Abstract
Hot water plumbing is an important conduit of microbes into the indoor environment and can increase risk of opportunistic pathogens (for example, Legionella pneumophila). We examined the combined effects of water heater temperature (39, 42, 48, 51 and 58 °C), pipe orientation (upward/downward), and water use frequency (21, 3 and 1 flush per week) on the microbial composition at the tap using a pilot-scale pipe rig. 16S rRNA gene amplicon sequencing indicated that bulk water and corresponding biofilm typically had distinct taxonomic compositions (R2Adonis=0.246, PAdonis=0.001), yet similar predicted functions based on PICRUSt analysis (R2Adonis=0.087, PAdonis=0.001). Although a prior study had identified 51 °C under low water use frequency to enrich Legionella at the tap, here we reveal that 51 °C is also a threshold above which there are marked effects of the combined influences of temperature, pipe orientation, and use frequency on taxonomic and functional composition. A positive association was noted between relative abundances of Legionella and mitochondrial DNA of Vermamoeba, a genus of amoebae that can enhance virulence and facilitate replication of some pathogens. This study takes a step towards intentional control of the plumbing microbiome and highlights the importance of microbial ecology in governing pathogen proliferation.
Introduction
There is growing interest in the microbiome inhabiting the built environment and its potential implications to human health (Hanski et al., 2012; Kelley and Gilbert, 2013; Oberauner et al., 2013). Building plumbing systems deliver water, along with its resident microbes, into homes and other buildings. Most attention has been directed towards opportunistic pathogens (OPs), such as Legionella pneumophila, which are particularly challenging to control as they are native to the fresh water environment. Collectively, OPs residing in building plumbing have become the top contributor of waterborne disease in the U.S. and other developed countries (Hilborn et al., 2013; Ashbolt, 2015; Beer et al., 2015), imposing an economic burden over $1 billion per year in the United States alone (Falkinham et al., 2015). Thus, understanding the composition of the microbiome at the tap, and its contribution of pathogens and general microbiota to built environment and human microbiomes is of great importance.
Hot water plumbing systems are of special interest as a typical route of OPs exposure is inhalation of aerosols and/or direct contact during showering (Feazel et al., 2009). Elevating the temperature of hot water storage tanks is known to be an effective measure to control L. pneumophila (Darelid et al., 2002; Buse and Ashbolt, 2011; ANSI/ASHRAE, 2015; Falkinham et al., 2015), and 60 °C has been demarcated as a regulatory threshold (VHA Directive 1061; Department of Veterans Affairs, 2014). However, motives to reduce energy usage and avoid scalding are in conflict with higher temperature settings (Lévesque et al., 2004; Rhoads et al., 2016a) and long-term effectiveness of thermal eradication has been questioned (Farhat et al. 2012). In contrast to a number of studies directly focusing on Legionella in hot water systems (Zietz et al., 2001; Borella et al., 2004; Zacheus and Martikainen, 2004; Mathys et al., 2008; Bargellini et al., 2011), very few have more broadly examined the microbiome as a whole. Prior studies included field surveys on microbial communities collected from shower head/hose biofilms (Feazel et al., 2009; Proctor et al., 2016) and bulk water samples from a hospital hot water system, where the effectiveness of monochloramine disinfection was examined (Baron et al., 2014). However, a systematic examination of the combined effects of water heater temperature and system design/operational conditions on the actual microbiome composition at the tap has been lacking.
Although hot water storage tanks may remain at elevated temperature settings, the reality at the distal tap is that the biofilm experiences only brief heat shock. Water use frequency at a given tap is thus of particular interest, as it dictates the frequency of heat shock as well as the length of time for the biofilm microbiota to recover from the perturbation, which could be further influenced by pipe orientation and associated convective mixing conditions (Rhoads et al., 2016b). In the bulk water phase, in-building stagnation was found to have a marked effect on the microbial composition of cold water lines (Lautenschlager et al., 2010; Ji et al., 2015). Moreover, warmer indoor temperature together with overnight stagnation could increase microbial metabolic activity level and alter the functional profile (Zhang et al., 2015). We thus hypothesized that different water use frequencies would alter the microbiome composition of both the biofilm and bulk water.
The relationship between the microbial composition of the biofilm and bulk water is also widely debated and likely to be influenced by operating conditions. Biofilm is often perceived to be a reservoir for OPs, where protist hosts can facilitate their replication and, along with the biofilm matrix, protect them from unfavorable conditions such as chlorine/monochloramine disinfection (Pryor et al., 2004; Cooper and Hanlon, 2010) and low nutrients (Lau and Ashbolt, 2009). However, microbes entrained in the biofilm must be released into the bulk water for relevance to human exposure, which can occur long after the initial entrapment/attachment of the microbe to the biofilm (Howe et al., 2002). Interestingly, OPs in the two phases do not necessarily respond the same way under a given condition. For example, at room temperature, stagnation predisposed bulk water to Legionella proliferation (Wang et al., 2012), but resulted in lowest Legionella concentrations in biofilm relative to those under turbulent and laminar flow conditions (Liu et al., 2006). Such questions call for a mechanistic understanding of the factors shaping the biofilm and bulk water microbiomes of domestic hot water systems.
To overcome complexities encountered in field studies, while maintaining relevance to real-world conditions, we constructed two identical recirculating hot water plumbing rigs with distal taps operated in triplicate under three water use scenarios (21, 3 and 1 flush per week) and two orientations (upward and downward flow). Water heater temperature was elevated in a step-wise fashion from 39 to 58 °C, while an identical control rig remained at 39 °C throughout the 15-month study period. Illumina amplicon sequencing of 16S rRNA genes was applied to map the bacterial and archaeal community composition of influent, recirculating line and distal taps with paired biofilm and first-flush bulk water samples. Metagenomic data was predicted using Phylogenetic Investigation of Communities by Reconstruction of Unobserved States (PICRUSt 1.0.0) to characterize hypothetical functionalities of resident microbes. Overall the study takes a step towards identifying to what extent building owners/operators may be able to intentionally control the microbiome composition at the tap through practical measures, and addresses a critical knowledge gap at the intersection of water infrastructure, energy saving and public health.
Materials and methods
System description
Two identical lab-scale hot water plumbing rigs were designed for paired comparison of the impact of water heater temperature and use frequency (Supplementary Figure 1), as reported previously in studies focused on the occurrence of Legionella (Rhoads et al., 2015, 2016b). After 5-month acclimation at 39 °C, the control rig was maintained at 39 °C, while the experimental rig was increased from 39 to 42, 48, 51 and 58 °C at intervals of 2–3 months. Both rigs were fed with Blacksburg (VA, USA) tap water pre-treated with Pentek GAC-BB granular activated carbon cartridges (Pentair Residential Filtration, Milwaukee, WI, USA). Factors examined within each rig included: water heater temperature setting (°C), time (5, 8, 10, 13 and 15 months), sample location (influent, recirculating line and distal tap), system (control rig and experimental rig), pipe orientation (upward and downward flow), phase (biofilm and bulk water), and water use frequency (high, medium, low: 21, 3, 1 flush/week respectively) in triplicate distal pipes. Notably, distal taps achieved room temperature within 25 min after flushing, in contrast to the recirculating line, which was consistently maintained at the temperature setting (Rhoads et al., 2015). If not specified, ‘temperature' denotes the water heater setting, while ‘effective' temperature indicates the average in the distal pipe during stagnation (that is, across the flush/stagnation cycle).
Sample collection and processing
Biofilm swab (~65 cm2) and first-flush bulk water (~500 ml) samples were collected after 2- to 3-month acclimation time at each condition, at the end of the stagnation cycle. For biofilm sampling, sterile cotton-tip applicators (Fisherbrand, distributed by Fisher Scientific Company LLC, Pittsburgh, PA, USA) were used to swab the interior surface of the influent, recirculating line, and ends of the distal tap pipes accessible from threaded union connections. Bulk water samples were concentrated onto 0.22 μm pore-size sterile mixed cellulose ester filters (Millipore, Billerica, MA, USA). Filters and swabs were transferred to lysis tubes and subject to DNA extraction using FastDNA Spin Kits (MP Biomedicals, Solon, OH, USA) according to manufacturer protocol. Illumina 16S rRNA amplicon sequencing was performed following the Earth Microbiome Project 16S rRNA Amplification Protocol Version 4_13 (http://www.earthmicrobiome.org/emp-standard-protocols/16s/). Minor changes were as previously described (Ji et al., 2015), including using Molecular Biology Grade Water (Quality Biological, Gaithersburg, MD, USA) and pooling 200 ng of amplicon from each sample before clean-up using QIAquick PCR Purification Kit (QIAGEN, Valencia, CA, USA). Universal primers 515f and barcoded 806r were used for PCR amplification. Illumina 16S rRNA amplicon sequencing (paired-end 250 bp, MiSeq platform) was performed by the Virginia Bioinformatics Institute (Blacksburg, VA, USA).
Microbiome data analyses
Paired-end reads were stitched using PANDAseq (Masella et al., 2012) and filtered based on quality score (>0.90, range 0–1) and sequence length (252–255 bp) following the EDAMAME course materials (https://edamame-course.github.io/, accessed 01/24/2016). A de novo Operational Taxonomic Unit (OTU) picking strategy was adopted in QIIME 1.8.0 (pick_de_novo_otus.py, Caporaso et al., 2010) at similarity cutoff of 0.97 (uclust; Edgar, 2010) using Greengenes 16S rRNA gene database v13_8 (McDonald et al., 2012). Mitochondrial and chlorophyll sequences were removed from both the OTU table and the phylogenetic tree before downstream analysis. After removal of singleton OTUs, the sequencing depth of the 427 samples ranged from 25 266 to 304 049 with a median value of 82 078, which translated into 62 494 OTUs in total. All samples were rarefied to the smallest sequencing depth (that is, 25 266) before downstream alpha and beta diversity analyses (rarefied 10 times and 100 times, respectively) in QIIME 1.8.0 (Caporaso et al., 2010). Shannon index was selected for the alpha diversity matrix to evaluate both evenness and richness components, which emphasizes richness and rare OTUs relative to the Simpson Index. Weighted UniFrac distance matrix was selected for beta diversity to capture phylogenetic distance while taking relative abundance into consideration (Lozupone and Knight, 2005). Principal Coordinate Analysis was further applied to the beta diversity and visualized in EMPeror (Vázquez-Baeza et al., 2013).
Metagenomic data was predicted from the OTU table generated by closed-reference OTU picking strategy (pick_closed_referfence_otus.py, uclust similarity cutoff at 0.97, Greengene v13_8) in QIIME 1.8.0 (Caporaso et al., 2010) following PICRUSt 1.0.0 online protocol (Langille et al., 2013). Bray-Curtis distance matrix was calculated from predicted functional profile in QIIME 1.8.0 (Caporaso et al., 2010). As PICRUSt predictions are based on sequenced reference bacterial genomes, weighted Nearest Sequenced Taxon Index (weighted NSTI) scores were applied to measure average phylogenetic similarities among the OTUs within a given sample and the reference bacterial genomes, with OTU abundance taken into consideration. In addition, the ratios of PICRUSt-compatible OTUs and total sequences were calculated to estimate the predictable proportion of microbial community. All factors were treated as non-numeric variables, and the relative contribution of each factor on the microbial phylogenetic and functional dissimilarities across samples were determined through Permutational Multivariate Analysis of Variance (Adonis, Anderson, 2001) among a priori defined groups in R studio (permutation=999, {vegan} version 2.3-0, Oksanen et al., 2016). Adonis is less sensitive to dispersion (that is, within-group variation) than alternative methods including ANOSIM (Oksanen et al., 2016). A complementary multivariate homogeneity of groups dispersions (variances) analysis (betadisper; Anderson, 2006) was applied to account for confounding dispersion effect.
To explore the relationship between biofilm and bulk water microbiota, we first examined if both phases trended similarly in response to various factors through Mantel and Procrustes tests (QIIME 1.8.0, Caporaso et al., 2010). Second, we explored to what extent biofilm and bulk water shared microbes via core OTU analysis on both sample groups and individual paired samples (169 pairs of bulk water- biofilm distal tap samples). Based on Levene's test of homogeneity of variances, one-way ANOVA with Tukey's honest significant differences (HSD) method for pair-wise comparisons, or Kruskal and Wallis test with Tukey and Kramer (Nemenyi) test was applied ({car} version 2.1-2, Fox and Weisberg, 2011; {stat} version 3.2-2; {PMCMR} version 4.1, Pohlert, 2014). A Venn diagram ({VennDiagram} version 1.6.16, Chen and Boutros, 2011) was constructed for the former. A complementary indicator microbes analysis ({indicspecies} version 1.7.5, De Caceres and Legendre, 2009) was performed to inform the identity of the shared/exclusive OTUs of interest with thresholds for both positive predictive value (A) and sensitivity (B) were set at 0.7 with a 95% confidence interval (P<0.05).
Associations between Legionella spp. and other microbial genera were characterized using Maximal Information-based Nonparametric Exploration (MINE) analysis ({minerva} version 1.4.1, Reshef et al., 2011) with a false detection rate correction at 0.05 (Benjamini and Hochberg, 1995). Mitochondrial sequences belonging to Acanthamoeba spp. and Vermamoeba vermiformis (née Hartmanella vermiformis) were detected. The ‘relative abundance' of mitochondrial V. vermiformis was calculated as the ratio of the number of mitochondrial sequences to the number of total sequences (no organelle or singletons), which was later incorporated into the taxonomy table at the genus-level before MINE analysis.
Data sharing
All sequences and metadata for this research are retrievable from QIITA Study ID 10502.
Results
Drivers of phylogenetic and functional microbiome dissimilarity
Phase (biofilm versus bulk water) was the major driver of phylogenetic dissimilarity across all samples (R2Adonis=0.246, PAdonis=0.001), followed by water heater temperature setting (R2Adonis=0.172, PAdonis= 0.001), sample location (influent, recirculating line, and distal tap; R2Adonis=0.124, PAdonis=0.001), and time (R2Adonis=0.100, PAdonis=0.001; Table 1). Water use frequency (R2Adonis=0.039, PAdonis=0.001) and pipe orientation (R2Adonis=0.013, PAdonis=0.001) had less pronounced effects on phylogenetic dissimilarity, although their impacts were apparent under certain conditions (for example, biofilm at T=58 °C, where lower water use frequency seemed to enhance the impact of pipe orientation, Supplementary Figure 2). Notably, elevated temperature and time together conferred a greater magnitude of impact on the phylogenetic dissimilarity relative to that of constant temperature and time (control rig) in both recirculating line (experimental rig, R2Adonis=0.759, PAdonis= 0.001; control rig; R2Adonis=0.444, PAdonis= 0.001) and distal tap (experimental rig, R2Adonis=0.249, PAdonis= 0.001; control rig; R2Adonis=0.112, PAdonis= 0.001) microbiomes, with the most striking contrast between the two rigs at 15 months when the experimental rig was at 58 °C (Supplementary Figure 3, distal tap). A statistically significant temporal trend was observed in influent samples (9.6–23.7 °C; R2Adonis=0.160, PAdonis=0.679), which was weaker than that in the control rig (consistently at 40 °C; R2Adonis=0.444, PAdonis=0.001) and experimental rig (incrementally increased from 40 °C; R2Adonis=0.759, PAdonis=0.001) recirculating lines. Temperature appeared to have exerted an influence on the microbiome composition over and above that of succession with time, given that there was no significant difference in the phylogenetic composition between the two rigs during the initial 5-month baseline at 39 °C (PAdonis=0.158).
Table 1. Relative impact of various factors on phylogenetic and functional microbiome dissimilarity across samples (Adonis and Betadisp, permutation=999).
| Factor |
Phylogenetic |
Functional |
Samples | ||||
|---|---|---|---|---|---|---|---|
| R2Adonis | PAdonis | PBetadisp | R2Adonis | PAdonis | PBetadisp | ||
| Phase | 0.246 | 0.001 | 0.001 | 0.087 | 0.001 | 0.001 | All samples (n=427) |
| Sample location | 0.124 | 0.001 | 0.001 | 0.055 | 0.001 | 0.003 | All samples (n=427) |
| t | 0.100 | 0.001 | 0.001 | 0.398 | 0.001 | 0.001 | All samples (n=427) |
| T | 0.172 | 0.001 | 0.003 | 0.194 | 0.001 | 0.003 | All samples (n=427), influent was set at 20°C. |
| Water use frequency | 0.039 | 0.001 | 0.039 | 0.021 | 0.005 | 0.040 | Distal tap samples (n=348) |
| Pipe orientation | 0.013 | 0.006 | 0.716 | 0.013 | 0.008 | 0.971 | Distal tap samples (n=348) |
| Phase | 0.311 | 0.001 | 0.001 | 0.104 | 0.001 | 0.001 | Distal tap samples (n=348) |
| Rig | 0.025 | 0.158 | 0.624 | 0.019 | 0.267 | 0.185 | Distal tap at 5 months, 39°C (n = 70) |
| t (at 39 °C) | 0.112 | 0.001 | 0.043 | 0.396 | 0.001 | 0.244 | Control rig distal tap samples (n=175) |
| Phase | 0.466 | 0.001 | 0.001 | 0.138 | 0.001 | 0.001 | Control rig distal tap samples (n=175) |
| t (with elevated T) | 0.249 | 0.001 | 0.015 | 0.513 | 0.001 | 0.398 | Experimental rig distal tap samples (n=173) |
| Phase | 0.256 | 0.001 | 0.001 | 0.084 | 0.001 | 0.001 | Experimental rig distal tap samples (n=173) |
| Phase | 0.126 | 0.001 | 0.098 | 0.104 | 0.003 | 0.069 | Recirc. Samples (n=45) |
| Rig | 0.124 | 0.001 | 0.003 | 0.072 | 0.016 | 0.962 | Recirc. Samples (n=45) |
| t (at 39 °C) | 0.444 | 0.001 | 0.730 | 0.681 | 0.001 | 0.519 | Control rig recirc samples (n=27) |
| t (with elevated T) | 0.759 | 0.001 | 0.687 | 0.608 | 0.001 | 0.807 | Experimental rig recirc samples (n=28) |
| t | 0.377 | 0.001 | 0.443 | 0.621 | 0.001 | 0.455 | Influent (n=24) |
| Rig | 0.041 | 0.001 | 0.039 | 0.012 | 0.011 | 0.053 | Distal tap samples (n=348) |
| T (<51 °C or ⩾51 °C) | 0.175 | 0.001 | 0.001 | 0.183 | 0.001 | 0.001 | Experimental rig distal tap samples (n=173) |
Both time (t) and water heater temperature setting (T) were treated as non-numeric factors. Statistical significance was set at 0.05: italic font indicates non-statistically significant.
When considering the microbiome composition from a functional standpoint, employing PICRUSt, there was no significant dissimilarity as a function of phase or sample location (Table 1). Instead, temperature and time were most influential, though the impact of elevated temperature on function was not as great as that which had been observed on phylogenetic profile (Table 1). For the influent water, the functional microbiome was found to be dissimilar with time (R2Adonis=0.621, PAdonis=0.001). Despite the low PICRUSt-compatible OTU ratio (16.2±6.6%), 81.3±6.3% sequences were PICRUSt-compatible across all samples (Supplementary Table 1). Although complex environmental samples pose a challenge to PICRUSt, accurate metagenome prediction was achieved in a prior study of soil, with NSTI values of 0.17±0.02 (mean±s.d.; Langille et al., 2013). NSTI scores aid in evaluating PICRUST accuracy, with a negative association with accurate metagenome prediction. Relatively low weighted NSTI scores in the present study indicated that PICRUSt was appropriate (mean±s.d.=0.10±0.02, Supplementary Table 1).
Effect of combined factors on microbiome composition
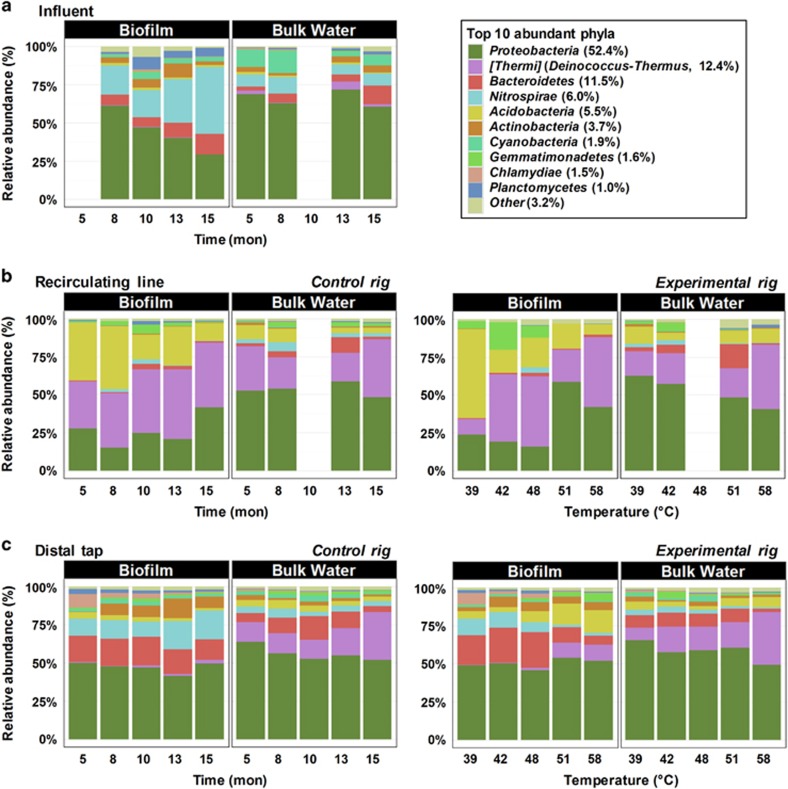
Grouping samples by phase, sample location, temperature and time resulted in 46 sample groups, which shared the same top abundant phyla (that is, phyla with over 1% of total sequences across all samples), but differed in their relative abundances (Figure 1). For instance, Nitrospirae favored the biofilm phase and were enriched with time in the influent and control rig distal taps, but diminished in the experimental rig distal taps with elevated temperature. In the recirculating line, Nitrospirae remained at a comparatively low relative abundance, with no obvious temporal trend in either rig. Almost all Nitrospirae sequences were classified as Nitrospira spp., which are nitrite-oxidizing bacteria with some members recently reported to be ‘comammox' (that is, complete ammonia oxidizer; Daims et al., 2015) and to have an optimal temperature range of 35–40 °C. In the recirculating line, higher temperatures (51 °C and 58 °C) were associated with smaller net decrease in ammonia concentration (Supplementary Table 2) and concurrently lower relative abundance of Nitrosomonas spp. in bulk water (Supplementary Table 3), which likely suggests inhibited ammonia oxidization given that influent ammonia level remained stable. Some other ammonia-oxidizing bacteria containing genera, including Nitrosospira (Burrell et al., 2001), were not found in the recirculating lines.
Figure 1.
Taxonomic profiles of sample groups at the phylum level: (a) influent, (b) recirculating line, (c) distal tap. Time is in month (mon) and temperature indicates water heater temperature setting. Top abundant phyla were defined as those that were >1% of total sequences across all samples combined (percentage shown in brackets); the remainder were combined and lumped in a category designated as ‘Other'. The three blank columns correspond to lost samples.
Biofilm versus bulk water: indicator OTUs
The indicator OTU concept provided a framework for examining the preference of individual microbes for the biofilm versus the bulk water. Ideally, an indicator OTU should meet two criteria: (1) 100% of the OTU sequences belong to the samples from the defined group (positive predictive value A=1.0) and (2) the OTU is detected in 100% of the samples from the defined group (sensitivity B=1.0; Cáceres and Legendre, 2009; Cáceres et al., 2010). With the confidence interval set at 95%, 8038 and 10 424 indicator OTUs were identified for biofilm and bulk water phase, respectively. Currently there is no consensus on the cutoff values for the positive predictive value (A) and sensitivity (B), whereas in general higher values imply stronger indicator OTUs. Applying 0.7 as an arbitrary threshold for both A and B, we observed that biofilm harbored less indicator OTUs (147) than did bulk water (232). Notably, OTUs within the same genus did not universally serve as indicators for the same phase or sample group. For instance, bulk water phase indicators included 11/631 Mycobacterium spp., 34/635 Meiothermus spp. and 1/188 Legionella spp. OTUs. Similarly, different OTUs within the same genus sometimes indicated different phases. Out of 1056 OTUs belonging to Nitrospira spp., 11 served as indicators for the biofilm phase while 7 others were indicators for the bulk water phase. This suggests that it may be inappropriate to assume that members of the same genus uniformly serve as indicators in domestic hot water systems.
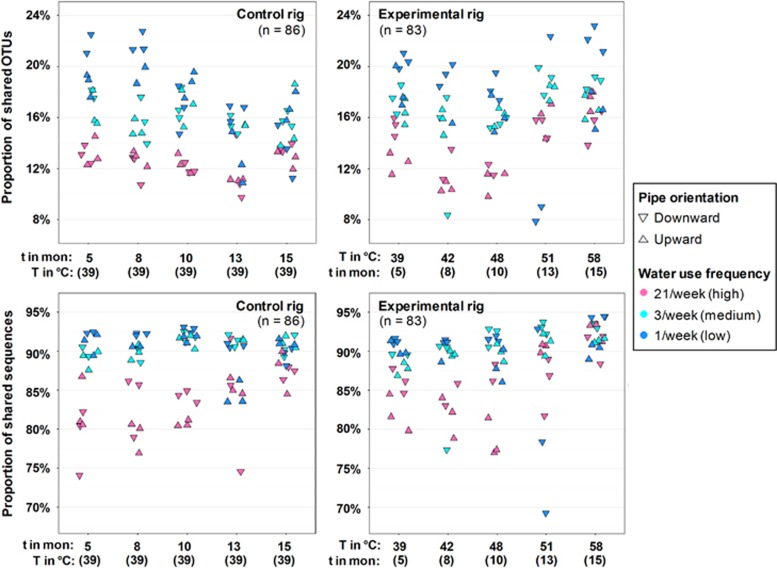
Bulk water and biofilm trended with each other in response to various factors including time and water heater temperature (Mantel: R2=0.608, P=0.001; Procrustes M2=0.698). In general, each bulk water and biofilm sample pair shared 8–24% OTUs, which comprised 68–95% of the total sequences between the pairs (Figure 2). Comparison between the two rigs suggested that elevated temperature (T⩾51 °C) induced a subtle increase in the proportion of shared OTUs between the biofilm and bulk water (Tukey HSD; adjusted P51–39°C=0.046, adjusted P58–39°C=0.005), although this trend was not significant when viewed as the proportion of shared sequences (adjusted P51–39°C=0.992, adjusted P58–39°C=0.258) (Supplementary Table 4). At T<51 °C, high water use frequency was associated with the fewest shared OTUs and sequences between paired bulk water and biofilm samples (Figure 2; adjusted P<0.001 for 21/week – 1/week and 3/week – 1/week, Supplementary Tables 5 and 6). Pipe orientation (upward and downward flow) had impact on the relationship between bulk water and biofilm in some situations. For instance, under high water use frequency (21/week), downward pipe harbored higher shared OTUs between phases, except at 10 and 15 months in the control rig (39 °C).
Figure 2.
Proportion of OTUs and sequences shared in paired bulk water and biofilm samples at distal taps. Proportion was calculated as either number of shared OTUs divided by number of total OTUs between the paired samples (membership); or the number of sequences belonging to shared OTUs divided by the number of total sequences between the paired samples (relative abundance) across time (t in month, mon) and temperature (T in °C). Color codes indicate water use frequency and shape indicates pipe orientation.
Influence of recirculating line microbes on those at the distal taps: core OTUs
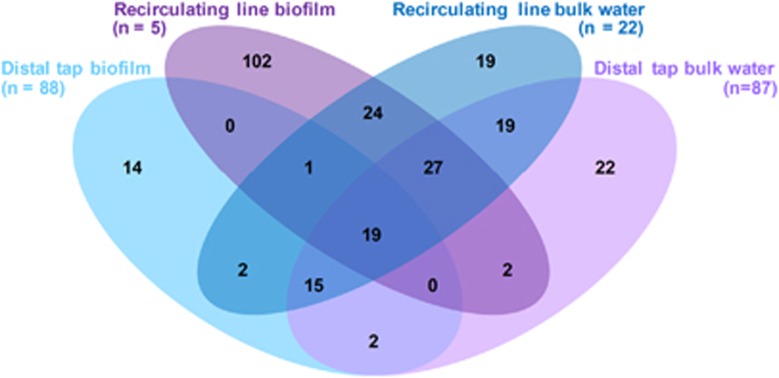
Beta diversity analysis indicated that the greatest contrast occurred between the influent and recirculating line, with the distal taps displaying noticeable overlap with both of these locations (Supplementary Figure 4). Of particular interest was the relative contribution of microbes from the influent and recirculating line to those observed at the tap, which can be conceptualized as survival of allochthonous microbes to the periodically heat-shocked distal tap environment. To facilitate this comparison, control rig samples were subject to core OTU analysis (that is, OTUs shared among all samples within the given group) corresponding to 4 groups including distal tap biofilm, recirculating line biofilm, distal tap bulk water, and recirculating line bulk water (Figure 3). There were 14, 102, 19 and 22 core OTUs confined to each of these four individual groups, respectively, with a total of 19 core OTUs common to all groups. The vast majority of distal tap bulk water core OTUs (82/106 OTUs) were shared with recirculating line core OTUs (both phases included). Of these, 34 OTUs were only shared with recirculating line bulk water and 2 were only shared with the recirculating line biofilm. Interestingly, 20 core OTUs that were shared between the two biofilm locations were also encompassed in recirculating line bulk water core OTUs. This suggests that significant migration occurs from the recirculating line to the tap via physical transport of bulk water. Biofilm at the tap is also a likely source of bulk water OTUs at the tap, with 36 core OTUs shared between these two phases and 2 of which confined to the tap.
Figure 3.
Venn diagram of core OTUs among each of the recirculating line and distal tap phases in the control rig across all sampling dates.
Negative association between alpha diversity (Shannon Index) and temperature
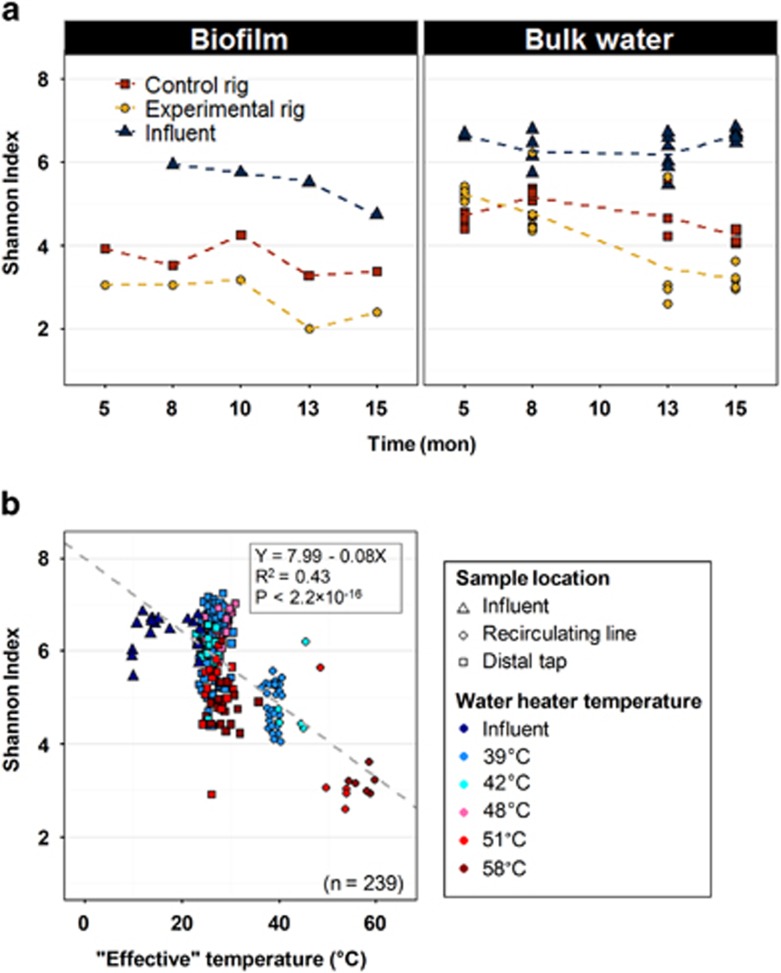
Alpha diversity trends were similar for both the biofilm and bulk water phases in that the influent was characterized by the highest Shannon Index, followed by the control rig recirculating line, and the experimental rig recirculating line (Figure 4a). In the biofilm phase, the experimental rig trended with the control rig with time; whereas in the bulk water phase, the experimental rig experienced a more drastic decrease concurrent with the elevated temperature. Linear regression ({stats} version 3.2.2, R Studio) further confirmed a negative association between Shannon Index and ‘effective' temperature (that is, average temperature across the flush/stagnation cycle) in bulk water samples (Figure 4b). Despite this negative association, influent samples, with ‘effective' temperatures below 15 °C, had lower alpha diversity than the fitted model, which contributed to a peak alpha diversity region around 20 to 25 °C. Distal tap samples did not vary much from each other in terms of ‘effective' temperature (26.8±2.2 °C), but displayed a wide range of alpha diversities. Generally, higher temperature (T⩾51 °C) appeared to be conducive to lower alpha diversity.
Figure 4.
(a) Alpha diversity (Shannon Index) across influent and recirculating lines in both rigs (note for experimental rig temperature was elevated with time); (b) negative association between alpha diversity and ‘effective' temperature in bulk water samples (gray line is linear correlation). Time is in month (mon) and ‘effective' temperature is in °C.
Patterns in Legionella occurrence and association with other microbes
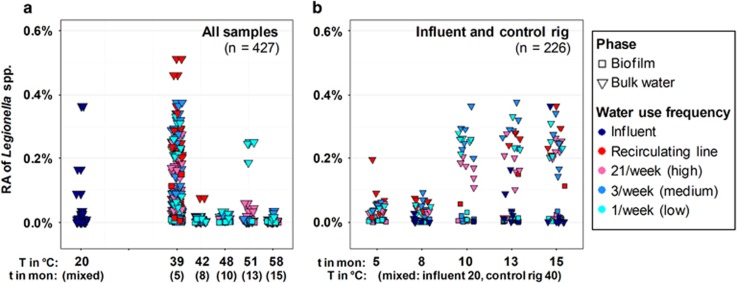
The relative abundance of Legionella spp. decreased with elevated temperature (Figure 5a), whereas it increased in the bulk water phase of the control rig with time (Figure 5b). Nevertheless, at T=51 °C, 3 out of 5 low water use frequency (1/week) bulk water samples displayed markedly high relative abundance of Legionella spp., 2 of which were from upward oriented pipes. Rhoads et al. (2015) also confirmed that this combination of 51 °C and low water use frequency uniquely selected for L. pneumophila at distal taps of the current system using quantitative PCR. Pipe orientation (upward and downward) alone did not show an obvious impact on enrichment of Legionella spp. (Supplementary Figure 5, paired upward/downward pipes).
Figure 5.
(a) Relative abundance (RA) of Legionella spp. in all samples under different water heater temperature settings (T in °C); (b) relative abundance (RA) of Legionella spp. in influent and control rig samples across time (t in month, mon). Relative abundance was calculated using Illumina 16S rRNA gene amplicon sequencing. Water use frequency and phase are indicated by color and shape, respectively. All influent samples were assigned a uniform temperature of 20 °C to facilitate comparison.
Mitochondrial sequences belonging to Acanthamoeba and Vermamoeba (V. vermiformis), which are known to serve as hosts for replication and enhanced virulence of L. pneumophila and other pathogens, were detected in 4 and 393 out of 427 samples, respectively. MINE analysis identified a total of 143 statistically significant associations between Legionella spp. and other microbial taxa under a false detection rate of 0.05. The highest maximal information coefficient (MIC) was with mitochondrial V. vermiformis (MIC=0.44). Legionella spp. was also found to be positively associated with Mycobacterium spp., Meiothermus spp., and Xanthomonadaceae family (MIC=0.35, 0.41, and 0.51, respectively); whereas negatively associated with Nitrospira spp. (MIC=0.28).
Discussion
This study utilized pilot-scale, replicated pipe rigs to unravel the complexity inherent in-building plumbing systems and gain insight into the impact of water heater temperature and water use frequency on the microbiome composition at the tap. The findings have important implications for informing selection of water heater settings as well as water system design, which are currently the targets for new design and optimization strategies for conserving energy and water. Such designs could inadvertently shape the numbers and kinds of microbes entering the built environment via tap water.
A total of 62 494 OTUs were identified across this study, about a quarter of which were accounted for by influent samples (15 478 OTUs, both bulk water and biofilm). This number was also approximately one order of magnitude higher than the maximum number of OTUs reported in prior drinking water microbiota surveys (6488 in Lautenschlager et al., 2013; Pinto et al., 2014; El-Chakhtoura et al., 2015; Lührig et al., 2015; Proctor and Hammes, 2015), which have focused exclusively on cold water. We found that elevated water heater temperature drove significant change with respect to both the phylogenetic composition and predicted functions of the microbiota at the tap. Interestingly, the bulk water composition shifted with a threshold response around 51 °C, whereas the biofilm displayed more of a continuous succession pattern as the temperature was gradually elevated. The findings are consistent with the expectation of the environment selecting microbial populations (the niche theory, Hutchinson, 1957), which could theoretically provide the basis for a prebiotic framework for selection of ‘beneficial' microbes in the built environment. In interpreting the results throughout, it is important to bear in mind that the analysis applied in this study was entirely DNA-based, and therefore cannot directly distinguish live, dead or viable but non-culturable microbes. However, systematic comparison across various interconnected locations of the pipe rigs through various operating conditions and time provided an indication of which microbes were likely most adaptive to each location/condition.
Although there appeared to be a sharp bifurcation between biofilm and bulk water microbial composition, as reported by others using culture and 16S rRNA gene based clone libraries (Martiny et al., 2005), highly similar functions between the two phases were predicted. This implies functional redundancy of the microbiota, or what has been dubbed as a ‘portfolio effect' in which positive and negative changes in individual taxa observed in the bulk water and biofilm essentially sum to zero (Allison and Martiny, 2008). Conceptually, the distinct dispersal limitations between planktonic (bulk water) and sessile (biofilm) states would allow for the development of functionally-equivalent, yet taxonomically diverse, species adapting to a similar set of environmental conditions (the neutral theory, Hubbell, 2001). In fact, neutral theory has been proposed in conceptualizing biofilms as microbial ‘landscapes' (Battin et al., 2007). Rather than a niche-neutral dichotomy, recent studies have recognized the importance of both selective and neutral processes in shaping gastrointestinal (Jeraldo et al., 2012), soil (Dumbrell et al., 2010) and wastewater (Ofiţeru et al., 2010) microbiomes.
Fundamental to this study is the fact that the distal tap environment is distinct from that of the recirculating line. Distal tap water samples essentially consisted of recirculating line inflow that had stagnated and cooled to room temperature (within ~25 min) since the previous flush of hot water. Stagnation has been reported to be a significant driving force for shift in potable cold water microbial communities (Lautenschlager et al., 2010; Ji et al., 2015). Within the control rig (T=39 °C), the substantial proportion of core OTUs unique to the distal tap bulk water (Figure 3) suggests that stagnation as short as 8 h (high water use frequency 21/week) is sufficient for a shift away from the core microbes defining the recirculating line bulk water or biofilm. It remains unclear, though, at what rate conversion from core to satellite microbiome generally occurs and whether elevated water heater temperature would influence the conversion time. The difference observed in as little as 8 h also demonstrated that the DNA-based approach applied in this study was able to capture microbial community shifts occurring on a short time scale.
The interrelationship between biofilm and bulk water microbes in building plumbing is one that is widely debated and of practical importance. For example, biofilm is widely considered to be a key environment for proliferation of OPs (Temmerman et al., 2006), but release to bulk water and conveyance via the tap is generally necessary to cause disease (Steinert et al., 2002). In this study, the high water use frequency was also characterized by the lowest proportion of shared OTUs and sequences between paired biofilm and bulk water sample (Figure 2, Supplementary Table 5). This suggests that the shorter stagnation time translates into less time for microbial interaction between phases to occur and take root. Microbes released from the biofilm to the bulk water would also be flushed out more frequently at a high use tap. Interestingly, the effect of higher water use frequency was diminished by elevated water heater temperature (T⩾51 °C, Supplementary Table 6). Elevated temperature (T⩾51 °C) was associated with a slightly increased proportion of shared OTUs between the two phases (Supplementary Table 4), which could be related to an overall decrease in total OTUs at the tap (Supplementary Figure 6). These findings have important implications for recommended sampling and monitoring protocols and whether biofilm or bulk water is most appropriately targeted over a range of conditions.
As noted above, the effect of gradually elevating the water heater temperature was not continuous, particularly in the bulk water. Notably, 51 °C appeared to be a critical threshold, with phylogenetic dissimilarity among distal tap biofilm samples, alpha diversity in distal tap water samples, ammonia oxidation in recirculating line bulk water, and interactive effects with water use frequency all bifurcating around this temperature. One possibility is that 51 °C demarcates a transition from a mesophilic to a thermophilic environment. For example, in anaerobic digestion, the mesophilic range is defined from 35–37 °C, while thermophilic digestion occurs from 50–57 °C, each imparting distinct dominant pathways for methanogenesis (Ali Shah et al., 2014). In sum, microbes must be biochemically adaptive, such that proteins, nucleic acids, ribosomes, and lipids can maintain function at hot temperatures. An earlier study of 204 complete genomes and proteomes reported increased purine load index (A+G content) in nucleotide composition as well as increased sum of fractions of IIe, Val, Tyr, Trp, Arg, Gly and Leu amino acids under elevated optimal growth temperatures (−10 to 110 °C, Zeldovich et al., 2007).
We considered whether the ‘effective' temperature profile (that is, the integral of instantaneous temperature over time between two sampling events) would provide a more meaningful predictor than the water heater temperature setting on observed phylogenetic dissimilarity patterns. Using this approach, distal tap sample compositions overlapped between those of the influent and recirculating samples (Supplementary Figure 4). Further, a negative association between ‘effective' temperature and microbial diversity (Shannon Index) was observed in distal tap and recirculating line bulk water samples. Although we could find no prior domestic hot water system studies for comparison, our observation is consistent with previous surveys of geothermal systems as a natural analog. A negative linear relationship between three temperature groups (62–87 °C) and the Shannon Index was reported for four sediment microbial communities in Great Boiling Spring (Cole et al., 2013). Similar negative association were documented along a temperature gradient of 52–75 °C in a Japanese hot spring (Everroad et al., 2012). Importantly, in the present study, the ‘peak' alpha diversity occurred around the ‘effective' temperature of 20–25 °C, which concurs with an extensive survey on 36 geothermal areas with 135 samples (Sharp et al., 2014), where they observed a bell-shape alpha diversity curve over a temperature range of 7.5–99 °C with peak value at 24 °C. Notably, the indoor built environment is commonly maintained from 20 to 25 °C.
Given that it is impossible to eradicate microbes from water systems, a ‘probiotic', or more specifically a ‘prebiotic' framework has been proposed for desirably manipulating the microbiome at the tap (Wang et al., 2013). Elevated temperature, in particular, can be highly effective for controlling OPs (Buse and Ashbolt, 2011; ANSI/ASHRAE, 2015). For instance, L. pneumophila has an optimal growth range of 25–42 °C (ASHRAE, 2000), with measurable deactivation at temperatures higher than 50 °C (WHO, 2007). In fact, a recent study has proposed a diagnostic approach to inform potential L. pneumophila risk through closely monitoring temperature profiles at critical control points within the hot water distribution system (Bédard et al., 2015). On the other hand, elevated temperature is not universally effective, particularly when only temporarily applied. Legionella have been observed to proliferate via necrotrophic growth on heat-killed cells following thermal disinfection (Temmerman et al., 2006). Thus, the present study makes it clear that temperature settings have much more broad sweeping effects on the microbial composition at the tap, likely exerting indirect microbial ecological controls on Legionella proliferation. Here we observed that water heater temperature settings from 39–58 °C were negatively associated with relative abundance of Legionella, but was less effective for Legionella control after cooling at the tap relative to the conditions of the consistently hot recirculating line. Further, at 51 °C, markedly high relative abundance of Legionella OTUs were observed in the bulk water phase at low-use distal taps, a measurement consistent with high L. pneumophila gene copy numbers determined by quantitative PCR (67 times higher on average than the inflow recirculating line water, Rhoads et al., 2015). It is speculated that, at 51 °C, the heat shock served to diminish microbes competitive with Legionella (that is, ‘probiotics'), creating an ecological ‘sweet spot' where Legionella could recover and proliferate with the extended stagnation (Rhoads et al., 2015). In fact, heat pre-treatment (50 °C for 30 min) is widely applied to increase the specificity for isolating Legionellae from hot water systems (Leoni and Legnani, 2001).
Mitochondrial Acanthamoeba spp. and Vermamoeba vermiformis sequences were detected, with the latter occurring in 92% of samples. Mitochondrial 16S-like ribosomal small subunit rRNA was reported to yield genotype-level identification and classification for Acanthamoeba comparable to that of 18S rRNA (Booton et al. 2001). A recent study (Fučíková and Lahr, 2016) using the complete mitochondrial genomes of Acanthamoeba castellanii and Vermamoeba vermiformis reported higher resolution of intraspecific phylogenetic divergences relative to nuclear rDNA. However, little is known about the primer (515F/barcoded 806R) specificity towards amoebal mitochondrial sequences. A major obstacle is the availability of sequenced whole amoebal mitochondrial genomes, for example, 2 for V. vermiformis in GenBank (accessed 11/14/2016): KT185627.1 and GU828005.1 (identical to NC_013986.1). Each of the two V. vermiformis mitochondrial genome sequences contains only one copy of 16S-like ribosomal small subunit rRNA. Nevertheless, as number of mitochondria per V. vermiformis cell vary as a function of metabolic status, the ‘relative abundance' for mitochondrial V. vermiformis is examined as a relative indicator of V. vermiforms cells.
Associations with Legionella spp. were examined to gain insight into potential antagonistic or synergistic relationships. In particular, a strong positive association was noted between the relative abundances of Legionella spp. and mitochondrial V. vermiformis. This finding suggests the likely prevalent existence of V. vermiformis in both bulk water and biofilm phases of the rigs and potentially a positive association between V. vermiformis and Legionella spp. in terms of abundance. From a microbial ecology perspective, some free-living amoebae are either obligate or facultative pathogens (Brieland et al., 1997; Lorenzo-Morales et al., 2007) and can serve as vehicles/reservoirs for L. pneumophila (Wadowsky et al., 1988; Kuiper et al., 2004). Positive correlations between V. vermiformis and Legionella have been reported in treated and distributed waters (Valster et al., 2011; Wang et al., 2012), yet noted as conditional relationships and hypothesized to be dependent on strain type (Buse and Ashbolt, 2011) and in-building plumbing temperature (Rhoads et al., 2015). Interestingly, Mycobacterium spp., a genus that contains some OP members, relative abundances were also found to be positively associated with those of Legionella spp. Previous studies have noted co-occurrence of M. avium complex and Legionella in water and/or biofilms in hot water systems ranging from 40 to 54 °C (Bukh and Roslev, 2014) as well as in a drinking water distribution system (Whiley et al., 2014). Positive correlation between Legionella spp. and Mycobacterium spp. gene copies measured by quantitative PCR has also been reported in a drinking water distribution system (Lu et al., 2016).
The design of the rig employed in this study uniquely served to simulate building plumbing conditions in a controlled, replicated fashion and isolate the effect of individual variables. The kind of insight gained would not be possible from a field study or simplistic bench-scale experiment alone, as the extreme complexities encountered from building to building make it impossible to identify causal factors. Nonetheless, it is important to note key differences in the design of the present study relative to the field. For instance, much longer distal pipes in the field would likely magnify impacts of cooling from water heater set points, pipe orientation and regrowth on levels of OPs in water. Real-world hot water systems will also have more complex water use patterns and employ a wide range of materials, which together will contribute to greater heterogeneity across distal tap microbiota. Overall this study demonstrates the possibility of implementing practical design and operation measures for intentionally controlling the microbiome composition at the tap, a topic of increasing interest not only for pathogen control, but also as linkages between the human microbiome and that of the built environment become more apparent.
Acknowledgments
This work was supported by The Alfred P. Sloan MoBE program, the National Science Foundation (CBET Award #1336650) and the Institute for Critical Technology and Applied Science (ICTAS) at Virginia Tech. We thank Brian Badgley for lab supplies, and Kelly Peterson, Arjan Ahluwalia, Daniel Logsdon for their laboratory assistance.
Footnotes
Supplementary Information accompanies this paper on The ISME Journal website (http://www.nature.com/ismej)
The authors declare no conflict of interest.
Supplementary Material
References
- Ali Shah F, Mahmood Q, Maroof Shah M, Pervez A, Ahmad Asad S. (2014). Microbial ecology of anaerobic digesters: the key players of anaerobiosis. Sci World J 204: 138752. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Allison SD, Martiny JBH. (2008). Resistance, resilience, and redundancy in microbial communities. Proc Natl Acad Sci USA 105: 11512–11519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson MJ. (2001). A new method for non-parametric multivariate analysis of variance. Aust Ecol 26: 32–46. [Google Scholar]
- Anderson MJ. (2006). Distance-based tests for homogeneity of multivariate dispersions. Biometrics 62: 245–253. [DOI] [PubMed] [Google Scholar]
- ANSI/ASHRAE. (2015) Standard 188 Legionellosis: Risk Management for Building Water Systems.
- Ashbolt NJ. (2015). Microbial contamination of drinking water and human health from community water systems. Curr Environ Health Rep 2: 95–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ASHRAE. (2000) ASHRAE Guideline 12, Minimizing the Risk of Legionellosis Associated with Building Water Systems.
- Bargellini A, Marchesi I, Righi E, Ferrari A, Cencetti S, Borella P et al. (2011). Parameters predictive of Legionella contamination in hot water systems: association with trace elements and heterotrophic plate counts. Water Res 45: 2315–2321. [DOI] [PubMed] [Google Scholar]
- Baron JL, VikramA, Duda S, Stout JE, Bibby K. (2014). Shift in the microbial ecology of a hospital hot water system following the introduction of an on-site monochloramine disinfection system. PLoS One 9: e102679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Battin TJ, Sloan WT, Kjelleberg S, Daims H, Head IM, Curtis TP et al. (2007). Microbial landscapes: new paths to biofilm research. Nat Rev Microbiol 5: 7–12. [DOI] [PubMed] [Google Scholar]
- Bédard E, Fey S, Charron D, Lalancette C, Cantin P, Dolcé P et al. (2015). Temperature diagnostic to identify high risk areas and optimize Legionella pneumophila surveillance in hot water distribution systems. Water Res 71: 244–256. [DOI] [PubMed] [Google Scholar]
- Beer KD, Gargano JW, Roberts VA, Hill VR, Garrison LE, Kutty PK et al. (2015). Surveillance for waterborne disease outbreaks associated with drinking water — United States, 2011–2012. Morb Mortal Wkly Rep 64: 842–848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. (1995). Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Statist Soc B 57: 289–300. [Google Scholar]
- Booton GC, Ledee DR, Awwad M, Sharma S, Nizsl I, Markus MM et al. (2001) Acanthamoeba mitochondrial 16S rDNA sequences: inferred phylogeny and support of nuclear ribosomal 18S rDNA. IXth International Meeting on the Biology and Pathogenicity of Free-Living Amoebae Proceedings. John Libbey Eurotext: Paris, France, pp 227–234. [Google Scholar]
- Borella P, Montagna MT, Romano-Spica V, Stampi S, Stancanelli G, Triassi M et al. (2004). Legionella infection risk from domestic hot water. Emerg Infect Dis 10: 457–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brieland JK, Fantone JC, Remick DG, LeGendre M, McClain M, Engleberg NC. (1997). The role of Legionella pneumophila-infected Hartmannella vermiformis as an infectious particle in a murine model of Legionnaires' disease. Infect Immun 65: 5330–5333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bukh AS, Roslev P. (2014). Mycobacterium avium complex in day care hot water systems, and persistence of live cells and DNA in hot water pipes. Curr Microbiol 68: 428–439. [DOI] [PubMed] [Google Scholar]
- Burrell PC, Phalen CM, Hovanec TA. (2001). Identification of bacteria responsible for ammonia oxidation in freshwater aquaria. Appl Environ Microb 67: 5791–5800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buse HY, Ashbolt NJ. (2011). Differential growth of Legionella pneumophila strains within a range of amoebae at various temperatures associated with in-premise plumbing. Lett Appl Microbiol 53: 217–224. [DOI] [PubMed] [Google Scholar]
- Cáceres MD, Legendre P. (2009). Associations between species and groups of sites: indices and statistical inference. Ecology 90: 3566–3574. [DOI] [PubMed] [Google Scholar]
- Cáceres MD, Legendre P, Moretti M. (2010). Improving indicator species analysis by combining groups of sites. Oikos 119: 1674–1684. [Google Scholar]
- Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK et al. (2010). QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7: 335–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Boutros PC. (2011). VennDiagram: a package for the generation of highly-customizable Venn and Euler diagrams in R. BMC Bioinformatics 12: 35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole JK, Peacock JP, Dodsworth JA, Williams AJ, Thompson DB, Dong H et al. (2013). Sediment microbial communities in Great Boiling Spring are controlled by temperature and distinct from water communities. ISME J 7: 718–729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper IR, Hanlon GW. (2010). Resistance of Legionella pneumophila serotype 1 biofilms to chlorine-based disinfection. J Hosp Infect 74: 152–159. [DOI] [PubMed] [Google Scholar]
- Daims H, Lebedeva EV, Pjevac P, Han P, Herbold C, Albertsen M et al. (2015). Complete nitrification by Nitrospira bacteria. Nature 528: 504–509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darelid J, Löfgren S, Malmvall BE. (2002). Control of nosocomial Legionnaires' disease by keeping the circulating hot water temperature above 55 degree C: experience from a 10-year surveillance programme in a district general hospital. J Hosp Infect 50: 213–219. [DOI] [PubMed] [Google Scholar]
- De Caceres M, Legendre P. (2009). Associations between species and groups of sites: indices and statistical inference. Ecology 90: 3566–3574. [DOI] [PubMed] [Google Scholar]
- Department of Veterans Affairs. (2014). Prevention of healthcare-associated Legionella disease and scald injury from potable water distribution systems. VHA Directive, 1061, pp A1–A7.
- Dumbrell AJ, Nelson M, Helgason T, Dytham C, Fitter AH. (2010). Relative roles of niche and neutral processes in structuring a soil microbial community. ISME J 4: 337–345. [DOI] [PubMed] [Google Scholar]
- Edgar RC. (2010). Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26: 2460–2461. [DOI] [PubMed] [Google Scholar]
- El-Chakhtoura J, Prest E, Saikaly P, van Loosdrecht M, Hammes F, Vrouwenvelder H. (2015). Dynamics of bacterial communities before and after distribution in a full-scale drinking water network. Water Res 74: 180–190. [DOI] [PubMed] [Google Scholar]
- Everroad RC, Otaki H, Matsuura K, Haruta S. (2012). Diversification of bacterial community composition along a temperature gradient at a thermal spring. Microb Environ 27: 374–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falkinham JO III, Pruden A, Edwards MA. (2015). Opportunistic premise plumbing pathogens: increasingly important pathogens in drinking water. Pathogens 4: 373–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farhat M, Moletta-Denat M, Frère J, Onillon S, Trouilhé M-C, Robine E. (2012). Effects of disinfection on Legionella spp., eukarya, and biofilms in a hot water system. Appl Environ Microb 78: 6850–6858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feazel LM, Baumgartner LK, Peterson KL, Frank DN, Harris JK, Pace NR. (2009). Opportunistic pathogens enriched in showerhead biofilms. Proc Natl Acad Sci USA 106: 16393–16399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox J, Weisberg S. (2011) An R Companion to Applied Regression Second EditionSAGE Publications: Thousand Oaks, California, USA. [Google Scholar]
- Fučíková K, Lahr DJG. (2016). Uncovering cryptic diversity in two Amoebazoan species using complete mitochondrial genome sequences. J Eukaryot Microbiol 63: 112–122. [DOI] [PubMed] [Google Scholar]
- Hanski I, von Hertzen L, Fyhrquist N, Koskinen K, Torppa K, Laatikainen T et al. (2012). Environmental biodiversity, human microbiota, and allergy are interrelated. Proc Natl Acad Sci USA 109: 8334–8339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilborn ED, Wade TJ, Hicks L, Garrison L, Carpenter J, Adam E et al. (2013). Surveillance for waterborne disease outbreaks associated with drinking water and other nonrecreational water—United States, 2009-2010. Morb Mortal Wkly Rep 62: 714–720. [PMC free article] [PubMed] [Google Scholar]
- Howe AD, Forster S, Morton S, Marshall R, Osborn KS, Wright P et al. (2002). Cryptosporidium oocysts in a water supply associated with a cryptosporidiosis outbreak. Emerg Infect Dis 8: 619–624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubbell SP. (2001) The Unified Neutral Theory of Biodiversity and Biogeography. Princeton University Press: Princeton, New Jersey, USA. [Google Scholar]
- Hutchinson GE. (1957). Concluding remarks. Cold Spring Harbor Symp Quant Biol 22: 415–427. [Google Scholar]
- Jeraldo P, Sipos M, Chia N, Brulc JM, Dhillon AS, Konkel ME et al. (2012). Quantification of the relative roles of niche and neutral processes in structuring gastrointestinal microbiomes. Proc Natl Acad Sci USA 109: 9692–9698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji P, Parks J, Edwards MA, Pruden A. (2015). Impact of water chemistry, pipe material and stagnation on the building plumbing microbiome. PLoS One 10: e0141087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley ST, Gilbert JA. (2013). Studying the microbiology of the indoor environment. Genome Biol 14: 202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuiper MW, Wullings BA, Akkermans ADL, Beumer RR, van der Kooij D. (2004). Intracellular proliferation of Legionella pneumophila in Hartmannella vermiformis in aquatic biofilms grown on plasticized polyvinyl chloride. Appl Environ Microb 70: 6826–6833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langille MGI, Zaneveld J, Caporaso JG, McDonald D, Knights D, Reyes JA et al. (2013). Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat Biotechnol 31: 814–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau HY, Ashbolt NJ. (2009). The role of biofilms and protozoa in Legionella pathogenesis: implications for drinking water. J Appl Microbiol 107: 368–378. [DOI] [PubMed] [Google Scholar]
- Lautenschlager K, Boon N, Wang Y, Egli T, Hammes F. (2010). Overnight stagnation of drinking water in household taps induces microbial growth and changes in community composition. Water Res 44: 4868–4877. [DOI] [PubMed] [Google Scholar]
- Lautenschlager K, Hwang C, Liu W-T, Boon N, Köster O, Vrouwenvelder H et al. (2013). A microbiology-based multi-parametric approach towards assessing biological stability in drinking water distribution networks. Water Res 47: 3015–3025. [DOI] [PubMed] [Google Scholar]
- Leoni E, Legnani PP. (2001). Comparison of selective procedures for isolation and enumeration of Legionella species from hot water systems. J Appl Microbiol 90: 27–33. [DOI] [PubMed] [Google Scholar]
- Lévesque B, Lavoie M, Joly J. (2004). Residential water heater temperature: 49 or 60 degrees Celsius? Can J Infect Dis 15: 11–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Lin YE, Stout JE, Hwang CC, Vidic RD, Yu VL. (2006). Effect of flow regimes on the presence of Legionella within the biofilm of a model plumbing system. J Appl Microbiol 101: 437–442. [DOI] [PubMed] [Google Scholar]
- Lorenzo-Morales J, Martínez-Carretero E, Batista N, Alvarez-Marín J, Bahaya Y, Walochnik J et al. (2007). Early diagnosis of amoebic keratitis due to a mixed infection with Acanthamoeba and Hartmannella. Parasitol Res 102: 167–169. [DOI] [PubMed] [Google Scholar]
- Lozupone C, Knight R. (2005). UniFrac: a new phylogenetic method for comparing microbial communities. Appl Environ Microb 71: 8228–8235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J, Struewing I, Vereen E, Kirby AE, Levy K, Moe C et al. (2016). Molecular detection of Legionella spp. and their associations with Mycobacterium spp., Pseudomonas aeruginosa and amoeba hosts in a drinking water distribution system. J Appl Microbiol 120: 509–521. [DOI] [PubMed] [Google Scholar]
- Lührig K, Canbäck B, Paul CJ, Johansson T, Persson KM, Rådström P. (2015). Bacterial community analysis of drinking water biofilms in southern Sweden. Microb Environ 30: 99–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martiny AC, Albrechtsen H-J, Arvin E, Molin S. (2005). Identification of bacteria in biofilm and bulk water samples from a non-chlorinated model drinking water distribution system: detection of a large nitrite-oxidizing population associated with Nitrospira spp. Appl Environ Microb 71: 8611–8617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masella AP, Bartram AK, Truszkowski JM, Brown DG, Neufeld JD. (2012). PANDAseq: PAired-eND Assembler for Illumina sequences. BMC Bioinformatics 13: 31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathys W, Stanke J, Harmuth M, Junge-Mathys E. (2008). Occurrence of Legionella in hot water systems of single-family residences in suburbs of two German cities with special reference to solar and district heating. Int J Hyg Environ Health 211: 179–185. [DOI] [PubMed] [Google Scholar]
- McDonald D, Price MN, Goodrich J, Nawrocki EP, DeSantis TZ, Probst A et al. (2012). An improved Greengenes taxonomy with explicit ranks for ecological and evolutionary analyses of bacteria and archaea. ISME J 6: 610–618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oberauner L, Zachow C, Lackner S, Högenauer C, Smolle K-H, Berg G. (2013). The ignored diversity: complex bacterial communities in intensive care units revealed by 16S pyrosequencing. Sci Rep 3: 1413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ofiţeru ID, Lunn M, Curtis TP, Wells GF, Criddle CS, Francis CA et al. (2010). Combined niche and neutral effects in a microbial wastewater treatment community. Proc Natl Acad Sci USA 107: 15345–15350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oksanen J, Blanchet FG, Kindt R, Legendre P, Minchin PR, O'Hara RB et al. (2016). Vegan: Community Ecology Package. R package version 2.3-5 https://cran.r-project.org/web/packages/vegan/vegan.pdf.
- Pinto AJ, Schroeder J, Lunn M, Sloan W, Raskin L. (2014). Spatial-temporal survey and occupancy-abundance modeling to predict bacterial community dynamics in the drinking water microbiome. mBio 5: e01135–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pohlert T. (2014), The Pairwise Multiple Comparison of Mean Ranks Package (PMCMR). R package http://CRAN.R-project.org/package=PMCMR.
- Proctor CR, Gächter M, Kötzsch S, Rölli F, Sigrist R, Walser J-C et al. (2016). Biofilms in shower hoses – choice of pipe material influences bacterial growth and communities. Environ Sci: Water Res Technol 2: 670–682. [Google Scholar]
- Proctor CR, Hammes F. (2015). Drinking water microbiology – from measurement to management. Curr Opin Biotechnol 33: 87–94. [DOI] [PubMed] [Google Scholar]
- Pryor M, Springthorpe S, Riffard S, Brooks T, Huo Y, Davis G et al. (2004). Investigation of opportunistic pathogens in municipal drinking water under different supply and treatment regimes. Water Sci Technol 50: 83–90. [PubMed] [Google Scholar]
- R Core Team. (2015) R: A language and environment for statistical computing. R Foundation for Statistical Computing: Vienna, Austria, https://www.R-project.org/ accessed on 02/29/2015. [Google Scholar]
- Reshef DN, Reshef YA, Finucane HK, Grossman SR, McVean G, Turnbaugh PJ et al. (2011). Detecting novel associations in large data sets. Science 334: 1518–1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhoads WJ, Ji P, Pruden A, Edwards MA. (2015). Water heater temperature set point and water use patterns influence Legionella pneumophila and associated microorganisms at the tap. Microbiome 3: 67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhoads WJ, Pruden A, Edwards MA. (2016. a). Surveys of green building water systems reveals elevated water age and water quality concerns. Environ Sci: Water Res Technol 2: 164–173. [Google Scholar]
- Rhoads WJ, Pruden A, Edwards MA. (2016. b). Convective mixing in distal pipes exacerbates Legionella pneumophila growth in hot water plumbing. Pathogens 5: E29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp CE, Brady AL, Sharp GH, Grasby SE, Stott MB, Dunfield PF. (2014). Humboldt's spa: microbial diversity is controlled by temperature in geothermal environments. ISME J 8: 1166–1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinert M, Hentschel U, Hacker J. (2002). Legionella pneumophila: an aquatic microbe goes astray. FEMS Microbiol Rev 26: 149–162. [DOI] [PubMed] [Google Scholar]
- Temmerman R, Vervaeren H, Noseda B, Boon N, Verstraete W. (2006). Necrotrophic growth of Legionella pneumophila. Appl Environ Microb 72: 4323–4328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valster RM, Wullings BA, van den Berg R, van der Kooij D. (2011). Relationships between free-living protozoa, cultivable Legionella spp., and water quality characteristics in three drinking water supplies in the Caribbean. Appl Environ Microb 77: 7321–7328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vázquez-Baeza Y, Pirrung M, Gonzalez A, Knight R. (2013). EMPeror: a tool for visualizing high-throughput microbial community data. Gigascience 2: 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wadowsky RM, Butler LJ, Cook MK, Verma SM, Paul MA, Fields BS et al. (1988). Growth-supporting activity for Legionella pneumophila in tap water cultures and implication of hartmannellid amoebae as growth factors. Appl Environ Microb 54: 2677–2682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Edwards MA, Falkinham JO III, Pruden A. (2012). Molecular survey of the occurrence of Legionella spp., Mycobacterium spp., Pseudomonas aeruginosa, and amoeba hosts in two chloraminated drinking water distribution systems. Appl Environ Microb 78: 6285–6294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Edwards MA, Falkinham JO III, Pruden A. (2013). Probiotic approach to pathogen control in premise plumbing systems? A review. Environ Sci Technol 47: 10117–10128. [DOI] [PubMed] [Google Scholar]
- Whiley H, Keegan A, Fallowfield H, Bentham R. (2014). Detection of LegionellaL. pneumophila and Mycobacterium Avium Complex (MAC) along potable water distribution pipelines. Int J Environ Res Public Health 11: 7393–7405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO. (2007) Legionella and the prevention of legionellosis http://www.who.int/water_sanitation_health/emerging/legionella_rel/en/ Accessed on July 1, 2016.
- Zacheus OM, Martikainen PJ. (2004). Occurrence of legionellae in hot water distribution systems of Finnish apartment buildings. Can J Microbiol 40: 993–999. [DOI] [PubMed] [Google Scholar]
- Zeldovich KB, Berezovsky IN, Shakhnovich EI. (2007). Protein and DNA sequences determinants of thermophilic adaptation. PLoS Comput Biol 3: e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H-H, Chen S-N, Huang T-L, Shang P-L, Yang X, Ma W-X. (2015). Indoor heating drives water bacterial growth and community metabolic profile changes in building tap pipes during the winter season. Int J Environ Res Public Health 12: 13649–13661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zietz B, Wiese J, Brengelmann F, Dunkelberg H. (2001). Presence of Legionellaceae in warm water supplies and typing of strains by polymerase chain reaction. Epidemiol Infect 126: 147–152. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.