Abstract
Herpes simplex virus 1 (HSV-1) infection manipulates distinct host DNA-damage responses to facilitate virus proliferation, but the molecular mechanisms remain to be elucidated. One possible HSV-1 target might be DNA damage-tolerance mechanisms, such as the translesion synthesis (TLS) pathway. In TLS, proliferating cell nuclear antigen (PCNA) is monoubiquitinated in response to DNA damage-caused replication fork stalling. Ubiquitinated PCNA then facilitates the error-prone DNA polymerase η (polη)-mediated TLS, allowing the fork to bypass damaged sites. Because of the involvement of PCNA ubiquitination in DNA-damage repair, we hypothesized that the function of PCNA might be altered by HSV-1. Here we show that PCNA is a substrate of the HSV-1 deubiquitinase UL36USP, which has previously been shown to be involved mainly in virus uptake and maturation. In HSV-1-infected cells, viral infection-associated UL36USP consistently reduced PCNA ubiquitination. The deubiquitination of PCNA inhibited the formation of polη foci and also increased cell sensitivity to DNA-damage agents. Moreover, the catalytically inactive mutant UL36C40A failed to deubiquitinate PCNA. Of note, the levels of virus marker genes increased strikingly in cells infected with wild-type HSV-1, but only moderately in UL36C40A mutant virus-infected cells, indicating that the UL36USP deubiquitinating activity supports HSV-1 virus replication during infection. These findings suggest a role of UL36USP in the DNA damage-response pathway.
Keywords: deubiquitylation (deubiquitination), DNA damage response, DNA polymerase, herpesvirus, proliferating cell nuclear antigen (PCNA), UL36USP, translesion synthesis
Introduction
During DNA replication, multiple environmental toxins and pathogen invasions cause DNA damages (1). High-fidelity DNA polymerase cannot accommodate these damaged bases, which leads to stalled and collapsed replication forks, resulting in DNA double-strand breaks (DSB)2 (2). DSB is a severe lethal damage that increases genome instability (3). To prevent DSB caused by collapsed replication forks, the translesion synthesis (TLS) pathway is activated to bypass DNA-damage sites, which is mediated by proliferating cell nuclear antigen (PCNA) monoubiquitination (2).
PCNA can form a homotrimer clamp that slides along the DNA chain to provide scaffold for replication complex assembly (4). Blockage of the replication fork results in prolonged single-stranded DNA that is rapidly coated by replication protein A (RPA). Then RPA triggers the recruitment of RAD6-RAD18 ubiquitination ligase to monoubiquitinate PCNA on lysine 164 (5). The monoubiquitinated PCNA then promotes the recruitment of polη that belongs to the error-prone Y-family polymerase used in TLS pathway. Polη contains three PCNA interaction protein (PIP) boxes and an ubiquitin-binding zinc finger domain. The PIP domains of polη are less efficient than canonical PIP domains in error-free DNA polymerase, which lead to a minor affinity between polη and PCNA during normal DNA replication. Polη has a flexible active site that can accommodate damaged DNA template, allowing replication complex to bypass the lesion sites, thus preventing replication fork collapse and leaving the damaged sites to be repaired by other error-free pathways (6, 7). These studies described a widely accepted model of the function of ubiquitinated PCNA in TLS. However, a debate on the role of PCNA ubiquitination in TLS arose from recent extensive quantitative studies by Hedglin et al. (8). Their results demonstrated that monoubiquitination of PCNA did not change the binding affinity between PCNA and polη, and the subsequent TLS across a DNA lesion was also independent of PCNA monoubiquitination. They proposed that PCNA monoubiquitination indirectly promotes DNA synthesis by increasing the residence time of polη within the damaged sites, likely through altering the chromatin structure around damaged sites, although further studies are needed to test this hypothesis (8).
Besides multiple chemical or physical stimuli that induce DNA damage, many pathogens also cause cellular DNA damage and even manipulate DNA-damage repair proteins to facilitate their own proliferation (9). HSV-1 is a large double-stranded DNA virus with an icosahedral capsid wrapped by an envelope. The tegument layer between the capsid and envelope contains proteins that are important for virus infection and capsid assembly. HSV-1 infection usually occurs at oral mucosa neurons. After primary infection, virus particles transport along the neuronal axons to the nucleus and establish a latent state. During the latent infection stage, virus DNA is packed into a repressed structure and most genes remain silent. Latent virus can be reactivated by multiple stimulus including UV exposure and transport from the neurons down to primary infection sites, resulting in herpetic stomatitis. The reactivation of HSV-1 can happen multiple times during one's lifetime. Besides the oral mucosa, HSV-1 also infects genitals and corneal tissues, causing genital herpes or herpes keratitis, respectively (10, 11).
The infection of HSV-1 activates a cellular defense system including DNA-damage response. Multi-virus proteins coordinate these DNA-damage response (DDR) pathways elegantly to facilitate virus DNA replication. It has been reported that the ATM signaling pathway is activated during HSV-1 infection, whereas ATR and DNA-PKCs pathways are inhibited (1, 12). In addition to function in the DDR signaling pathway, PCNA and several other DDR proteins including RPA, DNA-PKCs, Rad50, Ku86, Ku70, PARP1, and Mre11 have also been observed to aggregate at the HSV-1 virus replication compartment (13–15). However, the putative roles of PCNA in viral replication remain poorly documented.
UL36 (VP1/2) is the largest tegument protein of HSV-1 containing more than 3000 amino acids, which helps HSV-1 viruses enter cells (16, 17). When most tegument proteins are released into the cytoplasm, UL36 remains attached to capsid and facilitate viral DNA entering the nucleus. This direct attachment enables UL36 to play a central role in the whole tegument structure assembly and virions maturation (18–20). In addition, the N-terminal of UL36 cleaved from the full-length protein has been reported to possess deubiquitination activity (21). This fragment contains around 500 amino acids and named as UL36 ubiquitin-specific protease (UL36USP). Cysteine at position 40 in the HSV-1 F strain has been recognized as the active site for UL36USP deubiquitination activity (22). UL36USP can cleave K48 and K63 ubiquitin chains specifically and has no cleavage activity for ubiquitin-like protein modifications (23).
So far, two substrates of UL36USP have been identified (22, 24). Wang et al. (22) discovered that UL36USP inhibits the IFN-β pathway by deubiquitinating TRAF3. The O'Hare group (24) explored the effect of UL36 USP activity on its own stability. Moreover, HSV-1 infection has been found to regulate DNA-damage pathways, however, the detailed mechanisms remain unclear (1). Given that PCNA is observed at the HSV-1 replication center and knockdown of PCNA reduces viral replication and histone deposition, and PCNA monoubiquitination plays critical roles in TLS pathway, it is worth investigating the effect of HSV-1 virus infection on PCNA ubiquitination and the TLS pathway (25).
In this study, we demonstrated that in response to DNA damage, UL36USP deubiquitinated PCNA and inhibited polη recruitment, and cells with stably expressed UL36USP showed a significant decrease of cell viability. These results indicate that HSV-1 suppresses the TLS pathway through the deubiquitinating activity of UL36USP. Our findings provide new insight into DNA-repair pathways in HSV-1 infection.
Results
UL36USP deubiquitinates PCNA in vivo
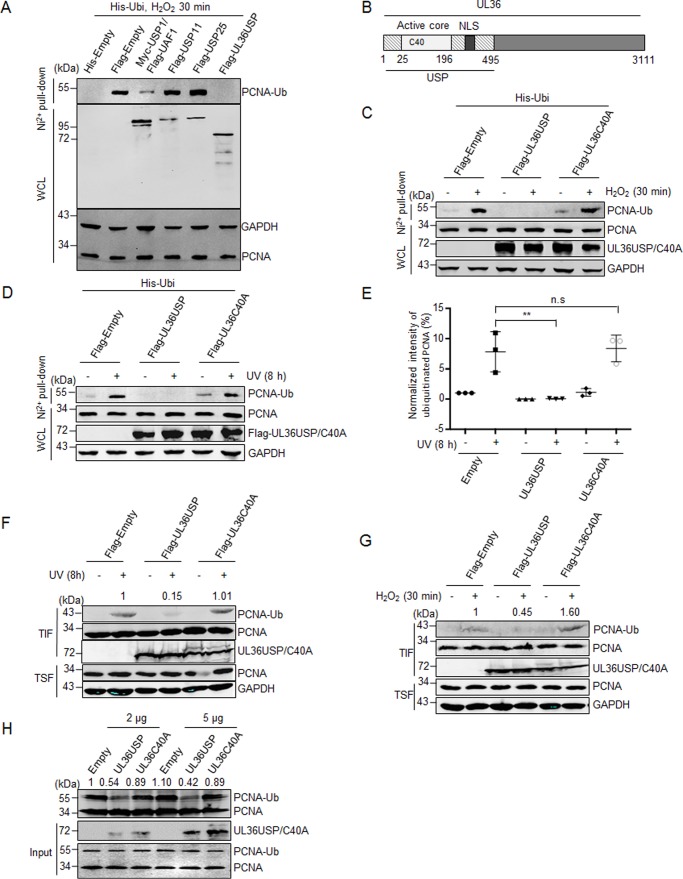
As a crucial member of cellular DDR pathways, the observation of PCNA in the HSV-1 virus DNA replication compartment suggests an active manipulation of PCNA by virus (13, 14). Because PCNA ubiquitination has been well studied in the DDR pathway, we wondered whether UL36USP, a deubiquitinase of the HSV-1 virus, targets monoubiquitinated PCNA in response to DNA damage. Different deubiquitinases were transfected into HEK293T cells together with His-ubiquitin, and PCNA ubiquitination was analyzed by His pulldown analysis. USP1/UAF1, a known deubiquitinase for PCNA was used as a positive control, whereas USP11 and USP25 were used as negative controls. In cells treated with H2O2, we found that UL36USP inhibited the level of PCNA ubiquitination more efficiently than USP1/UAF1 (Fig. 1A). Next, we investigated if the deubiquitinase activity of UL36USP is essential for its effect on PCNA during the DNA damage repair process. As shown in Fig. 1B, UL36USP is the N-terminal domain of HSV-1 large tegument protein UL36. Cysteine at position 40 is the key residue for the deubiquitination activity of UL36USP, and mutation of Cys at 40 to Ala (C40A) causes a significant loss of deubiquitination activity (22), we next explored whether the deubiquitination of PCNA by UL36USP was dependent on its deubiquitination activity. Cells were transfected with UL36USP or UL36C40A and the ubiquitination of PCNA was examined by a His pulldown assay after treatment with ultraviolet (UV) or H2O2. As expected, both UV and H2O2 promoted PCNA ubiquitination efficiently, and UL36USP inhibited PCNA ubiquitination completely, whereas the UL36C40A mutant exhibited no capability of deubiquitinating PCNA with or without DNA-damage stimuli (Fig. 1, C and D). The His pulldown experiment using UV as DNA-damage stimuli were repeated three times and quantified with Odyssey software. The intensity of ubiquitinated PCNA was normalized to that of the empty vector transfected but not stimulated group, and the deubiquitination effect of UL36USP on PCNA ubiquitination was significant with p value equaling 0.0076 (Fig. 1E).
Figure 1.
UL36USP deubiquitinates PCNA in vivo and in vitro. A, UL36USP reduces PCNA ubiquitination more efficiently than USP1/UAF1. HEK293T cells were cotransfected with His-ubiquitin and the indicated deubiquitinase plasmids, at 48 h after transfection, cells were treated with 100 μm H2O2 for 30 min and lysed. Cell lysates were subjected to His pulldown analysis, and the ubiquitinated PCNA was detected by Western blotting using anti-PCNA antibody. B, UL36USP is the N-terminal region of the HSV large tegument protein UL36. The cysteine at position 40 is the key amino acid of deubiquitinating activity. C–G, wild-type UL36USP, but not the C40A mutant, deubiquitinates PCNA in response to DNA damage induced by H2O2 or UV treatment. HEK293T cells were transfected with UL36USP or mutant UL36C40A that were defective in deubiquitination. At 40 h after transfection, cells were stimulated with or without H2O2 (100 μm, C) or UV (40 J/m2, D) and subjected to a His pulldown assay as described above. E, quantification of ubiquitinated PCNA normalized to the empty vector expressing group. Three independent His pulldown experiments were performed at the same conditions of D, the normalized intensities of PCNA ubiquitination was shown in scatter plots with standard deviation to represent error. F and G, HEK293T cells were transfected with UL36USP or UL36C40A for 40 h, and then treated with or without UV (F) or H2O2 (G). Cells were harvested, incubated with Triton X-100 buffer, and fractionated into chromatin-containing insoluble (TIF) and soluble (TSF) fractions. PCNA were concentrated in the insoluble fraction and probed by anti-PCNA antibody. The intensity of ubiquitinated PCNA was normalized to the total quantity of PCNA in TIF. H, in vitro deubiquitination assay shows UL36USP but not C40A deubiquitinates PCNA. HEK293T cells were transfected with His-ubiquitin and HA-PCNA plasmids. At 48 h after transfection, UV was used to promote PCNA ubiquitination. Ubiquitinated PCNA was precipitated by using Ni2+ beads and incubated with purified UL36USP or UL36C40A recombinant protein, as indicated, at 30 °C overnight, and then detected by Western blotting with antibody targeting PCNA. The intensity of ubiquitinated PCNA was normalized to the total quantity of PCNA calculated by adding ubiquitinated and unmodified PCNA. UL36USP and UL36C40A proteins were purified from E. coli. All the experiments described have been repeated more than 3 times. WCL, whole cell lysates; **, p value < 0.01; n.s., not significant.
To avoid the influence of ubiquitin overexpression in the His pulldown assay described above and confirm the deubiquitinating activity of UL36USP, we performed a chromatin fractionation assay. Cells transfected with UL36USP or -C40A were subjected to UV or H2O2 treatments, and endogenous chromatin-associated proteins were isolated by using Triton X-100 containing buffer. Monoubiquitinated PCNA should be concentrated in the Triton X-100-insoluble fraction (TIF), as it is associated with the chromatin. Indeed, UL36USP reduced accumulation of ubiquitinated PCNA induced by UV (Fig. 1F) or H2O2 treatments (Fig. 1G), whereas the UL36C40A mutant had no such effect. Interestingly, we also found an unexpected band that migrated slower than UL36C40A itself but not wild-type UL36USP (Fig. 1, F and G), which was similar to the results of ubiquitinated VP1–2 NT1 C65A in a previous report (24). This observation suggests that UL36C40A might be ubiquitinated, which will be discussed later. These results demonstrate that UL36USP deubiquitinates PCNA in vivo, whereas UL36C40A loses its deubiquitinating activity on PCNA, suggesting that viral protein UL36USP is involved in the TLS pathway.
UL36USP removes ubiquitin from PCNA in vitro
The in vivo experiments indicate that UL36USP deubiquitinates PCNA by its catalytic activity, to detect the direct deubiquitination of PCNA by UL36USP, we performed an in vitro deubiquitination experiment. Ubiquitinated PCNA was enriched by Ni2+ beads in Nonidet P-40 containing cell lysis buffer, and washed with deubiquitinating buffer three times. UL36USP or -C40A recombinant proteins purified from Escherichia coli were incubated with ubiquitinated PCNA at 30 °C following an experimental procedure reported by Huang et al. (26). As shown in Fig. 1H, UL36USP-deubiquitinated PCNA significantly, whereas the UL36C40A mutant did not. The result of the in vitro experiment confirms that UL36USP deubiquitinates PCNA directly by its catalytic activity but not through other proteins.
DNA damage stimulates the interaction between UL36USP and PCNA
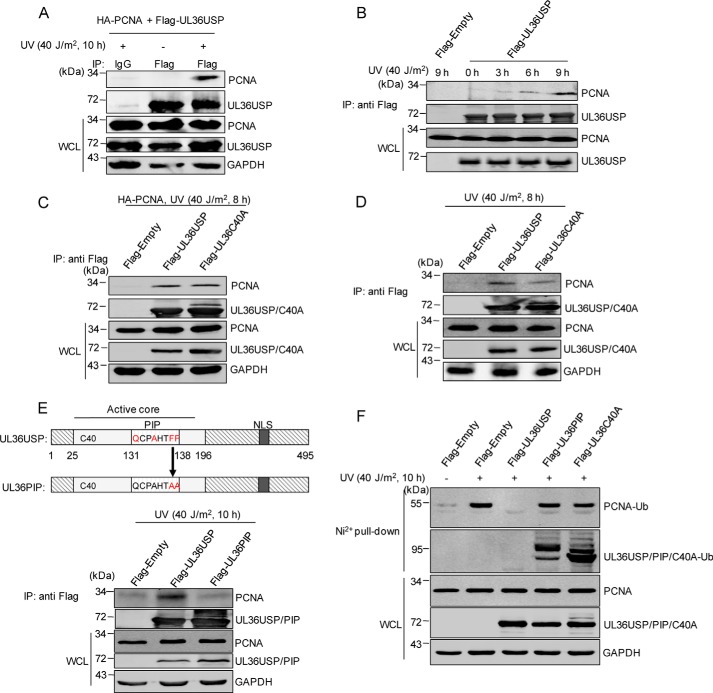
The observations that UL36USP-deubiquitinated PCNA spurred us to determine whether UL36USP interacted with PCNA, we then performed a co-immunoprecipitation (co-IP) assay to verify this hypothesis. HEK293T cells transfected with FLAG-UL36USP and HA-PCNA were treated with or without UV irradiation, cell lysates were immunoprecipitated using anti-FLAG antibody. As shown in Fig. 2A, the interaction between UL36USP and PCNA was detected in the presence of DNA-damage stimuli, suggesting that DNA damage stimulates the interaction of UL36USP with PCNA. We further investigated the interaction between UL36USP and endogenous PCNA after UV treatment for different times by using anti-FLAG® M2 magnetic beads (Fig. 2B). The interaction increasing with time prolongs and reached the maximum at 9 h after UV treatment, consistent with the result in Fig. 2A. This result indicates that UL36USP could respond to DNA damage by increasing its affinity with PCNA. Next, we investigated the interaction of both exogenous (Fig. 2C) and endogenous (Fig. 2D) PCNA to UL36USP and UL36C40A after DNA-damage stimuli. Both wild-type UL36USP and UL36C40A mutant associated with PCNA (Fig. 2, C and D), suggesting that the deubiquitinase activity of UL36USP has no effect on its binding to PCNA.
Figure 2.
UL36USP and UL36C40A interact with PCNA in response to DNA damage. A and B, UV treatment enhances the interaction between PCNA and UL36USP. HEK293T cells transfected with FLAG-UL36USP and HA-PCNA plasmids were treated with or without UV for the indicated time. After stimulation, a co-IP assay was performed to assess the interaction between PCNA with UL36USP or -C40A using the indicated antibodies. C, PCNA interacts with both UL36USP and UL36C40A. HEK293T cells transfected with HA-PCNA, FLAG-Empty, FLAG-UL36USP, or UL36C40A plasmids were treated with 40 J/m2 UV. At 8 h post-stimulation, cells were harvested and subjected to co-IP experiment. D, endogenous PCNA interacts with UL36USP and UL36C40A. HEK293T cells were transfected with FLAG-empty, FLAG-UL36USP, or UL36C40A plasmids and harvested as described in B. E, a schematic diagram of the UL36USP PIP domain and the UL36PIP mutation construct. The conserved amino acids of PIP domain are indicated by red, and the contiguous phenylalanines were mutated to alanines. The interaction between PCNA and UL36USP or UL36PIP was assessed by co-IP analysis using anti-FLAG® M2 magnetic beads, followed by Western blotting with anti-PCNA antibody. F, UL36PIP does not inhibit PCNA ubiquitination. HEK293T cells transfected with the indicated plasmids were subjected to 40 J/m2 UV stimuli for 10 h. And then a His pulldown assay was performed to examine ubiquitinated PCNA.
UL36USP interacts with PCNA by PIP
A PIP domain is found in UL36USP (Fig. 2E), which is conserved in herpes viruses (27). As the two conserved phenylalanines in the PIP domain play a crucial role in PIP-mediated PCNA interaction (27–31), we thus mutated these two residues to alanines (UL36PIP, Fig. 2E), and examined whether this mutant still interacted with PCNA. The result showed that mutation of the key residues in the PIP domain abolished its interaction with PCNA (Fig. 2E), suggesting that the PIP domain is critical in the UL36USP-PCNA interaction.
Moreover, to investigate whether mutation of the PIP domain will also affect the deubiquitination of PCNA by UL36USP, HEK293T cells transfected with FLAG-UL36USP or FLAG-UL36PIP or FLAG-UL36C40A mutants together with His-ubiquitin were subjected to UV stimulation, and then the levels of ubiquitinated PCNA were detected by a His pulldown assay (Fig. 2F). As expected, UV induced a significant increase of PCNA ubiquitination in the empty vector expressing group, whereas no ubiquitinated PCNA was detected in cells transfected with wild-type UL36USP. Similar to UL36C40A, the UL36PIP mutant could not inhibit PCNA ubiquitination. This result suggests that the PIP domain contributes to the deubiquitination process by promoting the interaction between UL36USP and PCNA.
UL36USP inhibits polη foci formation after DNA damage stimuli
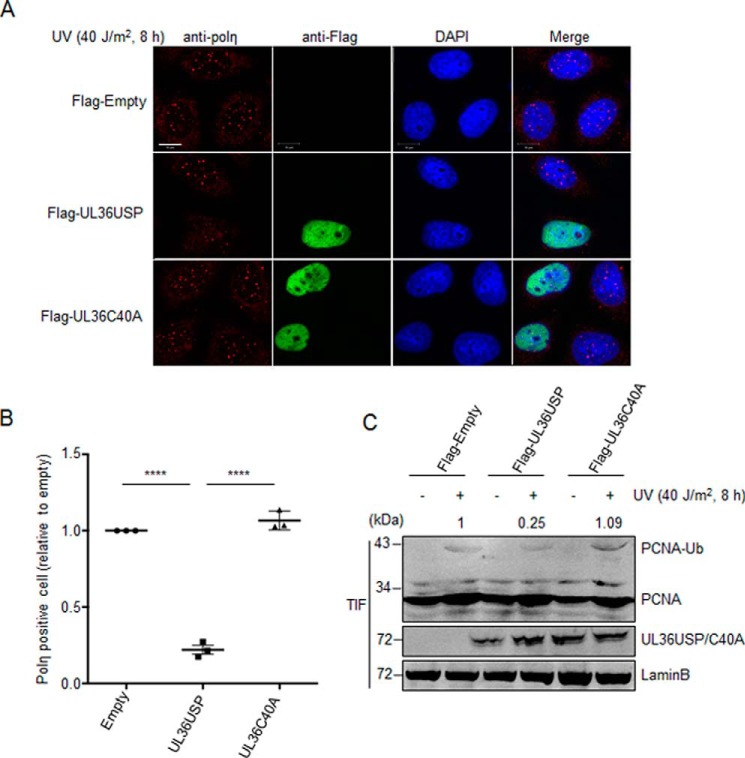
Because UL36USP deubiquitinated PCNA, we wondered whether it would compromise the accumulation of polη at replication stalling sites. HeLa cells transfected with FLAG-UL36USP, -C40A, or empty vector plasmids were treated with or without UV, and then subjected to immunofluorescence staining. Endogenous polη foci were detected by anti-polη antibody, whereas UL36USP or -C40A were labeled using anti-FLAG monoclonal antibody. Images were taken by Zeiss 710 confocal microscope. Cells with more than 10 polη focus were considered positive, and the percentage of polη foci-positive cells were counted, respectively, in three groups and then normalized to the empty vector group. This experiment was conducted three times and around 1000 cells for each group were counted and processed by using Bitplane Imaris software. Notice that due to low transfection efficiency in HeLa cells, only cells expressing UL36USP or -C40A (labeled green) were counted in each group (Fig. 3A). As expected, UL36USP decreased both the number of polη foci and percentage of polη foci-positive cells induced by DNA-damage stimuli. However, catalytically inactive UL36C40A expressing cells exhibited an equivalent percentage of polη foci-positive cells to that of control (Fig. 3, A and B). The expression of UL36USP or UL36C40A in cells that used immunofluorescence were detected at the same time. Quantification of ubiquitinated PCNA were normalized relative to Lamin B. Consistent with the reduced polη foci, a decreased ubiquitinated PCNA level was observed in UL36USP-expressing cells in comparison to the control or UL36C40A group (Fig. 3C). The relatively moderate deubiquitinating effect of UL36USP might be due to a restricted transfection efficiency in HeLa cells compared with that in HEK293T cells. These results indicate that UL36USP inhibits the TLS pathway via deubiquitinating PCNA and inhibiting polη foci formation.
Figure 3.
UL36USP inhibits polη foci formation in response to oxidative DNA damage. A, the number of polη foci was significantly decreases by UL36USP but not C40A. HeLa cells were transfected with UL36USP or -C40A plasmids and subjected to UV treatment. At 30 min after DNA-damage stimuli cells were fixed by cold methyl alcohol, immunofluorescence staining was performed using anti-polη and anti-FLAG antibodies. Endogenous polη were indicated by red fluorescence and UL36USP or UL36C40A by green. The scale bar represents 10 μm. B, quantification of polη foci-positive cells. Percentages of polη foci-positive cells (≥10 polη foci) were counted separately by Bitplane Imaris software. Three independent experiments were performed and ∼1000 cells were counted in each group. Error bars shows S.D. ****, p value < 0.0001. C, PCNA ubiquitination levels decreased in UL36USP-expressing cells compared with the control or UL36C40A group. HeLa cells transfected with UL36USP or -C40A plasmids were treated with or without UV. At 30 min after stimuli, cells were incubated with Triton X-100 buffer. Ubiquitinated PCNA in Triton X-100-soluble and -insoluble fractions was detected separately by Western blotting. The amount of ubiquitinated PCNA was normalized to Lamin B and indicated above the blot.
Viral infection-associated UL36USP deubiquitinates PCNA
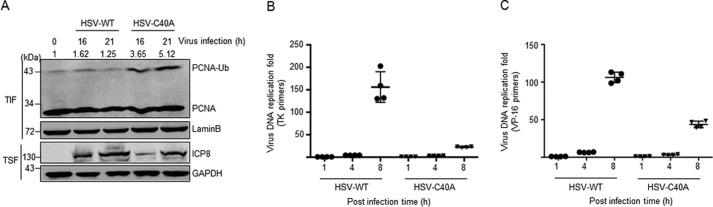
Because UL36USP transient expression caused significant deubiquitination of PCNA (Fig. 1), we further explored the effect of viral infection-associated UL36USP on PCNA during HSV-1 virus infection. We infected HEK293T cells with wild-type or UL36C40A HSV-1 virus and then chromatin-associated PCNA was enriched in TIF after chromatin fractionation (Fig. 4A). The infection of the catalytically inactive UL36C40A mutant virus resulted in a significant increase of PCNA ubiquitination, suggesting activation of the TLS pathway. On the contrary, cells infected with wild-type HSV-1 virus exhibited an equivalent level of PCNA ubiquitination to that of the mock infection group. These data suggest that wild-type HSV-1 virus infection also induces a DNA-damage response, which is compromised by the deubiquitination activity of virus infection-associated UL36USP.
Figure 4.
The deubiquitination activity of UL36USP is essential for viral DNA replication during HSV-1 infection. A, HSV-1 C40A mutant virus infection induced ubiquitination of PCNA, whereas wild-type virus exhibited an equivalent level compared with the control group. HEK293T cells were mock infected or infected with HSV-1 wild-type or UL36C40A mutant virus at 0.1 m.o.i. At 16 or 21 h post-infection, cells were fractionated into chromatin-containing fractions by incubating with Triton X-100 buffer. Western blotting was performed using the indicated antibodies. The intensity of ubiquitinated PCNA was normalized to Lamin B. B and C, HSV-1 wild-type virus DNA replication rate is higher than the UL36C40A mutant virus. Vero cells were infected with HSV-1 wild-type or UL36C40A mutant virus. Cells were harvested at the indicated time for quantitative real-time PCR analysis. The quantity of virus DNA was indicated by virus genes tk (B) and vp16 (C), respectively.
Because the level of ICP8 in cells infected by wild-type virus is higher than that in cells with HSV-1 UL36C40A mutant virus infection (Fig. 4A), we therefore assumed that HSV-1 facilitates its own replication through the deubiquitination activity of UL36USP. To assess this prediction, we performed quantitative RT-PCR to examine the replication rate of wild-type and UL36C40A mutant virus by determination of the levels of two virus DNA marker genes, thymidine kinase (tk) and vp16. The levels of both tk (Fig. 4B) and vp16 (Fig. 4C) increased strikingly in cells infected with wild-type HSV-1 but moderately in the UL36C40A mutant virus-infected group, indicating a higher replication rate of wild-type virus than C40A virus. These results demonstrate that the deubiquitinating activity of UL36USP facilitates HSV-1 virus DNA replication during infection.
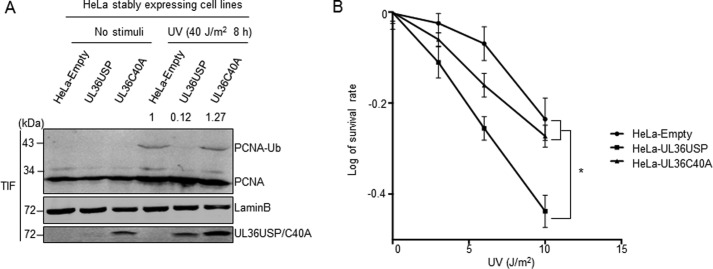
Stably expressed UL36USP decreases cellular viability after DNA damage
When suffering from DNA damage, cells with an activated TLS pathway would survive better than those with a deficient TLS pathway (6). To detect whether UL36USP affected cellular sensitivity to DNA damage, we constructed HeLa cells stably expressing FLAG-UL36USP or FLAG-UL36C40A using the plvx-IRES-zsGREEN lentivirus system as described under “Experimental procedures.” Monoclonal cells stably expressing UL36USP or -C40A were selected and stimulated with or without UV, and then the level of PCNA ubiquitination and cell viability were assessed. The UL36USP expressing cells showed a significant decrease of PCNA ubiquitination after exposure to UV irradiation, whereas UL36C40A expressing cells exhibited a much higher level of PCNA ubiquitination, which is similar to the control cells (Fig. 5A). Next, we examined the cell viability in these monoclonal cell lines treated with UV irradiation in different doses. 12 days later, the cell survival rates were calculated and normalized to that of cells without damage, respectively. DNA damage induced a decrease of cell numbers in all cell lines with the increase of UV intensity. However, UL36USP stably expressing cells exhibited a significantly lower survival rate compared with that of wild-type or UL36C40A-expressing cells (Fig. 5B). These results indicate that wild-type UL36USP leads to decreased cell viability by interrupting the cellular DNA damage repair ability.
Figure 5.
UL36USP stably expressing HeLa cells exhibited lower viability than that of wild-type or UL36C40A stably expressing cells after DNA damage. A, PCNA ubiquitination level decreased in UL36USP stably expressing HeLa cells compared with the control or UL36C40A group. HeLa cells stably expressing UL36USP or -C40A were treated with or without 40 J/m2 UV for 10 h and then incubated with Triton X-100. Triton-insoluble fractions were subjected to Western blotting to detect PCNA ubiquitination and UL36USP or -C40A expression. The intensity of ubiquitinated PCNA was normalized to Lamin B. B, UL36USP stably expressing HeLa cells showed decreased viability after DNA damage. UL36USP or UL36C40A stably expressing HeLa cells were treated with various ultraviolet radiation conditions as indicated. After 12 days, cell colonies were stained with crystal violet and counted; *, p value < 0.05.
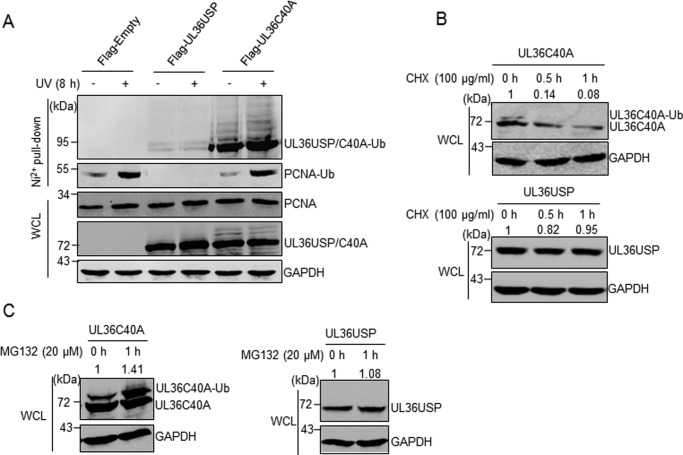
UL36USP deubiquitinates itself to enhance protein stability
Because the ubiquitin-proteasome system is a classic pathway for protein degradation, the unexpected band that migrated slower than UL36C40A itself observed in Figs. 1, F and G, and 2, C and D, prompted us to examine whether UL36USP or -C40A is ubiquitinated and degraded as an invading virus protein. The result of the His pulldown revealed multiple ubiquitinated bands of UL36C40A, whereas the level of ubiquitinated wild-type UL36USP was much lower. Also, the ubiquitination of UL36USP and -C40A was not affected by DNA damage stimulus (Fig. 6A). It is possible that ubiquitinated UL36USP could be deubiquitinated by itself, whereas the catalytic null mutant UL36C40A lost its deubiquitinating activity and maintained ubiquitin conjugates. Next, to further detect the degradation of UL36USP and -C40A, HEK293T cells stably expressing UL36USP or -C40A were treated with cycloheximide (CHX) for different times, and then the amount of UL36USP or -C40A was assessed. As shown in Fig. 6B, the level of UL36C40A decreased with a prolonged treatment time, whereas UL36USP abundance had no change (Fig. 6B). These results suggest that UL36C40A is degraded, whereas UL36USP stays stable after CHX treatment. Moreover, to explore whether the degradation of UL36C40A is executed by a ubiquitin-dependent proteasome system, UL36USP or -C40A stably expressing cells were treated with the proteasome-specific inhibitor MG132 and protein abundance was examined by Western blotting (Fig. 6C). UL36USP had no significant increase after 1 h of MG132 treatment, whereas ubiquitinated UL36C40A increased obviously (Fig. 6C). These results demonstrate that UL36USP deubiquitinates itself, whereas ubiquitinated UL36C40A is more sensitive to CHX and MG132 treatment, suggesting that the deubiquitination activity of UL36USP is required for its own stability.
Figure 6.
UL36USP and UL36C40A are ubiquitinated and degraded by proteasome, whereas UL36USP deubiquitinates itself to stabilize itself. A, UL36C40A had a stronger ubiquitination modification compared with UL36USP. HEK293T cells were transfected with His-ubiquitin and UL36USP or -C40A plasmids and treated with or without ultraviolet radiation. His pulldown assay was performed to examine the ubiquitination of UL36USP and UL36C40A using anti-FLAG antibody. B, UL36C40A degraded after CHX treatment but UL36USP does not. HEK293T cells stably expressing UL36USP or UL36C40A were treated with CHX to block protein synthesis for the indicated times. Whole cell lysates were detected by Western blotting. C, UL36C40A degradation was blocked by MG132 treatment. HEK293T cells stably expressing UL36USP or -C40A were treated with MG132 to block proteasome degradation. Whole cell lysates (WCL) were detected by Western blotting. The intensity of UL36USP and UL36C40A were normalized against those of GAPDH.
Discussion
As the largest tegument protein in HSV-1, the exploration of UL36 function has mainly focused on its role in viral maturation. Mounting evidence demonstrates that UL36 participates in virus uptake and the tegument assembly process (18, 32–36). Moreover, UL36USP, a deubiquitinating enzyme embedded in the N-terminal of UL36, could target both K48 and K63 polyubiquitin chains of TRAF3 and shows self-deubiquitinating activity (21, 22, 24). However, whether UL36USP manipulates other cellular pathways such as DDR remains unclear. In this study, we demonstrate PCNA as a new substrate of HSV-1 UL36USP. The DNA damage-induced PCNA ubiquitination is suppressed by wild-type UL36USP but not UL36C40A mutant, suggesting that PCNA is a cellular substrate of HSV-1 UL36USP.
DNA-damage response is used as a defense to viral infection. On the contrary, to inhibit the cellular antiviral defense, viruses will manipulate cellular pathways to change the hostile cellular environment to support its own replication. Previous studies indicate that HSV-1 regulates DDR pathways when the viral genome is transported to the cell nucleus. HSV-1 infection is found to increase single strand annealing and the Fanconi anemia (FA) pathway while decreasing homologous recombination (HR), non-homologous end joining (classic NHEJ), and microhomology-mediated end joining (MMEJ) pathways, although the detailed mechanisms remained to be elucidated (37, 38). In addition to these pathways, our data here indicates that the TLS pathway is also suppressed by HSV-1 through the deubiquitination activity of UL36USP, adding a new member into the cellular DDR network manipulated by HSV-1 infection. DDR pathways usually function coordinately with each other. For example, interstrand DNA cross-link repair requires cross-talk among TLS, FA network, and HR repair pathways. It is possible that TLS suppression by HSV-1 infection may coordinate with other DDR pathways. Therefore, additional studies are required to explore how UL36USP-mediated TLS correlates with other pathways including HR and FA in HSV-1-infected cells.
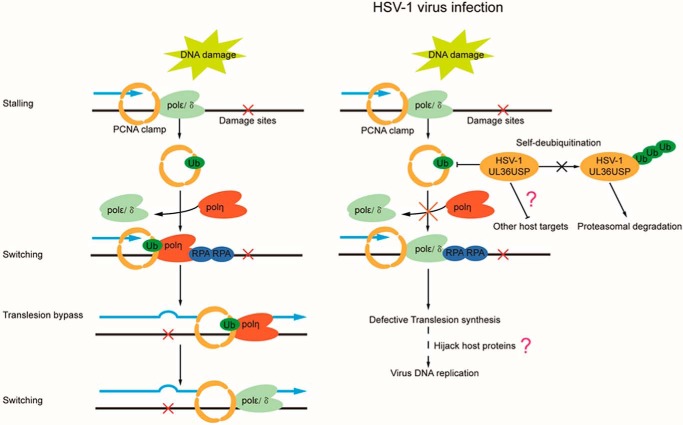
To overcome the cellular defense system and facilitate virus replication, as we found in this study, one possible mechanism is to inactivate the TLS pathway and hijack DNA-damage repair proteins to aid its own proliferation. In accordance with this scenario, during HSV-1 virus infection, certain cellular proteins including PCNA that interacts with ICP8 have been observed at the virus DNA replication center (13, 14). Besides, PCNA has been found to be important for HSV-1 virus DNA replication and histone deposition on the virus genome (25). In this study, we found that cells infected with the UL36C40A mutant virus showed a lower viral DNA replication rate than that with wild-type HSV-1. Consistently, we also found that cells infected by HSV-1 wild-type or UL36C40A mutant virus exhibited a significant difference in the accumulations of ubiquitinated PCNA on chromatin (Fig. 4). Our mechanistical studies revealed that in response to DNA damage, UL36USP interacts with and deubiquitinates PCNA, resulting in a reduced number of polη foci and decreased cell viability (Fig. 7). Although the role of PCNA ubiquitination in TLS is controversial, our findings here indicate that HSV-1 infection-associated UL36USP suppresses the cellular TLS pathway by deubiquitination of PCNA, and facilitates viral DNA replication as well, providing insight for understanding how viruses fight against the cellular defense system to benefit its own replication (Fig. 7). However, additional studies such as efficient detection of mutation frequency (39–42) will further help us to elucidate the effect of UL36USP on the function of polη in TLS. Moreover, as mentioned in our working model, whether deubiquitination of PCNA by UL36USP is directly associated with viral DNA replication or if UL36USP targets other host proteins, these questions remain to be elucidated in the future.
Figure 7.
HSV-1 UL36USP suppresses TLS pathway by deubiquitinating PCNA. DNA damage caused by UV and H2O2 induces PCNA monoubiquitination, which promotes polη foci formation and facilitates cell survival after DNA damage. HSV-1 inhibits the TLS pathway by deubiquiting PCNA through the deubiquitinase activity of the UL36USP protein, which results in a reduced number of polη foci, thus decreasing cell viability. Meanwhile, UL36USP deubiquitinates itself to increase protein stability.
The UL36USP catalytic motif is conserved among the herpesvirus family including Kaposi's sarcoma-associated herpesvirus (KSHV), Epstein-Barr virus (EBV), human cytomegalovirus, and pseudorabies virus (23). Similar to what we found here, BPLF1, the UL36USP homolog in EBV, has also been reported to deubiquitinate PCNA during EBV infection (27). The E3 ubiquitin ligase of PCNA, Rad18, is also a substrate of BPLF1, which is stabilized by BPLF1 deubiquitinating activity during EBV infection (43). It is interesting that both PCNA and its E3 ligase are substrates of the same deubiquitinase. Whether HSV-1 UL36USP could also deubiquitinate Rad18 is worthy to be tested. Different from UL36USP that only possesses deubiquitinating activity, BPLF1 also acts as a deneddylase in addition to its deubiquitinating activity, which inhibits culling-ring ligase activity and leads to a deregulation of the cell S phase that benefits virus replication (44). Besides, the deubiquitinating activity of UL48, a homogenous protein of UL36 in human cytomegalovirus, also plays a crucial role in virus replication (45). Together, these studies indicate that through their ubiquitin-specific protease, herpes viruses modulate DDR pathways to facilitate virus proliferation.
A previous report (24) indicated that loss of UL36 deubiquitinating activity resulted in a drastic reduction in the full-length protein level, which could be stabilized by proteasome inhibition, and studies of the stability of different HSV-1 UL36 truncations suggest that functional USP is required for stabilizing the fragments longer than the core USP domain. Consistently, here we also showed that UL36USP, which possesses deubiquitinating activity, could deubiquitinate and stabilize itself, whereas the strongly ubiquitinated UL36C40A is highly sensitive to CHX and MG132 treatment. These works together suggest that the deubiquitination activity of UL36USP contributes, at least partially, to its own stability. This work identified a new cellular substrate for UL36USP, demonstrated that the deubiquitinase of HSV-1 contributes to viral replication, and therefore enriched the knowledge of HSV-1 manipulation of cellular DNA damage pathways.
Experimental procedures
Cell lines, culture, and transfection
HEK293T, HeLa, and VERO cells were cultured in Dulbecco's modified Eagle's medium (DMEM, Gibco) supplemented with 10% fetal bovine serum (FBS, Gibco). Cells were grown at 37 °C in a humidified CO2 (5%) incubator, and transfected with polyethylenimine.
Plasmids and reagents
The 1485 bp encoding the deubiquitinase at the N terminus of UL36 protein was amplified by PCR and inserted into pcDNA3.0–3FLAG and pET28a vectors. UL36C40A and UL36PIP mutants were constructed by the point mutation method. The following antibodies were used in this study: anti-PCNA (BE0029, EASYBIO; 1:1000), anti-polη (BS6695, Bioworld; 1:100), mouse monoclonal anti-FLAG antibody (F3165, Sigma; 1:2000), anti-HA antibody (H9658, Sigma; 1:2000), mouse monoclonal anti-His antibody (D291–3, MBL, Nagoya, Japan; 1:2000), and anti-HSV-1 ICP8 antibody (A0914, Santa Cruz Biotechnology; 1:100).
HSV-1 virus infection and propagation
Wild-type HSV-1 F strain and UL36C40A mutant strain were propagated in VERO cells and cultured in DMEM containing 10% FBS. When cell density reached 90%, DMEM was removed and HSV-1 virus were added to the cells at a m.o.i. of 5–10, and then incubated at 37 °C. One h later, fresh DMEM containing 10% FBS was added and continuously cultured until a clear cytopathic effect was shown. Cells were collected in DMEM and placed at −80 °C for 1 day, and then thawed on ice and the freeze-thaw cycle was repeated for three times. The virus was stored at −80 °C after titration.
Co-immunoprecipitation
In Fig. 2A, HEK293T cells transfected with HA-PCNA and FLAG-UL36USP plasmids were lysed in Nonidet P-40 cell lysis buffer (50 mm Tris, 150 mm NaCl, 1% Nonidet P-40, 1 mm EDTA-2Na) and sonicated on ice using a Bioruptor Plus device. Immunoprecipitation and Western blotting were performed as described previously (46). In Fig. 2B, HEK293T cells transfected with HA-PCNA and FLAG-UL36USP or UL36C40A plasmids were lysed and sonicated as previously described. Cell lysates were centrifuged at 12,000 rpm for 15 min. FLAG-M2 beads were incubated with the supernatants for 3 h and then washed with cell lysis buffer 3 times. After being centrifuged at 3,000 rpm for 5 min, the supernatants were discarded, and the beads were rinsed with FLAG peptide for 3 h and centrifuged at 3,000 rpm for 5 min. The supernatants were collected and boiled with 6× SDS loading buffer. Then samples were subjected to SDS-PAGE and Western blot analyses.
His-ubiquitin pulldown assay
HEK293T cells were transfected with His-ubiquitin and FLAG-UL36/C40A plasmids, and then lysed with His pulldown lysis buffer containing 6 m guanidinium chloride, 0.1 m Na2HPO4/NaH2PO4, 0.01 m Tris-HCl, pH 8.0, 5 mm imidazole, and 10 mm β-mercaptoethanol. Cell lysates were incubated with nickel-nitrilotriacetic acid beads (Qiagen) for 4 h, and then washed with His pulldown wash buffer I (6 m guanidinium chloride, 0.1 m Na2HPO4/NaH2PO4, 0.01 m Tris-HCl, pH 8.0), buffer II (8 m urea, 0.1 m Na2HPO4/NaH2PO4, 0.01 m Tris-HCl, pH 8.0), buffer III (8 m urea, 0.1 m Na2HPO4/NaH2PO4, 0.01 m Tris-HCl, pH 6.3, 0.2% Triton X-100), and buffer IV (8 m urea, 0.1 m Na2HPO4/NaH2PO4, 0.01 m Tris-HCl, pH 6.3, 0.1% Triton X-100), respectively. The samples were centrifuged and the supernatant was denatured and loaded on a SDS-PAGE gel. Western blotting was performed using the indicated antibody. For detailed procedures see our previous article (47).
Chromatin fractionation assay
HEK293T cells were transfected with UL36USP or UL36C40A plasmids, at 42 h after transfection, cells were lysed with chromatin isolation buffer A containing 10 mm HEPES, pH 7.8, 10 mm KCl, 1.5 mm MgCl2, 0.34 m sucrose, 10% glycerol, 1 mm DTT, 0.1% Triton X-100, and 0.5% protease inhibitor, and incubated on ice for 30 min, and then centrifuged at 5,000 rpm for 5 min. The insoluble fraction was recovered and resuspended in buffer B (3 mm EDTA, 0.2 mm EGTA, 1 mm DTT, 0.5% protease inhibitor) and incubated on ice for 30 min. After centrifugation at 5,000 rpm for 5 min, the insoluble fraction was recovered and resuspended in buffer C (50 mm Tris, 150 mm NaCl, 2% Nonidet P-40, 1 mm EDTA-2Na), and incubated on ice for 10 min. Samples were denatured in 2× SDS loading buffer and separated by SDS-PAGE, and subjected to Western blot analysis.
In vitro deubiquitination assay
HEK293T cells were transfected with HA-PCNA and His-ubiquitin plasmids and cultured for 48 h. Cells were treated with 40 J/m2 for 8 h to promote the level of ubiquitinated PCNA, and lysed in cell lysis buffer using a ultrasonic cell disruption system. After centrifugation at 13,000 rpm for 10 min, the supernatant was collected and divided into six equal groups. The ubiquitinated PCNA was concentrated by incubating with 30 μl of nickel-nitrilotriacetic acid beads for 4 h. The beads were washed with deubiquitinating buffer (60 mm HEPES, 5 mm MgCl2, 4% glycerol, pH 7.6) three times and incubated with UL36USP or -C40A protein at 30 °C overnight (26). The reaction mixture was boiled in 2× SDS loading buffer and subjected to SDS-PAGE and Western blotting.
Immunofluorescence staining
HeLa cells cultured on coverslips were transfected with FLAG-UL36USP or -C40A plasmid and cultured for 42 h. Cells were treated with or without 40 J/m2 for 8 h and washed with cold PBS, and then fixed with methyl alcohol at −20 °C for 10 min. After washing with PBS, cells were blocked with 1% BSA in PBS, and then incubated with primary antibody at 37 °C for 1 h. Cells were washed with PBS and incubated with secondary antibody at 37 °C for 1 h. DAPI were used to stain the nucleus. Images were captured by Zeiss LSM710 and analyzed with Bitplane Imaris software.
Quantitative RT-PCR
VERO cells were infected with HSV-1 wild-type or C40A mutant virus, and incubated with lysis buffer containing proteinase K at a final concentration of 100 μg/ml at 37 °C overnight. DNA was extracted using phenol/chloroform and precipitated with ethanol, and resuspended with TE buffer. qRT-PCR was performed using Bio-Rad system. The primers used were: vp16-F, GCCGCCCCGTACCTCGTGAC; vp16-R, CAGCCCGCTCCGCTTGTCG; tk-F, GTATGATGACACAAACCCCG; tk-R, GAGTTTCACGCCACCAAGAT; GAPDH-F, TCTCTGCCCCCTCTGCTG; GADPH-R, ATGGTTCACACCCATGACGA.
Construction of cells lines stably expressing UL36USP or UL36C40A
HEK293T cells were transfected with pLVX-IRES-ZsGreen1 carrying FLAG-UL36USP or -C40A genes together with pLP2/R-1, pLP/VSVG, and pLP1/pRRE. The medium was changed at 6 and 24 h, respectively, after transfection. Another 10 ml of DMEM containing 5% FBS was added at 48 h, and the medium containing virus was collect 72 h after transfection. The medium containing virus were used to infect HeLa cells, at 48 h after infection, cells stably expressing UL36USP or -C40A were isolated with fluorescence-activated cell sorting (FACS).
Cell viability
HeLa cells stably expressing UL36USP or -C40A were seeded into six-well plates. 24 h later, cells were subjected to gradient UV stimulation and cultured for 12 days, and then stained with crystal violet. Colony numbers were counted.
Author contributions
X. D. D., J. H. G., and X. F. Z. designed the research, analyzed the data, and wrote the paper. X. D. D. performed the experiments, and C. F. Z. constructed the virus strains.
Acknowledgments
We thank Chuanzhen Yang for the help with co-IP assay, and Xiaochen Li and Liying Du of Core Facilities of Life Sciences, Peking University, for assistance with immunofluorescence staining and FACS.
This work was supported by National Science Foundation of China Grants 31470754 and 31670786, the National Key Research and Development Program of China Grant 2016YFC1302401, and Doctoral Fund of Ministry of Education of China Grant 20130001130003. The authors declare that they have no conflicts of interest with the contents of this article.
- DSB
- double-strand breaks
- TLS
- translesion synthesis
- PCNA
- proliferating cell nuclear antigen
- RPA
- replication protein A
- PIP
- PCNA interaction protein
- DDR
- DNA-damage response
- USP
- ubiquitin-specific protease
- TIF
- Triton X-100-insoluble fraction
- co-IP
- co-immunoprecipitation
- CHX
- cycloheximide
- FA
- Fanconi anemia
- HR
- homologous recombination
- EBV
- Epstein-Barr virus.
References
- 1. Luftig M. A. (2014) Viruses and the DNA damage response: activation and antagonism. Annu. Rev. Virol. 1, 605–625 [DOI] [PubMed] [Google Scholar]
- 2. Kannouche P. L., Wing J., and Lehmann A. R. (2004) Interaction of human DNA polymerase η with monoubiquitinated PCNA: a possible mechanism for the polymerase switch in response to DNA damage. Mol. Cell 14, 491–500 [DOI] [PubMed] [Google Scholar]
- 3. Chapman J. R., Taylor M. R., and Boulton S. J. (2012) Playing the end game: DNA double-strand break repair pathway choice. Mol. Cell 47, 497–510 [DOI] [PubMed] [Google Scholar]
- 4. Mailand N., Gibbs-Seymour I., and Bekker-Jensen S. (2013) Regulation of PCNA-protein interactions for genome stability. Nat. Rev. Mol. Cell Biol. 14, 269–282 [DOI] [PubMed] [Google Scholar]
- 5. Davies A. A., Huttner D., Daigaku Y., Chen S., and Ulrich H. D. (2008) Activation of ubiquitin-dependent DNA damage bypass is mediated by replication protein A. Mol. Cell 29, 625–636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Sale J. E., Lehmann A. R., and Woodgate R. (2012) Y-family DNA polymerases and their role in tolerance of cellular DNA damage. Nat. Rev. Mol. Cell Biol. 13, 141–152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Sale J. E. (2013) Translesion DNA synthesis and mutagenesis in eukaryotes. Cold Spring Harb. Perspect. Biol. 5, a012708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Hedglin M., Pandey B., and Benkovic S. J. (2016) Characterization of human translesion DNA synthesis across a UV-induced DNA lesion. Elife 5, e19788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Weitzman M. D., and Weitzman J. B. (2014) What's the damage? The impact of pathogens on pathways that maintain host genome integrity. Cell Host Microbe 15, 283–294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Whitley R. J., Kimberlin D. W., and Roizman B. (1998) Herpes simplex viruses. Clin. Infect. Dis. 26, 541–553; quiz 554–545 [DOI] [PubMed] [Google Scholar]
- 11. Weller S. K., and Coen D. M. (2012) Herpes simplex viruses: mechanisms of DNA replication. Cold Spring Harb. Perspect. Biol. 4, a013011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Chaurushiya M. S., and Weitzman M. D. (2009) Viral manipulation of DNA repair and cell cycle checkpoints. DNA Repair 8, 1166–1176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Wilcock D., and Lane D. P. (1991) Localization of P53, retinoblastoma and host replication proteins at sites of viral replication in herpes-infected cells. Nature 349, 429–431 [DOI] [PubMed] [Google Scholar]
- 14. Taylor T. J., and Knipe D. M. (2004) Proteomics of herpes simplex virus replication compartments: association of cellular DNA replication, repair, recombination, and chromatin remodeling proteins with ICP8. J. Virol. 78, 5856–5866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Lilley C. E., Carson C. T., Muotri A. R., Gage F. H., and Weitzman M. D. (2005) DNA repair proteins affect the lifecycle of herpes simplex virus 1. Proc. Natl. Acad. Sci. U.S.A. 102, 5844–5849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Shanda S. K., and Wilson D. W. (2008) UL36p is required for efficient transport of membrane-associated herpes simplex virus type 1 along microtubules. J. Virol. 82, 7388–7394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Zaichick S. V., Bohannon K. P., Hughes A., Sollars P. J., Pickard G. E., and Smith G. A. (2013) The herpesvirus VP1/2 protein is an effector of dynein-mediated capsid transport and neuroinvasion. Cell Host Microbe 13, 193–203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Cardone G., Newcomb W. W., Cheng N., Wingfield P. T., Trus B. L., Brown J. C., and Steven A. C. (2012) The UL36 tegument protein of herpes simplex virus 1 has a composite binding site at the capsid vertices. J. Virol. 86, 4058–4064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Lee J. I., Luxton G. W., and Smith G. A. (2006) Identification of an essential domain in the herpesvirus VP1/2 tegument protein: the carboxy terminus directs incorporation into capsid assemblons. J. Virol. 80, 12086–12094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Newcomb W. W., and Brown J. C. (2010) Structure and capsid association of the herpesvirus large tegument protein UL36. J. Virol. 84, 9408–9414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kattenhorn L. M., Korbel G. A., Kessler B. M., Spooner E., and Ploegh H. L. (2005) A deubiquitinating enzyme encoded by HSV-1 belongs to a family of cysteine proteases that is conserved across the family Herpesviridae. Mol. Cell 19, 547–557 [DOI] [PubMed] [Google Scholar]
- 22. Wang S., Wang K., Li J., and Zheng C. (2013) Herpes simplex virus 1 ubiquitin-specific protease UL36 inhibits β interferon production by deubiquitinating TRAF3. J. Virol. 87, 11851–11860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Schlieker C., Korbel G. A., Kattenhorn L. M., and Ploegh H. L. (2005) A deubiquitinating activity is conserved in the large tegument protein of the herpesviridae. J. Virol. 79, 15582–15585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Bolstad M., Abaitua F., Crump C. M., and O'Hare P. (2011) Autocatalytic activity of the ubiquitin-specific protease domain of herpes simplex virus 1 VP1–2. J. Virol. 85, 8738–8751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Sanders I., Boyer M., and Fraser N. W. (2015) Early nucleosome deposition on, and replication of, HSV DNA requires cell factor PCNA. J. Neurovirol. 21, 358–369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Huang T. T., Nijman S. M., Mirchandani K. D., Galardy P. J., Cohn M. A., Haas W., Gygi S. P., Ploegh H. L., Bernards R., and D'Andrea A. D. (2006) Regulation of monoubiquitinated PCNA by DUB autocleavage. Nat. Cell. Biol. 8, 339–347 [DOI] [PubMed] [Google Scholar]
- 27. Whitehurst C. B., Vaziri C., Shackelford J., and Pagano J. S. (2012) Epstein-Barr virus BPLF1 deubiquitinates PCNA and attenuates polymerase eta recruitment to DNA damage sites. J. Virol. 86, 8097–8106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Acharya N., Yoon J. H., Gali H., Unk I., Haracska L., Johnson R. E., Hurwitz J., Prakash L., and Prakash S. (2008) Roles of PCNA-binding and ubiquitin-binding domains in human DNA polymerase eta in translesion DNA synthesis. Proc. Natl. Acad. Sci. U.S.A. 105, 17724–17729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Haracska L., Acharya N., Unk I., Johnson R. E., Hurwitz J., Prakash L., and Prakash S. (2005) A single domain in human DNA polymerase ι mediates interaction with PCNA: implications for translesion DNA synthesis. Mol. Cell. Biol. 25, 1183–1190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Hishiki A., Hashimoto H., Hanafusa T., Kamei K., Ohashi E., Shimizu T., Ohmori H., and Sato M. (2009) Structural basis for novel interactions between human translesion synthesis polymerases and proliferating cell nuclear antigen. J. Biol. Chem. 284, 10552–10560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Vidal A. E., Kannouche P., Podust V. N., Yang W., Lehmann A. R., and Woodgate R. (2004) Proliferating cell nuclear antigen-dependent coordination of the biological functions of human DNA polymerase ι. J. Biol. Chem. 279, 48360–48368 [DOI] [PubMed] [Google Scholar]
- 32. Gibson W., and Roizman B. (1972) Proteins specified by herpes simplex virus: 8. characterization and composition of multiple capsid forms of subtypes 1 and 2. J. Virol. 10, 1044–1052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Zhou Z. H., Chen D. H., Jakana J., Rixon F. J., and Chiu W. (1999) Visualization of tegument-capsid interactions and DNA in intact herpes simplex virus type 1 virions. J. Virol. 73, 3210–3218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Bucks M. A., O'Regan K. J., Murphy M. A., Wills J. W., and Courtney R. J. (2007) Herpes simplex virus type 1 tegument proteins VP1/2 and UL37 are associated with intranuclear capsids. Virology 361, 316–324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Copeland A. M., Newcomb W. W., and Brown J. C. (2009) Herpes simplex virus replication: roles of viral proteins and nucleoporins in capsid-nucleus attachment. J. Virol. 83, 1660–1668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Roberts A. P., Abaitua F., O'Hare P., McNab D., Rixon F. J., and Pasdeloup D. (2009) Differing roles of inner tegument proteins pUL36 and pUL37 during entry of herpes simplex virus type 1. J. Virol. 83, 105–116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Schumacher A. J., Mohni K. N., Kan Y., Hendrickson E. A., Stark J. M., and Weller S. K. (2012) The HSV-1 exonuclease, UL12, stimulates recombination by a single strand annealing mechanism. PLoS Pathog. 8, e1002862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Karttunen H., Savas J. N., McKinney C., Chen Y. H., Yates J. R. 3rd, Hukkanen V., Huang T. T., and Mohr I. (2014) Co-opting the Fanconi anemia genomic stability pathway enables herpesvirus DNA synthesis and productive growth. Mol. Cell 55, 111–122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Han J., Liu T., Huen M. S., Hu L., Chen Z., and Huang J. (2014) SIVA1 directs the E3 ubiquitin ligase RAD18 for PCNA monoubiquitination. J. Cell Biol. 205, 811–827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Park J. M., Yang S. W., Yu K. R., Ka S. H., Lee S. W., Seol J. H., Jeon Y. J., and Chung C. H. (2014) Modification of PCNA by ISG15 plays a crucial role in termination of error-prone translesion DNA synthesis. Mol. Cell 54, 626–638 [DOI] [PubMed] [Google Scholar]
- 41. Zeman M. K., Lin J. R., Freire R., and Cimprich K. A. (2014) DNA damage-specific deubiquitination regulates Rad18 functions to suppress mutagenesis. J. Cell Biol. 206, 183–197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Kashiwaba S., Kanao R., Masuda Y., Kusumoto-Matsuo R., Hanaoka F., and Masutani C. (2015) USP7 is a suppressor of PCNA ubiquitination and oxidative-stress-induced mutagenesis in human cells. Cell Rep. 13, 2072–2080 [DOI] [PubMed] [Google Scholar]
- 43. Kumar R., Whitehurst C. B., and Pagano J. S. (2014) The Rad6/18 ubiquitin complex interacts with the Epstein-Barr virus deubiquitinating enzyme, BPLF1, and contributes to virus infectivity. J. Virol. 88, 6411–6422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Gastaldello S., Hildebrand S., Faridani O., Callegari S., Palmkvist M., Di Guglielmo C., and Masucci M. G. (2010) A deneddylase encoded by Epstein-Barr virus promotes viral DNA replication by regulating the activity of cullin-RING ligases. Nat. Cell. Biol. 12, 351–361 [DOI] [PubMed] [Google Scholar]
- 45. Kim E. T., Oh S. E., Lee Y. O., Gibson W., and Ahn J. H. (2009) Cleavage Specificity of the UL48 deubiquitinating protease activity of human cytomegalovirus and the growth of an active-site mutant virus in cultured cells. J. Virol. 83, 12046–12056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Hu B., Li S., Zhang X., and Zheng X. (2014) HSCARG, a novel regulator of H2A ubiquitination by downregulating PRC1 ubiquitin E3 ligase activity, is essential for cell proliferation. Nucleic Acids Res. 42, 5582–5593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Zhang J., Bai D., Ma X., Guan J., and Zheng X. (2014) hCINAP is a novel regulator of ribosomal protein-HDM2-p53 pathway by controlling NEDDylation of ribosomal protein S14. Oncogene 33, 246–254 [DOI] [PubMed] [Google Scholar]