Abstract
The existence of biogeographic patterns among most free-living microbial taxa has been well established, yet little is known about the underlying mechanisms that shape these patterns. Here, we examined soil bacterial β-diversity across different habitats in the drylands of northern China. We evaluated the relative importance of environmental factors versus geographic distance to a distance–decay relationship, which would be explained by the relative effect of basic ecological processes recognized as drivers of diversity patterns in macrobial theoretical models such as selection and dispersal. Although the similarity of bacterial communities significantly declined with increasing geographic distance, the distance–decay slope and the relative importance of factors driving distance–decay patterns varied across different habitats. A strong distance–decay relationship was observed in the alpine grassland, where the community similarity was influenced only by the environmental factors. In contrast, geographic distance was solely responsible for community similarity in the desert. Even the average compositional similarity among locations in the desert was distinctly lower compared with those in other habitats. We found no evidence that dispersal limitation strongly influenced the β-diversity of bacterial communities in the desert grassland and typical grassland. Together, our results provide robust evidence of habitat specificity for microbial diversity patterns and their underlying drivers. Our findings suggest that microorganisms also have multiple drivers of diversity patterns and some of which may be parallel to some fundamental processes for explaining biodiversity patterns in macroorganisms.
Introduction
Over the past few decades, spatial patterns in microbial biodiversity have been extensively investigated at regional (Green et al., 2004; Griffiths et al., 2011), continental (Fierer and Jackson, 2006; Lauber et al., 2009), and global scales (Fierer et al., 2009; Tedersoo et al., 2012). These studies provide strong evidence that most microbial taxa inhabiting different habitats exhibit biogeographic patterns that are similar to macroorganisms in spite of their microscopic size and hidden diversity (Green and Bohannan, 2006; Martiny et al., 2006). However, our understanding of the fundamental processes that underlie microbial biogeographic patterns remains limited (Nemergut et al., 2013; Wang et al., 2013). Such a knowledge gap impedes our ability to uncover the mechanisms underlying global variations in biodiversity and, ultimately, to predict ecosystem responses to current and future environmental changes.
The decline in community similarity with increasing geographic distance is a well-described pattern of biodiversity (Soininen et al., 2007; Morlon et al., 2008). Distance–decay curves offer a directional measure of the variation in community composition from site to site (that is, β-diversity; Anderson et al., 2011), and the slope of which can reflect the rate of species turnover over space. This commonly studied pattern has also been observed for microorganisms across a range of habitats at various taxonomic resolutions (Horner-Devine et al., 2004). Generally, two mechanisms could give rise to this pattern. First, environmental differences are likely to increase with increasing geographical distance. In this case, community composition thus becomes increasingly different with environmental changes as species are selected from local taxa pool based on their niche preferences (that is, niche-based or deterministic processes; Bell, 2010). Growing evidence supports the role of environmental factors, including soil pH, vegetation and aridity in structuring microbial communities (Chu et al., 2010; Maestre et al., 2015; Prober et al., 2015). Second, distance–decay patterns can also result from dispersal limitation, which is independent of niche differences. For example, neutral theory suggests that all individuals in the communities are ecologically equivalent, and species abundance and diversity are determined by stochastic events (that is, neutral or stochastic processes; Hubbell, 2001). In such a case, dispersal is limited such that individuals tend to disperse to nearby sites. Thus, closer sites will have more similar communities than between those further apart even without environmental influences (Bell, 2001). Evidence of distance–decay patterns driven by dispersal limitation in microorganisms has also been obtained in many studies (Cho and Tiedje, 2000; Peay et al., 2010). It is widely recognized that both processes are important for shaping the distance–decay relationship (Martiny et al., 2011), but the inconsistency of theoretical frameworks prevents more detailed profiles on the relative importance of the processes driving microbial diversity patterns.
In Vellend's (2010) conceptual synthesis, most mechanisms that contribute to patterns in the composition and diversity of species can be categorized into four classes of processes: selection, dispersal, drift and speciation. Selection is the outcome of environmental pressures causing variation in survival and reproduction within and among species; dispersal mainly refers to the movements of a species to a new location; drift reflects population sizes fluctuating in a location owing to inherent chance events; and speciation produces new species that are adapted to particular conditions. We attempt to use such a unified theoretical framework for identifying the processes underlying microbial biogeographic patterns. Although it is a fact that microbial taxa usually defined by molecular genetic methods (for example, sequence-based OTUs) are not the same as ‘species' in macroorganisms, it still makes sense to focus on the four processes for shaping microbial biogeography in some ways. For example, selection acts on multiple biological levels from genes to taxa (Lewontin, 1970). Hence, it allows us to consider selection generally for microbial taxa, which can be defined at different levels of gene sequence similarity. Sequence-based OTUs can offer valuable information about whether a microbial taxon has not dispersal at two sites rather than it has. Yet drift will, and so will speciation, not be easily evaluated for microbes if only sequence data is used because the population size is not measurable and the variation due to sequence or classification error will be just as great as true biological variation per se associated with sequence divergence, despite some detectable effects of drift on community assembly in microbial systems (Ofiteru et al., 2010; Stegen et al., 2013). Another reason is that this conceptual model relates many of the existing theories and models in community ecology to each of the four processes alone or their combinations. For instance, the idea of species ‘niches' (Chase and Leibold, 2003) is synonymous with selection while the full view of Hubbell's neutral theory (2001) represents the combined influence of drift, dispersal and speciation. Owing to intimate associations between micro- and macroorganisms and resulting impacts on each other's geographic distributions, this is a practical pathway to interpret biogeographic patterns displayed in microorganisms using a familiar and classic theoretical framework, which has been widely applied in macroorganisms (Martiny et al., 2006). In addition, although the four processes act in concert and quantitatively estimating the influences of these processes is rarely achieved, recent studies have provided evidence that some microorganisms are likely subjected to the same forces governing community assembly, similar to macroorganisms (Stegen et al., 2012; Nemergut et al., 2013). Until now, however, very little is known about whether these processes alone or in combination could be involved in observed microbial biogeographic patterns, and about what their relative importance is in determining the fundamental patterns of biodiversity. The distance–decay relationship is a well-known biodiversity pattern observed in communities from all domains of life. It is sensitive to critical ecological processes and thus often considered as a powerful tool for testing mechanistic ecological theories (Condit et al., 2002). It has been demonstrated that the relative role of the four processes can lead to a different observed distance–decay curve (Hanson et al., 2012). For instance, selection tends to produce a distance–decay relationship while dispersal counteracts it, and vice versa, such a curve may also provide potential insights for disentangling the relative importance of the four processes responsible for taxonomic diversity. For example, relative weak distance–decay relationship should be observed in habitats where dispersal is high as composition differentiation among locations will decrease with increasing newly established colonizers (Slatkin, 1987).
It is believed that the relative importance of these processes to β-diversity can vary across different spatial scales, habitat types and organisms, which thus results in the difference of the strength of distance–decay in ecological communities (that is, the slope of the distance–decay curve; Nekola and White, 1999). Many studies have shown that the relative importance of a particular mechanism that shapes the distance–decay pattern depends on the spatial scales, and such scale-dependent patterns have been observed in microorganisms (Bardgett and van der Putten, 2014; Wang et al., 2015). Furthermore, it is notable that the relative importance of different processes also likely varies across- and within-habitat types. For example, microbial community composition may differ substantially among habitat types owing to the effects of environmental gradients (Lozupone and Knight, 2007). Habitat differentiation would also affect the dispersal ability of hosting microbes, as highly aquatic substrates should allow for more dispersal than isolated habitats or solid substrates (Cermeno and Falkowski, 2009). A strong role of habitat specificity on microbial community assembly has been observed in Earth's major habitat types (Nemergut et al., 2011), such as desert (Andrew et al., 2012; Ronca et al., 2015), grassland (Zinger et al., 2011; Li et al., 2015), permafrost (Yergeau et al., 2007) and aquatic ecosystems (Zinger et al., 2014). Yet, few studies to date have focused on whether there are some associations between observed patterns in microbial diversity and processes driving them across different habitats.
To investigate the bacterial biogeographic patterns and the mechanisms underlying β-diversity in bacterial communities among habitat types, we conducted a 4000 km transect survey across four different habitats in China's drylands, including alpine grassland, desert, desert grassland and typical grassland (Supplementary Figure S1). This transect covered a continuous transition of four major habitat types according to the classification of Chinese terrestrial habitat types (Chinese Academy of Sciences, 2001; Supplementary Table S1). In total, 545 soil samples were collected and analyzed by sequencing 16S ribosomal RNA (rRNA) gene, the most commonly used indicator gene for bacterial diversity. We attempted to address the following two hypotheses. (i) The rate of distance decay (the slope of the distance–decay curve) will vary across different habitats. (ii) The relative importance of underlying factors (environmental variables or geographic distance) contributing to distance–decay relationships will differ across different habitats. Meanwhile, we predicted that the differences in such β-diversity patterns among different habitats would reflect some differences in relative importance of the ecological processes. For microorganisms, a detected distance–decay relationship and its underlying process may depend on taxonomic resolution; thus, we used the two commonly used groupings—97% and 99% sequence similarity—to define OTUs in our study.
Materials and methods
Study sites and experimental design
The study was conducted across a 4000 km transect of northern China's grasslands from the Xinjiang Uygur Autonomous Region to eastern Inner Mongolia in northern China (83.45° E to 120.36° E, 42.89° N to 49.19° N; Supplementary Figure S1). There are four types of habitats along this transect according to a vegetation map at a scale of 1:1 000 000 (Chinese Academy of Sciences, 2001), including alpine grassland, desert, desert grassland and typical grassland from west to east (Supplementary Table S1). The dominant species in the four habitats include Stipa spp. and Carex spp. (alpine grassland), Calligonum spp., Alhagi spp. and Ephedra spp. (desert and desert grassland), and Stipa spp., Leymus spp. and Agropyron spp. (typical grassland). Dominant soil types are classified as alpine steppe soil, gray desert and sandy soil, and brown pedocals.
Field sampling
We conducted this field sampling during July and August in 2012 near the period of highest plant aboveground biomass. We attempted to take the plant and soil samples at the same period of phenology, and therefore, we started to sample from the west to the east along this transect that had a decreasing trend of temperature in this direction. A total of 61 sites with an interval of 50~100 km were selected along the transect (Supplementary Figure S1). According to our whole sampling regime, there were only four sites in the alpine grassland because it had narrow spatial distribution relative to other habitat types across the transect. The minimum distance of sampling locations from human habitations was approximately 60 km, and each sampling site was representative of the local natural vegetation. Ten 1 m × 1 m quadrats were selected at each sampling site. Samples in bags that were broken were abandoned for use. Finally, we used only 545 samples for this study, with each site having 8–10 samples. In each quadrat, samples of living aboveground plants were clipped, sorted into species and stored in paper bags for biomass measurement and plant richness estimation (Supplementary Table S1). Soil samples per quadrat were collected from five soil cores (2.5 cm diameter × 10 cm depth) of the upper 10 cm of soil. Four soil cores were collected from four corners 10 cm away from the border line of the quadrat and one core was collected from the center of that quadrat. The five cores consisting of the soils from four corners and center of each quadrat were then mixed thoroughly. Composite soils were sieved through a 2.0 mm mesh to remove roots and rocks, homogenized by hand and separated into two parts: one was preserved for subsequent characterization of soil chemistry and the other was placed into a sterile plastic bag and immediately stored at −40 °C for later DNA extraction.
Climate data and geographic distance
Climate attributes, including the mean annual precipitation and mean annual temperature of each sampling site were obtained from the WorldClim global climate data set (Hijmans et al., 2005). Extracted data were processed in ArcGIS version 9.3 using Spatial Analysis tool (ESRI, Redlands, CA, USA). At each site, spatial geographical coordinates and elevations were recorded by a handheld GPS (eTrex Venture, Garmin, Olathe, Kansas, USA). The pairwise geographic distance between sites was calculated using the Imap package in R v.3.1.0 according to the GPS coordinates of each site. We then created a geographic distance matrix corresponding to sites distributed in each habitat type.
Soil physicochemical analysis
Total organic carbon and total nitrogen for soil samples from each quadrat were determined using wet oxidation and a modified Kjeldahl procedure (Wang et al., 2014). Total P was measured by colorimetric analysis with ammonium molybdate and persulfate oxidation (Kuo, 1996). Soil pH was measured after creating a 1: 2.5 (volume) fresh soil to water slurry. Soil moisture was determined gravimetrically after drying in an oven at 105 °C for 12 h.
Illumina sequencing analysis of 16S rRNA gene amplicons
Microbial genomic DNA was extracted from 0.5 g of well-mixed soil for each sample using the MoBio PowerSoil DNA isolation kit (MoBio Laboratories, Carlsbad, CA, USA) according to the manufacturer's protocol. The quality of the extracted DNA was assessed based on 260/280 nm and 260/230 nm absorbance ratios obtained using a NanoDrop ND-1000 Spectrophotometer (NanoDrop Technologies Inc., Wilmington, DE, USA). The final DNA concentration was quantified using a PicoGreen (Life Technologies, Grand Island, NY, USA) assay (Ahn et al., 1996) with a FLUOstar Optima (BMG Labtech, Jena, Germany), stored at −20 °C until use.
To determine the soil bacterial community composition and diversity in each soil sample, an amplicon survey of a portion of the 16S rRNA gene was performed. The primers 515F (5′-GTGCCAGCMGCCGCGGTAA-3′) and 806R (5′-GGACTACHVGGGTWTCTAAT-3′) targeting the V4 hypervariable regions of microbial 16S rRNA gene were selected (Caporaso et al., 2012). Both primers were tagged with adaptor, pad and linker sequences. The reverse primer contains a barcode sequence (12 mer) unique to each sample for pooling of multiple samples in one run of Miseq sequencing. All primers were synthesized by Eurofins/MWG (Huntsville, AL, USA).
PCR amplification was performed in triplicate using a Gene Amp PCR-System 9700 (Applied Biosystems, Foster City, CA, USA) in a 25 μl reaction volume, which contained 2.5 μl of 10 × PCR buffer II, 0.5 unit of AccuPrime Taq DNA Polymerase High Fidelity (Invitrogen, Carlsbad, CA, USA), 0.4 μm of each primer and 10 ng of purified template DNA. Thermal cycling conditions were as follows: an initial denaturation at 94 °C for 1 min followed by 30 cycles at 94 °C for 20 s, 53 °C for 25 s, 68 °C for 45 s and ended with a final extension step at 68 °C for 10 min.
Following amplification, 2 μl of the PCR product was used for agarose gel (1%) detection. The triplicate PCR reactions for each sample were combined and quantified with PicoGreen. From each sample, 200 ng of the PCR product was collected and pooled together with other samples in equimolar concentrations for one sequencing run. Primer and primer dimers were then separated out by electrophoresis on a 1% agarose gel. The pooled mixture was finally purified and recovered with a QIAquick gel extraction kit (QIAGEN Sciences, Valencia, CA, USA) and requantified with PicoGreen. Sequencing was conducted on an Illumina Miseq sequencer at the Institute for Environmental Genomics, University of Oklahoma.
Processing of sequencing data
After assigning each sequence to its sample according to its barcode, the sequences were quality trimmed using Btrim with threshold of average quality scores higher than 30 over a 5 bp window size and a minimum length of 100 bp (Kong, 2011). Paired-end reads with at least a 50 bp overlap and <5% mismatches were joined using FLASH v1.2.5 (Magoc and Salzberg, 2011). After removing the sequences with ambiguous bases, the sequences with lengths between 245 and 258 bp were subjected to chimera removal by U-Chime (Edgar et al., 2011) against 16S ‘Gold' database (reference database in the Broad Microbiome Utilities, version microbiomeutil-r20110519). Sequences were clustered into operational taxonomic units (OTUs) using the 97 and 99% sequence similarity threshold with UPARSE, respectively (Edgar, 2013) and singleton OTUs (with only one read) were removed. Final OTUs were generated based on the clustering results, and taxonomic annotations were assigned to each OTU's representative sequence by the Ribosomal Database Project (RDP) 16S Classifier (Wang et al., 2007). To correct for sampling effort (number of analyzed sequences per sample), the samples were rarefied at 11 612 sequences for the 97% resolution, and 19 992 sequences for the 99% resolution per sample for subsequent bacterial community analysis (Supplementary Figure S2). The above-mentioned steps were performed using an in-house pipeline that was built on the Galaxy platform at the Institute for Environmental Genomics, University of Oklahoma (http://zhoulab5.rccc.ou.edu:8080/).
Statistical analysis
We created four pairwise subsets of samples corresponding to the categorization of each habitat type. Each subset included matrices of the pairwise taxonomic distance (Bray–Curtis) and geographical distance that were constructed using the program R. The composite environmental distance matrix was generated with a normalized combination of the variables selected by the BioEnv (Clarke and Ainsworth, 1993). It is widely accepted that environmental variables usually covary with the changes of geographic distance. To disentangle their separate influences on the community composition, we used partial Mantel tests with 9999 permutations within the vegan R package (Oksanen et al., 2013) to examine the correlations between bacterial community similarity and geographic distance (Spearman correlation) while controlling for environmental distance, and between bacterial community similarity and environmental distance (Spearman correlation) while controlling for geographic distance in each subset (Martiny et al., 2006).
Patterns of community dissimilarity among samples were determined by nonmetric multidimensional scaling (Bray–Curtis distance, two dimensions; Kruskal, 1964). A dissimilarity test of the bacterial community composition was performed using nonparametric multivariate statistical tests and analysis of similarities (999 permutations; Clarke and Ainsworth, 1993). Both nonmetric multidimensional scaling and analysis of similarities were performed in the R software package using the vegan package. To investigate the differences of the bacterial taxonomic composition in different habitat types, we chose 24 dominant genera based on the taxonomic abundance data (average abundance >50 across all soil samples). Multivariate data analysis was then conducted with FactoMineR R package (Lê et al., 2008). The methods implemented in the package are conceptually similar with classical multivariate data analysis like principal component analysis or correspondence analysis, but this package can take into account different types of variables (quantitative or categorical) and different types of structure on the data (a partition on the variables and individuals, and a hierarchy on the variables). The graphical outputs including variables factor map and individuals factor map can thus display clearer distribution of the dominant bacterial taxa across different habitats than using the traditional principal component analysis method. To further exhibit the variation of the relative abundance of dominant bacterial genera across different habitats, we selected 12 most abundant genera based on the taxonomic abundance data and plotted all samples corresponding to each habitat with ggplot2 R package. Statistical analyses (one-way analysis of variance, P<0.05) were performed to test significance of group differences among the four habitat types.
To investigate the spatial structure of the habitats, integrated soil physicochemical parameters (total organic carbon, total nitrogen, TP, C/N ratio, N/P ratio, soil pH and moisture) and plant data (plant richness and aboveground net primary productivity) in each habitat were analyzed respectively using semivariograms (Rossi et al., 1992). We first converted the coordinate of longitude and latitude to UTM (Universal Transverse Mercartor Grid System) by setting the EPSG to 32748 for WGS 84, Northern Hemisphere with the rdgal R package. We then transformed environmental variables into vectors by principal components analysis. We only chose the first principal components for analyzing semivariograms, which explained 62.67 and 99.71% of variation in soil physicochemical parameters and plant data, respectively. Semivariograms for soil chemistry PC1 and plant PC1 were calculated using the geoR package in R and the appropriate model function was fit to the semivariograms. We found evident anisotropy in the directional semivariograms for both the soil chemistry PC1 and plant PC1. Therefore, models for the semivariograms were fitted using linear least-squares regression analysis. Generally, the nugget semivariance expressed as percentage of the total semivariance enables comparison of the relative nugget effect among soil properties (Trangmar et al., 1985). We therefore used this ratio to define distinct classes of spatial dependence for the soil chemistry PC1 and plant PC1 as follows: if the ratio was less than 25%, the variable had a strong spatial dependent; if the ratio was between 25 and 75%, the variable had a moderate spatial dependence; otherwise, the variable was considered random (pure nugget effect; Cambardella et al., 1994).
The rate of distance–decay of the bacterial communities was calculated as the slope of ordinary least-squares regression on the relationship between geographic distance (ln transformed) and community similarity (ln transformed). The significance of the relationship between community dissimilarity and geographical distance in each grassland type was assessed by using the Mantel test (Jackson and Somer, 1989) for each data subset. To test whether the slopes of the distance–decay curve (least squares) at the four grassland types were significantly different from zero or different from the slope of the overall distance–decay curve, we used matrix permutations to compare the slopes of each data subset against slopes in the randomized data sets based on 9999 permutations (Nekola and White, 1999). Owing to the scale dependence of distance–decay relationship and the existence for the differences in spatial scales among the four different habitats in our study, we chose two subsets of data (OTUs defined at the 97% resolution) from desert, desert grassland and typical grassland so as to further test our hypothesis about habitat-specific β-diversity patterns. For each data subset, there were also only four neighboring sites that had similar spatial scales with that in the alpine grassland. The rate of distance–decay of the bacterial communities and the significance test of such a relationship were also analyzed using the same method as above mentioned.
To determine the relative importance of geographic and local environmental factors in structuring bacterial communities, we conducted multiple regression analysis using multiple regression on matrices (MRM) approach, which can offer advantages over the traditional partial Mantel analysis to investigate linear, nonlinear or nonparametric relationships between a multivariate response distance matrix and any number of explanatory distance matrices (Legendre et al., 1994; Lichstein, 2007). Because there was strong collinearity among particular environmental factors, before applying MRM, we used variable clustering to assess the redundancy of the environmental variables by the varclus procedure in the Hmisc R package. The variables with higher correlation (Spearman's ρ2>0.7) were removed from the MRM analysis (for example, total nitrogen, total organic carbon, N/P, mean annual temperature; Supplementary Figure S3), but kept all other variables in the models. We then implemented a matrix randomization procedure with standardized predictor variables using ecodist R package (Goslee and Urban, 2007). To account for zero similarity values, bacterial community similarity (1 minus Bray–Curtis distance) was ln transformed and geographic distance was ln (x+1) transformed (Green et al., 2004; Talbot et al., 2014). To reduce the effect of spurious relationships between variables, we ran the MRM test twice. The first run was to remove the nonsignificant variables; we then reran the tests. We reported the model results from this second run.
Results
Across all the samples, we identified a total of 6 433 048 and 12 417 313 high-quality bacterial sequences, which were grouped into 30 792 and 124 024 OTUs using the 97 and 99% sequence similarity cutoff, respectively. Desert soils showed the lowest OTU richness (with an average of 1747±46 OTUs) compared with that in the alpine grassland (with an average of 1911±27), desert grassland (with an average of 2259±14) and typical grassland (with an average of 2335±15) at the 97% resolution (Supplementary Table S2). Pairwise community similarity between the samples was calculated based on the abundance of each OTU (defined at the 97% identity) using a rarefied Bray–Curtis index, which was highly correlated with the incidence-based Jaccard index (Mantel test: r=0.968, P=0.001).
The overall pattern of community composition across the transect is delineated on the first two coordinates of the nonmetric multidimensional scaling ordination based on the Bray–Curtis dissimilarity index. A distinctly different pattern was both observed among samples between groups for the 97 and 99% sequence identity (Supplementary Figure S4). This result was confirmed by the dissimilarity analysis of community composition, which showed that the bacterial community structures were significantly different (analysis of similarities: R=0.268 and 0.332, P<0.001, respectively). Within a single habitat (for example, desert grassland or typical grassland), the close clustering of samples indicated that there was similar community composition, whereas strong compositional variability was found among the desert samples (Supplementary Table S3).
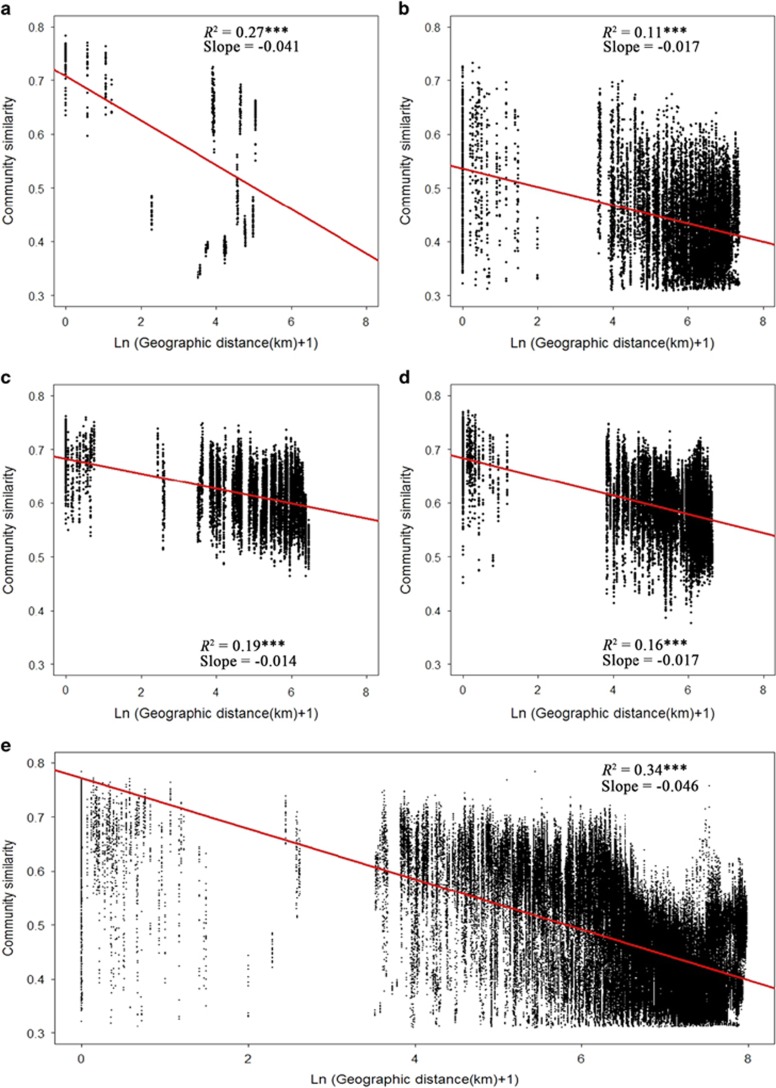
Community similarity versus geographic distance for each pairwise set of samples clearly displayed a significant distance–decay relationship for bacterial communities (at both the 97 and 99% resolutions) (Figure 1 and Supplementary Figure S5). However, the slopes of distance–decay that were estimated by linear regression models varied across different habitats. The slope in the alpine grassland was significantly steeper than those in the desert (slope=−0.017, P<0.001), desert grassland (slope=−0.014, P<0.001) and typical grassland (slope=−0.017, P<0.001; Supplementary Table S4). Compared with each habitat, the slope of the distance–decay curve over whole transect was highest (slope=−0.046), which was similar to that of the alpine grassland (slope=−0.041). In addition, overall lower similarity (average similarity=0.436) in bacterial community composition was found in the desert. We also found similar among-habitats differences of the distance–decay slopes for four dominant taxonomic groups (Actinobacteria, Acidobacteria, Alphaproteobacteria and Verrucomicrobia; Supplementary Figure S6,Supplementary Table S5). Although the estimated slopes for all sequences at the 99% resolution were not significantly different from those estimated at the 97%, the intercept (t=11.70, P<0.001) and average compositional similarity (t=2.69, P=0.05) at the 99% resolution were significantly lower than that of at the 97% resolution across all the habitats. Furthermore, the slopes of the distance–decay curve estimated using a couple of subset data from desert (slope=−0.014 and −0.016, P<0.0001, respectively), desert grassland (slope=−0.014 and −0.010, P<0.0001, respectively) and typical grassland (slope=−0.017 and −0.012, P<0.0001, respectively), which shared the similar spatial scales with the alpine grassland was similar to those estimated across the entire scales of the three habitats (Supplementary Table S6).
Figure 1.
Distance–decay curves of similarity for the bacterial communities (OTUs defined at the 97% sequence identity). The red lines denote the ordinary least squares linear regression across all samples in each habitat and the whole transect: (a) alpine grassland, (b) desert, (c) desert grassland, (d) typical grassland and (e) the whole transect. Statistics are derived from regression analysis (Supplementary Table S4). Asterisks represent significance of correlation (***P<0.0001).
The soil chemistry PC1 and plant PC1 in different habitats displayed difference in their spatial dependence as determined by linear semivariograms (Supplementary Figure S7, Supplementary Table S7). Semivariance calculated from the soil chemistry PC1 and plant PC1 significantly increased with increasing distance lags in the alpine grassland (R=0.655 and 0.840, P<0.001, respectively), desert grassland (R=0.479 and 0.305, P<0.001, respectively) and typical grassland (R=0.365 and 0.224, P<0.001, respectively), and showed strong (nugget, <25%) or moderate (nugget, 25∼75%) spatial dependence for environmental variables in these habitats. However, semivariance for both environmental vectors in the desert showed completely random changes with increasing lags (P>0.05), and thus the environmental variables were not spatially autocorrelated.
Across all the habitats, both environmental factors and geographic distance significantly influenced bacterial β-diversity (Table 1). Partial Mantel tests revealed that the similarity in bacterial community composition among samples (at both the 97% and 99% resolutions) was strongly correlated with environmental distance (ρ=−0.295 and −0.333, P=0.0001, respectively) and geographic distance (ρ=−0.289 and −0.305, P<0.001, respectively). Within each habitat type except the desert, bacterial community similarity was highly correlated with environmental distance, but was not or just weakly correlated with geographic distance for each pairwise set of samples. In contrast, community similarity was strongly correlated with geographic distance in the desert (ρ=−0.140 and −0.183, P=0.0001, respectively).
Table 1. The results of Spearman correlation between the bacterial community similarity (OTUs defined at both the 97% and the 99% sequence similarity) and geographic distance or environmental distance for all pairwise samples using partial Mantel test.
| Correlation between bacterial community similarity and: | Controlling for: |
Alpine grassland |
Desert |
Desert grassland |
Typical grassland |
The whole transect |
|||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| ρ | P | ρ | P | ρ | P | ρ | P | ρ | P | ||
| Environmental distance | Geographic distance | −0.785 (−0.812) | 0.0001 (0.0001) | −0.099 (−0.074) | 0.0067 (0.0189) | −0.430 (−0.555) | 0.0001 (0.0001) | −0.438 (−0.497) | 0.0001 (0.0001) | −0.295 (−0.333) | 0.0001 (0.0001) |
| Geographic distance | Environmental distance | −0.087 (−0.055) | 0.0882 (0.1207) | −0.140 (−0.183) | 0.0001 (0.0001) | −0.058 (−0.076) | 0.0237 (0.041) | −0.024 (−0.020) | 0.2002 (0.1744) | −0.289 (−0.305) | 0.0001 (0.0006) |
In brackets, we indicate the corresponding values at the 99% resolution. P-values are one-tailed tests based on 9999 permutations.
MRM was used to further identify the relative contributions of environmental factors versus geographic distance to bacterial community similarity (at both the 97 and 99% resolutions; Table 2). A large and significant proportion of the variability in bacterial community similarity can be explained by the MRM model in most habitat types except desert. In the alpine grassland, the MRM model explained up to 75% of the variability in community similarity (P<0.001), with soil moisture and plant species richness being the most important variables explaining community similarity (partial regression coefficient b=0.40 and 0.23, respectively, P<0.001). In contrast, only 5% of the variability in community similarity in the desert was explained by the MRM model (P<0.001), with geographic distance showing a sole effect on community similarity (b=0.11, P<0.001). In the typical grassland, soil pH and TP contributed the larger partial regression coefficient (b=0.10 and 0.06, P<0.001, respectively), with other factors such as geographic distance, mean annual precipitation, altitude and plant species richness contributing to smaller but significant partial regression coefficient (b=0.01–0.02, P<0.01). Over the whole transect, mean annual precipitation and geographic distance showed a strong effect on community similarity (b=0.41 and 0.12, respectively, P<0.001). Similar results were found when we used a taxon resolution of 99% sequence similarity.
Table 2. Results of the multiple regression analysis on matrices analysis (MRM) for each of the four habitats and the whole transect in the drylands of northern China.
| Alpine grassland | Desert | Desert grassland | Typical grassland | The whole transect | |
|---|---|---|---|---|---|
| R2=0.75 (0.78) P<0.001 | R2=0.05 (0.09) P<0.001 | R2=0.34 (0.39) P<0.001 | R2=0.46 (0.43) P<0.001 | R2=0.34 (0.40) P<0.001 | |
| Log(Geographic distance (km)+1) | NS (NS) | 0.11*** (0.14***) | 0.01* (NS) | 0.01 (0.01) | 0.12*** (0.19***) |
| MAP | NS (NS) | NS (NS) | 0.02*** (0.03**) | 0.02*** (0.02**) | 0.41*** (0.44***) |
| MAT | ND (NS) | ND (ND) | ND (ND) | ND (ND) | ND (ND) |
| Altitude | NS (NS) | ND (ND) | NS (NS) | 0.01* (NS) | ND (ND) |
| Soil total P | ND (ND) | NS (NS) | 0.03***(0.05***) | 0.06*** (0.06***) | 0.05* (0.06**) |
| C/N ratio | ND (ND) | NS (NS) | 0.01 (NS) | ND (ND) | 0.08** (0.05**) |
| Soil pH | ND (ND) | NS (NS) | 0.02** (0.03**) | 0.10*** (0.08***) | 0.07** (0.10***) |
| Soil moisture | 0.40*** (0.54***) | ND (ND) | 0.02*** (0.02*) | ND (ND) | ND (ND) |
| Plant richness | 0.23*** (0.35***) | ND (ND) | 0.02** (0.04***) | 0.02 (0.01*) | ND (ND) |
| ANPP | ND (ND) | NS (NS) | ND (ND) | ND (ND) | ND (ND) |
Abbreviations: ANPP, aboveground net primary productivity; MAP, mean annual precipitation; MAT, mean annual temperature; ND, not determined (removed by the varclus results); NS, not significant.
The variation (R2) of ln community similarity (1 minus Bray–Curtis distance) that is explained by the remaining variables. The partial regression coefficients (b) and associated P-values of the final model are reported from permutation test (nperm=9999) if its significance level is <0.05. *P⩽0.01, **P⩽0.001 and ***P⩽0.0001.
In brackets, we indicate the corresponding values at the 99% resolution.
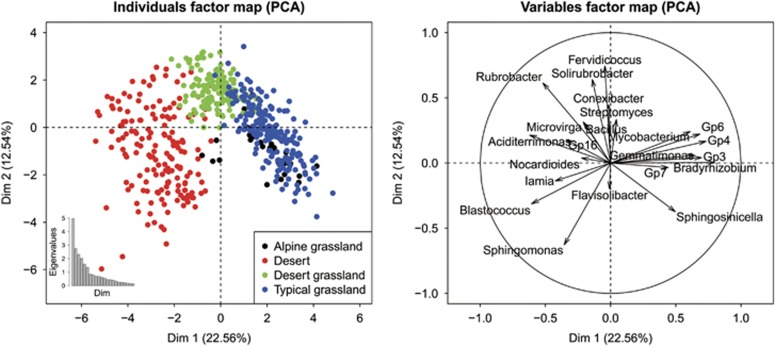
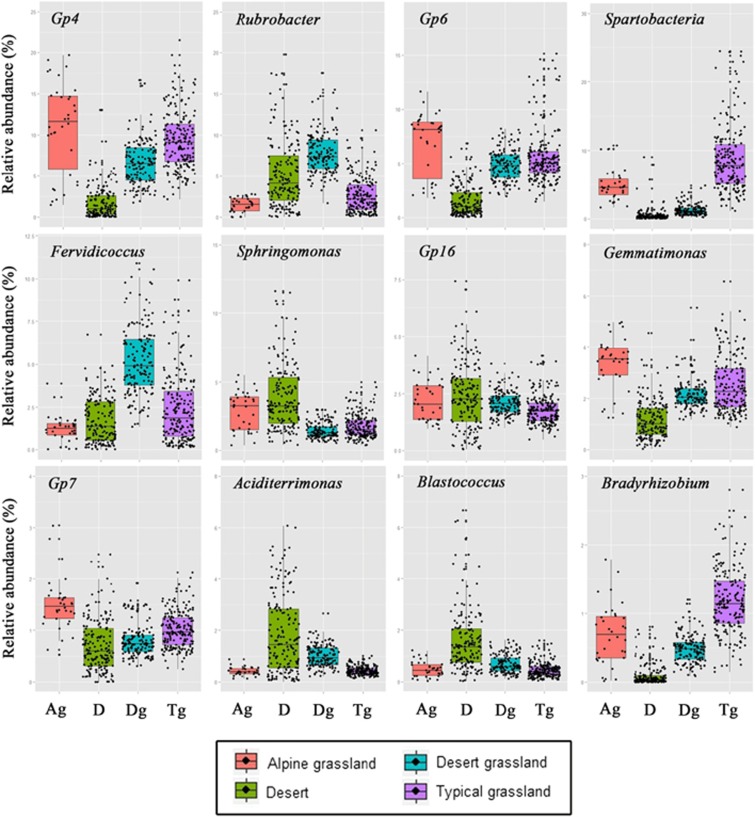
The distribution of dominant bacterial genera in the four different habitats showed that GP4, GP6, GP7, Gemmatimonas and so on were dominant in the alpine grassland, Aciditerrimonas, Blastococcus, Sphingomonas and so on were dominant in the desert, Fervidicoccus, Solirubrobacter, Rubrobacter and so on were dominant in the desert grassland, and GP4, GP6, Bradyrhizobium and so on were dominant in the typical grassland (Figure 2). We further investigated the changes of the relative abundance of dominant bacterial genera across different habitats (Figure 3), which all showed significant group differences among the four habitat types (one-way analysis of variance, P<0.001).
Figure 2.
Variables and individuals graph in principal component analysis using PCA function in FactoMineR package. We used taxonomic abundances data (OTUs defined at 97% sequence similarity) from 24 bacterial dominant genera (average abundance >50 across all soil samples) as quantitative variables, which were used to perform the PCA. Individuals were colored from the categorical variables, habitat types: black, red, green and blue plots represent samples from alpine grassland, desert, desert grassland and typical grassland, respectively. The percentage of variability explained by two dimensions was given: 22.56% for the first axis and 12.54% for the second axis. PCA, principal component analysis.
Figure 3.
Variation of relative abundance of the dominant bacterial genera in soils across different habitats. Ag, alpine grassland; D, desert; Dg, desert grassland; Tg, typical grassland. Points represent the samples in each habitat, and boxplot show quartile values for ecah taxon.
Discussion
We found robust evidence for distance–decay relationships of bacterial communities across each habitat, and most importantly, such a pattern of β-diversity was habitat specific. The data sets from those of the four dominant taxonomic groups further verified our findings (Supplementary Figure S6,Supplementary Table S5). We noted that the slopes of the distance–decay relationship (z-values) in our study ranged from 0.01 to 0.05, which is remarkably similar to those previously reported for bacteria (Horner-Devine et al., 2004). But the estimated slope z for bacteria reported here was much lower than that for macroorganisms (from ∼0.1 to 0.5), this may arise from the smaller body sizes of microbes and their higher dispersal rate relative to larger organisms (Woodcock et al., 2006). A steep distance–decay slope in the alpine grassland suggests that the turnover of bacterial communities within which may be higher than that in other habitats. Although such a difference of distance–decay slope among habitat types might result from the different spatial scales among habitats due to scale dependence of distance decay (Nekola and White, 1999), the subsets of data in other three habitats, which covered the similar spatial scales with alpine grassland provide further evidence for the existence of habitat-specific patterns, independent of spatial scales (Supplementary Table S6). It is worth pointing out that we did not observe an increase in slope z with increasing taxonomic resolution when we estimated it using a taxon resolution of 99% sequence similarity for both bacterial communities and dominant taxonomic groups (Supplementary Tables S4 and S5). This result is inconsistent with that observed for macroorganisms (Harcourt, 1999), possibly because of the differences of spatial scales or different methods adopted to define ‘species' among studies. Instead, the intercept and average compositional similarity of the distance–decay regression varied with taxonomic resolutions, decreasing with increasing taxonomic resolution for bacterial communities, suggesting the initial dissimilarity of bacterial communities would possibly increase at finer resolutions.
Given the spatial heterogeneity in different habitats, variation of distance–decay slopes may also strongly relate to the variability of spatial structure among habitats (Soininen et al., 2007). For example, the spatial configuration and context (for example, size or isolation of habitats) have important effects on the resistance to the movement of organisms, and therefore the dispersal abilities in different taxonomic groups (Tuomisto et al., 2003). Our result of linear semivariograms clearly showed spatial dependence for environmental variables such as soil and plant properties in the alpine grassland, desert grassland and typical grassland (Supplementary Figure S7, Supplementary Table S7). Meanwhile, such a linear relationship probably reflects large-scale continuous gradients over space (Ettema and Wardle, 2002), which is to some extent in accordance with our sampling schemes along this transect. All the sampling sites in each habitat were located in a relatively homogenous landscape, with few patches within these habitats. The result also suggests that spatial patterns of soil microorganisms observed in these habitats may be related to environmental heterogeneity caused primarily by soil and plant properties (Crist, 1998). In contrast, we found no significant increases of semivariance with increasing lags in the desert for both soil chemistry PC1 and plant PC1, suggesting that the variation of spatial structure at the sampling scales in this habitat is completely random (Cambardella et al., 1994). In this case, the spatially heterogeneous distributions of soil microorganisms might be mainly governed by stochastic events (for example, random changes in taxa abundance) rather than by environmental variables of spatially autocorrelated.
Our results showed that the relative importance of environmental factors versus geographic distance to community similarity also varied across different habitats (Tables 1 and 2). The environmental factors appear to have a sole effect on bacterial community similarity in the alpine grassland (ρ=−0.785 and −0.812, respectively). This finding was further supported by the result of the MRM model, in which variation in bacterial community similarity was well explained by the environmental factors such as soil moisture and plant species richness (R2=0.75 and 0.78, respectively). These results suggest that the variation in community composition in this habitat was probably driven by the spatial environmental heterogeneity, as displayed by semivariograms, which showed strong spatial autocorrelation for soil and plant properties (Supplementary Figure S7, Supplementary Table S7). Therefore, selection is likely to have dominant roles in driving bacterial β-diversity pattern in the alpine grassland. The process of selection represents the differences of relative fitness among taxa under surrounding environmental pressures (Chesson, 2000). It will differentiate microbial composition among locations, and thus produce a significant distance–decay relationship (Figure 1a). In this case, the compositional variation over space will become stronger with more environmental differences, which tends to strengthen such a relationship (that is, steepen the slope of the distance–decay curve). This is exactly coincident with the result of distance–decay relationship in this habitat, in which the estimated distance–decay slope was ~2.5 times higher than those in other habitats (Supplementary Tables S4 and S5).
Bacterial community similarity was weakly related to environment distance in the desert (ρ=−0.099 and −0.074, respectively), and was only influenced by the geographic distance according to the results of MRM model (Tables 1 and 2). One possible explanation is that we may have missed some spatially autocorrelated abiotic or biotic factors that strongly affect bacterial community composition (Martiny et al., 2011). The result of semivariograms in the desert also showed no spatial autocorrelation at sampling scales for measured environmental variables (Supplementary Figure S7, Supplementary Table S7). Actually, over such a broad-scale field survey, addressing the effect of all potentially important environmental variables is impractical, as the parameters that we measured only explained <10% of the variability in community similarity. Another possible explanation is that inherent stochastic processes such as drift and dispersal limitation may have a much greater role in driving bacterial β-diversity pattern in the desert (Legendre et al., 2009). Sole influences of geographic distance on the community composition suggest that bacterial dispersal in this area was limited probably because very few plants and animals can survive in such harsh environmental conditions, which thus reduce a chance for passive dispersal of microorganisms through their transportation. With a consequence of restricted dispersal, chance events would likely produce more compositionally similar communities between nearby locations than between those further apart. Thus, this spatial variation in composition leads to a negative relationship between community similarity and geographic distance (Morlon et al., 2008). Indeed, we found a significant distance–decay relationship for bacterial communities in this habitat (Figure 1 and Supplementary Figure S5). Furthermore, the height of the distance–decay curve in the desert was distinctly low (Figure 1,Supplementary Table S4). This result was consistent with that in the nonmetric multidimensional scaling analysis, implying great variability in the initial dissimilarity of bacterial communities.
There was no evidence that dispersal limitation strongly influenced the β-diversity of bacterial communities in the desert grassland and typical grassland owing to lack of the effect of geographic distance on the community similarity at both habitats (Tables 1 and 2). In addition, strong effects of environment on the bacterial community composition were found in most habitats, indicating that selection may have a prominent role in shaping microbial biogeographic patterns in drylands. This is consistent with many previous studies which have shown that environmental factors generally interact with taxa traits to determine which and how many taxa occur in different areas (Lawton, 1999; Hollister et al., 2010). In fact, the abundance and distribution of dominant taxonomic groups varied across habitats (Figures 2 and 3). Given the possible differences of biotic and abiotic characteristics among different habitats, the distribution of bacterial taxa structured by habitats should be associated with their ecological characteristics such as physiological capabilities or habitat preferences (Fierer et al., 2007; Pointing et al., 2009).
It is a challenge for ecologists and biogeographers to uncover and evaluate the processes that create and maintain the observed patterns in microbial diversity. It is also difficult to disentangle the relative importance of ecological processes by only analyzing distance–decay patterns. However, our study represents an important attempt to investigate microbial diversity patterns with a unique large-scale transect survey across continuous habitat types in the drylands and importantly, understand its intricate mechanism with basic processes in theoretical ecological models for macroorganisms.
Acknowledgments
We thank all the members of the Shenyang Sampling Campaign Team from the Institute of Applied Ecology, Chinese Academy of Sciences for their assistance during field sampling. We gratefully thank Jennifer Martiny for helpful suggestions and comments on the earlier version of the manuscript. We also greatly appreciate the three anonymous reviewers and the editors of this journal for giving important comments that significantly improved the quality of the paper. This work was financially supported by the Strategic Priority Research Program of the Chinese Academy of Sciences (XDB15010401), the General Program of the National Natural Science Foundation of China (31670457), the Office of the Vice President for Research at the University of Oklahoma, the Collaborative Innovation Center for Regional Environmental Quality.
Author contributions
XW, ZW, XL and XH designed the study, XW, XL performed the experiment, XW analyzed the data and XW, XL, JY, XH and JZ wrote the paper. All coauthors participated in discussions at the working group meetings and edited the manuscript.
Footnotes
Supplementary Information accompanies this paper on The ISME Journal website (http://www.nature.com/ismej)
The authors declare no conflict of interest.
Supplementary Material
References
- Ahn SJ, Costa J, Emanuel JR. (1996). PicoGreen quantitation of DNA: effective evaluation of samples pre- or post-PCR. Nucleic Acids Res 24: 2623–2625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson MJ, Crist TO, Chase JM, Vellend M, Inouye BD, Freestone AL et al. (2011). Navigating the multiple meanings of beta diversity: a roadmap for the practicing ecologist. Ecol Lett 14: 19–28. [DOI] [PubMed] [Google Scholar]
- Andrew DR, Fitak RR, Munguia-Vega A, Racolta A, Martinson VG, Dontsova K. (2012). Abiotic factors shape microbial diversity in Sonoran Desert soils. Appl Environ Microbiol 78: 7527–7537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bardgett RD, van der Putten WH. (2014). Belowground biodiversity and ecosystem functioning. Nature 515: 505–511. [DOI] [PubMed] [Google Scholar]
- Bell G. (2001). Ecology—Neutral macroecology. Science 293: 2413–2418. [DOI] [PubMed] [Google Scholar]
- Bell T. (2010). Experimental tests of the bacterial distance-decay relationship. ISME J 4: 1357–1365. [DOI] [PubMed] [Google Scholar]
- Cambardella CA, Moorman TB, Novak JM, Parkin TB, Karlen DL, Turco RF et al. (1994). Field-scale variability of soil properties in Central Iowa soils. Soil Sci Soc Am J 58: 1501–1511. [Google Scholar]
- Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Huntley J, Fierer N et al. (2012). Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J 6: 1621–1624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cermeno P, Falkowski PG. (2009). Controls on diatom biogeography in the ocean. Science 325: 1539–1541. [DOI] [PubMed] [Google Scholar]
- Chase JM, Leibold MA (2003). Ecological niches: linking classical and contemporary approaches. University of Chicago Press: Chicago, IL, USA. [Google Scholar]
- Chesson P. (2000). Mechanisms of maintenance of species diversity. Annu Rev Ecol Syst 31: 343–366. [Google Scholar]
- Cho JC, Tiedje JM. (2000). Biogeography and degree of endemicity of fluorescent Pseudomonas strains in soil. Appl Environ Microbiol 66: 5448–5456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu H, Fierer N, Lauber CL, Caporaso JG, Knight R, Grogan P. (2010). Soil bacterial diversity in the Arctic is not fundamentally different from that found in other biomes. Environ Microbiol 12: 2998–3006. [DOI] [PubMed] [Google Scholar]
- Clarke KR, Ainsworth M. (1993). A method of linking multivariate community structure to environmental variables. Mar Ecol Prog Ser 92: 205–219. [Google Scholar]
- Condit R, Pitman N, Leigh EG, Chave J, Terborgh J, Foster RB et al. (2002). Beta-diversity in tropical forest trees. Science 295: 666–669. [DOI] [PubMed] [Google Scholar]
- Crist TO. (1998). The spatial distribution of termites in shortgrass steppe—a geostatistical approach. Oecologia 114: 410–416. [DOI] [PubMed] [Google Scholar]
- Edgar RC, Haas BJ, Clemente JC, Quince C, Knight R. (2011). UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 27: 2194–2200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC. (2013). UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat Methods 10: 996–998. [DOI] [PubMed] [Google Scholar]
- Ettema CH, Wardle DA. (2002). Spatial soil ecology. Trends Ecol Evol 17: 177–183. [Google Scholar]
- Fierer N, Jackson RJ. (2006). From the Cover: the diversity and biogeography of soil bacterial communities. Proc Natl Acad Sci USA 103: 626–631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fierer N, Bradford MA, Jackson RB. (2007). Toward an ecological classification of soil bacteria. Ecology 88: 1354–1364. [DOI] [PubMed] [Google Scholar]
- Fierer N, Strickland MS, Liptzin D, Bradford MA, Cleveland CC. (2009). Global patterns in belowground communities. Ecol Lett 12: 1238–1249. [DOI] [PubMed] [Google Scholar]
- Goslee SC, Urban DL. (2007). The ecodist package for dissimilarity-based analysis of ecological data. J Stat Softw 22: 1–19. [Google Scholar]
- Green J, Bohannan BJ. (2006). Spatial scaling of microbial biodiversity. Trends Ecol Evol 21: 501–507. [DOI] [PubMed] [Google Scholar]
- Green JL, Holmes AJ, Westoby M, Oliver I, Briscoe D, Dangerfield M et al. (2004). Spatial scaling of microbial eukaryote diversity. Nature 432: 747–750. [DOI] [PubMed] [Google Scholar]
- Griffiths RI, Thomson BC, James P, Bell T, Bailey M, Whiteley AS. (2011). The bacterial biogeography of British soils. Environ Microbiol 13: 1642–1654. [DOI] [PubMed] [Google Scholar]
- Hanson CA, Fuhrman JA, Horner-Devine MC, Martiny JBH. (2012). Beyond biogeographic patterns: processes shaping the microbial landscape. Nat Rev Microbiol 10: 497–506. [DOI] [PubMed] [Google Scholar]
- Harcourt AH. (1999). Biogeographic relationships of primates on South-East Asian islands. Global Ecol Biogeogr 8: 55–61. [Google Scholar]
- Hijmans RJ, Cameron SE, Parra JL, Jones PG, Jarvis A. (2005). Very high resolution interpolated climate surfaces for global land areas. Int J Climatol 25: 1965–1978. [Google Scholar]
- Hollister EB, Engledow AS, Hammett AJM, Provin TL, Wilkinson HH, Gentry TJ. (2010). Shifts in microbial community structure along an ecological gradient of hypersaline soils and sediments. ISME J 4: 829–838. [DOI] [PubMed] [Google Scholar]
- Horner-Devine MC, Lage M, Hughes JB, Bohannan BJM. (2004). A taxa-area relationship for bacteria. Nature 432: 750–753. [DOI] [PubMed] [Google Scholar]
- Hubbell SP. (2001) The Unified Neutral Theory of Biodiversity and Biogeography (MPB-32). Princeton University Press: Princeton, NJ, USA. [Google Scholar]
- Jackson DA, Somer KM. (1989). Are probability estimates from the permutation model of Mantels test stable? Can J Zool 67: 766–769. [Google Scholar]
- Kong Y. (2011). Btrim: a fast, lightweight adapter and quality trimming program for next-generation sequencing technologies. Genomics 98: 152–153. [DOI] [PubMed] [Google Scholar]
- Kruskal JB. (1964). Nonmetric multidimensional scaling: a numerical method. Psychometrika 29: 115–129. [Google Scholar]
- Kuo S. (1996) Phosphorus. In: Sparks DL (ed). Methods of Soil Analysis. SSSA and ASA: Madison, WI, USA, pp 869–919. [Google Scholar]
- Lauber CL, Hamady M, Knight R, Fierer N. (2009). Pyrosequencing-based assessment of soil pH as a predictor of soil bacterial community structure at the continental scale. Appl Environ Microbiol 75: 5111–5120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawton JH. (1999). Are there general laws in ecology? Oikos 84: 177–192. [Google Scholar]
- Legendre P, Lapointe FJ, Casgrain P. (1994). Modeling brain evolution from behavior—a permutational regression approach. Evolution 48: 1487–1499. [DOI] [PubMed] [Google Scholar]
- Legendre P, Mi XC, Ren HB, Ma KP, Yu MJ, Sun IF et al. (2009). Partitioning beta diversity in a subtropical broad-leaved forest of China. Ecology 90: 663–674. [DOI] [PubMed] [Google Scholar]
- Lewontin RC. (1970). The units of selection. Annu Rev Ecol Syst 1: 1–18. [Google Scholar]
- Lê S, Julie J, Husson F. (2008). FactorMineR: an R package for multivariate analysis. J Stat Softw 25: 1–18. [Google Scholar]
- Li XL, Zhang JL, Gai JP, Cai XB, Christie P, Li XL. (2015). Contribution of arbuscular mycorrhizal fungi of sedges to soil aggregation along an altitudinal alpine grassland gradient on the Tibetan Plateau. Environ Microbiol 17: 2841–2857. [DOI] [PubMed] [Google Scholar]
- Lichstein JW. (2007). Multiple regression on distance matrices: a multivariate spatial analysis tool. Plant Ecol 188: 117–131. [Google Scholar]
- Lozupone CA, Knight R. (2007). Global patterns in bacterial diversity. Proc Natl Acad Sci USA 104: 11436–11440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maestre FT, Delgado-Baquerizo M, Jeffries TC, Eldridge DJ, Ochoa V, Gozalo B et al. (2015). Increasing aridity reduces soil microbial diversity and abundance in global drylands. Proc Natl Acad Sci USA 112: 15684–15689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magoc T, Salzberg SL. (2011). FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics 27: 2957–2963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martiny JBH, Bohannan BJM, Brown JH, Colwell RK, Fuhrman JA, Green JL et al. (2006). Microbial biogeography: putting microorganisms on the map. Nat Rev Microbiol 4: 102–112. [DOI] [PubMed] [Google Scholar]
- Martiny JBH, Eisen JA, Penn K, Allison SD, Horner-Devine MC. (2011). Drivers of bacterial beta-diversity depend on spatial scale. Proc Natl Acad Sci USA 108: 7850–7854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morlon H, Chuyong G, Condit R, Hubbell S, Kenfack D, Thomas D et al. (2008). A general framework for the distance-decay of similarity in ecological communities. Ecol Lett 11: 904–917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nekola JC, White PS. (1999). The distance decay of similarity in biogeography and ecology. J Biogeogr 26: 867–878. [Google Scholar]
- Nemergut DR, Costello EK, Hamady M, Lozupone C, Jiang L, Schmidt SK et al. (2011). Global patterns in the biogeography of bacterial taxa. Environ Microbiol 13: 135–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemergut DR, Schmidt SK, Fukami T, O'Neill SP, Bilinski TM, Stanish LF et al. (2013). Patterns and processes of microbial community assembly. Microbiol Mol Biol Rev 77: 342–356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ofiteru ID, Lunn M, Curtis TP, Wells GF, Criddle CS, Francis CA et al. (2010). Combined niche and neutral effects in a microbial wastewater treatment community. Proc Natl Acad Sci USA 107: 15345–15350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oksanen JF, Blanchet FG, Kindt R, Legendre P, Minchin PR, O'Hara RB et al (2013). vegan: Community Ecology Package, R package, Version 2.0-10 edn.
- Peay KG, Garbelotto M, Bruns TD. (2010). Evidence of dispersal limitation in soil microorganisms: isolation reduces species richness on mycorrhizal tree islands. Ecology 91: 3631–3640. [DOI] [PubMed] [Google Scholar]
- Pointing SB, Chan YK, Lacap DC, Lau MCY, Jurgens JA, Farrell RL. (2009). Highly specialized microbial diversity in hyper-arid polar desert. Proc Natl Acad Sci USA 106: 19964–19969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prober SM, Leff JW, Bates ST, Borer ET, Firn J, Harpole WS et al. (2015). Plant diversity predicts beta but not alpha diversity of soil microbes across grasslands worldwide. Ecol Lett 18: 85–95. [DOI] [PubMed] [Google Scholar]
- Ronca S, Ramond JB, Jones BE, Seely M, Cowan DA. (2015). Namib Desert dune/interdune transects exhibit habitat-specific edaphic bacterial communities. Front Microbiol 6: 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossi RE, Mulla DJ, Journel AG, Franz EH. (1992). Geostatistical tools for modeling and interpreting ecological spatial dependence. Ecol Monogr 62: 277–314. [Google Scholar]
- Slatkin M. (1987). Gene flow and the geographic structure of natural-populations. Science 236: 787–792. [DOI] [PubMed] [Google Scholar]
- Soininen J, McDonald R, Hillebrand H. (2007). The distance decay of similarity in ecological communities. Ecography 30: 3–12. [Google Scholar]
- Stegen JC, Lin XJ, Konopka AE, Fredrickson JK. (2012). Stochastic and deterministic assembly processes in subsurface microbial communities. ISME J 6: 1653–1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stegen JC, Lin XJ, Fredrickson JK, Chen XY, Kennedy DW, Murray CJ et al. (2013). Quantifying community assembly processes and identifying features that impose them. ISME J 7: 2069–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talbot JM, Bruns TD, Taylor JW, Smith DP, Branco S, Glassman SI et al. (2014). Endemism and functional convergence across the North American soil mycobiome. Proc Natl Acad Sci USA 111: 6341–6346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tedersoo L, Bahram M, Toots M, Diedhiou AG, Henkel TW, Kjoller R et al. (2012). Towards global patterns in the diversity and community structure of ectomycorrhizal fungi. Mol Ecol 21: 4160–4170. [DOI] [PubMed] [Google Scholar]
- Trangmar BB, Yost RS, Uehara G. (1985). Application of geostatistics to spatial studies of soil properties. Adv Agron 38: 45–94. [Google Scholar]
- Tuomisto H, Ruokolainen K, Yli-Halla M. (2003). Dispersal, environment, and floristic variation of western Amazonian forests. Science 299: 241–244. [DOI] [PubMed] [Google Scholar]
- Vellend M. (2010). Conceptual synthesis in community ecology. Q Rev Biol 85: 183–206. [DOI] [PubMed] [Google Scholar]
- Wang C, Wang X, Liu D, Wu H, Lv X, Fang Y et al. (2014). Aridity threshold in controlling ecosystem nitrogen cycling in arid and semi-arid grasslands. Nat Commun 5: 4799. [DOI] [PubMed] [Google Scholar]
- Wang J, Shen J, Wu Y, Tu C, Soininen J, Stegen JC et al. (2013). Phylogenetic beta diversity in bacterial assemblages across ecosystems: deterministic versus stochastic processes. ISME J 7: 1310–1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Garrity GM, Tiedje JM, Cole JR. (2007). Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol 73: 5261–5267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Van Nostrand JD, Deng Y, Lu X, Wang C, Zhou J et al (2015). Scale-dependent effects of climate and geographic distance on bacterial diversity patterns across northern China's grasslands. FEMS Microbiol Ecol 91: fiv133. [DOI] [PubMed] [Google Scholar]
- Woodcock S, Curtis TP, Head IM, Lunn M, Sloan WT. (2006). Taxa-area relationships for microbes: the unsampled and the unseen. Ecol Lett 9: 805–812. [DOI] [PubMed] [Google Scholar]
- Yergeau E, Newsham KK, Pearce DA, Kowalchuk GA. (2007). Patterns of bacterial diversity across a range of Antarctic terrestrial habitats. Environ Microbiol 9: 2670–2682. [DOI] [PubMed] [Google Scholar]
- Zinger L, DPH Lejon, Baptist F, Bouasria A, Aubert S, Geremia RA et al. (2011). Contrasting diversity patterns of Crenarchaeal, bacterial and fungal soil communities in an Alpine landscape. PLoS One 6: e19950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinger L, Boetius A, Ramette A. (2014). Bacterial taxa-area and distance-decay relationships in marine environments. Mol Ecol 23: 954–964. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.