Abstract
Marine Rhodobacteraceae (Alphaproteobacteria) are key players of biogeochemical cycling, comprise up to 30% of bacterial communities in pelagic environments and are often mutualists of eukaryotes. As ‘Roseobacter clade', these ‘roseobacters' are assumed to be monophyletic, but non-marine Rhodobacteraceae have not yet been included in phylogenomic analyses. Therefore, we analysed 106 genome sequences, particularly emphasizing gene sampling and its effect on phylogenetic stability, and investigated relationships between marine versus non-marine habitat, evolutionary origin and genomic adaptations. Our analyses, providing no unequivocal evidence for the monophyly of roseobacters, indicate several shifts between marine and non-marine habitats that occurred independently and were accompanied by characteristic changes in genomic content of orthologs, enzymes and metabolic pathways. Non-marine Rhodobacteraceae gained high-affinity transporters to cope with much lower sulphate concentrations and lost genes related to the reduced sodium chloride and organohalogen concentrations in their habitats. Marine Rhodobacteraceae gained genes required for fucoidan desulphonation and synthesis of the plant hormone indole 3-acetic acid and the compatible solutes ectoin and carnitin. However, neither plasmid composition, even though typical for the family, nor the degree of oligotrophy shows a systematic difference between marine and non-marine Rhodobacteraceae. We suggest the operational term ‘Roseobacter group' for the marine Rhodobacteraceae strains.
Introduction
Rhodobacteraceae (Garrity et al., 2005), one of the major subdivisions of Alphaproteobacteria, include >100 genera and >300 species with very diverse physiologies (Pujalte et al., 2014). Giovannoni and Rappé (2000) assigned most Rhodobacteraceae to the Roseobacter group (‘roseobacters'), which now includes >70 validly named genera, >170 validly named species (Pujalte et al., 2014) and numerous additional isolates and 16S rRNA phylotypes (http://www.arb-silva.de). Most roseobacters originate from marine habitats but some from (hyper-)saline lakes or soil (Pujalte et al., 2014). Other Rhodobacteraceae include only few strains of marine origin (Pujalte et al., 2014). Roseobacters show a very versatile physiology to dwell in greatly varying marine habitats (Buchan et al., 2005; Wagner-Döbler and Biebl, 2006; Brinkhoff et al., 2008; Newton et al., 2010; Sass et al., 2010; Laass et al., 2014; Luo and Moran, 2014; Collins et al., 2015) and account for large proportions of bacterioplankton communities (Selje et al., 2004; West et al., 2008; Alonso-Gutiérrez et al., 2009; Giebel et al., 2011; Buchan et al., 2014; Gifford et al., 2014; Wemheuer et al., 2015).
Genome plasticity might explain adaptability and diversity of roseobacters (Luo and Moran, 2014). Pelagic roseobacters show streamlined genomes (Voget et al., 2015) and possibly other adaptations to an oligotrophic lifestyle. Extrachromosomal replicons (ECRs) comprise chromids and genuine plasmids (Harrison et al., 2010; Petersen et al., 2013). Four replication systems (RepA, RepB, RepABC, DnaA-like) with about 20 phylogenetically distinguishable compatibility groups were identified in roseobacters (Petersen et al., 2013), but a comprehensive analysis of Rhodobacteraceae genome architecture is lacking. Whereas extensive studies of physiological, genetic and genomic features of roseobacters were performed, only scarce and unsystematic information is available on how these traits are distributed among marine and non-marine Rhodobacteraceae.
The Roseobacter group is presumed to be monophyletic and thus frequently called ‘Roseobacter clade' (Buchan et al., 2005; Newton et al., 2010; Luo et al., 2013; Pujalte et al., 2014). Strains within this group share >89% identity of the 16S rRNA gene (Brinkhoff et al., 2008; Luo and Moran, 2014) but a reliable delineation of this group from other Rhodobacteraceae cannot be carried out with this gene (Breider et al., 2014), as branch support and other rationales for suggested 16S rRNA gene-derived Rhodobacteraceae lineages (Pujalte et al., 2014) are lacking.
Even though more comprehensive phylogenetic analyses of the Roseobacter group have been conducted (Newton et al., 2010; Tang et al., 2010; Luo et al., 2012, 2013; Luo and Moran, 2014; Voget et al., 2015), an analysis of the phylogenetic affiliation of this group as a component of the entire Rhodobacteraceae has not yet been carried out. In the majority of phylogenomic analyses of roseobacters (Luo et al., 2012, 2014; Luo and Moran, 2014; Voget et al., 2015), non-roseobacter Rhodobacteraceae were missing, and a single set of selected genes was concatenated and analysed with a single inference method such as Maximum Likelihood (ML) assuming a single amino-acid substitution model for all genes. However, selections of genes, strains and other factors influence the resulting phylogenies (Jeffroy et al., 2006; Philippe et al., 2011; Salichos and Rokas, 2013; Breider et al., 2014). Even though evolutionary aspects of gene gains and losses of the Roseobacter group were analysed previously (Luo et al., 2013; Luo and Moran, 2014), these analyses did not consider the content of specific genomic features as adaptations to the environmental settings.
We carried out phylogenomic analyses of 106 sequenced Rhodobacteraceae genomes using distinct inference algorithms and distinct gene selections ranging from the analysis of the core genes to the ‘full' supermatrix, to address the following questions: (i) How is the Roseobacter group related to other Rhodobacteraceae? (ii) Is this relationship robust against variations in gene selection and phylogenetic inference? (iii) Do stable subgroups exist within this family that are supported by various phylogenomic approaches? and (iv) Do genomic characters correlate with marine or non-marine habitats?
Materials and methods
Genome-scale phylogenetic analysis
All applied methods are detailed in Supplementary File S1. Among the 106 genome-sequenced strains investigated, 13 strains of the Labrenzia/Stappia group taxonomically assigned to Rhodobacteraceae but rather placed within Rhizobiales in 16S rRNA gene analyses (Pujalte et al., 2014) were used as outgroup. An extended data set including 132 genomes was phylogenetically analysed using Genome BLAST Distance Phylogeny (Auch et al., 2006; Meier-Kolthoff et al., 2014). Digital DNA:DNA hybridization was used to check all species affiliations (Auch et al., 2010; Meier-Kolthoff et al., 2013a). Pairwise 16S rRNA gene similarities (Meier-Kolthoff et al., 2013b) were determined after extraction with RNAmmer version 1.2 (Lagesen and Hallin, 2007). The proteome sequences were phylogenetically investigated using the DSMZ phylogenomics pipeline (Anderson et al., 2011; Breider et al., 2014; Frank et al., 2014; Stackebrandt et al., 2014; Verbarg et al., 2014). Alignments were concatenated to three main supermatrices: (i) ‘core genes', alignments containing sequences from all proteomes; (ii) ‘full', alignments containing sequences from at least four proteomes; and (iii) ‘MARE', the full matrix filtered with that software (Meusemann et al., 2010). The core genes were further reduced to their 50, 100, 150 and 200 most conserved genes (up to 250 without outgroup). Long-branch extraction (Siddall and Whiting, 1999) to assess long-branch attraction artefacts (Bergsten, 2005) was conducted by removing the outgroup strains, generating the supermatrices anew and rooting the resulting trees with LSD version 0.2 (To et al., 2015).
ML and maximum parsimony (MP) phylogenetic trees were inferred as described (Andersson et al., 2011; Breider et al., 2014; Frank et al., 2014; Stackebrandt et al., 2014; Verbarg et al., 2014) but MP tree search was conducted with TNT version 1.1 (Goloboff et al., 2008). Additionally, best substitution models for each gene and ML phylogenies were calculated with ExaML version 3.0.7 (Stamatakis and Aberer, 2013). Ordinary and partition bootstrapping (Siddall, 2010) was conducted with 100 replicates except for full/ML for reasons of running time. To assess conflict between the genomic data and roseobacter monophyly, site-wise ML and MP scores were calculated from unconstrained and accordingly constrained best trees, optionally summed up per gene, and compared with Wilcoxon and T-tests (R Core Team, 2015) and, for ML, with the approximately unbiased test (Shimodaira and Hasegawa, 2001). Potential conflict with the 16S rRNA gene was measured using constraints derived from the supermatrix trees. Major sublineages were inferred from the phylogenomic trees non-arbitrarily as the maximally inclusive, maximally and consistently supported subtrees.
Analysis of character evolution
Phylogenetic correlations between pairs of binary characters (Pagel, 1994) were detected with BayesTraits version 2.0 (Pagel et al., 2004) in conjunction with the rooted ML phylogenies. Ratios of the estimated rates of change were calculated to verify the tendency toward marine or non-marine habitats. Three distinct genome samplings were used to detect an influence of only partially sequenced genomes; only results stable with respect to topology and genome sampling were considered further. Evolution of selected genomic characters was visualized using Mesquite v2.75 (Maddison and Maddison, 2011). Habitat assignments (Supplementary File S2) found in the literature only allowed for distinguishing marine and non-marine habitats but this distinction was fully supported by isolation location, physiology and environmental sequencing wherever available (Supplementary File S3). Habitats with a salt concentration comparable to the sea were considered marine (Hiraishi and Ueda, 1994; Brinkhoff and Muyzer, 1997).
The EnzymeDetector (Quester and Schomburg, 2011) was used for initial enzyme annotations of the genomes, improved by strain-specific information from BRENDA and AMENDA (Schomburg et al., 2013), BrEPS (Bannert et al., 2010) and BLAST (Altschul et al., 1990) search against UniProt (UniProt Consortium, 2013). To validate the completeness of each proteome, its proportion of enzymes was calculated (see Supplementary File S2). Enzymes were mapped on MetaCyc pathways (Caspi et al., 2012) as previously described (Chang et al., 2015). A pathway with ⩾75% of its enzymes present was initially assumed to be present too; for pathways discussed in detail, this was manually refined considering the enzymes essential for pathway functionality. The genomic clusters of orthologous groups (COGs) were taken from Integrated Microbial Genomes (Mavromatis et al., 2009) Prodigal annotations (Hyatt et al., 2010). Plasmid replication systems and compatibility groups were determined as described (Petersen et al., 2009, 2011), as were flagellar gene clusters and flagellar types (Frank et al., 2015a). For details, as well as for tests for phylogenetic inertia (Diniz-Filho et al., 1998) and quantification of oligotrophy (Lauro et al., 2009), see Supplementary File S1.
Results and discussion
Genome-scale phylogenetic analysis
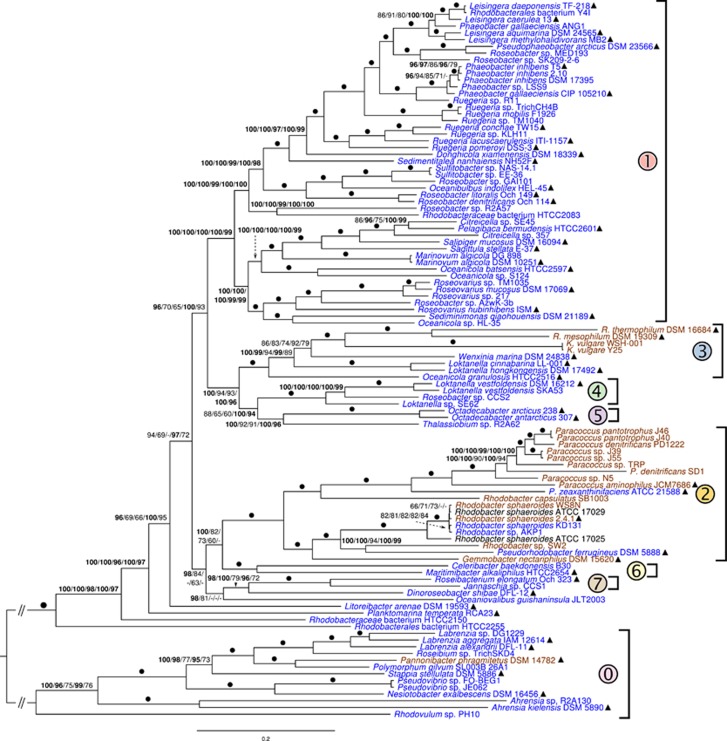
The core-genes analyses under LG (Le and Gascuel, 2008) as single ML model yielded seven maximally and consistently supported, maximally inclusive ingroup clades and four deeply branching strains (Figure 1, Supplementary File S3). Support was maximal for most branches in the distinct analyses (Supplementary File S3) but they differed regarding the backbone of the tree. The core-gene topology was almost identical to that of earlier phylogenomic analyses (Newton et al., 2010; Luo and Moran, 2014; Voget et al., 2015), provided the strains additionally included here were removed. Here the Roseobacter group appeared not monophyletic but paraphyletic, as clade 2 harbouring the non-roseobacter genera was nested within this group.
Figure 1.
ML tree inferred from the core-gene matrix (208 genes, 80 578 characters) under a single overall model of amino-acid evolution and rooted with the included outgroup strains. The branches are scaled in terms of the expected number of substitutions per site; double slashes indicate branches shortened by 90%. Numbers above branches (from left to right) are bootstrapping support values if >60% from (i) ordinary bootstrap under ML with a single overall model of amino-acid evolution; (ii) ordinary bootstrap under ML with one model per gene; (iii) partition bootstrap under ML; (iv) ordinary bootstrap under MP; and (v) partition bootstrap under MP. Values >95% are shown in bold; dots indicate branches with maximum support under all settings. The inferred major clades are indicated with numbers and colours; clade 2 comprises all ingroup strains not assigned to the Roseobacter group. Triangles indicate type strains. The colours of the tip labels indicate the habitat: blue, marine; brown, non-marine; uncoloured, unknown.
All core-gene inference and bootstrapping approaches showed 95% support for strains HTCC2255 and HTCC2150 branching first. Support for Planktomarina temperata RCA23T and Litoreibacter arenae DSM 18583T also branching before clade 2 was similarly high but collapsed under ML when partition bootstrapping or individual substitution models were applied (Figure 1). Average support was also lower under these settings; for details, see Supplementary File S3, which also provides revised genus and species affiliations. The extracted 16S rRNA gene sequences showed >89% similarity between all Rhodobacteraceae, not only between roseobacters (Supplementary File S3). The overall best 16S rRNA gene trees were not significantly better than the best constrained trees, confirming that the gene does not significantly support the Roseobacter clade.
Phylogenetic inference without outgroup and LSD rooting yielded an ML topology (Supplementary File S3) identical to the one obtained by pruning the outgroup from the tree depicted in Figure 1, irrespective of whether or not the gene set was renewed after strain removal. The core-gene matrices reduced to 200 genes again showed the same topology, albeit a slightly reduced support, whereas with ⩽150 genes the backbone was not supported any more and distinct between ML and MP (Supplementary File S3). The topologies different from that in Figure 1 showed a monophyletic yet statistically unsupported Roseobacter clade and included a cluster comprising HTCC2255, HTCC2155, P. temperata RCA23 and L. arenae in conflict with their deep branching (as in Figure 1) in earlier phylogenomic studies (Newton et al., 2010; Luo et al., 2012; Luo and Moran, 2014; Voget et al., 2015).
After removing the outgroup and re-estimating the root with LSD, all alternative trees showed an ingroup topology as in Figure 1. The six trees inferred from larger matrices (Supplementary File S3) that showed the Roseobacter group as monophyletic were also in conflict with earlier studies and, in the LSD test, showed a paraphyletic Roseobacter group as in Figure 1. Confirming Siddall (2010), conflicting branch support, if any, never originated from partition bootstrapping; analogously, conflict with the topology in Figure 1 assessed in paired-site tests was never significant when each gene was treated as a single character. Such alternative tests for topologies might help tackling incongruence in genome-scale phylogenetic analyses (Jeffroy et al., 2006). The topologies from distinct gene selections differed mainly regarding the placement of the outgroup, and distant outgroups are particularly frequently subject to long-branch attraction (Bergsten, 2005). Whereas the outgroup chosen here is more closely related to Rhodobacteraceae than Escherichia coli as used in Newton et al. (2010), we would recommend sampling even more and particularly non-marine Rhodobacteraceae genome sequences in future studies. However, outgroup removal and LSD rooting indicated that the alternative topologies (Supplementary File S3) rather than the one in Figure 1 might suffer from long-branch attraction to the outgroup. The Genome BLAST Distance Phylogeny analysis of the larger data set also showed HTCC2255 branching first within the ingroup with high support, followed by HTCC2150 (Supplementary File S3).
Our extensive phylogenomic analyses hardly support a monophyletic Roseobacter group. Particularly the core-genes analysis, methodologically most similar to earlier studies, shows paraphyletic roseobacters. This finding is in conflict with literature claims but not actually with the underlying analyses (Buchan et al., 2005; Newton et al., 2010; Luo et al., 2012; Luo and Moran, 2014; Pujalte et al., 2014; Voget et al., 2015) because it is just due to increased strain sampling, particularly of Paracoccus and Rhodobacter. The tree shown in Figure 1 is not in conflict with previous analyses, whereas the topologies observed with some other gene sets are.
The Roseobacter group has been considered as the marine Rhodobacteraceae (Giovannoni and Rappé, 2000; Buchan et al., 2005; Brinkhoff et al., 2008; Luo et al., 2012) but also includes non-marine genera such as Rubellimicrobium and Ketogulonicigenium (Buchan et al., 2005; Brinkhoff et al., 2008; Pujalte et al., 2014). Ketogulonicigenium, however, also comprises marine ribotypes (Gifford et al., 2014). Paracoccus and Rhodobacter, which form clade 2 in our analysis (Figure 1), are no roseobacters and mainly non-marine, whereas P. zeaxanthinifaciens ATCC 21588T (Berry et al., 2003) and R. sphaeroides KD131 (Lim et al., 2009) dwell in the sea. P. denitrificans has marine ribotypes (Gifford et al., 2014), whereas strain PD1222 was derived from PD1001 (de Vries et al., 1989), isolated from garden soil (Nokhal and Schlegel, 1983). Obviously, some Paracoccus and Rhodobacter strains became secondarily marine.
As a conclusion, definitions and uses of the terms ‘Roseobacter group' and ‘Roseobacter clade' need to be reconsidered. The Roseobacter clade is neither unambiguously supported by our in-depth analysis nor by previous studies, which either lacked branch support or sufficient strain sampling. We suggest using the term ‘Roseobacter group', not ‘clade', for the marine Rhodobacteraceae. This operational definition is consistent with the current general use of the name of the group and avoids overinterpreting the phylogenetic evidence. Our analysis further shows that a transition from marine to non-marine habitats occurred several times independently within Rhodobacteraceae. An equivalent step occurred only once in the evolution of the SAR11 clade, another prominent lineage of marine but strictly pelagic Alphaproteobacteria (Luo et al., 2015). Rhodobacteraceae genomes often contain many transposable elements (Vollmers et al., 2013), ECRs (Petersen et al., 2013) and gene-transfer agents (Zhao et al., 2009; Luo and Moran, 2014). The lack of these traits in the streamlined genomes of the SAR11 clade might explain just one transition between marine and freshwater habitats (Luo et al., 2015). The more frequent habitat transitions within Rhodobacteraceae call for an analysis of the underlying genomic adaptations.
Genomic traits of marine versus non-marine Rhodobacteraceae
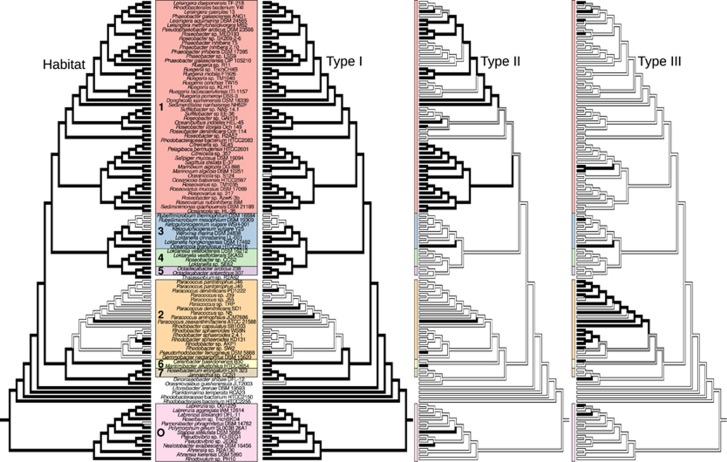
Altogether 1835 enzymes with distinct EC numbers were predicted, 106 in all strains and 419 in >90%. Their proportion in each proteome ranged between 11.58% (Paracoccus sp. J55) and 24.05% (Oceaniovalibus guishaninsula JLT2003; for further details, see Supplementary File S2). The enzymes indicated 322 pathways overall, 18 present in all strains. The Integrated Microbial Genomes annotations yielded 4873 COGs overall, 345 present in all strains. Of the 106 strains, 85 were assigned to a marine or saline habitat, 19 to a non-marine one and for 2 a habitat assignment was not possible (Figure 1). Apparently, the marine state was lost and regained several times in evolution, even though it is phylogenetically conserved (Figure 2, Supplementary File S3). This result supports the claims of a predominantly marine roseobacter group, as the habitat can apparently be well predicted from the phylogenetic position of a strain and thus indirectly supports the reliability of our habitat assignments.
Figure 2.
Ancestral character-state reconstruction under ordered MP for the presence (black) or absence (white) of H, marine or equivalent habitat; I, (S)-2-haloacid dehalogenase (EC 3.8.1.2); II, ectoine synthase (EC 4.2.1.108); and III, 6-phosphofructokinase (EC 2.7.1.11). The tree topology and the number and colour codes for the major clades are as in Figure 1. Grey shading indicates uncertainties in character-state assignment. The major types of phylogenetic distributions represented by the three genomic characters are: I, losses predominantly in non-marine strains; II, gains mainly in marine strains; and III, gains predominantly in non-marine strains.
In contrast to χ2 tests, considering phylogenies in comparative biology has a high chance to avoid false positives and false negatives (Pagel, 1994; Pagel et al., 2004). The BayesTraits software estimates rates of changes in a phylogeny and, for a pair of discrete characters, statistically compares a model with independent rates of change to one with the changes in one character dependent on the states of the other. BayesTraits analyses allow for detecting genomic adaptations that accompany phylogenetically independent switches in habitat preferences. As the inferred tree topologies were not in agreement throughout (Figure 1, Supplementary File S3), only those BayesTraits results were considered further that were significant under all tested topologies and thus independent, for example, of a monophyletic or a paraphyletic Roseobacter group.
Up to 90 pathways were significantly habitat correlated (59 identified by all analyses; Supplementary File S4), 391 (255) enzymes (Supplementary File S4) and 563 (386) COGs (Supplementary File S4). As judged from the estimated rates of changes as well as MP reconstructions of ancestral character states on the major distinct topologies, significant characters exhibited three major types of phylogenetic distributions (Table 1, Figure 2, Supplementary File S3): (i) inheritance from the most recent common ancestor and loss in non-marine strains; (ii) absence in the ancestor, gain in marine strains; and (iii) absence in the ancestor, gain in non-marine strains. In few cases, ambiguity in character-state reconstruction prevented the distinction between types (i) and (ii).
Table 1. Selected genomic characters that were significantly (α=0.01) habitat-correlated in the BayesTraits tests under all conditions (for sulphoacetaldehyde acetyltransferase (EC 2.3.3.15) under most conditions), along with their overall type of evolution (as in Figure 2), sum of evolutionary rates estimated by BayesTraits indicating co-occurrence divided by overall sum of rates and percentage occurrences (on which the test is not based) in marine and non-marine strains.
| Feature | ID | Rate quotient | Marine | Non-marine |
|---|---|---|---|---|
| I. Lost in non-marine habitats | ||||
| Chloride channel protein | COG0038 | 0.834 | 96 | 32 |
| Ca2+/Na+ antiporter | COG0530 | 0.783 | 99 | 42 |
| NhaP-type Na+/H+ and K+/H+ antiporters | COG0025 | 0.829 | 76 | 21 |
| (S)-2-haloacid dehalogenase | EC 3.8.1.2 | 0.751 | 94 | 21 |
| Mercury (Hg) II reductase | EC 1.16.1.1 | 0.887 | 98 | 26 |
| Carbon monoxide dehydrogenase | EC 1.2.99.2 | 0.654 | 93 | 42 |
| Precorrin-8X methylmutase | EC 5.4.1.2 | 0.445 | 96 | 63 |
| Precorrin-4 C11-methyltransferase | EC 2.1.1.133 | 0.687 | 95 | 68 |
| Precorrin-6B methylase 2 | COG2242 | 0.794 | 85 | 37 |
| Predicted cobalamin binding protein | COG5012 | 0.596 | 90 | 42 |
| Cobalamin biosynthesis protein CbiD | COG1903 | 0.774 | 38 | 5 |
| Choline-glycine betaine transporter | COG1292 | 0.733 | 98 | 69 |
| II. Gained in marine habitats | ||||
| Ectoine synthase | EC 4.2.1.108 | 0.979 | 41 | 0 |
| Ectoine biosynthesis pathway | P101-PWY | 0.954 | 38 | 0 |
| Betaine-homocysteine S-methyltransferase | EC 2.1.1.5 | 0.763 | 25 | 0 |
| Glycine betaine degradation | PWY-3661 | 0.883 | 94 | 74 |
| Gamma butyrobetaine dioxygenase | EC 1.14.11.1 | 0.969 | 48 | 0 |
| Trimethylamine-corrinoid protein Co-methyltransferase | EC 2.1.1.250 | 0.493 | 62 | 11 |
| Trimethylamine-N-oxide reductase | EC 1.6.6.9 | 0.891 | 12 | 5 |
| Probable taurine catabolism dioxygenase | COG2175 | 0.859 | 52 | 16 |
| Nitrile hydratase | EC 4.2.1.84 | 0.749 | 62 | 0 |
| Arylsulphatase | EC 3.1.6.1 | 0.752 | 61 | 11 |
| Precorrin-3B synthase | EC 1.14.13.83 | 0.596 | 47 | 37 |
| Unclear whether lost or gained | ||||
| ABC-type tungstate transport system, periplasmic component | COG4662 | 0.567 | 70 | 5 |
| ABC-type tungstate transport system, permease component | COG2998 | 0.676 | 69 | 5 |
| Sulphoacetaldehyde acetyltransferase | EC 2.3.3.15 | 0.434 | 69 | 53 |
| Trimethylamine-N-oxide reductase (cytochrome c) | EC 1.7.2.3 | 0.763 | 34 | 26 |
| III. Gained in non-marine habitats | ||||
| ABC-type sulphate transport system, permease component | COG4208 | 0.128 | 12 | 95 |
| ABC-type sulphate transport system, periplasmic component | COG1613 | 0.129 | 10 | 95 |
| ABC-type sulphate/molybdate transport systems, ATPase component | COG1118 | 0.13 | 10 | 95 |
| Formaldehyde oxidation pathway II (glutathione-dependent) | PWY-1801 | 0.412 | 8 | 58 |
| S-(hydroxymethyl)glutathione synthase | EC 4.4.1.22 | 0.372 | 8 | 63 |
| Taurine dehydrogenase | EC 1.4.99.2 | 0.154 | 4 | 37 |
| 6-Phosphofructokinase | EC 2.7.1.11 | 0.398 | 16 | 58 |
P-values differ depending on the underlying tree and strain sampling and are provided in the according sments. The characters are presence and absence of enzymes (as indicated by their EC number), COGs (as indicated by their COG number) and pathways (as indicated by their Metacyc IDs). Ancestral character-state reconstructions of the exemplary occurrences of (S)-2-haloacid dehalogenase (EC 3.8.1.2) for type I, ectoine synthase (EC 4.2.1.108) for type II and 6-phosphofructokinase (EC 2.7.1.11) for type III are depicted in Figure 2.
Genomic traits predominantly lost in non-marine strains
The transition into non-marine habitats is reflected by adaptations to their different ecology and biogeochemistry such as losses (and lack of gains) of genetic traits directly related to the strongly reduced NaCl concentration from ~1.015 M in the sea to <10 mM in soil and freshwater habitats (Table 1,Figure 2). Marine bacteria require Na+ for growth, and some use a Na+ circuit for various functions (Kogure, 1998). The significant trend to evolutionary losses (and no gains) of Na+ antiporters and a Cl− channel protein in non-marine Rhodobacteraceae and thus their probably reduced capability to transport Na+ and Cl− across the cell membrane is consistent with the strongly reduced NaCl concentration in non-marine habitats and the fact that most Paracoccus and Rhodobacter strains do not require Na+ for growth (Pujalte et al., 2014).
The gene encoding (S)-2-haloacid dehalogenase showed a significant trend to be missing in non-marine genomes (Table 1). The great majority of organohalogens is produced by macroalgae, sponges, corals, tunicates, polychaetes and other marine organisms, even though terrestrial and freshwater cyanobacteria, fungi and bacteria can also produce them (Fielman et al., 2001; Gribble, 2003, 2012). Particularly the halogenated metabolites produced by macroalgae are toxic for various animals and bacteria, such as Vibrio sp. and Acinetobacter sp. (Paul et al., 2006; Cabrita et al., 2010; Gribble, 2012). As many marine Rhodobacteraceae not only live in close association with microalgae and macroalgae, sponges and corals (Buchan et al., 2005; Brinkhoff et al., 2008; Raina et al., 2009; Lachnit et al., 2011; Webster et al., 2011) but may also be exposed to particulate detrital and dissolved organohalogens, they presumably use dehalogenases for detoxifying and utilizing these compounds as substrates (Novak et al., 2013). Conversely, the lack of (S)-2-haloacid dehalogenase could reflect an adaptation to a reduced exposure to such toxic compounds.
The significantly correlated mercury-II reductase gene is missing in many non-marine but present in almost all marine strains (Table 1). Bacterial detoxification of mercury (Hg) by reducing oxidized Hg(I) to volatile Hg(0) is widely distributed in habitats with Hg(I) or Hg(II) (Barkay et al., 2010). Under a reduced redox potential, mercury is present as Hg(0), and bacteria dwelling under these conditions often lack Hg reductase. The occurrence of non-marine Rhodobacteraceae such as Paracoccus in soil, compost or sewage under reduced or zero-oxygen conditions and of Rhodobacter in anaerobic freshwater conditions (Pujalte et al., 2014) thus may explain their tendency to lose the ability to detoxify Hg(I/II).
The lack of the carbon monoxide dehydrogenase (CODH) gene was also significantly correlated with non-marine habitats (Table 1). It is often part of the bacterial enzyme complex to oxidize CO to CO2 (King and Weber, 2007). The large subunit of the CODH complex (coxL) gene occurs in two forms. Nearly all marine Rhodobacteraceae harbour form II but only those which perform CO oxidation harbour form I (Cunliffe, 2011). Oxidation of this secondary green-house gas is an important process in pelagic marine ecosystems (Tolli et al., 2006; Moran and Miller, 2007; Dong et al., 2014), but the exact function of coxL form II is still unclear. Comparison with published sequences (King, 2003) showed that form I and form II were distinct clusters of orthologues in our data set; the BayesTraits test was always significant for form I, the one for form II only for half of the assessed combinations of trees and strain samplings (Supplementary File S4). Therefore, CO oxidation, an adaptation of a complementary mode of energy acquisition under nutrient-depleted marine conditions, appears unsuitable in non-marine lineages.
Absences of five genes encoding enzymes operating on cobalamin and precorrin and thus biosynthesis and binding of vitamin B12 were correlated with non-marine habitats (Table 1). The vitamin B12 biosynthetic pathway has previously been demonstrated in roseobacters (Newton et al., 2010; Luo and Moran, 2014). In contrast to other major groups of marine bacteria such as the SAR11 clade and Bacteroidetes (Sañudo-Wilhelmy et al., 2014), marine Rhodobacteraceae are major vitamin suppliers for B12-auxotrophic prokaryotes and eukaryotic primary producers, such as chlorophytes, diatoms, dinoflagellates, coccolithophores and brown algae (Croft et al., 2005; Wagner-Döbler et al., 2010; Helliwell et al., 2011; Bertand and Allen, 2012; Sañudo-Wilhelmy et al., 2014). The loss of the ability to produce vitamin B12 in non-marine Rhodobacteraceae could be due to more dominant bacteria, adapted to lakes and soil for an evolutionary longer period and major other producers of this vitamin. In fact, it has been shown that the ability to produce vitamin B12 can be lost rapidly in a freshwater alga when exposed to a continuous supply of B12 (Helliwell et al., 2015). This scenario appears to be applicable to the non-marine Rhodobacteraceae. Soil Rhizobiales are known to produce vitamin B12 (Kazamia et al., 2012; Sañudo-Wilhelmy et al., 2014), whereas Rhodobacter often dwells in eutrophic lakes or biofilms where cyanobacteria as vitamin-B12 producers are abundant. Macrophytes not requiring vitamin B12 (Helliwell et al., 2011) often dominate as primary producers in soil and freshwater ecosystems, which may also result in its reduced demand.
Gains and losses of both permease and periplasmic component of the ATP-binding cassette (ABC)-type tungstate transport system were significantly correlated to the habitat, with ambiguous MP reconstructions indicating either losses in non-marine Rhodobacteraceae or gains by marine ones (Table 1). Oxyanions of molybdenum (MoO42−) and tungsten (WO42−) are the main sources of these essential trace metals in bacterial cells (Johnson et al., 1996; Hille, 2002). The ubiquitous Mo-containing enzymes have important roles in the global cycles of nitrogen, carbon and sulphur (Kisker et al., 1997). High-affinity molybdate/tungstate ABC-type transporters (Schwarz et al., 2007) allow bacteria to scavenge these oxyanions in the presence of sulphate, whose concentration in seawater is ca. 105 times higher than that of molybdate and ca. 108 times higher than that of tungstate (Bruland, 1983). In terrestrial ecosystems, the concentration of tungstate is much higher (Senesi et al., 1988), and owing to its similarity to molybdate, both may be transported by the same carrier (Bevers et al., 2006; Taveirne et al., 2009; Gisin et al., 2010). Therefore, loss of this highly specific transport system appears to be an adaptation to non-marine habitats.
Genomic traits predominantly gained in marine strains
The ectoine biosynthesis pathway and ectoine synthase distributions were significantly habitat-correlated; in contrast to the characters discussed above, MP reconstructions indicated the absence of ectoine synthesis in the common ancestor and gains predominantly in marine genomes (Table 1). Ectoine is a compatible solute that helps surviving extreme osmotic stress by acting as an osmolyte and is synthesized by a wide range of bacteria (Ventosa et al., 1998; Reshetnikov et al., 2004; Trotsenko et al., 2007). Independent gains by several marine lineages (Figure 2) indicate that, whereas osmolytes are mandatory in the ocean, several alternatives can be used, as confirmed by the results for the following characters.
Carnitine, glycine betaine and proline are other osmolytes widespread in prokaryotes (Welsh, 2000; Hoffmann and Bremer, 2011). The enzyme mediating the last step in the biosynthesis of carnitine, gamma butyrobetaine dioxygenase, shows the same type of evolutionary changes as the ectoine-related characters (Table 1). Carnitine can also be taken up by a betaine/carnitine/choline transporter (Lidbury et al., 2014), which was present in the ancestor and predominantly lost by non-marine strains (Table 1). Thus marine Rhodobacteraceae originally relied on carnitine uptake and tended to additionally gain the ability to synthesize it.
Carnitine can be decomposed by various ways to glycine betaine (Kleber, 1997; Welsh, 2000; Wargo and Hogan, 2009). Catabolism of glycine betaine and the enzyme mediating its first step, betaine-homocysteine S-methyltransferase, show gains by marine strains (Table 1). In contrast, almost all genomes encoded ABC transporters for proline/glycine betaine such as COG4176, which do not show a significant relationship to the habitat (Supplementary File S4). As biosynthetic traits for the osmolytes ectoine and carnitine were lost and gained several times independently by marine strains, they might be exposed to distinct magnitudes of osmotic stress, as typical for coastal areas.
Genes encoding enzymes involved in the reduction of trimethylamine-N-oxide (TMAO) and demethylation of trimethylamine (TMA) were significantly correlated with the habitat, with gains mainly occurring in the ocean (Table 1). TMAO is well known as terminal electron acceptor in anaerobic microbial respiration (Arata et al., 1992; Gon et al., 2001) and as osmolyte in a variety of marine biota (Seibel and Walsh, 2002; Gibb and Hatton, 2004; Treberg et al., 2006). A TMAO-specific ABC transporter and genes encoding TMAO decomposition via TMA, dimethylamine and monomethylamine are widespread in marine metagenomic libraries and bacteria, including roseobacters and the SAR11 clade (Lidbury et al., 2014, 2015). Most marine Rhodobacteraceae can use TMA and TMAO as sole N source and probably oxidize the methyl groups to CO2 as a complementary pathway to conserve energy (Lidbury et al., 2015), whereas Roseovarius can even use TMA and TMAO as sole C source (Chen, 2012). Such traits are unknown from aerobic freshwater environments (Treberg et al., 2006).
The probable dioxygenase as a key enzyme in taurine catabolism was gained significantly often during transitions to marine habitats (Table 1). Taurine is an important organosulphur compound and compatible solute in marine and freshwater invertebrates and fish (Treberg et al., 2006; Lidbury et al., 2015). Freshwater and soil bacteria use taurine predominantly as S but not as C source, whereas marine bacteria, including Rhodobacteraceae, can use taurine both as S and as C and energy source (King and Quinn, 1997; Kertesz, 2000). In fact, taurine ABC transporters are major transport systems in marine bacterial communities comprising large proportions of Alphaproteobacteria, including Rhodobacteraceae (Gifford et al., 2012; Williams et al., 2012). Taurine as S source requires an oxygenation with taurine dioxygenase as key enzyme and cleavage to aminoacetaldehyde and sulphite (Kertesz, 2000). This pathway might be encoded as COG2175 (probable taurine catabolism dioxygenase). However, taurine dioxygenase (EC 1.14.11.17) was not significantly habitat-correlated (Supplementary File S4). Taurine as C and energy source can be metabolized via two pathways (Kertesz, 2000; Denger et al., 2009). One involves taurine-pyruvate aminotransferase (EC 2.6.1.77) as key enzyme to generate alanine and sulphoacetaldehyde. This pathway was encoded in most genomes, and its gains and losses showed no significant dependency on the habitat (Supplementary File S4). In an alternative pathway, taurine is directly deaminated to sulphoacetaldehyde by a dehydrogenase, which was significantly habitat-correlated but more frequent in non-marine strains (Table 1). In both pathways, sulphoacetaldehyde is cleaved to acetylphosphate and sulphite by a sulphoacetaldehyde acetyltransferase, which was significantly habitat-correlated in the majority of the analyses and more frequently occurred in marine Rhodobacteraceae (Table 1). Thus taurine seems more important as C and energy source than as a general S source, with significant differences in the metabolic pathways between marine and non-marine strains.
An arylsulphatase was gained significantly more frequently in the sea (Table 1). Arylsulphatases cleave sulphate from phenolic compounds and, in a fungus, from breakdown products of the polysaccharide fucoidan, that is, sulphated fucose as monomers or oligomers (Shvetsova et al., 2015). Fucoidan is a major component of marine macroalgae, including Fucus, Laminaria and Macrocystis (Deniaud-Bouët et al., 2014), but not known from freshwater or soil. Marine Rhodobacteraceae are major colonizers of Fucus and other macroalgae and can grow on fucoidan (Lachnit et al., 2011; Bengtsson et al., 2011). If their arylsulphatase could also target fucoidan, marine Rhodobacteraceae would apparently benefit directly on macroalgae or detrital particles and colloids by utilizing breakdown products of fucoidan after cleaving the sulphate group.
Gains of nitrile hydratase were also correlated with marine habitats (Table 1). This enzyme is involved not only in acrylonitrile degradation (Kato et al., 2000) but also in the indole-3-acetonitrile pathway for the biosynthesis of indole-3-acetic acid (IAA), a plant hormone. Three pathways can lead to IAA, which is synthesized by Rhizobiaceae (Ghosh et al., 2011) and by a marine Sulfitobacter strain, enhancing growth of the abundant diatom Pseudonitzschia multiseries (Amin et al., 2015). In marine metatranscriptomic data sets from the Pacific, Roseobacter group-specific transcripts of all three IAA biosynthetis pathways were detected, mainly from the indole-3-acetonitrile pathway (Amin et al., 2015). This is consistent with IAA biosynthesis being significantly gained by marine Rhodobacteraceae, and its potential role in their symbiosis with phytoplankton and possibly also macroalgae, corresponding to the role of Rhizobiaceae for higher plants.
Genomic traits predominantly gained in non-marine strains
Similar to the characters discussed above, the following ones were significantly phylogenetically correlated with the habitat but tended to be gained by non-marine Rhodobacteraceae (Table 1), as revealed by the estimated rates of changes and the MP reconstructions (Figure 2). These genomic traits must be interpreted particularly carefully because most Rhodobacteraceae are marine and thus the non-marine ones are less representative for the multitude of non-marine bacteria than the ones dwelling in the ocean for the plethora of marine strains.
Distinct genes encoding ABC sulphate transporters showed a significant and positive relationship with non-marine habitats. This markedly differs from a sulphate permease of the SulP superfamily (COG0659) encoded in almost all genomes, which showed no habitat correlation (Supplementary File S4) and is also widespread among other bacteria (Aguilar-Barajas et al., 2011). Sulphate concentrations in marine waters are around 28 mM, whereas in common freshwater systems they are in the submillimolar range (Wetzel, 2001). A marine Rhodobacter strain can take up sulphate at concentrations ranging from 50 μM to 2 mM, obviously applying two transport systems with different affinities (Imhoff et al., 1983; Warthmann and Cypionka, 1996). This is in line with the finding that non-marine rather than marine Rhodobacteraceae harbour high-affinity sulphate uptake systems in addition to sulphate permease to cope with the low sulphate concentrations in freshwaters. For many marine Rhodobacteraceae, sulphate is probably not the major S source because uptake of dimethylsulphoniopropionate, occurring at concentrations in the nanomolar range, meets most of their sulphur demand (Simo et al., 2002; Malmstrom et al., 2004; Moran et al., 2012). Marine dimethylsulphoniopropionate is the major global source of organic sulphur, including the climatically relevant dimethylsulphide, whereas this and other organic S compounds are also produced in freshwater and terrestrial systems but to much lower extent (Schäfer et al., 2010). Accordingly, we did not find a significant dependency of the occurrence of genes encoding dimethylsulphoniopropionate decomposition and the habitat (Supplementary File S4).
Gains of S-(hydroxymethyl)glutathione synthase were also significantly related to non-marine habitats (Table 1). This enzyme catalyses the first of the three reactions of the equally significant glutathione-dependent formaldehyde oxidation II pathway for detoxifying formaldehyde (Gonzalez et al., 2006). For methylotrophic bacteria such as Paracoccus or Rhodobacter, formaldehyde is also a central intermediate for oxidizing methanol or methylamine (Ras et al., 1995; Harms et al., 1996; Barber et al., 1996; Chistoserdova, 2011). Although the enzymes catalysing the second and third reaction were encoded in almost all genomes and showed no dependency on the habitat (Supplementary File S4), S-(hydroxymethyl)glutathione synthase has mainly been gained in Paracoccus and Rhodobacter. The first reaction of the formaldehyde oxidation pathway II is spontaneous in vivo but can be accelerated by the enzyme (Goenrich et al., 2002). It might be present only in strains that produce and consume large amounts of intracellular formaldehyde, whereas the spontaneous reaction could be sufficient for detoxifying exogenous formaldehyde (Goenrich et al., 2002). Roseobacters might use methanol as a supplementary energy rather than C source (Chen, 2012), hence the significant correlation with non-marine habitats might be due to the specific physiologies of Paracoccus and Rhodobacter.
Non-marine habitats were significantly related to gains of the gene encoding 6-phosphofructokinase (Table 1), the key enzyme of glucose metabolism via the Embden–Meyerhof–Parnas pathway (EMPP) (Flamholz et al., 2013). Marine Rhodobacteraceae, Gammaproteobacteria and Flavobacteria lack the EMPP and catabolize glucose exclusively via the Entner–Doudoroff pathway (EDP; Klingner et al., 2015), thus yielding slightly less ATP but, in contrast to the EMP, also NADPH (Flamholz et al., 2013). Klingner et al. (2015) showed that, even when the EMPP was additionally encoded in the genome, glucose was completely catabolized via the EDP. Rhodobacter sphaeroides behaves similarly (Fuhrer et al., 2005). Use of the EDP was interpreted as protection against oxidative stress, which is of major importance in near-surface marine habitats (Klingner et al., 2015). The EDP is evolutionary older (Romano and Conway, 1996), in accordance with its wide distribution in Proteobacteria (Flamholz et al., 2013) and its presence in ancestral Rhodobacteraceae as shown here. Apparently 6-phosphofructokinase was acquired several times independently by Rhodobacteraceae, most prominently in lineages with non-marine strains (Figure 2). Thus our analysis fully supports previous physiological studies and highlights the evolutionary importance of the EDP for the breakdown of glucose in the sea.
Genomic traits unrelated to habitats
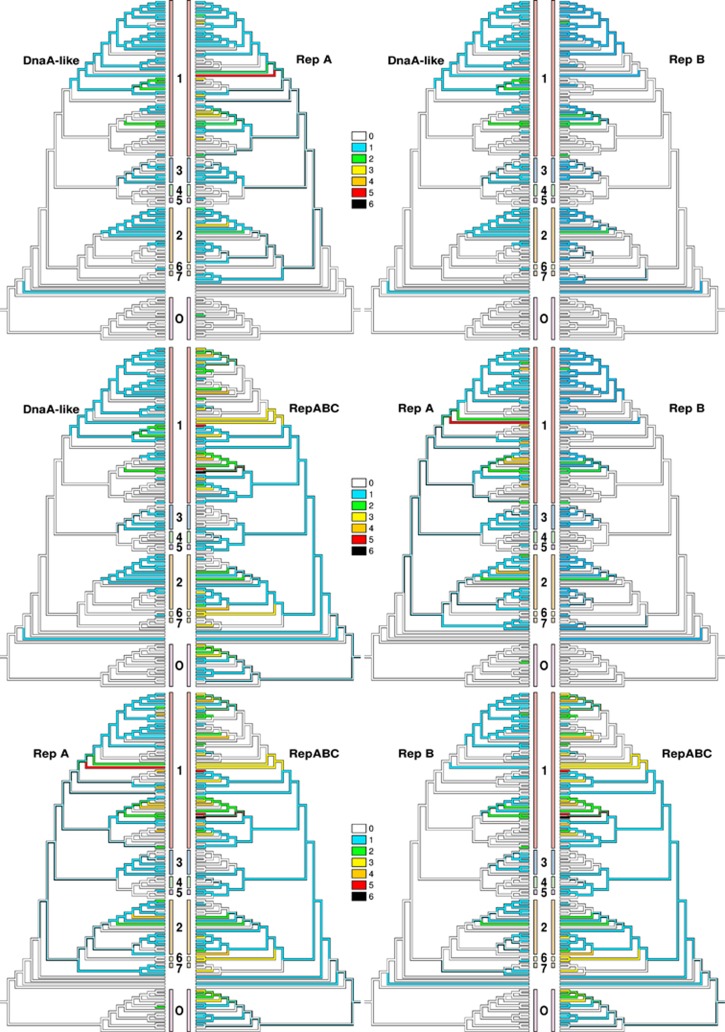
The comparison of habitat-related characters with genomic traits such as ECRs that show other distributional types provides additional valuable insights. The current study represents the most comprehensive comparison of ECRs in Rhodobacteraceae and revealed up to 12 replicons (chromosome, chromids, plasmids; Harrison et al., 2010) in a single bacterium (Supplementary File S2), in agreement with previous results (Pradella et al., 2004, 2010; Frank et al., 2015a), as well as 53 DnaA-like, 79 RepA, 52 RepB and 140 RepABC replication modules. Their phylogeny-based classification indicated distinct compatibility groups in the outgroup (Supplementary File S2), whereas BayesTraits analysis showed no correlation between ECRs and habitat (Supplementary File S5). The replication systems were not distinct between marine and non-marine Rhodobacteraceae (Petersen et al., 2009), in line with our phylogenetic findings (Figure 1).
Occurrences of ECR types DnaA-like, RepA and RepB but not RepABC significantly depended on each other (Figure 3, Supplementary File S5). Only RepB and DnaA-like showed a significant phylogenetic inertia under all conditions (Supplementary File S3). An explanation might be that RepABC-type ECRs do not frequently occur on chromids (Harrison et al., 2010) but on genuine plasmids (Dogs et al., 2013; Frank et al., 2014), which often contain type-IV secretion systems enabling horizontal transfer via conjugation (Petersen et al., 2013). Significant positive correlations between ECRs and type-IV secretion systems were indeed found (Supplementary File S2). The significant positive correlation between the dTDP-L-rhamnose biosynthesis pathway and RepA (Supplementary File S6) is in agreement with previous studies (Frank et al., 2015b; Michael et al., 2016), much like the one between the archetypal type-1 flagellum and TDP-L-rhamnose biosynthesis (Frank et al., 2015a, 2015b). L-rhamnose is essential for proteobacterial surface polysaccharides (Giraud and Naismith, 2000) and the typical swim-or-stick lifestyle of surface-associated roseobacters (Belas et al., 2009; Michael et al., 2016).
Figure 3.
Ancestral character-state reconstruction under ordered MP for the number of ECR replicases of the distinct types DnaA, RepA, RepB and RepABC and according pairwise phylogenetic cross-comparisons of their abundances in each genome. For the labels of the tips, see Figure 2, where the same tree topology is depicted in exactly the same layout. The number and colour codes between the trees refer to the clades as indicated in Figure 1 (O=outgroup). The colours depicted on the topologies indicate the number of replicases of each type as follows: white, 0; blue, 1; green, 2; yellow, 3; orange, 4; red, 5; and black, 6. Presences and absences alone are correlated between DnaA, RepA and RepB but not between RepABC and the others.
The oligotrophy index neither exhibited a habitat-specific distribution nor phylogenetic inertia (Supplementary File S2), in line with the distinct positions of oligotrophic, genome-streamlined strains such as Planktomarina temperata RCA23T, HTCC2255, HTCC2150, Oceanicola batsensis HTCC2597T, HTCC2083 and Sulfitobacter sp. NAS-14.1 (Figure 1). Evolutionary plasticity regarding the oligotrophic lifestyle might have helped Rhodobacteraceae to dwell in diverse habitats. Genome size and the number of replication systems were not correlated with oligotrophy, but its variance decreased for large genomes (Supplementary File S3). This result does not support the finding of Lauro et al. (2009), possibly because not only oligotrophy but also eutrophic and constant environments can cause genome streamlining (van de Guchte et al., 2006; Giovannoni et al., 2014).
Conclusion
Our comprehensive analysis provides new insights into phylogenomics and the evolutionary adaptation of Rhodobacteraceae to marine and non-marine habitats. It builds on previous phylogenomic analyses of the Roseobacter group, extends them to the entire Rhodobacteraceae and elucidates that evolutionary adaptations to marine and non-marine habitats of the most recent common ancestor were accompanied by distinct gains and losses of genes (summarized in Figure 4), obviously improving fitness of these lineages in these habitats. Of course, not all genomic features of relevance could be addressed here. For instance, quite a few roseobacters are well known to carry out aerobic anoxygenic photosynthesis (Wagner-Döbler and Biebl, 2006; Voget et al., 2015). However, non-marine Rhodobacteraceae do not carry out this form of energy acquisition (Koblizek, 2015), and Rhodobacter is even an archetypal anaerobic anoxygenic photosynthetic bacterium (Koblizek, 2015). Therefore, this trait was beyond the scope of our study. Nevertheless, our comprehensive analysis forms a profound and stimulating basis for further and refined research on specific aspects of these adaptations.
Figure 4.
A summary of the interpretation of the genetic traits of the most recent marine ancestor of Rhodobacteraceae significantly gained or lost during adaptation to non-marine and gained during better adaptation to marine habitats.
Acknowledgments
We thank Oliver Frank and Palani Kandavel Kannan for help with the BayesTraits analysis and Gerrit Wienhausen for help with design of Figure 4. This work was supported by Deutsche Forschungsgemeinschaft within TRR 51.
Footnotes
Supplementary Information accompanies this paper on The ISME Journal website (http://www.nature.com/ismej)
The authors declare no conflict of interest.
Supplementary Material
References
- Aguilar-Barajas E, Díaz-Pérez C, Ramírez-Díaz MI, Riveros-Rosas H, Cervantes C. (2011). Bacterial transport of sulfate, molybdate, and related oxyanions. BioMetals 24: 687–707. [DOI] [PubMed] [Google Scholar]
- Alonso-Gutiérrez J, Lekunberri I, Teira E, Gasol JM, Figueras A, Novoa B. (2009). Bacterioplankton composition of the coastal upwelling system of ‘Ría de Vigo', NW Spain. FEMS Microbiol Ecol 70: 493–505. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. (1990). Basic local alignment search tool. J Mol Biol 215: 403–410. [DOI] [PubMed] [Google Scholar]
- Amin SA, Hmelo LR, van Tol HM, Durham BP, Carlson LT, Heal KR et al. (2015). Interaction and signalling between a cosmopolitan phytoplankton and associated bacteria. Nature 522: 98–101. [DOI] [PubMed] [Google Scholar]
- Anderson I, Scheuner C, Göker M, Mavromatis K, Hooper SD, Porat I et al. (2011). Novel insights into the diversity of catabolic metabolism from ten haloarchaeal genomes. PLoS One 6: e20237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arata H, Shimizu M, Takamiya K. (1992). Purification and properties of trimethylamine N-oxide reductase from aerobic photosynthetic bacterium Roseobacter denitrificans. J Biochem 112: 470–475. [DOI] [PubMed] [Google Scholar]
- Auch AF, Henz SR, Holland BR, Göker M. (2006). Genome BLAST distance phylogenies inferred from whole plastid and whole mitochondrion genome sequences. BMC Bioinformatics 7: 350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auch AF, Klenk H-P, Göker M. (2010). Standard operating procedure for calculating genome-to-genome distances based on high-scoring segment pairs. Stand Genomic Sci 2: 142–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bannert C, Welfle A, Aus dem Spring C, Schomburg D. (2010). BrEPS: a flexible and automatic protocol to compute enzyme-specific sequence profiles for functional annotation. BMC Bioinformatics 11: 589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barber RD, Rott MA, Donohue TJ. (1996). Characterization of a glutathione-dependent formaldehyde dehydrogenase from Rhodobacter sphaeroides. J Bacteriol 178: 1386–1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barkay T, Kritee K, Boyd E, Geesey G. (2010). A thermophilic bacterial origin and subsequent constraints by redox, light and salinity on the evolution of the microbial mercuric reductase. Environ Microbiol 12: 2904–2917. [DOI] [PubMed] [Google Scholar]
- Belas R, Horikawa E, Aizawa SI, Suvanasuthi R. (2009). Genetic determinants of Silicibacter sp. TM1040 motility. J Bacteriol 191: 4502–4512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bengtsson MM, Sjøtun K, Storesund JE, Øvreas L. (2011). Utilization of kelp-derived carbon sources by kelp surface-associated bacteria. Aquat Microb Ecol 62: 191–199. [Google Scholar]
- Bergsten JA. (2005). A review of long-branch attraction. Cladistics 21: 163–193. [DOI] [PubMed] [Google Scholar]
- Berry A, Janssens D, Hümbelin M, Jore JPM, Hoste B, Cleenwerck I et al. (2003). Paracoccus zeaxanthinifaciens sp. nov., a zeaxanthin-producing bacterium. Int J Syst Evol Microbiol 53: 231–238. [DOI] [PubMed] [Google Scholar]
- Bertand EM, Allen AE. (2012). Influence of vitamin B auxotrophy on nitrogen metabolism in eukaryotic phytoplankton. Front Microbiol 3: 375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bevers LE, Hagedoorn PL, Krijger GC, Hagen WR. (2006). Tungsten transport protein A (WtpA) in Pyrococcus furiosus: the first member of a new class of tungstate and molybdate transporters. J Bacteriol 188: 6498–6505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breider S, Scheuner C, Schumann P, Fiebig A, Petersen J, Pradella S et al. (2014). Genome-scale data suggest reclassifications in the Leisingera-Phaeobacter cluster including proposals for Sedimentitalea gen. nov. and Pseudophaeobacter gen. nov. Front Microbiol 5: 416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brinkhoff T, Giebel HA, Simon M. (2008). Diversity, ecology, and genomics of the Roseobacter clade: a short overview. Arch Microbiol 189: 531–539. [DOI] [PubMed] [Google Scholar]
- Brinkhoff T, Muyzer G. (1997). Increased species diversity and extended habitat range of sulfur-oxidizing Thiomicrospira spp. Appl Environ Microbiol 63: 3789–3796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruland KW. (1983) Trace elements in sea water. In: Riley JP, Chester R (eds), Chemical Oceanography vol. 8. Academic Press: London, UK, p 157. [Google Scholar]
- Buchan A, González JM, Moran MA. (2005). Overview of the marine Roseobacter lineage. Appl Environ Microbiol 71: 5665–5677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchan A, LeCleir GR, Gulvik Ca, González JM. (2014). Master recyclers: features and functions of bacteria associated with phytoplankton blooms. Nat Rev Microbiol 12: 686–698. [DOI] [PubMed] [Google Scholar]
- Cabrita MT, Vale C, Rauter AP. (2010). Halogenated compounds from marine algae. Mar Drugs 8: 2301–2317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caspi R, Altman T, Dreher K, Fulcher CA, Subhraveti P, Keseler IM et al. (2012). The MetaCyc database of metabolic pathways and enzymes and the BioCyc collection of pathway/genome databases. Nucleic Acids Res 40: D742–D753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang A, Schomburg I, Placzek S, Jeske L, Ulbrich M, Xiao M et al. (2015). BRENDA in 2015: exciting developments in its 25th year of existence. Nucleic Acids Res 43: D439–D446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y. (2012). Comparative genomics of methylated amine utilization by marine Roseobacter clade bacteria and development of functional gene markers (tmm, gmaS). Environ Microbiol 14: 2308–2322. [DOI] [PubMed] [Google Scholar]
- Chistoserdova L. (2011). Modularity of methylotrophy, revisited. Environ Microbiol 13: 2603–2622. [DOI] [PubMed] [Google Scholar]
- Collins AJ, Fullmer MS, Gogarten JP, Nyholm SV. (2015). Comparative genomics of Roseobacter clade bacteria isolated from the accessory nidamental gland of Euprymna scolopes. Front Microbiol 6: 123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croft MT, Lawrence AD, Raux-Deery E, Warren MJ, Smith AG. (2005). Algae acquire vitamin B12 through a symbiotic relationship with bacteria. Nature 438: 90–93. [DOI] [PubMed] [Google Scholar]
- Cunliffe M. (2011). Correlating carbon monoxide oxidation with cox genes in the abundant marine Roseobacter clade. ISME J 5: 685–691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Vries GE, Harms N, Hoogendijk J, Stouthamer AH. (1989). Isolation and characterization of Paracoccus denitrificans mutants with increased conjugation frequencies and pleiotropic loss of a (nGATCn) DNA-modifying property. Arch Microbiol 152: 52–57. [Google Scholar]
- Denger K, Mayer J, Buhmann M, Weinitschke S, Smits THM, Cook AM. (2009). Bifurcated degradative pathway of 3-sulfolactate in Roseovarius nubinhibens ISM via sulfoacetaldehyde acetyltransferase and (S)-cysteate sulfolyase. J Bacteriol 191: 5648–5656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deniaud-Bouët E, Kervarec N, Gurvant M, Tonon T, Kloareg B, Hervé C. (2014). Chemical and enzymatic fractionation of cell walls from Fucales: insights into the structure of the extracellular matrix of brown algae. Ann Bot 114: 1203–1216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diniz-Filho JAF, De Sant'Ana CER, Bini LM. (1998). An eigenvector method for estimating phylogenetic inertia. Evolution 52: 1247–1262. [DOI] [PubMed] [Google Scholar]
- Dogs M, Voget S, Teshima H, Petersen J, Davenport K, Dalingault H et al. (2013). Genome sequence of Phaeobacter inhibens type strain (T5T), a secondary metabolite producing representative of the marine Roseobacter clade, and emendation of the species description of Phaeobacter inhibens. Stand Genomic Sci 9: 334–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong H, Hong Y, Lu S, Xie L. (2014). Metaproteomics reveals the major microbial players and their biogeochemical functions in a productive coastal system in the northern South China Sea. Environ Microbiol Rep 6: 683–695. [DOI] [PubMed] [Google Scholar]
- Fielman KT, Woodin SA, Lincoln DE. (2001). Polychaete indicator species as a source of natural halogenated organic compounds in marine sediments. Environ Toxicol Chem 20: 738–747. [PubMed] [Google Scholar]
- Flamholz A, Noor E, Bar-Even A, Liebermeister W, Milo R. (2013). Glycolytic strategy as a tradeoff between energy yield and protein cost. Proc Natl Acad Sci USA 110: 10039–10044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank O, Göker M, Pradella S, Petersen J. (2015. a). Ocean's twelve: flagellar and biofilm chromids in the multipartite genome of Marinovum algicola DG898 exemplify functional compartmentalization in Proteobacteria. Environ Microbiol 17: 4019–4034. [DOI] [PubMed] [Google Scholar]
- Frank O, Michael V, Päuker O, Boedeker C, Jogler C, Rohde M et al. (2015. b). Plasmid curing and the loss of grip – The 65-kb replicon of Phaeobacter inhibens DSM 17395 is required for biofilm formation, motility and the colonization of marine algae. Syst Appl Microbiol 38: 120–127. [DOI] [PubMed] [Google Scholar]
- Frank O, Pradella S, Rohde M, Scheuner C, Klenk H-P et al. (2014). Complete genome sequence of the Phaeobacter gallaeciensis type strain CIP 105210T (= DSM 26640T=BS107T. Stand Genomic Sci 9: 914–932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuhrer T, Fischer E, Sauer U. (2005). Experimental identification and quantification of glucose metabolism in seven bacterial species. J Bacteriol 187: 1581–1590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrity GM, Bell JA, Lilburn T, Class I. (2005) Alphaproteobacteria class. nov. In: Brenner DJ, Krieg NR, Stanley JT, Garrity GM (eds), Bergey's Manual of Sytematic Bacteriology, 2nd edn. Vol. 2 The Proteobacteria), part C The Alpha -, Beta -, Delta -, and Epsilonproteobacteria Springer: New York, USA, p 1. [Google Scholar]
- Ghosh S, Ghosh P, Maiti TK. (2011). Production and metabolism of indole acetic acid (IAA) by root nodule bacteria (Rhizobium): a review. J Pure Appl Microbiol 5: 523–540. [Google Scholar]
- Gibb SW, Hatton AD. (2004). The occurrence and distribution of trimethylamine-N-oxide in Antarctic coastal waters. Mar Chem 91: 65–75. [Google Scholar]
- Giebel H-A, Kalhoefer D, Lemke A, Thole S, Gahl-Janssen R, Simon M et al. (2011). Distribution of Roseobacter RCA and SAR11 lineages in the North Sea and characteristics of an abundant RCA isolate. ISME J 5: 8–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gifford SM, Sharma S, Booth M, Moran MA. (2012). Expression patterns reveal niche diversification in a marine microbial assemblage. ISME J 7: 281–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gifford SM, Sharma S, Moran MA. (2014). Linking activity and function to ecosystem dynamics in a coastal bacterioplankton community. Front Microbiol 5: 185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giovannoni SJ, Rappé MS. (2000) Evolution, diversity and molecular ecology of marine prokaryotes. In: Kirchman DL (ed). Microbial Ecology of the Oceans. Wiley: New York, USA, pp 47–84. [Google Scholar]
- Giovannoni SJ, Thrash JC, Temperton B. (2014). Implications of streamlining theory for microbial ecology. ISME J 8: 1553–1565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giraud M-F, Naismith JH. (2000). The rhamnose pathway. Curr Opin Struct Biol 10: 687–696. [DOI] [PubMed] [Google Scholar]
- Gisin J, Mueller A, Pfaender Y, Leimkuehler S, Narberhuas F. (2010). A Rhodobacter capsulatus member of a universal permease family imports molybdate and other oxyanions. J Bacteriol 192: 5943–5952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goenrich M, Bartoschek S, Hagemeier CH, Griesinger C, Vorholt JA. (2002). A glutathione-dependent formaldehyde-activating enzyme (Gfa) from Paracoccus denitrificans detected and purified via two-dimensional proton exchange NMR spectroscopy. J Biol Chem 277: 3069–3072. [DOI] [PubMed] [Google Scholar]
- Goloboff PA, Farris JS, Nixon KC. (2008). TNT, a free program for phylogenetic analysis. Cladistics 25: 774–786. [Google Scholar]
- Gon S, Giudici-Orticoni MT, Méjean V, Iobbi-Nivol C. (2001). Electron transfer and binding of the c-type cytochrome TorC to the trimethylamine N-oxide reductase in Escherichia coli. J Biol Chem 276: 11545–11551. [DOI] [PubMed] [Google Scholar]
- Gonzalez CF, Proudfoot M, Brown G, Korniyenko Y, Mori H, Savchenko AV et al. (2006). Molecular basis of formaldehyde detoxification: characterization of two S-formylglutathione hydrolases from Escherichia coli, FrmB and YeiG. J Biol Chem 281: 14514–14522. [DOI] [PubMed] [Google Scholar]
- Gribble GW. (2003). The diversity of naturally produced organohalogens. Chemosphere 52: 289–297. [DOI] [PubMed] [Google Scholar]
- Gribble GW. (2012). A recent survey of naturally occurring organohalogen compounds. Environ Chem 12: 396–405. [Google Scholar]
- Harms N, Ras J, Reijnders WNM, Van Spanning RJM, Stouthamer AH. (1996). S-formylglutathione hydrolase of Paracoccus denitrificans is homologous to human esterase D: a universal pathway for formaldehyde detoxification? J Bacteriol 178: 6296–6299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison PW, Lower RPJ, Kim NKD, Young JPW. (2010). Introducing the bacterial 'chromid': not a chromosome, not a plasmid. Trends Microbiol 18: 141–148. [DOI] [PubMed] [Google Scholar]
- Helliwell KE, Wheeler GL, Leptos KC, Goldstein RE, Smith AG. (2011). Insights into the evolution of vitamin B12 auxotrophy from sequenced algal genomes. Mol Biol Evol 28: 2921–2933. [DOI] [PubMed] [Google Scholar]
- Helliwell KE, Collins S, Kazamia E, Purton S, Wheeler GL, Smith AG. (2015). Fundamental shift in vitamin B-12 eco-physiology of a model alga demonstrated by experimental evolution. ISME J 9: 1446–1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hille R. (2002). Molybdenum and tungsten in biology. Trends Biochem Sci 27: 360–367. [DOI] [PubMed] [Google Scholar]
- Hiraishi A, Ueda Y. (1994). Intrageneric structure of the genus Rhodobacter: transfer of Rhodobacter sulfidophilus and related marine species to the genus Rhodovulum gen. nov. Int J Syst Bacteriol 44: 15–23. [Google Scholar]
- Hoffmann T, Bremer E. (2011). Protection of Bacillus subtilis against cold stress via compatible-solute acquisition. J Bacteriol 193: 1552–1562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyatt D, Chen G-L, Locascio PF, Land ML, Larimer FW, Hauser LJ. (2010). Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics 11: 119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imhoff JF, Kramer M, Trüper HG. (1983). Sulfate assimilation in Rhodopseudomonas sulfidophila. Arch Microbiol 136: 96–101. [Google Scholar]
- Jeffroy O, Brinkmann H, Delsuc F, Philippe H. (2006). Phylogenomics: the beginning of incongruence? Trends Genet 22: 225–231. [DOI] [PubMed] [Google Scholar]
- Johnson MK, Rees DC, Adams MWW. (1996). Tungstoenzymes. Chem Rev 96: 2817–2840. [DOI] [PubMed] [Google Scholar]
- Kato Y, Ryoko O, Asano Y. (2000). Distribution of aldoxime dehydratase in microorganisms. Appl Environ Microbiol 66: 2290–2296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazamia E, Czesnick H, Nguyen TTV, Croft MT, Sherwood E, Sasso S et al. (2012). Mutualistic interactions between vitamin B12-dependent algae and heterotrophic bacteria exhibit regulation. Environ Microbiol 14: 1466–1476. [DOI] [PubMed] [Google Scholar]
- Kertesz MA. (2000). Riding the sulfur cycle - metabolism of sulfonates and sulfate esters in Gram-negative bacteria. FEMS Microbiol Rev 24: 135–175. [DOI] [PubMed] [Google Scholar]
- King GM. (2003). Molecular and culture-based analyses of aerobic carbon monoxide oxidizer diversity. Appl Environ Microbiol 69: 7257–7265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King GM, Weber CF. (2007). Distribution, diversity and ecology of aerobic CO-oxidizing bacteria. Nat Rev Microbiol 5: 107–118. [DOI] [PubMed] [Google Scholar]
- King JE, Quinn JP. (1997). The utilization of organosulphonates by soil and freshwater bacteria. Lett Appl Microbiol 24: 474–478. [Google Scholar]
- Kisker C, Schindelin H, Rees DC. (1997). Molybdenum-cofactor-containing enzymes: structure and mechanism. Annu Rev Biochem 66: 233–267. [DOI] [PubMed] [Google Scholar]
- Kleber H-P. (1997). Bacterial carnitine metabolism. FEMS Microbiol Lett 147: 1–9. [DOI] [PubMed] [Google Scholar]
- Klingner A, Bartsch A, Dogs M, Wagner-Döbler I, Jahn D, Simon M et al. (2015). Large-scale 13 C-flux profiling reveals conservation of the Entner-Doudoroff pathway as glycolytic strategy among glucose-using marine bacteria. Appl Environ Microbiol 81: 2408–2422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kogure K. (1998). Bioenergetics of marine bacteria. Curr Opin Biotechnol 9: 278–282. [DOI] [PubMed] [Google Scholar]
- Koblizek M. (2015). Ecology of aerobic anoxygenic phototrophs in aquatic environments. FEMS Microbiol Rev 39: 854–870. [DOI] [PubMed] [Google Scholar]
- Lachnit T, Meske D, Wahl M, Harder T, Schmitz R. (2011). Epibacterial community patterns on marine macroalgae are host-specific but temporally variable. Environ Microbiol 13: 655–665. [DOI] [PubMed] [Google Scholar]
- Lagesen K, Hallin P. (2007). RNAmmer: consistent and rapid annotation of ribosomal RNA genes. Nucleic Acids Res 35: 3100–3108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laass S, Kleist S, Bill N, Drueppel K, Kossmehl S, Woehlbrand L et al. (2014). Gene regulatory and metabolic adaptation processes of Dinoroseobacter shibae DFL12T during oxygen depletion. J Biol Chem 289: 13219–13231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauro FM, McDougald D, Thomas T, Williams TJ, Egan S, Rice S et al. (2009). The genomic basis of trophic strategy in marine bacteria. Proc Natl Acad Sci USA 106: 15527–15533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le SQ, Gascuel O. (2008). An improved general amino acid replacement matrix. Mol Biol Evol 25: 1307–1320. [DOI] [PubMed] [Google Scholar]
- Lidbury I, Murrell JC, Chen Y. (2014). Trimethylamine N-oxide metabolism by abundant marine heterotrophic bacteria. Proc Natl Acad Sci USA 111: 2710–2715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lidbury ID, Murrell JC, Chen Y. (2015). Trimethylamine and trimethylamine N-oxide are supplementary energy sources for a marine heterotrophic bacterium: implications for marine carbon and nitrogen cycling. ISME J 9: 760–769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim S-K, Kim SJ, Cha SH, Oh Y-K, Rhee H-J, Kim M-S et al. (2009). Complete genome sequence of Rhodobacter sphaeroides KD131. J Bacteriol 191: 1118–1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo H, Csuros M, Hughes AL, Moran MA. (2013). Evolution of divergent life history strategies in marine alphaproteobacteria. mBio 4: e00373–e003713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo H, Löytynoja A, Moran MA. (2012). Genome content of uncultivated marine Roseobacters in the surface ocean. Environ Microbiol 14: 41–51. [DOI] [PubMed] [Google Scholar]
- Luo H, Moran MA. (2014). Evolutionary ecology of the marine Roseobacter clade. Microbiol Mol Biol Rev 78: 573–587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo H, Swan BK, Stepanauskas R, Hughes AL, Moran MA. (2014). Evolutionary analysis of a streamlined lineage of surface ocean Roseobacters. ISME J 8: 1428–1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo H, Thompson LR, Stingl U, Hughes AL. (2015). Selection maintains low genomic GC content in marine SAR11 lineages. Mol Biol Evol 32: 2738–2748. [DOI] [PubMed] [Google Scholar]
- Maddison WP, Maddison DR. (2011), Mesquite: a modular system for evolutionary analysis. Version 2.75. Mesquite website http://mesquiteproject.org.
- Malmstrom RR, Kiene RP, Kirchman DL. (2004). Identification and enumeration of bacteria assimilating dimethylsulfoniopropionate (DMSP) in the North Atlantic and Gulf of Mexico. Limnol Oceanogr 49: 597–606. [Google Scholar]
- Mavromatis K, Ivanova NN, Chen I-M a, Szeto E, Markowitz VM, Kyrpides NC. (2009). The DOE-JGI standard operating procedure for the annotations of microbial genomes. Stand Genomic Sci 1: 63–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meier-Kolthoff JP, Auch AF, Klenk H-P, Göker M. (2013. a). Genome sequence-based species delimitation with confidence intervals and improved distance functions. BMC Bioinformatics 14: 60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meier-Kolthoff JP, Göker M, Spröer C, Klenk H-P. (2013. b). When should a DDH experiment be mandatory in microbial taxonomy? Arch Microbiol 195: 413–418. [DOI] [PubMed] [Google Scholar]
- Meier-Kolthoff JP, Auch AF, Klenk H-P, Göker M. (2014). Highly parallelized inference of large genome-based phylogenies. Concurr Comp Pract Exp 26SI: 1715–1729. [Google Scholar]
- Meusemann K, von Reumont BM, Simon S, Roeding F, Strauss S, Kück P et al. (2010). A phylogenomic approach to resolve the arthropod tree of life. Mol Biol Evol 27: 2451–2464. [DOI] [PubMed] [Google Scholar]
- Michael V, Frank O, Bartling P, Scheuner C, Göker M, Brinkmann H et al. (2016). Biofilm-plasmids with a rhamnose operon are essential determinants of the 'swim-or-stick' lifestyle in roseobacters. ISME J 10: 2498–2513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moran MA, Miller WL. (2007). Resourceful heterotrophs make the most of light in the coastal ocean. Nat Rev Microbiol 5: 792–800. [DOI] [PubMed] [Google Scholar]
- Moran MA, Reisch CR, Kiene RP, Whitman WB. (2012). Genomic insights into bacterial DMSP transformations. Ann Rev Mar Sci 4: 523–542. [DOI] [PubMed] [Google Scholar]
- Newton RJ, Griffin LE, Bowles KM, Meile C, Gifford S, Givens CE et al. (2010). Genome characteristics of a generalist marine bacterial lineage. ISME J 4: 784–798. [DOI] [PubMed] [Google Scholar]
- Nokhal T-H, Schlegel HG. (1983). Taxonomic study of Paracoccus denitrificans. Int J Syst Bacteriol 33: 26–37. [Google Scholar]
- Novak HR, Sayer C, Isupov MN, Paszkiewicz K, Gotz D, Spragg AM et al. (2013). Marine Rhodobacteraceae l-haloacid dehalogenase contains a novel His/Glu dyad that could activate the catalytic water. FEBS J 280: 1664–1680. [DOI] [PubMed] [Google Scholar]
- Pagel M, Meade A, Barker D. (2004). Bayesian estimation of ancestral character states on phylogenies. Syst Biol 53: 673–684. [DOI] [PubMed] [Google Scholar]
- Pagel M. (1994). Detecting correlated evolution on phylogenies: a general method for the comparative analysis of discrete characters. Proc R Soc B Biol Sci 255: 37–45. [Google Scholar]
- Paul NA, De Nys R, Steinberg PD. (2006). Chemical defence against bacteria in the red alga Asparagopsis armata: linking structure with function. Mar Ecol Prog Ser 306: 87–101. [Google Scholar]
- Petersen J, Brinkmann H, Berger M, Brinkhoff T, Päuker O, Pradella S. (2011). Origin and evolution of a novel DnaA-like plasmid replication type in Rhodobacterales. Mol Biol Evol 28: 1229–1240. [DOI] [PubMed] [Google Scholar]
- Petersen J, Brinkmann H, Pradella S. (2009). Diversity and evolution of repABC type plasmids in Rhodobacterales. Environ Microbiol 11: 2627–2638. [DOI] [PubMed] [Google Scholar]
- Petersen J, Frank O, Göker M, Pradella S. (2013). Extrachromosomal, extraordinary and essential - the plasmids of the Roseobacter clade. Appl Microbiol Biotechnol 97: 2805–2815. [DOI] [PubMed] [Google Scholar]
- Petersen J. (2011). Phylogeny and compatibility: plasmid classification in the genomics era. Arch Microbiol 193: 313–321. [DOI] [PubMed] [Google Scholar]
- Philippe H, Brinkmann H, Lavrov DV, Littlewood DTJ, Manuel M, Wörheide G et al. (2011). Resolving difficult phylogenetic questions: why more sequences are not enough. PLoS Biol 9: e1000602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pradella S, Allgaier M, Hoch C, Päuker O, Stackebrandt E, Wagner-Döbler I. (2004). Genome organization and localization of the pufLMg of the photosynthesis reaction center in phylogenetically diverse marine Alphaproteobacteria. Appl Environ Microbiol 70: 3360–3369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pradella S, Päuker O, Petersen J. (2010). Genome organisation of the marine Roseobacter clade member Marinovum algicola. Arch Microbiol 192: 115–126. [DOI] [PubMed] [Google Scholar]
- Pujalte MJ, Lucena T, Ruvira MA, Arahal DR, Macián MC. (2014) The family Rhodobacteraceae. In: Rosenberg E, DeLong EF, Lory S, Stackebrandt E, Thompson F (eds), The Prokaryotes – Alphaproteobacteria and Betaproteobacteria, 4th edn. Springer: Berlin, Germany, pp 545–577. [Google Scholar]
- Quester S, Schomburg D. (2011). EnzymeDetector: an integrated enzyme function prediction tool and database. BMC Bioinformatics 12: 376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team.. (2015). R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing: Vienna, Austria.
- Raina JB, Tapiolas D, Willis BL, Bourne DG. (2009). Coral-associated bacteria and their role in the biogeochemical cycling of sulfur. Appl Environ Microbiol 75: 3492–3501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ras J, Van Ophem PW, Reijnders WNM, Van Spanning RJM, Duine JA, Stouthamer AH et al. (1995). Isolation, sequencing, and mutagenesis of the gene encoding NAD- and glutathione-dependent formaldehyde dehydrogenase (GD-FALDH) from Paracoccus denitrificans, in which GD-FALDH is essential for methylotrophic growth. J Bacteriol 177: 247–251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reshetnikov AS, Khmelenina VN, Trotsenko YA. (2004). Detection of ectoin biosynthesis genes in halotolerant aerobic methylotrophic bacteria. Dokl Biochem Biophys 396: 200–202. [DOI] [PubMed] [Google Scholar]
- Romano AH, Conway T. (1996). Evolution of carbohydrate metabolic pathways. Res Microbiol 147: 448–455. [DOI] [PubMed] [Google Scholar]
- Salichos L, Rokas A. (2013). Inferring ancient divergences requires genes with strong phylogenetic signals. Nature 497: 327–331. [DOI] [PubMed] [Google Scholar]
- Sass H, Koepke B, Ruetters H, Feuerlein T, Droege S, Cypionka H et al. (2010). Tateyamaria pelophila sp nov., a facultatively anaerobic alphaproteobacterium isolated from tidal-flat sediment, and emended descriptions of the genus Tateyamaria and of Tateyamaria omphalii. Int J Syst Evol Microbiol 60: 1770–1777. [DOI] [PubMed] [Google Scholar]
- Sañudo-Wilhelmy SA, Gómez-Consarnau L, Suffridge C, Webb EA. (2014). The role of B vitamins in marine biogeochemistry. Ann Rev Mar Sci 6: 339–367. [DOI] [PubMed] [Google Scholar]
- Schäfer H, Myronova N, Boden R. (2010). Microbial degradation of dimethylsulphide and related C1-sulphur compounds: organisms and pathways controlling fluxes of sulphur in the biosphere. J Exp Bot 61: 315–334. [DOI] [PubMed] [Google Scholar]
- Schomburg I, Chang A, Placzek S, Söhngen C, Rother M, Lang M et al. (2013). BRENDA in 2013: integrated reactions, kinetic data, enzyme function data, improved disease classification: New options and contents in BRENDA. Nucleic Acids Res 41: D764–D772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz G, Hagedoorn P-L, Fischer K. (2007) Molybdate and tungstate: uptake, homeostasis, cofactors, and enzymes. In: Nies D, Silver S (eds), Molecular Microbiology of Heavy Metals. Springer: Heidelberg, Germany, pp 421–451. [Google Scholar]
- Seibel BA, Walsh PJ. (2002). Trimethylamine oxide accumulation in marine animals: relationship to acylglycerol storage. J Exp Biol 205: 297–306. [DOI] [PubMed] [Google Scholar]
- Selje N, Simon M, Brinkhoff T. (2004). A newly discovered Roseobacter cluster in temperate and polar oceans. Nature 427: 445–448. [DOI] [PubMed] [Google Scholar]
- Senesi N, Padovano G, Brunetti G. (1988). Scandium, titanium, tungsten and zirconium content in commercial inorganic fertilizers and their contribution to soil. Environ Technol Lett 9: 1011–1020. [Google Scholar]
- Shimodaira H, Hasegawa M. (2001). CONSEL: for assessing the confidence of phylogenetic tree selection. BMC Bioinformatics 17: 1246–1247. [DOI] [PubMed] [Google Scholar]
- Shvetsova SV, Zhurishkina EV, Bobrov KS, Ronzhina NL, Lapina IM, Ivanen DR et al. (2015). The novel strain Fusarium proliferatum LE1 (RCAM02409) produces α-L-fucosidase and arylsulfatase during the growth on fucoidan. J Basic Microbiol 55: 471–479. [DOI] [PubMed] [Google Scholar]
- Siddall ME, Whiting MF. (1999). Long-branch abstractions. Cladistics 15: 9–24. [Google Scholar]
- Siddall ME. (2010). Unringing a bell: metazoan phylogenomics and the partition bootstrap. Cladistics 26: 444–452. [DOI] [PubMed] [Google Scholar]
- Simo R, Archer SD, Pedrós-Alió C, Gilpin L, Stelfox-Widdicombe CE. (2002). Coupled dynamics of dimethylsulfoniopropionate and dimethylsulfide cycling and the microbial food web in surface waters of the North Atlantic. Limnol Oceanogr 41: 53–61. [Google Scholar]
- Stackebrandt E, Scheuner C, Göker M, Schumann P. (2014) The family Intrasporangiaceae. In: Rosenberg E, DeLong EF, Lory S, Stackebrandt E, Thompson F (eds), The Prokaryotes – Actinobacteria. 4th edn. Springer: Heidelberg, Germany, pp 397–424. [Google Scholar]
- Stamatakis A, Aberer AJ. (2013). Novel Parallelization Schemes for Large-Scale Likelihood-Based Phylogenetic Inference. Proceedings - IEEE 27th International Parallel and Distributed Processing Symposium: Boston, MA, USA, pp 1195–1204.
- Taveirne ME, Sikes ML, Olson JW. (2009). Molybdenum and tungsten in Campylobacter jejuni: their physiological role and identification of separate transporters regulated by a single ModE-like protein. Mol Microbiol 74: 758–771. [DOI] [PubMed] [Google Scholar]
- Tang K, Huang H, Jiao N, Wu CH. (2010). Phylogenomic analysis of marine Roseobacters. PLoS One 5: e11604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- UniProt Consortium. (2013). Update on activities at the Universal Protein Resource (UniProt) in 2013. Nucleic Acids Res 41: D43–D47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- To TH, Jung M, Lycett S, Gascuel O. (2015). Fast dating using least-squares criteria and algorithms. Syst Biol 65: 82–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tolli JD, Sievert SM, Taylor CD. (2006). Unexpected diversity of bacteria capable of carbon monoxide oxidation in a coastal marine environment, and contribution of the Roseobacter-associated clade to total CO oxidation. Appl Environ Microbiol 72: 1966–1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treberg JR, Speers-Roesch B, Piermarini PM, Ip YK, Ballantyne JS, Driedzic WR. (2006). The accumulation of methylamine counteracting solutes in elasmobranchs with differing levels of urea: a comparison of marine and freshwater species. J Exp Biol 209: 860–870. [DOI] [PubMed] [Google Scholar]
- Trotsenko IA, Doronina NV, Li TD, Reshetnikov AS. (2007). Moderately haloalkaliphilic aerobic methylobacteria. Mikrobiologiia 76: 293–305. [PubMed] [Google Scholar]
- van de Guchte M, Penaud S, Grimaldi C, Barbe V, Bryson K, Nicolas P et al. (2006). The complete genome sequence of Lactobacillus bulgaricus reveals extensive and ongoing reductive evolution. Proc Natl Acad Sci USA 103: 9274–9279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ventosa A, Nieto JJ, Oren A. (1998). Biology of moderately halophilic aerobic bacteria. Microbiol Mol Biol Rev 62: 504–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verbarg S, Göker M, Scheuner C, Schumann P, Stackebrandt E. (2014) The families Erysipelotrichaceae emend., Coprobacillaceae fam. nov., and Turicibacteraceae fam. nov. In: Rosenberg E, DeLong EF, Lory S, Stackebrandt E, Thompson F (eds), The Prokaryotes – Firmicutes and Tenericutes. 4th edn. Springer: Heidelberg, Germany, pp 79–105. [Google Scholar]
- Voget S, Wemheuer B, Brinkhoff T, Vollmers J, Dietrich S, Giebel H-A et al. (2015). Adaptation of an abundant Roseobacter RCA organism to pelagic systems revealed by genomic and transcriptomic analyses. ISME J 9: 371–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vollmers J, Voget S, Dietrich S, Gollnow K, Smits M, Meyer K et al. (2013). Poles apart: extreme genome plasticity and a new xanthorhodopsin-like gene family in the genomes of Octadecabacter arcticus 238 and Octadecabacter antarcticus 307. PLoS One 8: e63422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner-Döbler I, Ballhausen B, Berger M, Brinkhoff T, Buchholz I, Bunk B et al. (2010). The complete genome sequence of the algal symbiont Dinoroseobacter shibae: a hitchhiker's guide to life in the sea. ISME J 4: 61–77. [DOI] [PubMed] [Google Scholar]
- Wagner-Döbler I, Biebl H. (2006). Environmental biology of the marine Roseobacter lineage. Ann Rev Microbiol 60: 255–280. [DOI] [PubMed] [Google Scholar]
- Wargo MJ, Hogan DA. (2009). Identification of genes required for Pseudomonas aeruginosa carnitine catabolism. Microbiology 155: 2411–2419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warthmann R, Cypionka H. (1996). Characteristics of assimilatory sulfate transport in Rhodobacter sulfidophilus. FEMS Microbiol Lett 142: 243–246. [Google Scholar]
- Webster NS, Botté ES, Soo RM, Whalan S. (2011). The larval sponge holobiont exhibits high thermal tolerance. Environ Microbiol Rep 3: 756–762. [DOI] [PubMed] [Google Scholar]
- Welsh DT. (2000). Ecological significance of compatible solute accumulation by micro-organisms: from single cells to global climate. FEMS Microbiol Rev 24: 263–290. [DOI] [PubMed] [Google Scholar]
- Wemheuer B, Wemheuer F, Hollensteiner J, Meyer F, Voget S, Daniel R. (2015). The green impact: bacterioplankton response towards a phytoplankton spring bloom in the southern North Sea assessed by comparative metagenomic and metatranscriptomic approaches. Front Microbiol 6: 805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West NJ, Obernosterer I, Zemb O, Lebaron P. (2008). Major differences of bacterial diversity and activity inside and outside of a natural iron-fertilized phytoplankton bloom in the Southern Ocean. Environ Microbiol 10: 738–756. [DOI] [PubMed] [Google Scholar]
- Wetzel RG. (2001) Limnology: Lake and River Ecosystems. 3rd edn. Academic Press: San Diego, CA, USA. [Google Scholar]
- Williams TJ, Long E, Evans F, DeMaere MZ, Lauro FM, Raftery MJ et al. (2012). A metaproteomic assessment of winter and summer bacterioplankton from Antarctic Peninsula coastal surface waters. ISME J 6: 1883–1900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Wang K, Budinoff C, Buchan A, Lang A, Jao N et al. (2009). Gene transfer agent (GTA) genes reveal diverse and dynamic Roseobacter and Rhodobacter populations in the Chesapeake Bay. ISME J 3: 364–373. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.