Abstract
Collodaria are heterotrophic marine protists that exist either as large colonies composed of hundreds of cells or as large solitary cells. All described species so far harbour intracellular microalgae as photosymbionts. Although recent environmental diversity surveys based on molecular methods demonstrated their consistently high contribution to planktonic communities and their worldwide occurrence, our understanding of their diversity and biogeography is still very limited. Here we estimated the 18S ribosomal DNA (rDNA) gene copies per collodarian cell for solitary (5770±1960 small subunit (SSU) rDNA copies) and colonial specimens (37 474±17 799 SSU rDNA copies, for each individual cell within a colony) using single-specimen quantitative PCR. We then investigated the environmental diversity of Collodaria within the photic zone through the metabarcoding survey from the Tara Oceans expedition and found that the two collodarian families Collosphaeridae and Sphaerozoidae contributed the most to the collodarian diversity and encompassed mostly cosmopolitan taxa. Although the biogeographical patterns were homogeneous within each biogeochemical biome considered, we observed that coastal biomes were consistently less diverse than oceanic biomes and were dominated by the Sphaerozoidae while the Collosphaeridae were dominant in the open oceans. The significant relationships with six environmental variables suggest that collodarian diversity is influenced by the trophic status of oceanic provinces and increased towards more oligotrophic regions.
Introduction
Radiolarians are skeleton-bearing marine heterotrophic protists belonging to the eukaryotic phylum Retaria, which is included within the super-group Rhizaria (Nikolaev et al., 2004; Adl et al., 2005; Moreira et al., 2007). Radiolaria encompass >700 extant species, classified in five well-established orders, among which the Acantharia possess a skeleton made of strontium sulfate and the Taxopodia, Collodaria, Nassellaria and Spumellaria a skeleton made of opaline silica (Suzuki and Not, 2015). As it is currently impossible to maintain them alive in culture, most of our knowledge on radiolarians come from paleontological studies (Wever et al., 2002) and less is known about their actual diversity and ecology in modern oceans (Suzuki and Not, 2015; Caron, 2016).
Among radiolarians, Collodaria are a poorly studied group in both paleontological and biological research. Each collodarian colony is composed of hundreds to thousands of collodarian cells embedded in a gelatinous matrix while the solitary specimens are composed of a single cell. The size of collodarian colonies ranges from a few micrometres to a maximum recorded of three metres (for a colonial specimen; Swanberg and Harbison, 1979). Some collodarian species harbour silicified structures (either as needle-like spines or skeleton) while other typically lack any mineral structures (naked collodarian; Biard et al., 2015). Although our knowledge on their life cycle is still limited, the releases of small (2–10 μm) flagellate cells (reproductive swarmers) have been reported on several occasions (Anderson, 1983). Also, very little is known with respect to the feeding behaviour of Collodaria, and all species reported so far harbour numerous symbiotic microalgae (photosymbionts), mostly identified as the dinoflagellate Brandtodinium nutricula (Hollande and Enjumet, 1953; Probert et al., 2014). The taxonomical delineation of the different collodarian species is challenging due to the limited number of morphological criteria available (Brandt, 1885; Haeckel, 1887). However, an integrative morpho-molecular approach recently allowed to better understand the diversity of Collodaria and clearly distinguished 3 monophyletic families (Collosphaeridae, Collophidiidae and Sphaerozoidae) among Collodaria and 20 clades merging solitary and colonial species (Biard et al., 2015).
So far, only a few studies have described the geographical distribution of collodarian taxa throughout the world ocean and reported that Collodaria are globally distributed across a large variety of marine environments (Pavshtiks and Pan'kova, 1966; Strelkov and Reshetnyak, 1971; Swanberg, 1979). They preferentially inhabit the near surface of oligotrophic waters where they can be locally abundant, from 30 to exceptionally 20 000 colonies m−3 (Khmeleva, 1967; Caron and Swanberg, 1990; Dennett et al., 2002). Recent in situ estimation of collodarian abundances highlighted that Collodaria were the main contributors to the rhizarian biomass and represent a substantial fraction of the global carbon standing stock in the upper 100 m of the oceans (Biard et al., 2016). Though recent studies unravelled the importance of Collodaria in marine ecosystems (Biard et al., 2016; Guidi et al., 2016), our understanding of the collodarian biodiversity, its extent and distribution, is still very limited.
In the past decade, environmental molecular diversity surveys based on the 18S ribosomal RNA (rRNA) gene regularly highlighted a high diversity and a relative important contribution of radiolarians to planktonic communities in marine ecosystems (Countway et al., 2007; Not et al., 2007; Edgcomb et al., 2011) and, in particular, of the Collodaria, from photic layers to the bathypelagic regions of the oceans (de Vargas et al., 2015; Pernice et al., 2015). Similar molecular approaches applied to the analyses of protist communities collected by sediment traps also highlighted the important contribution of Collodaria in the particle export to the deep ocean (Amacher et al., 2009; Fontanez et al., 2015). Yet these studies generally lacked taxonomic resolution, as no reliable reference database for the 18S rDNA was available for detailed assignation of Collodaria, and did not consider the quantification of the collodarian rDNA copy number in each cell, a parameter largely variable among marine protists (Zhu et al., 2005; Godhe et al., 2008).
In this study, we investigated the global biogeography of the Collodaria across the variety of marine ecosystems sampled during the Tara Oceans expedition (Pesant et al., 2015). We also investigated the relationships between the distribution of collodarian diversity across a variety of biogeochemical biomes and a set of 15 environmental variables.
Materials and methods
Real-time quantitative PCR (qPCR) analysis of single-specimen collodarian
Colonial and solitary collodarian specimens were collected in the bay of Villefranche-sur-Mer (France, 43°41′10″ N, 7°18′50″ E) using a Regent Net (680-μm mesh size) or a hand net (Supplementary Figure S1) for surface and vertical (0–75 m) net tows. Each specimen was micropipette isolated, cleaned into 0.2 μm filtered seawater and imaged under a binocular microscope. DNA from each specimen was extracted using the MasterPure Complete DNA Purification Kit (Epicentre, Le Perray, France) following the manufacturer's instructions. The 18S rRNA gene was amplified and sequenced using the set of primers S32col/V9R as previously described (Biard et al., 2015). All sequences have been deposited in the GenBank database under accession numbers KY263810, KY263812–KY263827. Two colonial species, Sphaerozoum fuscum and Collozoum pelagicum, and one solitary species Procyttarium primordialis, were identified based on morphological and molecular identity according to the latest classification of Collodaria (Biard et al., 2015).
In order to avoid eukaryote contaminations from preys or microalgal photosymbionts, we designed two collodarian-specific primers, Col-961-1 F (5′-CARCTAGGGGTTGGCAAAT-3′) and Col-1075 R (5′-CACATCTTGTGGTGCCCTT-3′). Primers were designed and optimized using a reference alignment of 38 Sphaerozoidae 18S rDNA sequences and using PrimaClade (Gadberry et al., 2005) and the OligoAnalyzer 3.1 software program (Integrated DNA Technologies, Leuven, Belgium). The specificity of the newly designed primers was evaluated by PCR using genomic DNA from Acantharia, Nassellaria, Spumellaria and the collodarian photosymbiont Brandtodinium nutricula. PCR was performed as previously described (Biard et al., 2015).
We used the Col-961-1 F/Col-1075 R primer set to PCR amplify a 114-bp fragment of a Collozoum inerme (accession no. KR058247) to be used as standard for qPCR assays. The amplicon was cloned into Escherichia coli. Plasmid DNA was extracted using the NucleoSpin Plasmid (NoLid) Kit (Macherey-Nagel, Hœrdt, France) and newly constructed plasmids were linearized using NotI enzyme. Linearized plasmids were analysed by electrophoresis in 1% agarose gel and concentration measured using a Qubit Fluorometer (Fischer Scientific, Illkirch, France). The number of copies in the standard was calculated as previously described (Zhu et al., 2005). A serial 10-fold dilutions (10−1–10−6) were used to obtain standard curves.
All reactions were performed in technical duplicate with a LightCycler 480 Real-Time PCR System (Roche, Boulogne-Billancourt, France), using the LightCycler 480 SYBR Green I Master Kit (Roche). Reactions were performed by denaturing at 95 °C for 5 min, followed by 45 cycles of denaturation at 95 °C for 10 s, annealing at 60 °C for 15 s and extension at 72 °C for 15 s, respectively. Data were retrieved at the extension step. A melting curve analysis was added at the end of each run to ensure a specific amplification.
Metabarcoding sample acquisition and processing
The environmental diversity of Collodaria was explored in 653 samples (including 4 size fractions, 0.8–5, 5–20, 20–180 and 180–2 000 μm) collected at the surface and the depth of the chlorophyll maximum (DCM) at 113 sampling stations during the Tara Oceans expedition (Supplementary Figure S2; Pesant et al., 2015). Detailed descriptions of samples used in the present study and their environmental context are published in open access at PANGAEA (Biard et al., 2017). The V9-18S rDNA metabarcodes (from the Tara Oceans expedition (2009–2012; published in open access at PANGAEA (de Vargas et al., 2017a, b), and at the European Nucleotide Archive under project accession number PRJEB6610) were extracted for each sample and processed with a bioinformatics pipeline previously described (de Vargas et al., 2015). Briefly, the pipeline consisted of: (1) quality checking, (2) filtering (metabarcodes present in <2 samples and with <3 reads were removed) and, (3) clustering into operational taxonomic units (OTUs) using the Swarm software (Mahé et al., 2014). For the present analysis, we only considered OTUs with ⩾1 000 sequences present in at least two different samples. This threshold led to the removal of 3% of all sequences included in the initial data set, while it removed 99% of unique collodarian OTUs. From this, the OTUs were assigned by comparison to the Protist Ribosomal Reference (PR2) database (Guillou et al., 2013) modified with the inclusion of new collodarian reference sequences (Biard et al., 2015). For OTUs having contentious assignation (for example, several matches with different reference sequences or low hit score), they were classified as uncertain. We finally eliminated the sampling stations having <2 samples to homogenize the final data set.
To determine the most relevant identity threshold to analyse the collodarian biodiversity, we calculated pairwise identity values for the full-length 18S rRNA gene and its hypervariable regions V4 and V9 (Supplementary Figure S3), using the seqidentity function implemented in the bio3d package (Grant et al., 2006). The reference alignment used for this analysis comprised 81 collodarian 18S rDNA sequences available on the GenBank database, representing the most exhaustive collodarian data set up to date. We consequently considered for the present study only the collodarian OTUs with ⩾80% identity to a reference sequence.
Data analyses
All data analyses and statistics described below were performed using R 3.2.0. (R Core Team, 2015) and the ggplot2 (Wickham, 2009), Hmisc 3.16-0 (Harrell, 2015), and vegan 2.3-0 (Oksanen et al., 2015) packages as well as custom scripts. For each sampling station, we did not find significant differences in OTU composition between the four different size fractions nor the two depths and consequently pooled all the sequences from different samples collected in the same sampling station for statistical analyses of the OTU composition and richness.
We created an environmental data set composed of 15 environmental variables available online at PANGAEA (Biard et al., 2017), recorded at the surface and the DCM, to investigate their relationships with the collodarian biodiversity. For each sampling stations, we calculated the average value of the environmental variables recorded between the surface water and the DCM. Collodarian diversity (expressed here as the OTU richness), as well as other environmental variables, were log-transformed to normalize them, and we calculated the Pearson correlation coefficient between pairs of variables. Because of the increased risk of a type I error when several tests of significance are performed simultaneously (Legendre and Legendre, 2012), we used the sequential Bonferroni adjustment to test the significance of the correlation coefficients (Rice, 1989).
Results
Quantification of 18S rRNA gene copy number in colonial and solitary Collodaria
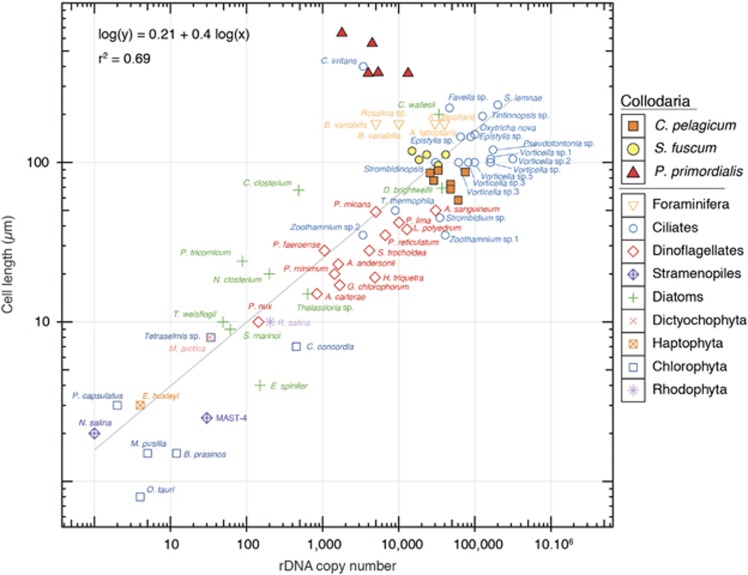
We quantified the number of 18S rRNA gene copies of two colonial and one solitary collodarian species (Table 1). Overall, when normalized by the number of cells found within a colony (that is, estimated by the number of central capsules forming the colony), colonial specimens showed about sevenfold more copies than solitary specimens, 37 474±17 799 (mean±s.e.m.) and 5770±1960, respectively. The rDNA copy number between the two different colonial species appeared lower in Sphaerozoum punctatum (26 189±4849) than in C. pelagicum (45 534±6767). We then compared the estimates for collodarian 18S rRNA gene copy numbers with previous estimates for a broad range of marine protists, extracted from the literature (Supplementary Table S1). The number of rDNA copies was significantly correlated with cell length (F=163, R2adj=0.69, P<0.001; Figure 1). Although all colonial specimens matched the general pattern, the solitary specimens (P. primordialis) display relatively low rDNA copy numbers compared with their large cell size.
Table 1. Quantification of rDNA copy numbers in colonial and solitary Collodaria with quantitative PCR.
| Organism (ID no.) | Clade affiliation | Accession no. (SSU) | Organism length (mm) | Central capsules (n)a | Central capsule size (μm)b | Copies per central capsule (n) | s.d. |
|---|---|---|---|---|---|---|---|
| Colonial collodarians | |||||||
| Sphaerozoum fuscum (1) | C2 | KY263810 | 8.25 | 225 | 118±23 | 14 933 | 667 |
| Sphaerozoum fuscum (2) | C2 | KY263812 | 5.77 | 200 | 112±13 | 23 275 | 575 |
| Sphaerozoum fuscum (3) | C2 | KY263813 | 3.67 | 114 | 96±14 | 32 895 | 1316 |
| Sphaerozoum fuscum (4) | C2 | KY263814 | 4.09 | 134 | 104±10 | 18 470 | 37 |
| Sphaerozoum fuscum (5) | C2 | KY263815 | 4.49 | 131 | 112±9 | 41 374 | 3740 |
| Mean | — | — | — | — | — | 26 189 | 4849 |
| Collozoum pelagicum (1) | C8 | KY263816 | 12.17 | 173 | 89±10 | 32 977 | 2572 |
| Collozoum pelagicum (2) | C8 | KY263817 | 8.10 | 151 | 87±12 | 74 834 | 0 |
| Collozoum pelagicum (3) | C8 | KY263818 | 3.44 | 100 | 68±9 | 47 950 | 1150 |
| Collozoum pelagicum (4) | C8 | KY263819 | 10.29 | 245 | 77±8 | 28 755 | 633 |
| Collozoum pelagicum (5) | C8 | KY263820 | 7.28 | 111 | 85±11 | 60 450 | 991 |
| Collozoum pelagicum (6) | C8 | KY263821 | 8.20 | 196 | 86±10 | 25 765 | 153 |
| Collozoum pelagicum (7) | C8 | KY263822 | 4.82 | 108 | 73±13 | 48 009 | 1806 |
| Mean | — | — | — | — | — | 45 534 | 6767 |
| Mean for all colonial collodarians | — | — | — | — | — | 37 474 | 17 799 |
| Solitary collodarians | |||||||
| Procyttarium primordialis (1) | C10 | KY263823 | 4.96 | 1 | 661 | 1790 | 90 |
| Procyttarium primordialis (2) | C10 | KY263824 | 2.95 | 1 | 374 | 5355 | 275 |
| Procyttarium primordialis (3) | C10 | KY263825 | 3.94 | 1 | 370 | 13 250 | 450 |
| Procyttarium primordialis (4) | C10 | KY263826 | 2.64 | 1 | 370 | 3980 | 80 |
| Procyttarium primordialis (5) | C10 | KY263827 | 4.69 | 1 | 570 | 4475 | 75 |
| Mean | — | — | — | — | — | 5770 | 1960 |
Abbreviation: SSU, small subunit.
Central capsules are proxies for the number of collodarian cells.
For colonial collodarian, the central capsule size is the average size (±s.d.) estimated from the measurement of 15 randomly selected central capsules.
Figure 1.
Correlation between the rDNA copy number per cell estimated by quantitative PCR and the cell length across eukaryotic marine protists including the three different collodarian species (filled circles). Detailed measurements are provided in Supplementary Table S1.
Biogeography of Collodaria in the global ocean
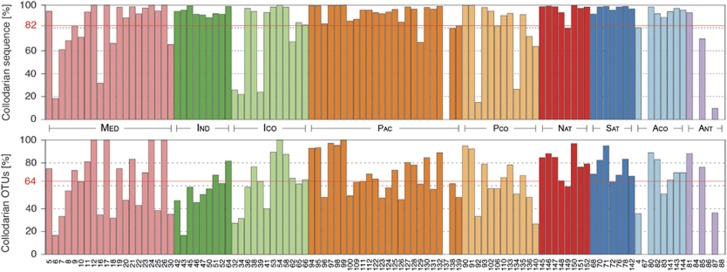
After filtering, a total of 147 078 627 quality-checked V9 rDNA gene sequences were assigned to the Rhizaria and further clustered in 517 OTUs. Out of these sequences, Collodaria accounted for a total of 133 301 730 sequences and 230 OTUs. Based on relative sequence abundances, we estimated that Collodaria contributed for an average of 82% of the total rhizarian sequences and at least 64% of the rhizarians OTUs, considering that we applied a strict cutoff excluding rare OTU in this study (Figure 2). Collodaria were the dominant lineage within the Rhizaria within all biomes, except in the Antarctic biome, where three sampling stations completely lacked Collodaria. Overall, variations of collodarian contribution to the rhizarian sequences matched very well their contribution to rhizarian OTUs, except for a few sampling stations (for example, station 43 in the Indian Ocean) where the collodarian contribution to rhizarian OTUs was lower than their contribution to rhizarian sequences.
Figure 2.
Contribution of Collodaria to the Rhizaria lineage across the Tara Oceans expedition sampling stations. Upper panel: Contribution of Collodaria to the total number of Rhizaria sequences. Lower panel: Contribution of Collodaria to the total number of Rhizaria OTUs. The red lines display the mean contributions for each data set. Contributions are geographically divided according to Longhurst's Biogeochemical Biomes by (Longhurst, 2010). A full colour version of this figure is available at the ISME journal online.
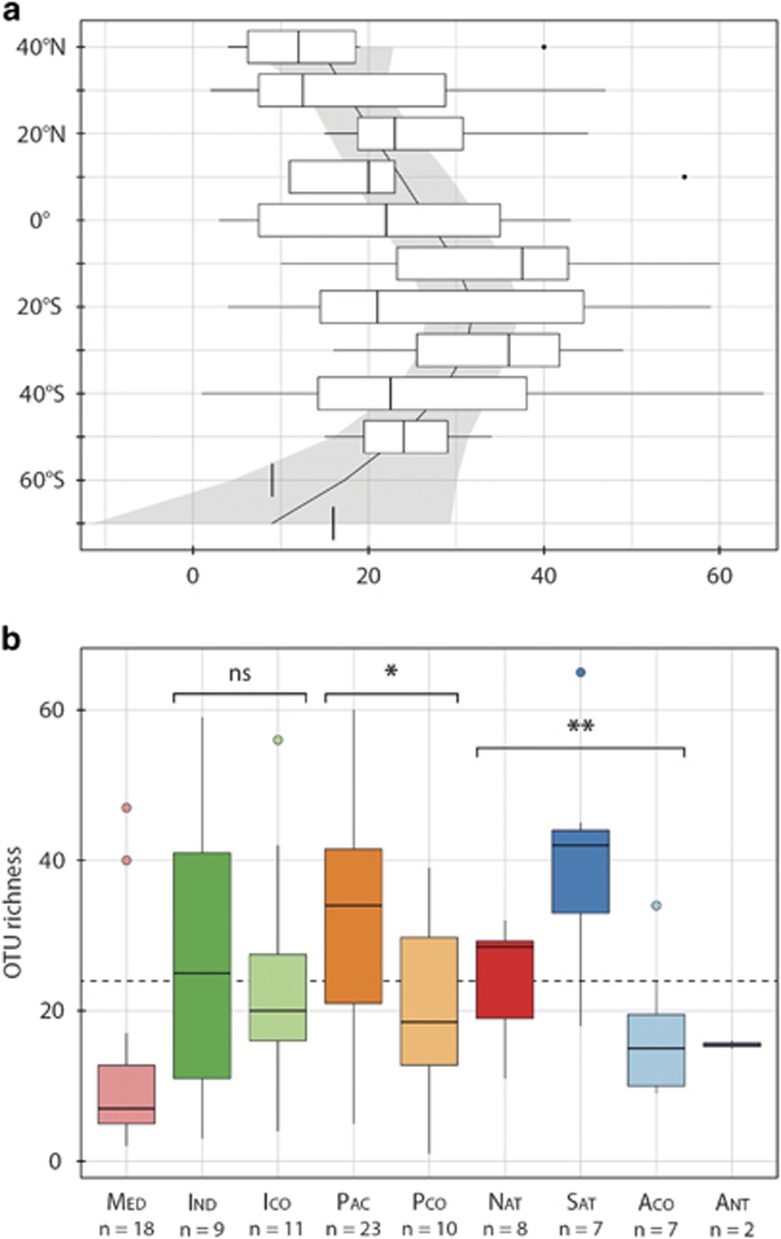
The latitudinal variations of collodarian OTU richness revealed a hump-shaped relationship centred between 10 and 30° S and exhibiting higher diversity at low latitudes (Figure 3a). Additionally, the collodarian richness greatly varied within and between the different biogeochemical biomes, reaching the highest average value (40 OTUs) in the Atlantic Trade Wind Biome (Sat; Figure 3b). The Mediterranean Sea (Med) was the least diverse biome, with an average richness of 12 OTUs. Overall, coastal biomes (that is, Med–Ico–Pco–Aco) had a significantly lower OTU richness (mean=17.26, s.d.=13.28) compared with the open-ocean biomes (that is, Ind–Pac–Nat–Sat–Ant) (mean=30.29, s.d.=15.05), t(93)=4.48, P<0.001. For each of the three oceanic basins, Indian Ocean, Pacific Ocean and Atlantic Ocean (North and South), we compared the OTU richness between coastal and oceanic biomes (Figure 3b). Although we observed a small level of statistical difference (P<0.05 and P<0.01, respectively) between the coastal and oceanic sampling stations for the Pacific and Atlantic Oceans, we did not observe any significant difference within the Indian Ocean.
Figure 3.
Variation in collodarian diversity across oceanic basins and latitudes. (a) Latitudinal distribution of collodarian OTU richness. Loess regression with polynomial fitting was computed to illustrate the latitudinal pattern. Pale colour area displays a 0.95 confidence interval around the trend. (b) Variation of OTU richness across Longhurst's Biomes. Sample size (n) for each biome is indicated along the x axis. The dashed line represents the overall mean OTU richness (S=24). P-value (NS: P>0.05, *P<0.05, **P<0.01) for Welch test show the significant decrease in OTU richness between coastal and oceanic biomes. A full colour version of this figure is available at the ISME journal online.
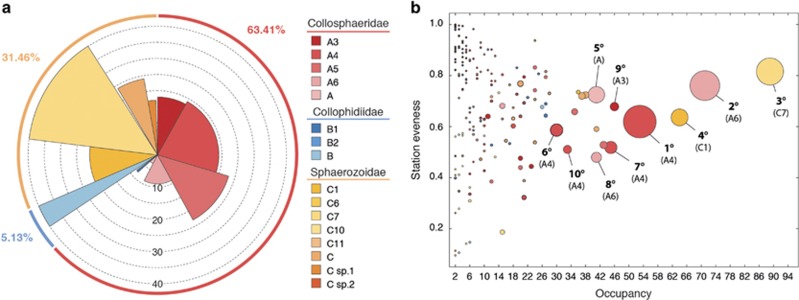
The environmental diversity of OTUs assigned to the Collodaria covered all the currently recognized collodarian families. For 89 OTUs having uncertain assignations with the procedure used for the entire data set, we identified the closest environmental and cultured match using the NCBI BLASTN tool and assigned them to the Collophidiidae (41 OTUs), the Sphaerozoidae (37) and the Collosphaeridae (11) according to the established collodarian reference database (Supplementary Table S2; Biard et al., 2015). When considering all the OTUs together, the Collosphaeridae (Clade A) was the most sequence-abundant family, accounting for an average of 63.41% of all collodarian sequences (Figure 4a). Although it was less abundant (31.46% of all sequences), the Sphaerozoidae (Clade C) was the most diverse family encompassing almost half of all collodarian OTUs (109 OTUs), while the Collosphaeridae only gathered 72 OTUs (Figure 4a). The third family, the Collophidiidae (Clade B), showed an overall low relative abundance (5.13%) and was fairly diverse (49 OTUs; Figure 4a). We further distinguished 13 clades among the three families, clades A4, B1 and C7 being the most abundant and, clades A5, B1 and C7 being the most diverse clades within each family (Figure 4a). Overall, the clade A4 was the most abundant with 21.59% of all collodarian sequences.
Figure 4.
Diversity of the Collodaria. (a) Contribution of the different clades to the total collodarian OTUs (height=number of OTUs) and the total number of collodarian sequence (radius=% contribution) (b) Occupancy and spatial evenness of each collodarian V9 rDNA OTU identified across stations sampled. A low value of spatial evenness indicates that the number of metabarcodes within an OTU is not equally distributed over the sampling stations where the OTU is present and vice versa. Each circle represents one of the collodarian V9 rDNA OTU with its size being proportional to the total read abundance. The 10 most abundant OTUs are labelled according to their rank and taxonomic clade affiliation. A full colour version of this figure is available at the ISME journal online.
We observed that the most abundant OTUs were widely spread across the sampling stations (Figure 4b) and that each of the 15 most abundant were distributed at least in 32 sampling stations among a total of 95 (that is, 34%). A few other ‘rare' OTUs occurred in a limited number of sampling locations, typically being observed in two or three sampling stations.
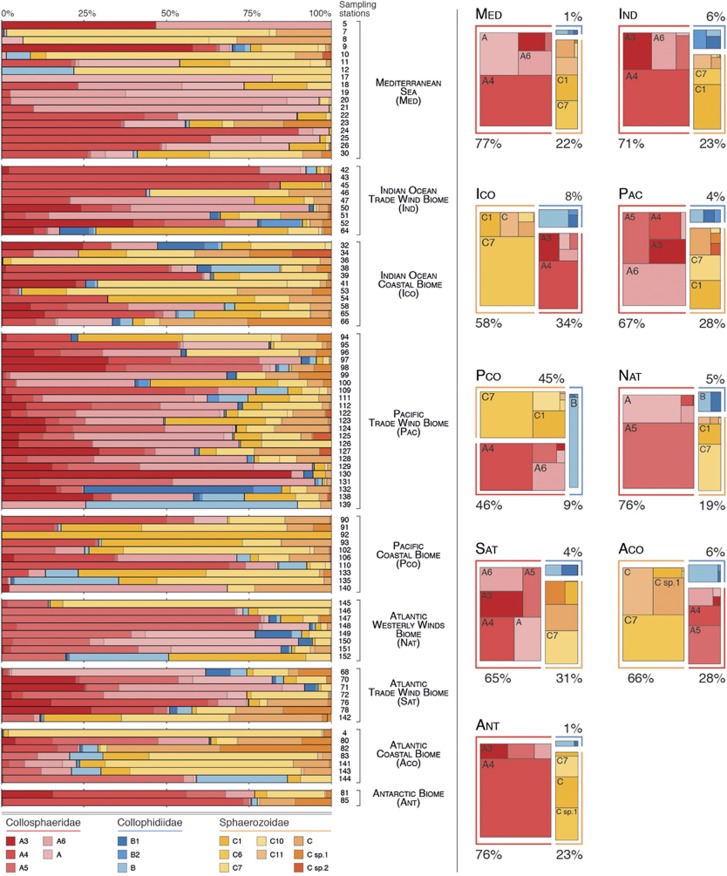
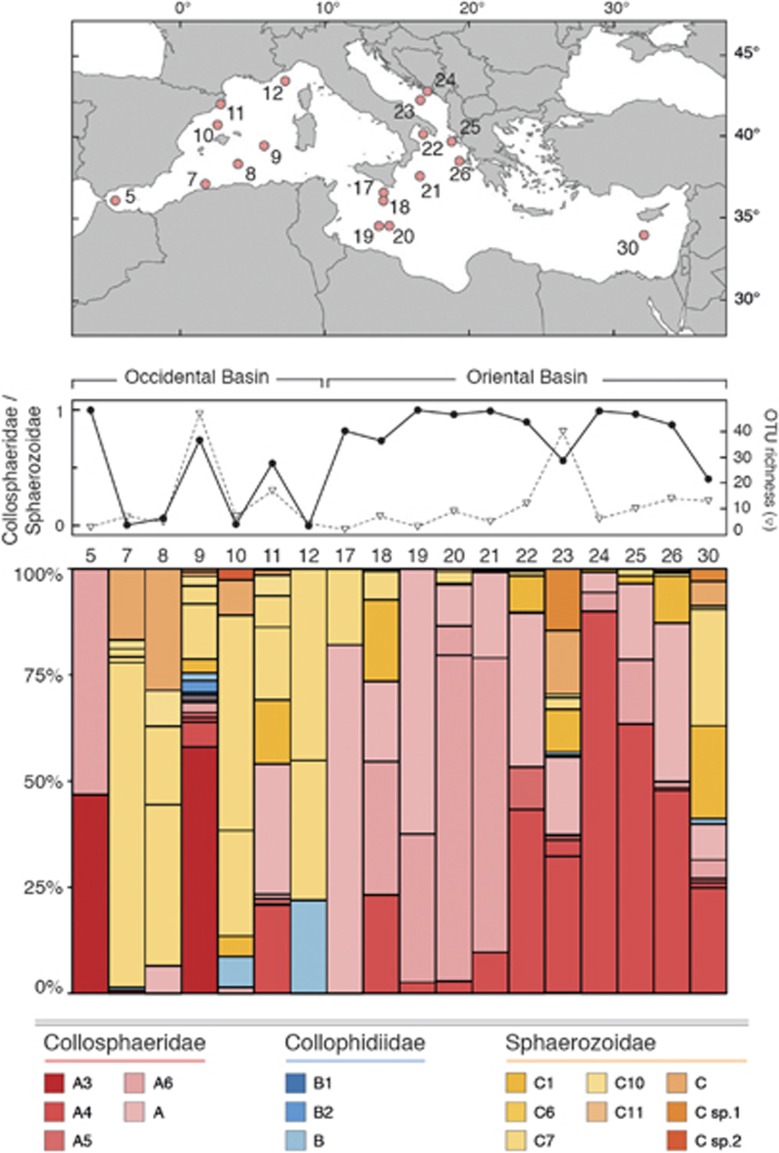
The geographical distribution of the different collodarian families and clades among nine different biogeochemical regions revealed a rather homogeneous distribution, with a few clades (one or two) prevailing at each sampling station (Figure 5). On average, the Collosphaeridae appeared dominant (65–76%) in open-ocean waters (that is, Ind, Pac, Nat, Sat and Ant), while we observed a rather clear domination (45–66%) of the Sphaerozoidae at coastal biomes (that is, Ico, Pco and Aco), with the exception of the Mediterranean Sea (Med) with an overall domination of the Collosphaeridae (77% Figure 5). Collophidiidae were rare, accounting for <10% of the collodarian sequences in all biomes, with the exception of a few sampling stations (for example, 132 and 139, both in the Pacific Trade Wind Biome) where they displayed significantly higher contribution (Figure 5). Within each biome, the distribution patterns were rather homogeneous across sampling stations except for the Mediterranean Sea where a clear shift appeared in clade composition when going from the occidental basin (sampling stations 5–12) to the oriental basin (17–30), the Sphaerozoidae being dominant in the western part of the sea (48% of collodarian sequences) and replaced by Collosphaeridae in the eastern part (84% of collodarian sequences) (Figure 6). Although the collodarian diversity was low throughout the two basins, higher diversity was observed locally in three sampling stations (for example, 9, 11 and 23). This higher collodarian diversity consistently occurred with more even contribution of the two main collodarian family (ratio Collosphaeridae/Sphaerozoidae; Figure 6).
Figure 5.
Relative abundance of rDNA metabarcodes assigned to the different collodarian families and clades defined previously (Biard et al., 2015) and across the Tara Oceans expedition sampling stations. Relative contributions are geographically divided according to Longhurst's Biomes (Longhurst, 2010). For each biome, average contributions of the different collodarian families are shown on the right panel and the five most abundant clades are labelled according to their affiliation. Detailed numerical values are shown in Supplementary Table S3.
Figure 6.
Diversity pattern of the Collodaria across the Mediterranean Sea. Upper panel: location of the sampling stations. Middle panel: (in filled black circles) variation of ratio between the relative abundances of Collosphaeridae and Sphaerozoidae (a high value of ratio meaning a domination of Collosphaeridae, a low ratio, a domination of the Sphaerozoidae) and (with inverted triangle) the collodarian diversity (expressed as the OTUs richness). Lower panel: relative abundance of rDNA metabarcodes across sampling stations.
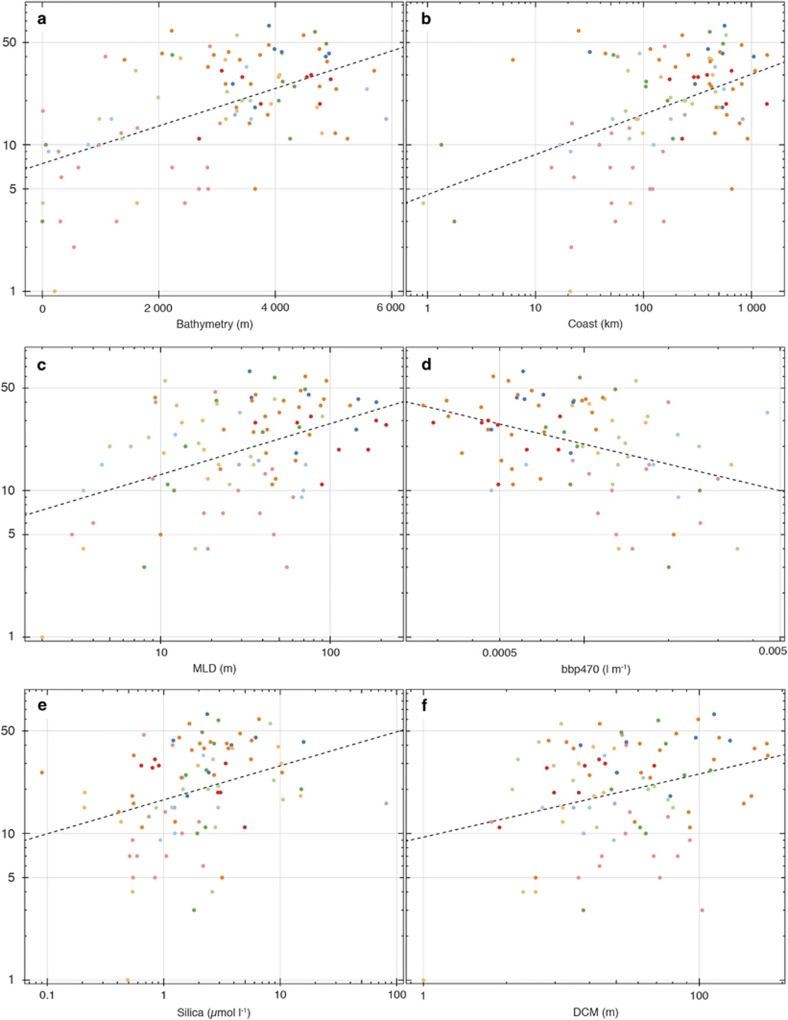
We then investigated the variation in collodarian diversity with regards to a set of environmental variables available for the samples considered (Supplementary Table S4). Whether considering the 16 clades or the 3 families illustrated in the data set, their relationships with environmental variables were not statistically significant, yet we observed increasing or decreasing trends for a number of different variables (Supplementary Figure S4). Instead of the taxonomic groups, when considering the log-transformed collodarian OTU richness, we found significant relationships with six environmental variables, the bathymetry (that is, bottom depth), the distance to the coast, the depth of the mixed layer, the backscattering coefficent of particles, the silica concentration and the DCM (Figure 7 and Supplementary Table S4). The Pearson correlation coefficient between collodarian diversity and the bathymetry of each sampling station was the highest, r(93)=0.5456, P<0.001 (Figure 7a) and explained 29% of the total variation in collodarian diversity. Although the data were rather scattered on the plots, the OTU richness showed significant increases with the following log-transformed variables: the distance to the coast, r(92)=0.4706, P<0.001 (Figure 7b), the depth of the mixed layer, r(90)=0.4360, P<0.001 (Figure 7c), the silica concentration, r(86)=0.3223, P<0.01 (Figure 7e), and the depth of the DCM, r(79)=0.2973, P<0.01 (Figure 7f). The OTU richness showed a significant decrease with the log-transformed backscattering coefficient of particles, a proxy for the particulate organic carbon, r(80)=−0.3933, P<0.001 (Figure 7d).
Figure 7.
Correlations between the collodarian diversity (log of OTU richness) and mean environmental variables across the Tara Oceans sampling stations (linear regression lines are displayed). Each sampling station (dot) is coloured according to one of the Longhurst's Biomes. (a) Relation with the bathymetry (m). (b) Relation with the log of the distance to the coast (km). (c) Relation with the log of the MLD (m). (d) Relation with the log of the backscattering coefficient of particules, 470 nm (bbp470) (l m−1). (e) Relation with the log of the silica concentration (μmol m−1). (f) Relation with the log of the DCM (m).
Discussion
Quantifying the rDNA copy numbers
Using specifically designed qPCR primers, we determined the number of 18S rRNA gene copies for 17 collodarian specimens, encompassing three different species (Table 1). The 7 times higher 18S rRNA gene copy number observed between the colonial compared with the solitary specimens could be explained by the presence of multiple nuclei, estimated from 4 to 100 nuclei per cells in colonial forms (Suzuki et al., 2009), while solitary specimens typically possess a single nucleus (Huth, 1913; Anderson, 1976; Suzuki et al., 2009). Other hypotheses, such as the difference in ploidy between the two forms, the solitary being a haploid stage of the colonial specimens (Biard et al., 2015), cannot be ruled out.
The number of rDNA copies weighted by the number of central capsules (that is, actual number of cells) showed a distribution consistent with that of other marine protists (Figure 1) and similar compared with that of the Foraminifera (5000–40 000 18S rRNA gene copies; Weber and Pawlowski, 2013), their closest relative in the overall comparison in the rDNA copy content in marine protists (Supplementary Table S1). Our results confirm and fit the previously reported correlation between size and number of rDNA copies for marine protists (Zhu et al., 2005; Godhe et al., 2008). When considering the full specimen (that is, the entire colony), colonial collodarian displayed the highest rDNA copy number ever recorded in any marine protist. Among the 12 colonial specimens we analysed here, the highest number of rDNA copies (11 300 000 rDNA copies) was recorded for a C. pelagicum colony measuring 8 × 2 mm2 (~14 mm2). Such high content is similar to the number of rDNA copies estimated in the multicellular crustacean copepod Mesocyclops edax (Supplementary Figure S5; Wyngaard et al., 1995). As colonial collodarian often display sizes larger than several centimetres, up to a few metres (Swanberg and Harbison, 1979), we estimate that a 400 × 2 mm2 cylindrical colony (that is, 2512 mm2, the size of a colony reported in Swanberg and Harbison (1979)) with a central capsule density of 9.12 capsule mm−2 (Dennett et al., 2002) could possess almost one billion rDNA copies. These high values illustrate the difficulty to quantitatively appreciate the real significance of Collodaria abundance inferred from metabarcoding surveys, as they can potentially lead to an overestimation of their importance, depending on the care with which the samples have been collected. Indeed, colonial Collodaria are easily broken upon collection with plankton net or during filtration procedures, and DNA signature along with the high rDNA copy content could propagate to smaller size fractions (for example, 0.8–5 μm), where the high diversity and contribution of Collodaria have been reported on several occasions (Not et al., 2007; Sauvadet et al., 2010; Edgcomb et al., 2011; Massana, 2011). Although we cannot rule out the impact of cell breakage in the propagation of collodarian DNA, the lack of differences in collodarian OTU composition among the four size fractions collected during the Tara Oceans expedition could also be due to the presence of different collodarian life stages (for example, flagellate reproductive swarmers; Anderson, 1983), which could represent collodarian DNA also in the small size fractions.
Refining the data set accuracy
Interpretations of data from metabarcoding diversity surveys depend on a number of methodological and analytical processes such as PCR artefacts and sequencing errors, which are known to inflate diversity estimates (Kunin et al., 2010; Lee et al., 2012), quality-control procedures of the sequences generated (Huse et al., 2010; Quince et al., 2011) and intragenomic polymorphism, recently demonstrated in several rhizarian taxa (that is, Foraminifera, Acantharia), including the Nassellaria, the closest relative to Collodaria in molecular phylogenies (Pillet et al., 2012; Decelle et al., 2014). Using interspecific variability estimates across collodarian families (Supplementary Figure S3) and a conservative abundance threshold of 1000 sequences, we defined a total of 230 OTUs. Our estimates, based on a global survey, are 26 times lower than previous estimates (that is,~6000 OTUs) for a subset of sampling stations from the same Tara Oceans expedition (de Vargas et al., 2015), yet twice more than the number of extant collodarian species described to date (that is, 95 species; Dr N Suzuki, personal communication). Additionally, further efforts will be needed to fully assess the actual collodarian diversity, such as the requirement for taxon-specific delineation of sequence abundance or the use of clustering thresholds to limit potential diversity overestimations from environmental survey analyses (Brown et al., 2015). In the case of Collodaria, qPCR estimations of 18S rRNA gene copy number for three species (two colonial and one solitary) suggested a major difference between solitary and colonial forms (Table 1), two forms that cannot currently be differentiated based on the 18S and 28S rRNA taxonomic gene marker resolution (Biard et al., 2015). In this regards, estimations of central capsule abundance from sequence abundance will be biased until we are able to separate solitary from colonial forms based on molecular signature, which could potentially be resolved using more variable marker such as mitochondrial gene marker (Leray et al., 2013).
Insight into the biodiversity and ecology of collodarian
Taxonomic assignment of the 230 OTUs based on a recent morpho-molecular reference framework for Collodaria (Biard et al., 2015) did not reveal new lineages represented by environmental sequences only at the family level. At finer taxonomic resolution, a few dominant OTUs, assigned mostly to the Collosphaeridae and Sphaerozoidae, were consistently dominant throughout the different sampling stations, suggesting that, besides Collophidiidae, Collodaria as a whole are likely to be cosmopolitan organisms (Figures 4b and 5). These observations are consistent with a previous examination of collodarian biodiversity distribution assessed from diverse locations, suggesting that collodarian species can be divided into three groups: (1) widely distributed species typically belonging to the Collosphaeridae clades A4, A5 and A6 or the Sphaerozoidae clade C7, (2) tropical distributed species such as clades A3, B1 or C9, confined to the surface waters, and (3) endemics species, absent from any existing morpho-molecular databases (Strelkov and Reshetnyak, 1971; Biard et al., 2015). The clades included in the first category were consistently found being the most abundant throughout the different sampling stations of the present data set (Figures 4b and 5). We did not find strong evidence suggesting the presence of endemic clades even though we recorded several ‘rare' OTUs being present in few sampling stations (Figure 4b). Despite our extensive geographical coverage, most of the stations sampled were located in tropical and temperate zones, with only a few coastal conditions and a limited number of polar sampling stations (Supplementary Figure S2). In this respect, we cannot exclude that our sampling might have missed a number of ecological niches where additional collodarian taxa could have been found.
The Collophidiidae family, with its unique genus Collophidium (Biard et al., 2015), was rather rarely encountered across the sampling stations investigated in our study (Figures 4a and 5), which were all restricted to the shallow photic layers of the oceans (Pesant et al., 2015). This sampling strategy might have introduced a bias against this family as assignation of environmental sequences, mostly obtained from previous clone libraries surveys, showed that the Collophidiidae were consistently the dominant collodarian family in deep-water samples (Biard et al., 2015). This observation has been confirmed by a recent study from which a substantial number of metabarcodes assigned to the genus Collophidium were extracted from deep-water samples (3000–4000 m) acquired worldwide (Pernice et al., 2015). Exploring further this deep-dwelling collodarian community might change our understanding of the ecology and diversity of Collodaria in the global ocean.
The observed decrease of collodarian richness towards high latitudes (Figure 3a) is not only consistent with previous analyses (Strelkov and Reshetnyak, 1971) but also with other latitudinal trends observed for radiolarians (Boltovskoy et al., 2010) or different marine organisms, such as Tintinnids or copepods (Dolan et al., 2006; Rombouts et al., 2009). When considering biogeochemical regions defined by Longhurst (2010), we consistently observed a higher contribution of sequences affiliated to Sphaerozoidae (species mostly lacking silicified structure; Biard et al., 2015) in low-diversity coastal biomes, whereas Collosphaeridae (skeleton-bearing taxa; Biard et al., 2015) were the dominant collodarians in the more diverse open-ocean biomes (Figures 3b and 5). Similar to our observations, numerous surface sediment records suggested that Collosphaeridae are likely to be encountered in open-ocean waters (Boltovskoy et al., 2010), but the lack of fossil record for spicule-bearing and naked Collodaria prevent accurate testing of this distribution pattern.
Links between collodarian diversity and environmental variables
Testing the relationships between the collodarian OTU richness, within each clade or family, with the 14 environmental variables available did not provide any significant linear correlations (Supplementary Figure S4), but significant relationships were found when considering the total collodarian OTU richness instead (Figure 7). The observed patterns of increasing or decreasing diversity suggest that all three families (with the exception of the Collophidiidae largely under-represented) displayed the same variation for the variables considered and that none of the significant variability affected preferentially one of the families.
Overall, both the bathymetry and the distance from the coast of each sampling station revealed the highest correlation with OTU richness (Figures 7a and b), indicating that the collodarian diversity is likely to increase towards more oceanic conditions, as it was previously suggested (Figure 3b; Swanberg, 1979). We also found a positive correlation between the water column stratification (estimated via the mixed-layer depth), suggesting that the collodarian diversity increase with a deeper stratification of the ocean (Figure 7c). Although the extent of the mixed layer depth (MLD) is variable over seasons, with deeper stratification in winter and a shallow MLD in summer, its extent in tropical regions is rather stable over time, with a deep MLD (de Boyer Montégut et al., 2004). In the present study, most of the samples were collected in tropical regions (Supplementary Figure S2) with deep MLD suggesting an increasing diversity toward tropical and oligotrophic regions where Collodaria are known to be more abundant (Dennett et al., 2002; Biard et al., 2016). Such a pattern was supported by the significant increase of diversity with decreasing particulate organic carbon and increasing DCM. We found no significant correlation between OTU richness and sea temperature (Supplementary Table S4), whereas it has been suggested earlier that collodarian diversity may increase towards warmer waters (Strelkov and Reshetnyak, 1971). However, our sampling coverage provided only a narrow range of temperature (interquartile range=7 °C, between 18 and 25 °C) and thus could have limited the significance of temperature in explaining the diversity patterns observed.
Although the silica concentrations were rather low across the sampling stations and typical of tropical waters, the overall positive correlation between silica concentration and collodarian diversity (Figure 7e) was unexpected as most Collodaria have reduced silicified structures (spines or skeleton) or even lack these structures (Biard et al., 2015). Additionally, while we could have expected a domination of naked Sphaerozoidae (for example, Collozoum spp.) in low silica concentration waters, the analysis of OTU composition did not reveal such a domination but rather suggested that the diversity of the three collodarian families increased with increasing silica concentration (Supplementary Figure S4). Although the decrease of radiolarian skeleton weight with decreasing silica availability in surface waters has been previously suggested through analysis of marine sediments from the Cenozoic (that is, <60 million years; Lazarus et al., 2009), the present pattern could, however, indicate that, with increasing silica concentration, a large morphological diversity, in particular, silicified collodarian (that is, spicules and skeleton-bearing species), could be encountered.
The cosmopolitan distribution of collodarian families and clades observed in the present study provide insights into the ecology of Collodaria and suggested that these protists have adapted to various environmental conditions. Although we recovered six significant relationships with different environmental parameters, other processes might be considered to fully understand the distribution of collodarian diversity, such as the influence of temporal scales, as it has been proved to have potential impacts on plankton diversity (Rombouts et al., 2010; Egge et al., 2015), or including biotic variables such as predation (Shurin, 2001) or photosymbiosis (Biard et al., 2016), known to affect the distribution of plankton communities.
Acknowledgments
This work was supported by the PhD fellowship Emergence-UPMC ‘DESIR' from the Université Pierre et Marie Curie. We are grateful to the Biogenouest Genomics and Genomer plateforme core facility for its technical support and to the Dr John Dolan (Villefranche-sur-Mer) for providing us sampling facilities. We would like to thank Dr Ian Probert for insightful comments and English proof reading. We thank the French Governement 'Investissement d'Avenir' program OCEANOMICS (ANR-11-BTBR-0008) and the ANR project POSEIDON (ANR-09-BLAN-0348). This article is contribution number 50 of Tara Oceans. For the Tara Oceans expedition, we thank the commitment of the CNRS (in particular, Groupement de Recherche GDR3280), European Molecular Biology Laboratory (EMBL), Genoscope/CEA, VIB, Stazione Zoologica Anton Dohrn, UNIMIB, Fund for Scientific Research—Flanders, Rega Institute, KU Leuven, The French Ministry of Research. We also thank the support and commitment of Agnès b. and Etienne Bourgois, the Veolia Environment Foundation, Région Bretagne, Lorient Agglomération, World Courier, Illumina, the EDF Foundation, FRB, the Prince Albert II de Monaco Foundation, the Tara schooner and its captains and crew. We are also grateful to the French Ministry of Foreign Affairs for supporting the expedition and to the countries who graciously granted sampling permissions. Tara Oceans would not exist without continuous support from 23 institutes (http://oceans.taraexpeditions.org). We further declare that all data reported herein are fully and freely available from the date of publication, with no restrictions, and that all of the samples, analyses, publications, and ownership of data are free from legal entanglement or restriction of any sort by the various nations whose waters the Tara Oceans expedition sampled in.
Footnotes
Supplementary Information accompanies this paper on The ISME Journal website (http://www.nature.com/ismej)
The authors declare no conflict of interest.
Supplementary Material
References
- Adl SM, Simpson AGB, Farmer MA, Andersen RA, Anderson OR, Barta JR et al. (2005). The new higher level classification of eukaryotes with emphasis on the taxonomy of protists. J Eukaryot Microbiol 52: 399–451. [DOI] [PubMed] [Google Scholar]
- Amacher J, Neuer S, Anderson I, Massana R. (2009). Molecular approach to determine contributions of the protist community to particle flux. Deep-Sea Res Pt I 56: 2206–2215. [Google Scholar]
- Anderson OR. (1976). A cytoplasmic fine-structure study of two spumellarian Radiolaria and their symbionts. Mar Micropaleontol 1: 81–99. [Google Scholar]
- Anderson OR. (1983) Radiolaria. Springer-Verlag: New York, USA, pp 355. [Google Scholar]
- Biard T, Bigeard E, Audic S, Poulain J, Gutierrez-Rodriguez A, Stemmann L et al. (2017). Abundances of metabarcodes and OTUs, and contextual data of samples selected for a study of the biogeography and diversity of Collodaria (Radiolaria) in the global ocean. doi:10.1594/PANGAEA.872436. [DOI] [PMC free article] [PubMed]
- Biard T, Pillet L, Decelle J, Poirier C, Suzuki N, Not F. (2015). Towards an integrative morpho-molecular classification of the collodaria (Polycystinea, Radiolaria). Protist 166: 374–388. [DOI] [PubMed] [Google Scholar]
- Biard T, Stemmann L, Picheral M, Mayot N, Vandromme P, Hauss H et al. (2016). In situ imaging reveals the biomass of giant protists in the global ocean. Nature 532: 504–507. [DOI] [PubMed] [Google Scholar]
- Boltovskoy D, Kling SA, Takahashi K, Bjørklund K. (2010). World atlas of distribution of recent polycystina (Radiolaria). Palaeontol Electron 13: 230. [Google Scholar]
- de Boyer Montégut C, Madec G, Fischer AS, Lazar A, Iudicone D. (2004). Mixed layer depth over the global ocean: an examination of profile data and a profile-based climatology. J Geophys Res Oceans 109: C12003. [Google Scholar]
- Brandt KAH. (1885) Die Koloniebildenden Radiolarien (Sphaerozoeen) des Golfes von Neapel und der Angrenzenden Meeresabschnitte. R Friedlander & Sohn: Berlin, Germany. [Google Scholar]
- Brown EA, Chain FJJ, Crease TJ, MacIsaac HJ, Cristescu ME. (2015). Divergence thresholds and divergent biodiversity estimates: can metabarcoding reliably describe zooplankton communities? Ecol Evol 5: 2234–2251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caron DA. (2016). The rise of Rhizaria. Nature 532: 444–445. [DOI] [PubMed] [Google Scholar]
- Caron DA, Swanberg NR. (1990). The ecology of planktonic sarcodines. Rev Aquat Sci 3: 147–180. [Google Scholar]
- Countway PD, Gast RJ, Dennett MR, Savai P, Rose JM, Caron DA. (2007). Distinct protistan assemblages characterize the euphotic zone and deep sea (2500 m) of the western North Atlantic (Sargasso Sea and Gulf Stream). Environ Microbiol 9: 1219–1232. [DOI] [PubMed] [Google Scholar]
- de Vargas C, Audic S, Tara Oceans Consortium C, Tara Oceans Expedition P. (2017. a). Total V9 rDNA information organized at the metabarcode level for the Tara Oceans Expedition (2009-2012); doi:10.1594/PANGAEA.873277.
- de Vargas C, Audic S, Tara Oceans Consortium C, Tara Oceans Expedition P. (2017. b). Total V9 rDNA information organized at the OTU level for the Tara Oceans Expedition (2009-2012); doi:10.1594/PANGAEA.873275.
- Decelle J, Romac S, Sasaki E, Not F, Mahé F. (2014). Intracellular diversity of the V4 and V9 regions of the 18S rRNA in marine protists (Radiolarians) assessed by high-throughput sequencing. PLoS One 9: e104297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennett MR, Caron DA, Michaels AF, Gallager SM, Davis CS. (2002). Video plankton recorder reveals high abundances of colonial Radiolaria in surface waters of the central North Pacific. J Plankton Res 24: 797–805. [Google Scholar]
- Dolan JR, Lemée R, Gasparini S, Mousseau L, Heyndrickx C. (2006). Probing diversity in the plankton: using patterns in tintinnids (planktonic marine ciliates) to identify mechanisms. Hydrobiologia 555: 143–157. [Google Scholar]
- Edgcomb V, Orsi W, Bunge J, Jeon S, Christen R, Leslin C et al. (2011). Protistan microbial observatory in the Cariaco Basin, Caribbean. I. Pyrosequencing vs Sanger insights into species richness. ISME J 5: 1344–1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egge ES, Johannessen TV, Andersen T, Eikrem W, Bittner L, Larsen A et al. (2015). Seasonal diversity and dynamics of haptophytes in the Skagerrak, Norway, explored by high-throughput sequencing. Mol Ecol 24: 3026–3042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontanez KM, Eppley JM, Samo TJ, Karl DM, DeLong EF. (2015). Microbial community structure and function on sinking particles in the North Pacific Subtropical Gyre. Aquat Microbiol 6: 469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gadberry MD, Malcomber ST, Doust AN, Kellogg EA. (2005). Primaclade - a flexible tool to find conserved PCR primers across multiple species. Bioinformatics 21: 1263–1264. [DOI] [PubMed] [Google Scholar]
- Godhe A, Asplund ME, Härnström K, Saravanan V, Tyagi A, Karunasagar I. (2008). Quantification of diatom and dinoflagellate biomasses in coastal marine seawater samples by real-time PCR. Appl Environ Microbiol 74: 7174–7182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant BJ, Rodrigues APC, ElSawy KM, McCammon JA, Caves LSD. (2006). Bio3d: an R package for the comparative analysis of protein structures. Bioinformatics 22: 2695–2696. [DOI] [PubMed] [Google Scholar]
- Guidi L, Chaffron S, Bittner L, Eveillard D, Larhlimi A, Roux S et al. (2016). Plankton networks driving carbon export in the oligotrophic ocean. Nature 532: 465–470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillou L, Bachar D, Audic S, Bass D, Berney C, Bittner L et al. (2013). The Protist Ribosomal Reference database (PR2): a catalog of unicellular eukaryote small sub-unit rRNA sequences with curated taxonomy. Nucleic Acids Res 41: D597–D604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haeckel E. (1887). Report on the Radiolaria collected by H. M.S. Challenger during the years 1873–1876. Zoology 18: 1–1803. [Google Scholar]
- Harrell FE. (2015) Hmisc: Harrell Miscellaneous. R package version 3.15-0. Available at: http://CRAN.R-project.org/package=Hmisc.
- Hollande A, Enjumet M. (1953). Contribution à l'étude biologique des Sphaerocollides (Radiolaires Collodaires et Radiolaires polycyttaires) et de leurs parasites. Ann Sci Nat Zool 15: 99–183. [Google Scholar]
- Huse S, Welch D, Morrison H, Sogin M. (2010). Ironing out the wrinkles in the rare biosphere through improved OTU clustering. Environ Microbiol 12: 1889–1898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huth W. (1913). Zur entwicklungsgeschichte der thalassicollen. Arch Für Protistenkd 30: 1–124. [Google Scholar]
- Khmeleva N. (1967). Role of radiolarians in the estimation of the primary production in the Red Sea and the Gulf of Aden. Dokl Akad Nauk SSSR 172: 1430–1433. [Google Scholar]
- Kunin V, Engelbrektson A, Ochman H, Hugenholtz P. (2010). Wrinkles in the rare biosphere: pyrosequencing errors can lead to artificial inflation of diversity estimates. Environ Microbiol 12: 118–123. [DOI] [PubMed] [Google Scholar]
- Lazarus D, Kotrc B, Wulf G, Schmidt D. (2009). Radiolarians decreased silicification as an evolutionary response to reduced Cenozoic ocean silica availability. Proc Natl Acad Sci USA 106: 9333–9338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee CK, Herbold CW, Polson SW, Wommack KE, Williamson SJ, McDonald IR et al. (2012). Groundtruthing next-gen sequencing for microbial ecology–biases and errors in community structure estimates from PCR amplicon pyrosequencing. PLoS One 7: e44224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legendre P, Legendre L. (2012) Numerical Ecology 3rd English edn Elsevier: Amsterdam, The Netherlands. [Google Scholar]
- Leray M, Yang JY, Meyer CP, Mills SC, Agudelo N, Ranwez V et al. (2013). A new versatile primer set targeting a short fragment of the mitochondrial COI region for metabarcoding metazoan diversity: application for characterizing coral reef fish gut contents. Front Zool 10: 34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longhurst AR. (2010) Ecological Geography of the Sea. Academic Press: London, UK. [Google Scholar]
- Mahé F, Rognes T, Quince C, de Vargas C, Dunthorn M. (2014). Swarm: robust and fast clustering method for amplicon-based studies. PeerJ 2: e593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massana R. (2011). Eukaryotic picoplankton in surface oceans. Annu Rev Microbiol 65: 91–110. [DOI] [PubMed] [Google Scholar]
- Moreira D, von der Heyden S, Bass D, López-García P, Chao E, Cavalier-Smith T. (2007). Global eukaryote phylogeny: combined small- and large-subunit ribosomal DNA trees support monophyly of Rhizaria, Retaria and Excavata. Mol Phylogenet Evol 44: 255–266. [DOI] [PubMed] [Google Scholar]
- Nikolaev SI, Berney C, Fahrni JF, Bolivar I, Polet S, Mylnikov AP et al. (2004). The twilight of Heliozoa and rise of Rhizaria, an emerging supergroup of amoeboid eukaryotes. Proc Natl Acad Sci USA 101: 8066–8071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Not F, Gausling R, Azam F, Heidelberg JF, Worden AZ. (2007). Vertical distribution of picoeukaryotic diversity in the Sargasso Sea. Environ Microbiol 9: 1233–1252. [DOI] [PubMed] [Google Scholar]
- Oksanen J, Blanchet GF, Kindt R, Legendre P, Minchin PR, O'Hara RB et al. (2015). Vegan: Community Ecology Package. R package vegan, vers.2.2-1.
- Pavshtiks EA, Pan'kova LA. (1966). O pitanii pelagicheskoi molodi morskikh okunei roda Sebastes planktonon v Devisovom prolive. Mater Nauchnoi Sesii Polyarnago Nauchnoissledovatelskogo Instituta Morskogo Rybn Khozyaistva Okeanogr 6: 1–87. [Google Scholar]
- Pernice MC, Giner CR, Logares R, Perera-Bel J, Acinas SG, Duarte CM et al. (2015). Large variability of bathypelagic microbial eukaryotic communities across the world's oceans. ISME J 10: 945–958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pesant S, Not F, Picheral M, Kandels-Lewis S, Bescot NL, Gorsky G et al. (2015). Open science resources for the discovery and analysis of Tara Oceans data. Sci Data 2: 150023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pillet L, Fontaine D, Pawlowski J. (2012). Intra-genomic ribosomal RNA polymorphism and morphological variation in Elphidium macellum suggests inter-specific hybridization in Foraminifera. PLoS One 7: e32373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Probert I, Siano R, Poirier C, Decelle J, Biard T, Tuji A et al. (2014). Brandtodinium gen. nov. and B. nutricula comb. Nov. (Dinophyceae), a dinoflagellate commonly found in symbiosis with polycystine radiolarians. J Phycol 50: 388–399. [DOI] [PubMed] [Google Scholar]
- Quince C, Lanzen A, Davenport RJ, Turnbaugh PJ. (2011). Removing noise from pyrosequenced amplicons. BMC Bioinformatics 12: 38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team.. (2015). R: A Language and Environment for Statistical Computing http://www.R-project.org/.
- Rice WR. (1989). Analyzing tables of statistical tests. Evolution 43: 223–225. [DOI] [PubMed] [Google Scholar]
- Rombouts I, Beaugrand G, Ibañez F, Gasparini S, Chiba S, Legendre L. (2010). A multivariate approach to large-scale variation in marine planktonic copepod diversity and its environmental correlates. Limnol Oceanogr 55: 2219–2229. [Google Scholar]
- Rombouts I, Beaugrand G, Ibanez F, Gasparini S, Chiba S, Legendre L. (2009). Global latitudinal variations in marine copepod diversity and environmental factors. Proc R Soc B Biol Sci 276: 3053–3062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauvadet A-L, Gobet A, Guillou L. (2010). Comparative analysis between protist communities from the deep-sea pelagic ecosystem and specific deep hydrothermal habitats: protist associated with hydrothermal environments. Environ Microbiol 12: 2946–2964. [DOI] [PubMed] [Google Scholar]
- Shurin JB. (2001). Interactive effects of predation and dispersal on zooplankton communities. Ecology 82: 3404–3416. [Google Scholar]
- Strelkov A, Reshetnyak V. (1971). Colonial spumellarian radiolarians of the world ocean. Acad Sci USSR Explor Fauna Seas 9: 295–418. [Google Scholar]
- Suzuki N, Not F. (2015) Biology and ecology of Radiolaria. In: Ohtsuka S, Suzaki T, Horiguchi T, Suzuki N, Not F (eds). Marine Protists. Springer: Japan, pp 179–222. [Google Scholar]
- Suzuki N, Ogane K, Aita Y, Kato M, Sakai S, Kurihara T et al. (2009). Distribution patterns of the radiolarian nuclei and symbionts using DAPI-fluorescence. Bull Natl Mus Nat Sci Ser B 35: 169–182. [Google Scholar]
- Swanberg NR. (1979) The Ecology of Colonial Radiolarians: Their Colony Morphology, Trophic Interactions and Associations, Behavior, Distribution, and the Photosynthesis of their Symbionts. Massachusetts Institute of Technology and Woods Hole Oceanographic Institution. [Google Scholar]
- Swanberg NR, Harbison GR. (1979). The ecology of Collozoum longiforme, sp. nov., a new colonial radiolarian from the equatorial Atlantic Ocean. Deep Sea Res Part Oceanogr Res Pap 27: 715–732. [Google Scholar]
- de Vargas C, Audic S, Henry N, Decelle J, Mahé F, Logares R et al. (2015). Eukaryotic plankton diversity in the sunlit ocean. Science 348: 1261605. [DOI] [PubMed] [Google Scholar]
- Weber AA-T, Pawlowski J. (2013). Can abundance of protists be inferred from sequence data: a case study of Foraminifera. PLoS One 8: e56739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wever PD, Dumitrica P, Caulet JP, Nigrini C, Caridroit M. (2002) Radiolarians in the Sedimentary Record. CRC Press. [Google Scholar]
- Wickham H. (2009) ggplot2: Elegant Graphics for Data Analysis. Springer Science & Business Media. [Google Scholar]
- Wyngaard GA, McLaren IA, White MM, Sévigny J-M. (1995). Unusually high numbers of ribosomal RNA genes in copepods (Arthropoda: Crustacea) and their relationship to genome size. Genome 38: 97–104. [DOI] [PubMed] [Google Scholar]
- Zhu F, Massana R, Not F, Marie D, Vaulot D. (2005). Mapping of picoeucaryotes in marine ecosystems with quantitative PCR of the 18S rRNA gene. FEMS Microbiol Ecol 52: 79–92. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.