosis. Biochemistry 33, 12487–12494 [DOI] [PubMed] [Google Scholar]
- 14. Kumano-Kuramochi M., Shimozu Y., Wakita C., Ohnishi-Kameyama M., Shibata T., Matsunaga S., Takano-Ishikawa Y., Watanabe J., Goto M., Xie Q., Komba S., Uchida K., and Machida S. (2012) Identification of 4-hydroxy-2-nonenal-histidine adducts that serve as ligands for human lectin-like oxidized LDL receptor-1. Biochem. J. 442, 171–180 [DOI] [PubMed] [Google Scholar]
- 15. Schneider C., Tallman K. A., Porter N. A., and Brash A. R. (2001) Two distinct pathways of formation of 4-hydroxynonenal. Mechanisms of nonenzymatic transformation of the 9- and 13-hydroperoxides of linoleic acid to 4-hydroxyalkenals. J. Biol. Chem. 276, 20831–20838 [DOI] [PubMed] [Google Scholar]
- 16. Baggio B., Gambaro G., Zambon S., Marchini F., Bassi A., Bordin L., Clari G., and Manzato E. (1996) Anomalous phospholipid n-6 polyunsaturated fatty acid composition in idiopathic calcium nephrolithiasis. J. Am. Soc. Nephrol. 7, 613–620 [DOI] [PubMed] [Google Scholar]
- 17. Steinbrecher U. P., Witztum J. L., Parthasarathy S., and Steinberg D. (1987) Decrease in reactive amino groups during oxidation or endothelial cell modification of LDL. Correlation with changes in receptor-mediated catabolism. Arteriosclerosis 7, 135–143 [DOI] [PubMed] [Google Scholar]
- 18. Tertov V. V., Kaplun V. V., Dvoryantsev S. N., and Orekhov A. N. (1995) Apolipoprotein B-bound lipids as a marker for evaluation of low density lipoprotein oxidation in vivo. Biochem. Biophys. Res. Commun. 214, 608–613 [DOI] [PubMed] [Google Scholar]
- 19. Itabe H., Takeshima E., Iwasaki H., Kimura J., Yoshida Y., Imanaka T., and Takano T. (1994) A monoclonal antibody against oxidized lipoprotein recognizes foam cells in atherosclerotic lesions. Complex formation of oxidized phosphatidylcholines and polypeptides. J. Biol. Chem. 269, 15274–15279 [PubMed] [Google Scholar]
- 20. Gillotte K. L., Hörkkö S., Witztum J. L., and Steinberg D. (2000) Oxidized phospholipids, linked to apolipoprotein B of oxidized LDL, are ligands for macrophage scavenger receptors. J. Lipid Res. 41, 824–833 [PubMed] [Google Scholar]
- 21. Hoppe G., Ravandi A., Herrera D., Kuksis A., and Hoff H. F. (1997) Oxidation products of cholesteryl linoleate are resistant to hydrolysis in macrophages, form complexes with proteins, and are present in human atherosclerotic lesions. J. Lipid Res. 38, 1347–1360 [PubMed] [Google Scholar]
- 22. Kawai Y., Saito A., Shibata N., Kobayashi M., Yamada S., Osawa T., and Uchida K. (2003) Covalent binding of oxidized cholesteryl esters to protein: implications for oxidative modification of low density lipoprotein and atherosclerosis. J. Biol. Chem. 278, 21040–21049 [DOI] [PubMed] [Google Scholar]
- 23. LaRosa J. C. (2001) Prevention and treatment of coronary heart disease: who benefits? Circulation 104, 1688–1692 [DOI] [PubMed] [Google Scholar]
- 24. Alwaili K., Alrasadi K., Awan Z., and Genest J. (2009) Approach to the diagnosis and management of lipoprotein disorders. Curr. Opin. Endocrinol. Diabetes Obes. 16, 132–140 [DOI] [PubMed] [Google Scholar]
- 25. Albertini R., Moratti R., and De Luca G. (2002) Oxidation of low-density lipoprotein in atherosclerosis from basic biochemistry to clinical studies. Curr. Mol. Med. 2, 579–592 [DOI] [PubMed] [Google Scholar]
- 26. Fujiwara M., Sato T., Tazaki H., Yamamoto I., Kawasumi K., and Arai T. (2013) Changes in plasma fatty acid composition in hyperlipidemia dogs. Asian J. Anim. Vet. Adv. 8, 639–646 [Google Scholar]
- 27. Dalcanale E., and Montanari F. (1986) Selective oxidation of aldehydes to carboxylic acids with sodium chlorite-hydrogen peroxide. J. Org. Chem. 51, 567–569 [Google Scholar]