Abstract
Members of the CAP superfamily (cysteine-rich secretory proteins, antigen 5, and pathogenesis-related 1 proteins), also known as SCP superfamily (sperm-coating proteins), have been implicated in many physiological processes, including immune defenses, venom toxicity, and sperm maturation. Their mode of action, however, remains poorly understood. Three proteins of the CAP superfamily, Pry1, -2, and -3 (pathogen related in yeast), are encoded in the Saccharomyces cerevisiae genome. We have shown previously that Pry1 binds cholesterol in vitro and that Pry function is required for sterol secretion in yeast cells, indicating that members of this superfamily may generally bind sterols or related small hydrophobic compounds. On the other hand, tablysin-15, a CAP protein from the horsefly Tabanus yao, has been shown to bind leukotrienes and free fatty acids in vitro. Therefore, here we assessed whether the yeast Pry1 protein binds fatty acids. Computational modeling and site-directed mutagenesis indicated that the mode of fatty acid binding is conserved between tablysin-15 and Pry1. Pry1 bound fatty acids with micromolar affinity in vitro, and its function was essential for fatty acid export in cells lacking the acyl-CoA synthetases Faa1 and Faa4. Fatty acid binding of Pry1 is independent of its capacity to bind sterols, and the two sterol- and fatty acid-binding sites are nonoverlapping. These results indicate that some CAP family members, such as Pry1, can bind different lipids, particularly sterols and fatty acids, at distinct binding sites, suggesting that the CAP domain may serve as a stable, secreted protein domain that can accommodate multiple ligand-binding sites.
Keywords: cholesterol, fatty acid, fatty acid binding protein, fatty acid transport, Saccharomyces cerevisiae, secretion, sterol, CAP protein family, acyl-CoA synthetase
Introduction
Proteins belonging to the CAP superfamily (pfam PF00188), also known as SCP (sperm-coating proteins), are present in all kingdoms of life, and they have been implicated in a number of different physiological processes, including immune defense in mammals and plants, pathogen virulence, sperm maturation and fertilization, venom toxicity, and prostate and brain cancer. Named after the three founding members of this protein superfamily, the mammalian cysteine-rich secretory protein, the insect venom allergen antigen 5, and the plant pathogen-induced pathogenesis-related 1 protein, this superfamily comprises more than 8,400 members in over 1,850 species. Most CAP proteins are secreted glycoproteins, and they all share a common CAP domain of ∼150 amino acids, which adopts a unique α-β-α sandwich fold. The structural conservation of this domain suggests that CAP proteins exert a fundamentally similar function. Their precise molecular mode of action, however, has remained elusive (1, 2).
Several CAP family members have been shown to bind lipids, suggesting that lipid binding may constitute a conserved mode of action of these proteins. First, glioma pathogenesis-related 2 protein (GLIPR-2/RTVP-1/GAPR-1) is the smallest of the mammalian CAPs and the one most closely related to yeast Pry1 (3, 4). It is remarkable because it is thus far the only CAP protein that is not secreted (5). Instead, the protein is myristoylated and associates with the cytosolic surface of the Golgi membrane (6). GLIPR-2 is highly expressed in glioblastoma, which accounts for more than 65% of all human primary brain tumors (7–9). The nonmyristoylated protein binds to the surface of liposomes containing negatively charged lipids (10).
Second, tablysin-15, a salivary venom allergen of the blood-feeding horsefly Tabanus yao, acts as a potent inhibitor of platelet aggregation through binding integrins (11). In addition, tablysin-15 scavenges eicosanoids as well as free fatty acids through binding these ligands in a hydrophobic channel, thereby inhibiting the proinflammatory action of cysteinyl leukotrienes that are released from mast cells in the area of the insect bite (12).
Third, Pry proteins (pathogen related in yeast), CAP superfamily members from Saccharomyces cerevisiae, are required for the export of sterols in vivo and purified Pry1 binds sterols in vitro (13). The sterol-binding and export function of yeast Pry proteins appears to be a conserved function of members of the CAP protein superfamily because expression of the human CAP protein CRISP2 complements the defect in sterol export of a yeast mutant lacking Pry function, and purified CRISP2 binds sterols in vitro (13, 14). The capacity to bind sterols is also conserved in SmVal4, a CAP protein from the human parasite Schistosoma mansoni, as well as in PR-1, the founding member of the CAP superfamily from plants (15, 16). Consistent with the proposed defense function of PR-1, sterol sequestration by PR-1 inhibits growth of plant pathogenic oomycetes, which are auxotrophic for sterols (16). Sterol binding by Pry1 requires a flexible loop containing aromatic amino acids, termed the caveolin-binding motif (17). The CAP domain of the yeast Pry1 forms dimers in vitro, and sterol binding is Mg2+-dependent and requires the presence of the aliphatic isooctane side chain on the steroid ring (4, 18).
The observation that tabylsin-15 binds leukotrienes and free fatty acids in vitro prompted us to assess whether the yeast Pry proteins also binds free fatty acids and if so, whether this fatty acid binding is of physiological significance in yeast. Here, we provide evidence that the lipid-binding pocket described for tablysin-15 is conserved in the yeast Pry1 and that Pry1 has two independent lipid-binding sites: one for sterols and one for free fatty acids. Binding and export of free fatty acids by Pry1 is important for the survival of cells that accumulate high intracellular levels of free fatty acids as is the case in double mutants lacking the two acyl-CoA synthetases Faa1 and Faa4. These results indicate that lipid binding and sequestration constitutes a conserved function of different CAP family members and thus may constitute a uniform mode of action of these proteins.
Results and discussion
The fatty acid-binding channel of tablysin-15 is conserved in Pry1
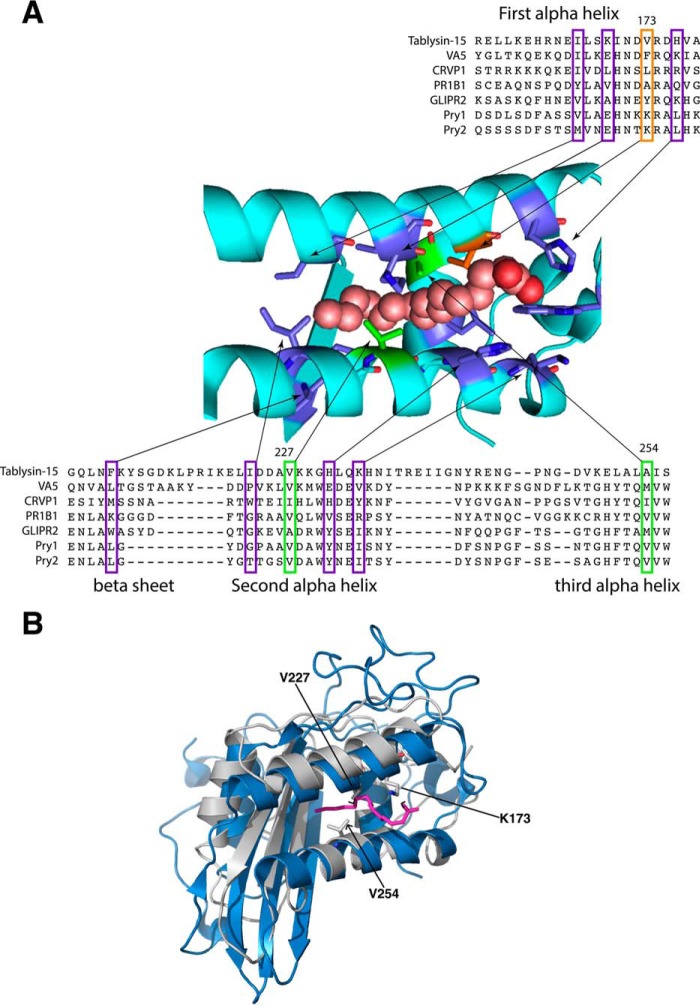
Tablysin-15 binds leukotrienes as well as free palmitic acid by a lipid-binding channel formed between the two parallel running helices, α1 and α3, and closed at the bottom by a much shorter helix α4, which runs perpendicular to helices α1 and α3 (12). To examine whether yeast Pry1 could also bind fatty acids, we first compared the sequence of tablysin-15 to that of other CAP proteins, including yeast Pry1 and Pry2. Tablysin-15 and Pry1 only share 23% sequence identity. However, key residues that constitute this lipid-binding pocket, but not their spacing, are conserved in the analyzed CAP family members (Fig. 1A). In particular, valine 227 and valine 254 of Pry1 are predicted to lay in close proximity to the hydrocarbon portion of the bound palmitate and to contact the ligand from two opposite sites. The structure of tablysin-15 and Pry1, although displaying a similar fold, do not superimpose well with root mean square deviation (r.m.s.d.)2 of 4.597 Å for all main-atoms. The palmitate binding cavity of both proteins, however, superimposes reasonably well and reveals a conserved position and spacing of the two helices, α1 and α3, which together form the previously identified fatty acid-binding pocket (12) (Fig. 1B).
Figure 1.
The palmitate-binding site of tablysin-15 is conserved in yeast Pry1. A, a multiple sequence alignment limited to the residues important for palmitic acid binding of tablysin-15 with taxonomically diverse CAP family members is shown. Sequences from the following CAP family members with their UniProt identifier given in the parentheses were aligned: horsefly tablysin-15 (F8QQG5), wasp venom allergen 5 (VA5, Q05110), Naja atra natrin-1 (CRVP1, Q7T1K6), tomato PR1B1 (P04284), human GLIPR2/GAPR1 (Q9H4G4), yeast Pry1 (P47032), yeast Pry2 (P36110). Key residues shown in colors are those forming the fatty acid-binding site in tablysin (12). Shown in green and orange are those that were mutated. B, comparison of the structure of the fatty acid-binding pocket of tablysin-15 to that of yeast Pry1. The structures of the two proteins around the fatty acid-binding pocket are superimposed with tablysin-15 in blue and Pry1 in gray with the bound palmitic acid shown in magenta. Key residues lysine 173, valine 227, and valine 254 are indicated.
Yeast Pry1 binds fatty acids
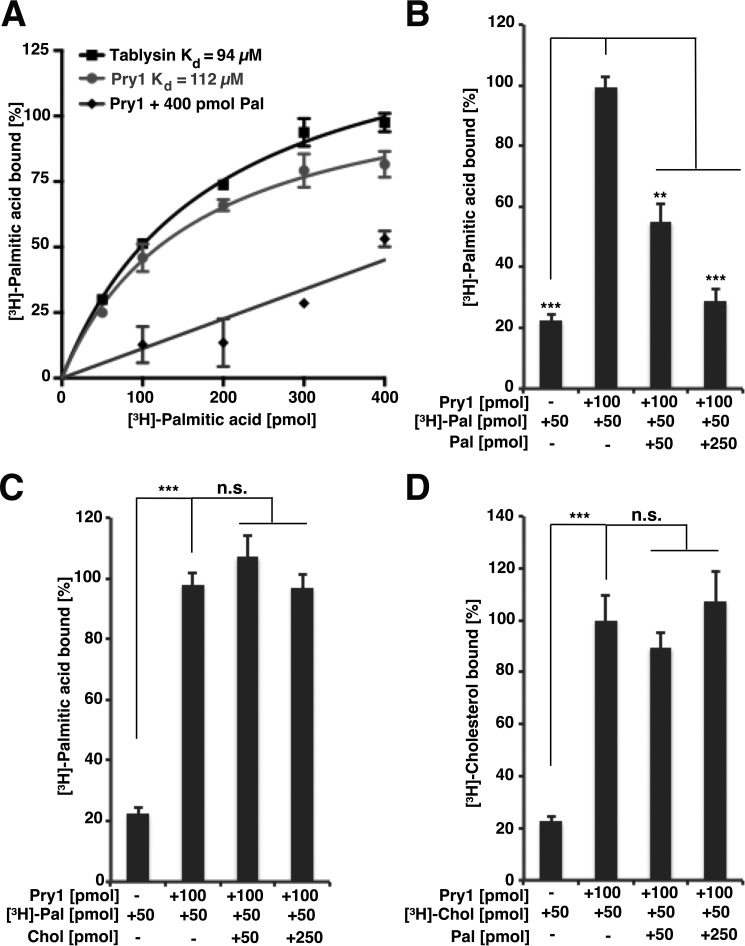
Given that the fatty acid-binding pocket in Pry1 is conserved at both the sequence and the structural level, we tested whether Pry1 indeed binds fatty acids in vitro. Therefore, purified hexahistidine-tagged Pry1 was incubated with increasing concentrations of [3H]-palmitic acid, the protein was separated from unbound ligand by ion exchange chromatography, and the protein-bound ligand was quantified. This analysis revealed that Pry1 bound palmitic acid with a saturable binding kinetics and an apparent Kd of 112 μm, which is comparable with that of tablysin-15 (Kd of 94 μm) (Fig. 2A). Palmitate binding by Pry1 was specific as it could be competed for by the addition of unlabeled palmitate (Fig. 2, A and B), but not by addition of unlabeled cholesterol (Fig. 2C). Correspondingly, binding of [3H]-cholesterol to Pry1 could not be competed for by the addition of unlabeled palmitic acid (Fig. 2D).
Figure 2.
Yeast Pry1 binds palmitic acid. A, comparison of the in vitro palmitic acid binding affinities of tablysin-15 and Pry1. Purified tablysin-15 and Pry1 (100 pmol) were incubated with the indicated concentration of [3H]-palmitic acid in the presence (Pry1 + 400 pmol Pal) or absence of unlabeled competitor ligand (Tablysin, Pry1). The protein was separated from the unbound ligand by adsorption to an anion-exchange matrix and the protein-bound radioligand was quantified by scintillation counting. B, unlabeled palmitic acid competes with [3H]-palmitic acid for binding to Pry1. Binding of [3H]-palmitic acid (50 pmol) to Pry1 (100 pmol) was assessed in the presence of the indicated concentration of unlabeled palmitic acid (Pal). The first bar shows background values obtained in the absence of added protein. C, cholesterol does not compete with [3H]-palmitic acid for binding to Pry1. Binding of [3H]-palmitic acid (50 pmol) to Pry1 (100 pmol) was assessed in the presence of the indicated concentration of unlabeled cholesterol (Chol). D, palmitic acid does not compete with [3H]-cholesterol for binding to Pry1. Binding of [3H]-cholesterol (50 pmol) to Pry1 (100 pmol) was assessed in the presence of the indicated concentration of unlabeled palmitic acid (Pal). Values represent means ± S.D. of three independent determinations. Asterisks denote statistical significance (**, p < 0.001; ***, p < 0.0001). n.s., nonsignificant.
Unlike Pry1, tablysin-15 did not bind free cholesterol in vitro (Fig. 3A). To test whether the lack of sterol-binding of tablysin-15 observed in vitro could also be seen in vivo, we expressed tablysin-15 in yeast and assayed its capacity to export sterols, particularly cholesteryl acetate (13, 17). Therefore, cells were labeled with [14C]-cholesterol, lipids were isolated from the cell pellet and the culture supernatant, separated by thin layer chromatography (TLC), and the ratio between intracellular and extracellular cholesteryl acetate was quantified and blotted as an export index. Expression of tablysin-15 in cells lacking Pry function failed to complement the block in sterol export, thus confirming the results of the in vitro binding assay (Fig. 3, B and C).
Figure 3.
Tablysin-15 does not bind sterols in vitro and fails to complement the sterol export defect of cells lacking Pry function. A, tablysin-15 does not bind sterols in vitro. The indicated amount of purified Pry1 or tablysin-15 was incubated with [3H]-cholesterol (50 pmol) and ligand binding was quantified. B, expression of tablysin-15 in pry mutant yeast cells does not rescue the block in sterol export. Heme-deficient cells of the indicated genotype expressing tablysin-15 were radiolabeled with [14C]-cholesterol. Cells were washed and chased with media containing unlabeled cholesterol, lipids were extracted from the cell pellet (P) and the culture supernatant (S) and separated by TLC. The positions of free cholesterol (FC), cholesteryl acetate (CA), and steryl esters (STE) are indicated. The asterisk marks the position of an unidentified cholesterol-derivative. C, quantification of the export of cholesteryl acetate. The export index indicates the relative percentages of cholesteryl acetate that are exported. Data represent mean ± S.D. of three independent experiments. Asterisks denote statistical significance (***, p < 0.0001).
Taken together, these results indicate that Pry1 has at least two independent lipid-binding sites, the caveolin-binding motif, which is required for binding free sterols, and the hydrophobic binding channel between helices α1 and α3, which constitutes the binding site for free fatty acids and leukotrienes. These two lipid-binding sites are not connected with each other in the structure of the CAP domain of Pry1 (4). In addition, the results indicate that sterol binding is not a universally conserved function of CAP family members, as tablysin-15 is the first CAP protein found that does not bind sterols in both the in vitro and the in vivo assay. Similarly, fatty acid binding may not be a universally conserved feature of CAP family members, as suggested by the fact that the venom antigen Ves v 5 from wasp, does not show a hydrophobic channel in its structure (12, 19).
Mutations in the fatty acid-binding pocket of Pry1 affect palmitate binding
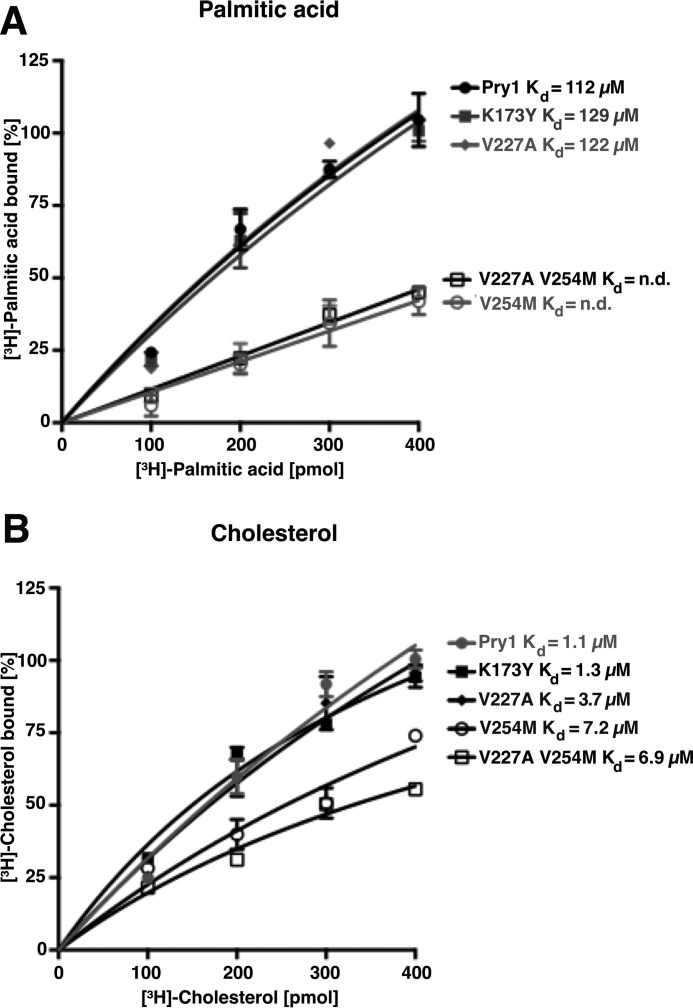
To confirm that the hydrophobic pocket formed by helices α1 and α3 is indeed the palmitate-binding site of Pry1, we generated point-mutant versions in which key residues of the binding pocket were mutated. As expected from structural modeling, substitution of lysine 173 by tyrosine or of valine 227 by alanine (as found in GAPR1) had only a slight impact on fatty acid binding (Fig. 4A). Instead, the exchange of valine 254 by methionine and the double substitution of both valine 227 by alanine and valine 254 by methionine strongly reduced palmitic acid binding of Pry1. The fact that the double mutation does not result in a further decrease of palmitate binding compared with the single substitution of valine 254 by methionine indicates that the valine at position 254 is most crucial for fatty acid binding.
Figure 4.
Mutations within the fatty acid-binding pocket of Pry1 abrogate binding of fatty acids but not that of sterols. A, valine 254 is crucial for in vitro binding of palmitic acid by Pry1. The indicated residues in the fatty acid-binding pocket of Pry1 were mutated, and in vitro binding of [3H]-palmitic acid (100–400 pmol) to the wild-type (wt) and mutant versions of Pry1 was assessed. No Kd values can be determined for the V254M single and the V227A,V254M double mutant (n.d.). B, mutations in the fatty acid-binding pocket do not strongly affect sterol binding by Pry1. The indicated point-mutant versions of Pry1 (100 pmol) were incubated with [3H]-cholesterol (100–400 pmol) and ligand binding was quantified. Values represent means ± S.D. of three independent determinations.
To examine whether these substitutions in the fatty acid-binding pocket, which are only a few residues away from the flexible loop that is required for cholesterol binding, the caveolin-binding motif contained in residues 236–248 of Pry1, we examined in vitro cholesterol binding by these mutant versions of Pry1. Cholesterol binding was not affected by the substitutions at position 173 and only slightly affected by the mutations at residue 227 (Fig. 4B). In contrast, the exchange of valine 254 by methionine and the double substitution at positions 227 and 254 reduced the cholesterol-binding affinity of Pry1 by more than 6-fold. Because valine 254 is buried in the core of the protein, its substitution by methionine might impact the overall stability of the protein and this might explain the observed slight reduction in sterol binding.
Mutations in the sterol-binding motif of Pry1 do not affect palmitate binding
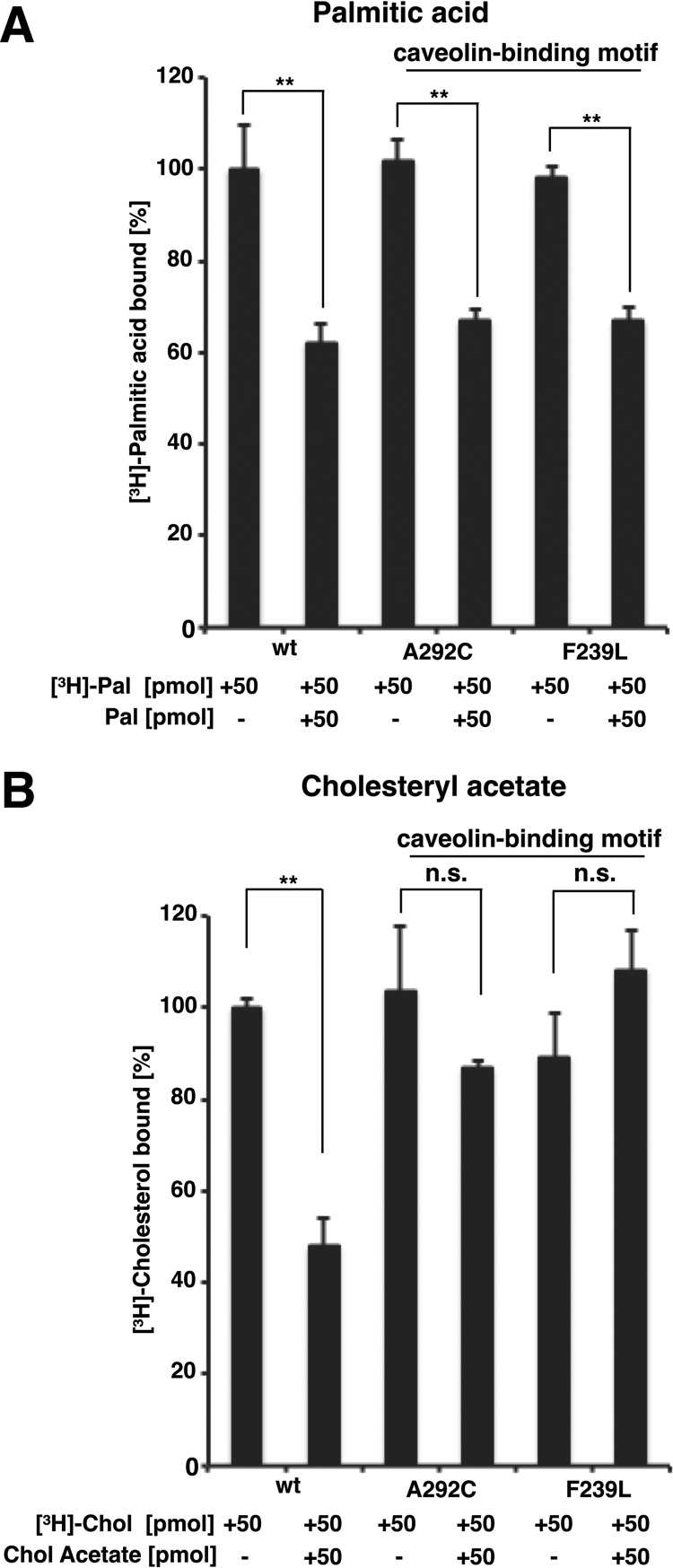
To confirm that the two lipid-binding sites on Pry1 are distinct, we tested whether mutations in the caveolin-binding motif, which affect binding of cholesteryl acetate, would also affect the binding of palmitic acid. Mutant versions of Pry1 containing an exchange of alanine at position 292 by cysteine or that of phenylalanine at position 239 by leucine, which have been shown previously to affect cholesteryl acetate binding (Fig. 5B) (17), still bind palmitic acid with wild-type efficiency and binding could be competed for by the addition of unlabeled palmitic acid (Fig. 5A). These data thus support the notion that the two lipid-binding sites on Pry1 are distinct and that they act independent of each other.
Figure 5.
Mutations in sterol-binding loop of Pry1, the caveolin-binding motif, do not affect in vitro binding of fatty acids. A, Pry1 wild-type (wt) and mutant proteins bearing point mutations in the caveolin-binding motif, A292C and F239L (100 pmol each), were incubated with [3H]-palmitic acid (50 pmol) in the presence or absence of unlabeled palmitic acid (50 pmol) and ligand binding was quantified. B, Pry1 wild-type (wt) and mutant proteins bearing point mutations in the caveolin-binding motif (100 pmol each) were incubated with [3H]-cholesterol (50 pmol) in the presence or absence of unlabeled cholesteryl acetate (50 pmol) and binding of the radioligand was quantified. Values represent means ± S.D. of three independent determinations. Asterisks denote statistical significance (**, p < 0.001).
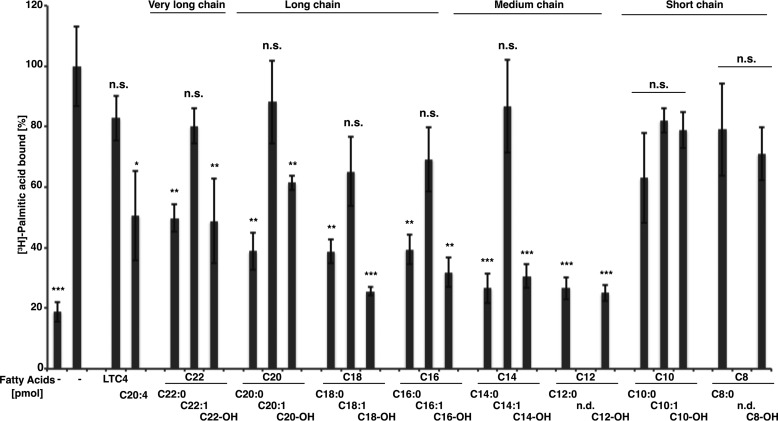
Pry1 preferentially binds saturated long-chain fatty acids
To probe the substrate specificity of the fatty acid-binding pocket, we performed in vitro competition binding assays with different types of fatty acids. Oleic (C18:1), palmitoleic (C16:1), or myristoleic acid (C14:1) competed less well for binding than did their saturated counterparts, stearic (C18:0), palmitic (C16:0), or myristic acid (C14:0), indicating that binding of saturated acyl chains is preferred over that of Δ9-unsaturated ones (Fig. 6). Moreover, binding of medium- (C12 and C14) and long-chain saturated fatty acids (C16, C18, C20, and C22) was preferred over that of short-chain fatty acids (C8 and C10) because both caprilic (C8) and decanoic (C10) fatty acids essentially failed to compete with [3H]-palmitic acid for binding to Pry1, whereas the medium-chain (C12, C14), the long-chain (C16, C18, C20), and even the very long-chain fatty acids (C22) competed efficiently (Fig. 6).
Figure 6.
Pry1 preferentially binds saturated medium- and long-chain fatty acids and fatty alcohols. Ligand binding specificity was tested by a competition binding assay. Pry1 (100 pmol) was incubated with [3H]-palmitic acid (50 pmol) in the presence or absence of the indicated unlabeled fatty acid (50 pmol), arachidonic acid (C20:4), or the cysteinyl leukotriene LTC4, and binding of the radioligand was quantified. The chain length and degree of unsaturation of the tested fatty acids and the fatty alcohols (-OH) is indicated. Values represent means ± S.D. of three independent determinations. Asterisks denote statistical significance (*, p < 0.01; **, p < 0.001; ***, p < 0.0001). n.s., nonsignificant (p > 0.02); n.d., not determined.
To assess the importance of the carboxyl group, we also tested binding of the corresponding fatty alcohols. Except for the short-chain alcohols (C8, and C10), all the tested medium-, long-, and very long-chain alcohols competed efficiently with [3H]-palmitic acid for binding to Pry1 (Fig. 6). These data thus indicate that fatty acid binding requires a minimal chain length of more than 10 carbon atoms, that kinks in the chain, as introduced by a single cis-double bond, are not tolerated, and that the carboxyl group is less important and can be replaced by a primary alcohol.
Given that tablysin-15 binds arachidonic acid (C20:4) and leukotrienes, we also tested whether the addition of arachidonic acid (C20:4) or that of the cysteinyl leukotriene LTC4 would compete for palmitate binding. Whereas the polyunsaturated arachidonic acid effectively competed for binding to Pry1, LTC4 did not (Fig. 6). These results thus indicate that Pry1 can bind polyunsaturated fatty acids, such as arachidonic acids (C20-Δ5,8,11,14), which have its second double bond at position 8 of the acyl chain. Monounsaturated fatty acids, however, contain their double bond at position 9 of the acyl chain and do not bind to Pry1. LTC4, on the other hand, is composed of an arachidonic acid that contains a glutathione-modified cysteinyl group. The observation that arachidonic acid, but not LTC4, binds to Pry1 suggests that the presence of the glutathione-modified cysteinyl group interferes with its binding to Pry1.
Pry1 is required for export of fatty acids in vivo
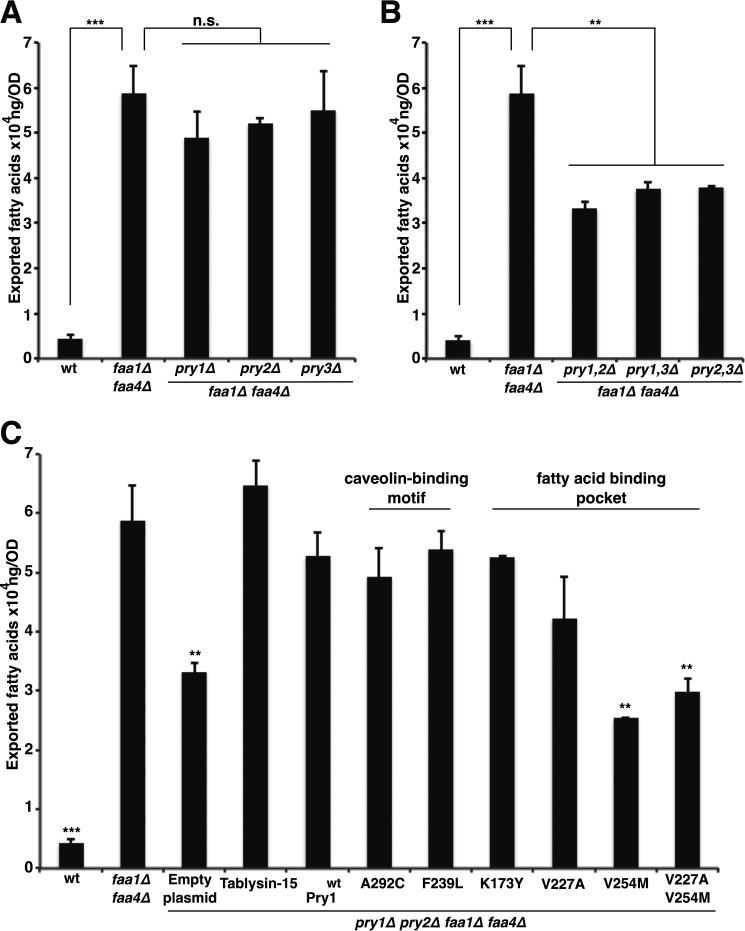
To examine whether fatty acid binding by Pry1 is of physiological significance, we tested whether Pry1 is required for the export of fatty acids from cells that accumulate high levels of free fatty acids. Faa1 and Faa4 are two of at least five different acyl-CoA synthetases present in yeast, and they are required for CoA activation of exogenous fatty acids (20). Mutants lacking the two acyl-CoA synthetases Faa1 and Faa4 have been shown previously to secrete free fatty acids into the culture medium (21, 22). To test whether Pry proteins are required for the export of fatty acids from faa1Δ faa4Δ double mutant cells, we deleted each one of the three PRY genes in an faa1Δ faa4Δ double mutant background and quantified the levels of free fatty acids secreted from these cells into the culture medium. The results of this analysis confirm that faa1Δ faa4Δ double mutant cells have greatly elevated levels of fatty acids in the medium when compared with wild-type cells (Fig. 7A). Deletion of any one of the three Pry proteins, however, did not affect the levels of the secreted fatty acids. In contrast, double deletion of two of the Pry proteins resulted in a marked decrease of the levels of the exported fatty acids, indicating that Pry proteins function redundantly in the export of fatty acids as they do in the export of cholesteryl acetate (Fig. 7B) (13).
Figure 7.
Fatty acid binding by Pry and tablysin-15 is required for the in vivo export of fatty acids from mutant cells lacking the acyl-CoA synthetases Faa1 and Faa4. A, faaΔ1 faa4Δ double mutant cells export fatty acids even in the absence of one of the three yeast Pry proteins. Wild-type (wt) and faa1Δ faa4Δ double mutant cells lacking any one of the three Pry proteins were cultivated in SC media to 2 optical density. Cells were pelleted and the free fatty acids present in the culture supernatant were analyzed and quantified by GC-MS. B, Pry double mutant cells have a block in export of fatty acids. Cells of the indicated genotype were cultivated in SC media and levels of fatty acid exported into the culture supernatant were quantified. C, mutations in the fatty acid-binding pocket of Pry1 abrogate the in vivo export of fatty acids. Quadruple mutant cells (pry1Δ pry2Δ faa1Δ faa4Δ) carrying a plasmid-borne wild-type copy of tablysin-15, Pry1, or one of the indicated point-mutant versions were cultivated in SC media, and fatty acids exported into the culture supernatant were quantified. Values represent means ± S.D. of three independent determinations. Asterisks denote statistical significance (**, p < 0.001; ***, p < 0.0001).
Next, we tested whether mutations in the caveolin-binding motif or those in the fatty acid-binding pocket affect the export of fatty acids. In agreement with the in vitro binding results, the mutations in the caveolin-binding motif did not affect the export of fatty acids by Pry1 (Fig. 7C). The mutations in the fatty acid-binding pocket, V254M, however, reduced the levels of exported fatty acids back to background levels, i.e. levels observed in cells lacking Pry function.
Pry function is essential for the viability of cells that accumulate free fatty acids
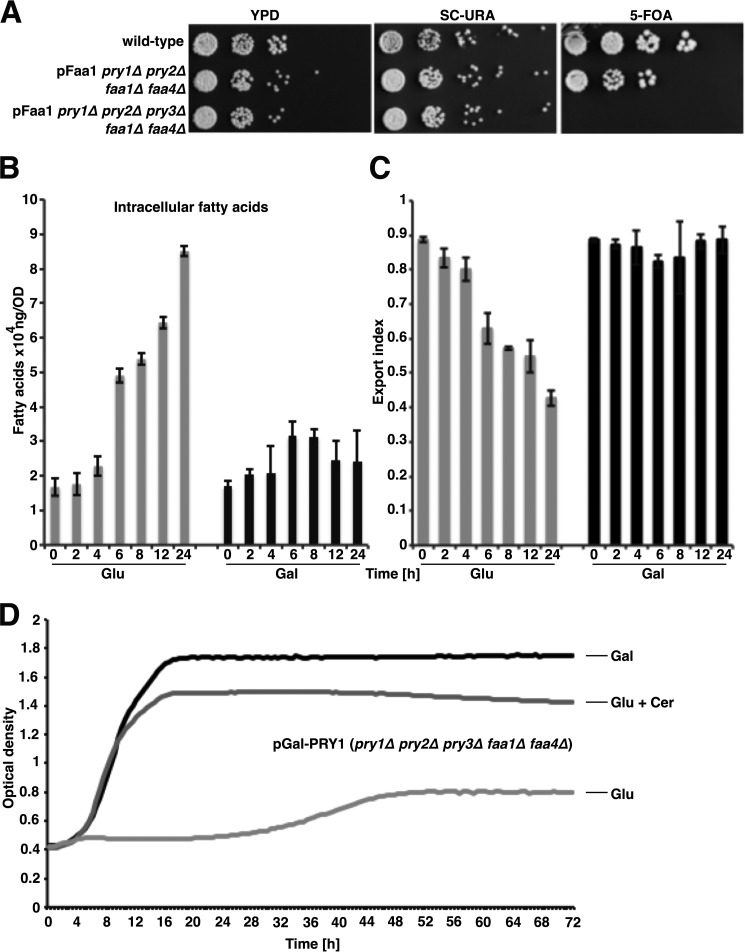
Given that the deletion of two of the three Pry proteins greatly reduced free fatty acid exported in faa1Δ faa4Δ mutant cells, but did not completely block fatty acid export, we wondered whether deletion of the third PRY gene would further reduce levels of exported fatty acids or whether it would render cells nonviable. To test this, we generated faa1Δ faa4Δ double mutant cells, carrying a rescuing plasmid containing FAA1, and lacking either two or all three of the PRY genes. We then tested whether the rescuing pFaa1 plasmid, which was marked by URA3, could be lost by strains lacking two of the PRY genes (pry1Δ pry2Δ) or all three of them (pry1Δ pry2Δ pry3Δ). Although cells lacking only two of the three PRY genes could lose the plasmid-borne copy of FAA1, as indicated by their growth on media containing 5-fluoroorotic acid (5-FOA), which selects for cells that are uracil auxotroph, cells lacking all three PRY genes failed to grow on 5-FOA medium, indicating that PRY function becomes essential in cells lacking the acyl-CoA synthetases Faa1 and Faa4 (Fig. 8A).
Figure 8.
Pry function is essential in cells that export fatty acids. A, wild-type, quadruple (pry1Δ pry2Δ faa1Δ faa4Δ), and quintuple (pry1Δ pry2Δ pry3Δ faa1Δ faa4Δ) mutant cells bearing a plasmid-borne copy of FAA1 (pFaa1) were serially diluted 10-fold and stamped on YPD, SC-URA, or 5-fluoroorotic acid (5-FOA) containing solid media. Plates were incubated at 30 °C for 3 days. B, transcriptional shut off of PRY1 results in a time-dependent increase of intracellular fatty acids. Quintuple mutant cells (pry1Δ pry2Δ pry3Δ faa1Δ faa4Δ) bearing a plasmid-borne copy of PRY1 under the transcriptional control of a galactose-inducible promoter (pGal-PRY1) were cultivated in galactose-containing media and diluted into fresh media containing either glucose (Glu) or galactose (Gal). Cells were cultivated for the indicated period of time and intracellular fatty acid levels were quantified by GC-MS. C, export of fatty acids is arrested upon shut off of PRY1 transcription. Cells were cultivated as described for panel B and the ratio of extracellular fatty acids to that of the sum of both intracellular and extracellular fatty acid is shown as an export index. D, blocking fatty acid synthesis rescues growth of cells lacking Pry function. Quintuple mutant cells (pry1Δ pry2Δ pry3Δ faa1Δ faa4Δ) bearing the glucose repressible pGal-PRY1 were cultivated in media containing galactose (Gal), glucose (Glu), or glucose and the fatty acid synthase inhibitor cerulenin (Cer) (0.1 μg/ml). Cell density over time was recorded using a bioimager. The growth curves shown are representative of three independent experiments. Values represent means ± S.D. of three independent determinations.
Because Pry1 binds fatty acids in vitro and Pry function is required for the export of fatty acids in cells that are impaired in CoA activation of free fatty acids, we wondered whether Pry function is required for the export of these free fatty acids from cells. If this were the case, one would expect that the levels of intracellular fatty acids would increase upon depletion of Pry function. To test this, we generated a strain in which the transcription of PRY1 could be shut off through a switch in carbon sources. Cultivation of this strain, bearing a plasmid-borne copy of PRY1 under control of a galactose-inducible and glucose-repressible promoter, in a background lacking genomic Pry function as well as the two acyl-CoA synthetases (pGAL-PRY1 pry1Δ pry2Δ pry3Δ faa1Δ faa4Δ), in glucose media resulted in a time-dependent increase of intracellular free fatty acids (Fig. 8B). When cultivated in galactose-containing media, on the other hand, the levels of intracellular fatty acids remained on a constant low level. Under these conditions, about 90% of the free fatty acids were exported from the cells, whereas under repressible conditions, that is in glucose media, the fatty acid export index (the fraction of exported fatty acid divided by the sum of extracellular and intracellular fatty acids) displayed a time-dependent decline, reaching a minimum of 40% of fatty acids exported after cultivation for 24 h in glucose media (Fig. 8C). These data thus indicate that the levels of exported fatty acids correlate with the steady-state levels of Pry1 protein and, given that Pry1 is a secreted glycoprotein (13), they support the notion that fatty acids are exported from cells as complexes bound to Pry1.
Because high levels of intracellular fatty acids are toxic (23–26), the marked increase of intracellular fatty acids in cells depleted of Pry1 is likely to explain the nonviability of cells lacking all Pry function in a faa1Δ faa4Δ mutant background. Consistent with this notion, growth of these cells was rescued by sublethal doses of cerulenin, an inhibitor of fatty acid synthesis (27) (Fig. 8D).
Taken together, the data presented in this study indicate that the yeast CAP superfamily member Pry1, and possibly many other CAP family members, can bind and thereby scavenge at least two different types of lipid, sterols, and fatty acids. In yeast, both these lipid-binding functions of the Pry proteins are of physiological importance because they become essential under specific growth conditions. Sterol-binding is important to secrete modified sterols, such as cholesteryl acetate, and required for growth in the presence of eugenol, a plant-derived antimicrobial compound that binds to the caveolin-binding motif of Pry1 and whose toxic action is thereby neutralized (13, 18). For the antimicrobial defense of plants, on the other hand, scavenging of sterols by PR-1 is likely important to inhibit the growth of sterol auxotrophic pathogens, such as oomycetes (16). Fatty acid binding, however, is required for the growth of yeast cells under conditions of elevated levels of intracellular free fatty acids, as occurs in mutants lacking major acyl-CoA synthetase, but is presumably also important in cells having reduced synthesis or uptake of CoA itself, or those displaying enhanced lipid remodeling induced by membrane-perturbing agents. Insect venom antigens of the CAP superfamily may employ this hydrophobic binding channel to scavenge and sequester immune modulators of their hosts, as exemplified by the leukotriene binding of tablysin-15 (12). The key question, of course, is what the uniform function of the conserved CAP domain is. Is it the three-dimensional structure of the domain that is of functional importance, as is commonly the case for proteins? Alternatively, the CAP domain, with its unique fold, might serve as a stable and secreted scaffold into which different types of hydrophobic binding pockets can be implemented. The secreted CAP domain containing proteins could thus act to scavenge and neutralize the action of small hydrophobic compounds that these proteins encounter in their specific physiological setting. This ligand-binding CAP domain could then be further functionalized by the addition of auxiliary modules, such as the arginine-glycine-aspartic acid (RGD) motif, which is present in tablysin-15 and binds integrins (11), or the potassium ion channel inhibitor-like fold, which is present in the cysteine-rich domain of stecrisp and the mouse testis specific protein 1 (Tpx-1) (28, 29).
Experimental procedures
Yeast strains, growth conditions and epitope tagging, and site-directed mutagenesis
Yeast mutant strains were cultivated either in rich media, YPD (containing 1% Bacto yeast extract, 2% Bacto peptone, and 2% glucose) (US Biological, Salem, MA), in YPGal containing 2% galactose instead of glucose, or in minimal defined media (containing 0.67% yeast nitrogen base without amino acids (US Biological), 0.73 g/liter amino acids, and 2% glucose). Media supplemented with sterols and fatty acids contained 0.05 mg/ml Tween 80 and 20 μg/ml cholesterol (Sigma). To bypass heme deficiency, cells were grown in media supplemented with 10 μg/ml δ-aminolevulinic acid. To counterselect for the presence of URA3-containing plasmids, 5-FOA (bts Biotech Trade & Service Gmbh, St. Leon-Rot, Germany) was added to solid media at 1 g/liter. Mutant strains were generated by gene disruption using PCR deletion cassettes and a marker rescue strategy (30). Cerulenin (Sigma Aldrich) was diluted in DMSO and used from a 1,000× stock. For growth analysis, strains were diluted 10-fold and diluted samples were spotted onto plates supplemented with the appropriate medium. Plates were grown for 3 days at 30 °C and were then photographed. Growth curves were recorded using a Bioscreen C MBR (Oy Growth Curves Ab Ltd., Helsinki, Finland).
Expression and purification of tablysin-15 and Pry1
For in vivo lipid export assays, DNA encoding tablysin-15 was synthesized (GenScript, Piscataway, NJ), PCR amplified, and cloned into a pGREG506-based plasmid in which the GAL promoter was replaced by an ADH promoter and the proteins were fused to the Pry1 ER signal sequence, amino acids 1–19 (13). Wild-type and mutant versions of Pry1 were PCR amplified and cloned into the NcoI and XhoI sites of pET22b vector (Novagen, Merck, Darmstadt, Germany), which contains a PelB signal sequence to direct the secretion of the expressed protein into the periplasmic space. Similarly, plasmids were transformed into Escherichia coli BL21, and the proteins were expressed as a polyhistidine-tagged fusion after lactose induction and overnight growth of the bacteria at 24 °C. Cells were harvested, lysed, and incubated with Ni-NTA beads (Qiagen, Hilden, Germany) as per instructions of the manufacturer. Beads were washed and proteins were eluted with imidazole, concentrated, and quantified. Protein concentration was determined by Lowry assay using Folin reagent and BSA as standard.
Lipid labeling and analysis
Acetylation and export of sterols into the culture media were examined as described previously (31). Yeast mutants deficient in heme biosynthesis (hem1Δ) were cultivated in presence of cholesterol/Tween-80 and labeled with 0.025 μCi/ml of [14C]-cholesterol (American Radiolabeled Chemicals Inc., St. Louis, MO). Cells were harvested by centrifugation, washed with synthetic complete (SC) media, diluted to an A600 of 1 into fresh media containing nonradiolabeled cholesterol, and grown overnight. Cells were centrifuged and lipids were extracted from the cell pellet and the culture supernatant using chloroform/methanol (1:1; v/v). Samples were dried and separated by TLC on silica gel 60 plates (Merck, Darmstadt, Germany) using the solvent system, petroleum ether/diethylether/acetic acid (70:30:2; per volume). TLC plates were exposed to phosphorimager screens and radiolabeled lipids were visualized and quantified using a phosphorimager (GE Healthcare).
Fatty acid quantification
Fatty acid methyl esters (FAMEs) were prepared from 5 A600 nm units of yeast cells. Wild-type and mutant cells were grown in SC medium at 30 °C. Cells were washed once in cold water, and a fatty acid standard (C17:0; 50 μg) (Sigma) was added. Cells were disrupted using glass beads, and lipids were extracted using chloroform/methanol (1:1; per volume) for cellular lipids and chloroform/methanol/concentrated HCl (1:2:0.03; per volume) for extracellular fatty acids. FAMEs were produced using boron trifluoride at 100 °C for 45 min and were recovered by extraction with petrol ether. After evaporation, extracts were resuspended in hexane. FAMEs were separated with an Agilent 7890A gas chromatograph (GC) equipped with a DB-23 capillary column (30 m × 0.25 mm × 0.25 μm) (Agilent Technologies, Santa Clara, CA). The temperature of the injection port was set to 250 °C, its pressure to 26.24 p.s.i. (average velocity, 48.17 cm/s), and the septum purge flow to 3 ml/min. Split injections occurred through an Agilent 7693A automated liquid sampler. The initial oven temperature (100 °C; held for 2 min) was increased to 160 °C at the rate of 25 °C/min and was then increased again to 250 °C at 8 °C/min. The final oven temperature was held for an additional 4 min. FAMEs were detected with a flame ionization detector (Agilent Technologies) set at 270 °C with H2, air, and helium flows set at 30, 400, and 27.7 ml/min, respectively. FAMEs were quantified relative to the internal standard, and the relative response factor for each FAME was determined from a four-level calibration curve (r2, 0.999).
In vitro lipid binding assay
The radioligand binding assay was performed as described previously (13, 32). Purified protein (100 pmol) in binding buffer (20 mm Tris, pH 7.5, 30 mm NaCl, 0.05% Triton X-100) was incubated with [3H]-cholesterol (50 pmol) or [3H]-palmitic acid for 1 h at 30 °C. The protein was then separated from the unbound ligand by adsorption to Q-Sepharose beads (GE Healthcare), the beads were washed, and the protein-bound radioligand was quantified by scintillation counting. For competition binding assays, unlabeled cholesterol, palmitic acid (50 pmol), saturated and monounsaturated fatty acids, fatty alcohols, arachidonic acid (all from Sigma), or LTC4 (Avanti Polar Lipids, Alabaster, AL) were included in the binding reaction. To determine nonspecific binding, the binding assay was performed without the addition of the protein. Statistical significance of data was analyzed by a multiple Student's t test (Prism, GraphPad Software, La Jolla, CA).
Author contributions
R. D. conducted most of the experiments, analyzed the results, and generated most of the figures. L. M.-S. helped with fatty acid analysis and data processing. D. G. and O. A. A. performed the molecular modeling, based on which the mutants were generated. R. D., D. G., O. A. A., and R. S. conceived the idea for the project and analyzed the data. R. S. wrote the paper.
Acknowledgments
We thank Dr. Xueqing Xu and Dr. John Andersen (National Institute of Allergy and Infectious Diseases, National Institutes of Health, Rockville, MD) for recombinant tablysin-15, members of the lab for helpful discussions, and Stéphanie Cottier for comments on the manuscript.
This work was supported by the Swiss National Science Foundation, 31003A_153416. The authors declare that they have no conflict of interest with the contents of this article.
- r.m.s.d.
- root mean square deviation of atomic positions
- FAMEs
- fatty acid methyl esters
- TLC
- thin-layer chromatography
- 5-FOA
- 5-fluoroorotic acid
- SC
- synthetic complete
- GC
- gas chromatograph.
References
- 1. Cantacessi C., Campbell B. E., Visser A., Geldhof P., Nolan M. J., Nisbet A. J., Matthews J. B., Loukas A., Hofmann A., Otranto D., Sternberg P. W., and Gasser R. B. (2009) A portrait of the “SCP/TAPS” proteins of eukaryotes—developing a framework for fundamental research and biotechnological outcomes. Biotechnol. Adv. 27, 376–388 [DOI] [PubMed] [Google Scholar]
- 2. Gibbs G. M., Roelants K., and O'Bryan M. K. (2008) The CAP superfamily: cysteine-rich secretory proteins, antigen 5, and pathogenesis-related 1 proteins—roles in reproduction, cancer, and immune defense. Endocr. Rev. 29, 865–897 [DOI] [PubMed] [Google Scholar]
- 3. Miosga T., Schaaff-Gerstenschläger I., Chalwatzis N., Baur A., Boles E., Fournier C., Schmitt S., Velten C., Wilhelm N., and Zimmermann F. K. (1995) Sequence analysis of a 33.1 kb fragment from the left arm of Saccharomyces cerevisiae chromosome X, including putative proteins with leucine zippers, a fungal Zn(II)2-Cys6 binuclear cluster domain and a putative α2-SCB-α2 binding site. Yeast 11, 681–689 [DOI] [PubMed] [Google Scholar]
- 4. Darwiche R., Kelleher A., Hudspeth E. M., Schneiter R., and Asojo O. A. (2016) Structural and functional characterization of the CAP domain of pathogen-related yeast 1 (Pry1) protein. Sci. Rep. 6, 28838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Serrano R. L., Kuhn A., Hendricks A., Helms J. B., Sinning I., and Groves M. R. (2004) Structural analysis of the human Golgi-associated plant pathogenesis related protein GAPR-1 implicates dimerization as a regulatory mechanism. J. Mol. Biol. 339, 173–183 [DOI] [PubMed] [Google Scholar]
- 6. Eberle H. B., Serrano R. L., Füllekrug J., Schlosser A., Lehmann W. D., Lottspeich F., Kaloyanova D., Wieland F. T., and Helms J. B. (2002) Identification and characterization of a novel human plant pathogenesis-related protein that localizes to lipid-enriched microdomains in the Golgi complex. J. Cell Sci. 115, 827–838 [DOI] [PubMed] [Google Scholar]
- 7. Murphy E. V., Zhang Y., Zhu W., and Biggs J. (1995) The human glioma pathogenesis-related protein is structurally related to plant pathogenesis-related proteins and its gene is expressed specifically in brain tumors. Gene 159, 131–135 [DOI] [PubMed] [Google Scholar]
- 8. Rich T., Chen P., Furman F., Huynh N., and Israel M. A. (1996) RTVP-1, a novel human gene with sequence similarity to genes of diverse species, is expressed in tumor cell lines of glial but not neuronal origin. Gene 180, 125–130 [DOI] [PubMed] [Google Scholar]
- 9. Thompson T. C. (2010) Glioma pathogenesis-related protein 1: tumor-suppressor activities and therapeutic potential. Yonsei Med. J. 51, 479–483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Van Galen J., Van Balkom B. W., Serrano R. L., Kaloyanova D., Eerland R., Stüven E., and Helms J. B. (2010) Binding of GAPR-1 to negatively charged phospholipid membranes: unusual binding characteristics to phosphatidylinositol. Mol. Membr. Biol. 27, 81–91 [DOI] [PubMed] [Google Scholar]
- 11. Ma D., Gao L., An S., Song Y., Wu J., Xu X., and Lai R. (2010) A horsefly saliva antigen 5-like protein containing RTS motif is an angiogenesis inhibitor. Toxicon 55, 45–51 [DOI] [PubMed] [Google Scholar]
- 12. Xu X., Francischetti I. M., Lai R., Ribeiro J. M., and Andersen J. F. (2012) Structure of protein having inhibitory disintegrin and leukotriene scavenging functions contained in single domain. J. Biol. Chem. 287, 10967–10976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Choudhary V., and Schneiter R. (2012) Pathogen-related yeast (PRY) proteins and members of the CAP superfamily are secreted sterol-binding proteins. Proc. Natl. Acad. Sci. U.S.A. 109, 16882–16887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Schneiter R., and Di Pietro A. (2013) The CAP protein superfamily: function in sterol export and fungal virulence. Biomol. Concepts 4, 519–525 [DOI] [PubMed] [Google Scholar]
- 15. Kelleher A., Darwiche R., Rezende W. C., Farias L. P., Leite L. C., Schneiter R., and Asojo O. A. (2014) Schistosoma mansoni venom allergen-like protein 4 (SmVAL4) is a novel lipid-binding SCP/TAPS protein that lacks the prototypical CAP motifs. Acta Crystallogr. D Biol. Crystallogr. 70, 2186–2196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Gamir J., Darwiche R., Van't Hof P., Choudhary V., Stumpe M., Schneiter R., and Mauch F. (2017) The sterol-binding activity of pathogenesis-related protein 1 reveals the mode of action of an antimicrobial protein. Plant J. 89, 502–509 [DOI] [PubMed] [Google Scholar]
- 17. Choudhary V., Darwiche R., Gfeller D., Zoete V., Michielin O., and Schneiter R. (2014) The caveolin-binding motif of the pathogen-related yeast protein Pry1, a member of the CAP protein superfamily, is required for in vivo export of cholesteryl acetate. J. Lipid Res. 55, 883–894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Darwiche R., and Schneiter R. (2016) Cholesterol-binding by the yeast CAP family member Pry1 requires the presence of an aliphatic side chain on cholesterol. J. Steroids Horm. Sci. 7, 172 [Google Scholar]
- 19. Henriksen A., King T. P., Mirza O., Monsalve R. I., Meno K., Ipsen H., Larsen J. N., Gajhede M., and Spangfort M. D. (2001) Major venom allergen of yellow jackets, Ves v 5: structural characterization of a pathogenesis-related protein superfamily. Proteins 45, 438–448 [DOI] [PubMed] [Google Scholar]
- 20. Johnson D. R., Knoll L. J., Levin D. E., and Gordon J. I. (1994) Saccharomyces cerevisiae contains four fatty acid activation (FAA) genes: an assessment of their role in regulating protein N-myristoylation and cellular lipid metabolism. J. Cell Biol. 127, 751–762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Michinaka Y., Shimauchi T., Aki T., Nakajima T., Kawamoto S., Shigeta S., Suzuki O., and Ono K. (2003) Extracellular secretion of free fatty acids by disruption of a fatty acyl-CoA synthetase gene in Saccharomyces cerevisiae. J. Biosci. Bioeng. 95, 435–440 [DOI] [PubMed] [Google Scholar]
- 22. Scharnewski M., Pongdontri P., Mora G., Hoppert M., and Fulda M. (2008) Mutants of Saccharomyces cerevisiae deficient in acyl-CoA synthetases secrete fatty acids due to interrupted fatty acid recycling. FEBS J. 275, 2765–2778 [DOI] [PubMed] [Google Scholar]
- 23. Garbarino J., Padamsee M., Wilcox L., Oelkers P. M., D'Ambrosio D., Ruggles K. V., Ramsey N., Jabado O., Turkish A., and Sturley S. L. (2009) Sterol and diacylglycerol acyltransferase deficiency triggers fatty acid-mediated cell death. J. Biol. Chem. 284, 30994–31005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Petschnigg J., Wolinski H., Kolb D., Zellnig G., Kurat C. F., Natter K., and Kohlwein S. D. (2009) Good fat, essential cellular requirements for triacylglycerol synthesis to maintain membrane homeostasis in yeast. J. Biol. Chem. 284, 30981–30993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Connerth M., Czabany T., Wagner A., Zellnig G., Leitner E., Steyrer E., and Daum G. (2010) Oleate inhibits steryl ester synthesis and causes liposensitivity in yeast. J. Biol. Chem. 285, 26832–26841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Fakas S., Qiu Y., Dixon J. L., Han G. S., Ruggles K. V., Garbarino J., Sturley S. L., and Carman G. M. (2011) Phosphatidate phosphatase activity plays key role in protection against fatty acid-induced toxicity in yeast. J. Biol. Chem. 286, 29074–29085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Omura S. (1981) Cerulenin. Methods Enzymol. 72, 520–532 [PubMed] [Google Scholar]
- 28. Guo M., Teng M., Niu L., Liu Q., Huang Q., and Hao Q. (2005) Crystal structure of the cysteine-rich secretory protein stecrisp reveals that the cysteine-rich domain has a K+ channel inhibitor-like fold. J. Biol. Chem. 280, 12405–12412 [DOI] [PubMed] [Google Scholar]
- 29. Gibbs G. M., Scanlon M. J., Swarbrick J., Curtis S., Gallant E., Dulhunty A. F., and O'Bryan M. K. (2006) The cysteine-rich secretory protein domain of Tpx-1 is related to ion channel toxins and regulates ryanodine receptor Ca2+ signaling. J. Biol. Chem. 281, 4156–4163 [DOI] [PubMed] [Google Scholar]
- 30. Gueldener U., Heinisch J., Koehler G. J., Voss D., and Hegemann J. H. (2002) A second set of loxP marker cassettes for Cre-mediated multiple gene knockouts in budding yeast. Nucleic Acids Res. 30, e23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Tiwari R., Köffel R., and Schneiter R. (2007) An acetylation/deacetylation cycle controls the export of sterols and steroids from S. cerevisiae. EMBO J. 26, 5109–5119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Im Y. J., Raychaudhuri S., Prinz W. A., and Hurley J. H. (2005) Structural mechanism for sterol sensing and transport by OSBP-related proteins. Nature 437, 154–158 [DOI] [PMC free article] [PubMed] [Google Scholar]