Abstract
Marine microbes exhibit seasonal cycles in community composition, yet the key drivers of these patterns and microbial population fidelity to specific environmental conditions remain to be determined. To begin addressing these questions, we characterized microbial dynamics weekly for 3 years at a temperate, coastal site with dramatic environmental seasonality. This high-resolution time series reveals that changes in microbial community composition are not continuous; over the duration of the time series, the community instead resolves into distinct summer and winter profiles with rapid spring and fall transitions between these states. Here, we show that these community shifts involve switching between closely related strains that exhibit either summer or winter preferences. Moreover, taxa repeat this process annually in both this and another temperate coastal time series, suggesting that this phenomenon may be widespread in marine ecosystems. To address potential biogeochemical impacts of these community changes, PICRUSt-based metagenomes predict seasonality in transporters, photosynthetic proteins, peptidases and carbohydrate metabolic pathways in spite of closely related summer- and winter-associated taxa. Thus, even small temperature shifts, such as those predicted by climate change models, could affect both the structure and function of marine ecosystems.
Introduction
Planktonic microbes dominate the biomass and biogeochemical cycles in the world's oceans. Although a number of environmental variables have been shown to affect microbial communities, the field lacks consensus on how strongly these environmental factors influence community assembly (Nemergut et al., 2013). Moreover, quantifying the relative importance of environmental selection, stochasticity, dispersal, evolution and predation in shaping microbial community composition is essential to predicting ecosystem responses to environmental alterations including climate change (Doney et al., 2012). Here, we focus on the impact of environmental selection on marine microbiomes. Yet, identifying key variables through observational sampling is complicated by a number of factors, including covariation of environmental variables (for example, temperature, light and day length; Gilbert et al., 2012; Yung et al., 2015), stochastic effects (Baltar et al., 2015), geographic differences in communities and responses and ecological divergence in closely related microbes (Johnson et al., 2006; Hunt et al., 2008; Yung et al., 2015). Despite these challenges, there is general agreement that bacterioplankton respond to physical conditions (for example, temperature, salinity), resource availability (for example, nutrients, organic matter) and interactions with other organisms (Fuhrman et al., 2006; Steele et al., 2011; Gilbert et al., 2012; Gifford et al., 2014; Hunt and Ward 2015; Salter et al., 2015; Yung et al., 2015). However, differentiating between potential community drivers requires well-resolved environment and microbial community characterization to tease apart relationships between phylogeny, community and environment.
With the growing evidence of global climate change's impacts on marine ecosystems, there is increasing urgency to mechanistically link the impact of environmental changes on microbial populations and their biogeochemical processes. Culture-based and modeling studies have primarily focused on potential shifts in the types, geographic range and productivity of phytoplankton, due to their importance in biogeochemical cycling (Thomas et al., 2012; Dutkiewicz et al., 2013). Yet, to fully understand the ecological and biogeochemical implications of climate change, we must consider climatic influences on the diversity and activity of the entire marine microbiome. Compared with phytoplankton, predicting bacterial responses to environmental changes is complicated by the wide range of substrates used by heterotrophic bacteria and different functional roles for even very closely related bacteria (Kashtan et al., 2014; Yawata et al., 2014). To gain insight into how future climate conditions might affect microbial community structure and function, we characterize the microbiome of a coastal marine ecosystem with large seasonal changes in temperature and other environmental parameters.
High-resolution, long-term time series can track microbial responses to environmental changes as they span the range of seasonal and episodic environmental conditions. In contrast with monthly oceanographic measurements or single research cruises, they can better follow both environmental variables and microbial community composition over time to capture time-lagged responses, resolve within-season dynamics, identify rare events and situate these disturbances into annual cycles and long-term climatic trends (Gilbert et al., 2012; Needham et al., 2013; El-Swais et al., 2015; Hunt and Ward, 2015; Lindh et al., 2015; Teeling et al., 2016). Here, we examine weekly changes in the bacterioplankton community at a temperate coastal site to identify the key environmental drivers that shape microbial communities. With both high-resolution (weekly) sampling and several years of observations, this data set is well-positioned to examine the drivers and mechanisms underlying annual cycles in bacterial community composition.
Materials and methods
Environmental sampling
The samples were collected at the Pivers Island Coastal Observatory (PICO) site (34.7181 °N 76.6707 °W) near the Beaufort Inlet (US East Coast; Supplementary Figure S1A) weekly from January 2011 to December 2013. Seawater was collected at 1030 h local time using a 5 liter Niskin bottle centered at 1 m or a peristaltic pump with the tubing open at 1 m and processed within 1 h. Methods for determination of surface water temperature, pH, salinity, dissolved inorganic nutrient concentrations, chlorophyll a concentration and bacterioplankton and phytoplankton abundances were described previously (Hunt et al., 2013; Johnson et al., 2013).
DNA extraction and sequencing
Microbial biomass was collected by filtering ~1 liter of seawater through a 0.22-micron Sterivex filter (Millipore, Darmstadt, Germany) and the filters were stored at −80 °C until extraction. Nucleic acids were extracted as described previously (Massana et al., 1997), with some modifications. In brief, the cells were lysed by bead-beating on ice three times for 30 s in lysis solution (0.75 m sucrose, 40 mm EDTA, 50 mm Tris pH 8.0), followed by consecutive incubations with lysozyme (60 mg ml−1; 37 °C) and SDS (1% 55 °C). DNA was purified by phenol–chloroform extraction, RNase treatment, isopropanol precipitation and PCR inhibitor removal (Zymo, Irvine, CA, USA). DNA concentration was measured using a NanoDrop ND-1000.
Microbial communities were characterized using a dual index amplicon library approach targeting the 16S rRNA gene V3–V4 region (Kozich et al., 2013; Yung et al., 2016). PCR reactions contained 20 ng of template gDNA and 0.4 U of Q5 DNA polymerase (NEB, Ipswich, MA, USA) as well as a final concentration of 200 μm dNTPs, 2 mm MgCl2 and 0.5 μm of each primer. PCR reactions were thermocycled using the following protocol: 98 °C for 30 s; and 28 cycles at 98 °C for 10 s, 55 °C for 30 s and 72 °C for 30 s; with a final extension at 72 °C for 2 min. Triplicate reactions per sample were pooled and gel-purified. In total, 151 libraries were paired-end (2 × 250 bp) sequenced on the MiSeq (Illumina, San Diego, CA, USA) at Duke's Genome Sequencing and Analysis Core Facility.
Sequence processing
Sequences were demultiplexed and assigned to corresponding samples using CASAVA (Illumina). Sequences were processed using USEARCH v7 (Edgar, 2010). Briefly, low-quality sequence ends were trimmed at Phred quality (Q) of 30 using a 10 bp running window. Paired-end reads were merged when reads had a ⩾10 bp overlap with no mismatches; the resulting joined sequences were then filtered to remove reads with expected errors >1 or a length <400 nt. At this point, singleton sequences were excluded and the remaining sequences were assigned to operational taxonomic units (OTUs) of 98.5% pairwise identity using the centroid-based UPARSE-OTU clustering algorithm (Edgar, 2013), resulting in OTUs of at least 97% similarity. Chimeras were removed at the OTU clustering step and using ChimeraSlayer in UCHIME (Edgar et al., 2011). OTUs occurring less than five times in the entire data set were removed, yielding a total of 10 290 OTUs and 3 032 382 sequences in the weekly data set (Supplementary Table S1). Libraries were sub-sampled to 20 082 reads per library and corrected for rRNA gene operon number using rrnDB (Stoddard et al., 2014), then normalized by the total prokaryotic cell counts as measured using flow cytometry to account for seasonal patterns in cell abundances (Alonso-Sáez et al., 2015). Although libraries contain plastid sequences that are not included in the cell counts, these sequences comprise an average of 12% of total reads and, therefore, their inclusion is unlikely to substantially alter the observed patterns in microbial community dynamics. The inverse Simpson's diversity index was computed using the ‘vegan' R package (Oksanen et al., 2013), and a Loess-smoothed curve (span=0.75) was used to show trends in diversity.
Characterization of the microbial community
Canonical correspondence analysis (CCA; ter Braak and Verdonschot, 1995) was used to visualize the temporal dynamics of the microbial community and to identify the environmental and biological factors that were most closely associated with compositional changes. Although the environmental variable data set was largely complete, missing data points (<1% of metadata) were replaced with the average value observed in the time series. Microbial communities were related to a constrained set of environmental and biological variables using CCA implemented in the vegan package (Oksanen et al., 2013). The constrained parameter set was determined by performing a stepwise selection (Akaike information criterion, 999 permutations per step) using the ‘step' function in vegan. Synchronous dynamics in the 100 most abundant OTUs, which comprise 68% of total reads, were revealed using soft clustering with Mfuzz (Kumar and Futschik, 2007). The number of clusters was set to 10 and the fuzzifier variable was set to 1.08. Of the 100 most abundant OTUs, 99 had a membership value of at least 0.7 and thus were assigned to clusters, and no OTUs were assigned to multiple clusters. As bacterial taxa may have delayed responses to different environmental drivers, we cross-correlated the 100 most abundant OTUs with select environmental variables (temperature, no-sky projected daily insolation, salinity, chlorophyll a and ammonium) for a range of lag times using the CCF function in R.
Construction of the phylogenetic tree
The most common sequence in each of the 100 most abundant OTUs was automatically aligned using PyNAST, followed by manual refinement and filtering to remove poorly aligned hypervariable regions using a lane mask, to yield a 402 bp alignment of the partial 16S rRNA gene. We then used PhyML 3.0 to construct a maximum-likelihood tree with automatic model selection by SMS to find the best-fit substitution model and transition/transversion ratio (Guindon et al., 2010). The following parameter settings were used: DNA substitution was modeled using the GTR parameter; estimated proportion of invariable nucleotide sites was 0.342; the gamma shape parameter was set to 1.228; six gamma rate categories were used and a BIONJ tree was initially used and improved using nearest-neighbor interchange with 1000 bootstrap replicates. The taxonomies of representative sequences were classified using the RDP naive Bayesian classifier using the Greengenes version 13.5 database.
Partitioning ubiquitous OTUs into sub-OTUs
We used the minimum entropy decomposition pipeline version 2.1 (Eren et al., 2015) to partition the 35 ubiquitous OTUs into sub-OTUs. In brief, minimum entropy decomposition uses nucleotide entropy along the length of the sequence reads to differentiate genetic variation from sequence error. The sequence reads are partitioned into sequentially refined ‘nodes' based on the nucleotide present in the position with the highest entropy. This iterative process continues until nodes achieve a minimum entropy threshold, which was set to 0.0965. We set the minimum substantive abundance of an oligotype to 0.02% of the total reads to reduce noise from rare sequences, allowed within-node variation of four nucleotides and used default criteria for all other parameters.
PICRUSt predictions of marine metagenomes
The metagenome of PICO communities was predicted based on the 16S rRNA gene library composition using PICRUSt (Langille et al., 2013). In brief, after the removal of OTUs annotated as chloroplasts or mitochondria, the 16S rRNA gene sequences were used for closed-reference OTU picking based on the Greengenes v. 13.5 database using MacQIIME v.1.9.1 (Caporaso et al., 2010). The resulting OTU table was then rarified to 19 506 sequences per sample and used to predict the genes and gene families present in the seawater samples using PICRUSt v.1.0.0. We calculated the weighted nearest sequenced taxon index scores for each sample, representing the average branch length separating an OTU from a reference OTU (mean=0.12, s.d.=0.02). We then collapsed the predicted KEGG Orthology into KEGG pathways and KEGG categories and used one-way analysis of variance with Tukey's honest significant difference post hoc to test the associations between relative abundance of metagenomic functions (at both the KEGG pathway and KEGG category level) and seasonality (summer, winter and transitional periods). To define summer, winter and transition periods, we first use local regression (LOESS) to fit smooth curves to temperature data and then calculated the rate of temperature change. We found that most rapid temperature changes occurred between 15 and 22.5 °C. Therefore, we define seasons based on this range, that is, summer >22.5 °C, winter <15 °C, with sampling points between 22.5 and 15 °C comprising the transition periods. As temperature fluctuates from week to week, we differentiated summer and winter seasons when the temperature remained in a new temperature regime for two consecutive weeks.
Re-analysis of the Western English Channel microbial community
The quality-filtered 16S rRNA gene sequence data set generated by the Plymouth Marine Lab time series from the Western English Channel (Gilbert et al., 2012) was downloaded from the MG-RAST server. Sequence clustering was performed as described above, with OTU assignment to 98.5% pairwise identity and sequence libraries were rarefied to 4584 reads per library. As described previously, Mfuzz was used for assigning the 100 most abundant OTUs to soft clusters by setting number of clusters to 10 and the fuzzifier variable to 1.1. OTUs were assigned to clusters to which they had a membership value of at least 0.7; 96 OTUs met this threshold and no OTUs were assigned to multiple clusters.
Results and Discussion
To identify the key factors in microbial community assembly, bacterial community composition and a suite of environmental variables were measured weekly for 3 years (January 2011–December 2013) as part of the PICO (Supplementary Figure S1). The PICO time series, located adjacent to the Beaufort Inlet, Beaufort NC USA (34.7 °N 76.7 °W), captures the dynamics of the temperate, coastal ocean including large seasonal changes in environmental variables such as light, temperature and chlorophyll a (Figure 1a; Johnson et al., 2013). This site encompasses a larger range in environmental parameters than many long-term marine microbial time series, which are located farther from the coast and in more consistent environments (for example, North Atlantic [HOT], Sargasso Sea [BATS], Coastal California [SPOT], Western English Channel). Although our site is at the mouth of an estuary and in relatively shallow water (depth ~4.5 m), environmental conditions including salinity (mean=32.69 ppt; s.d.=2.16) reflect a strong coastal ocean, rather than estuarine signature. Nevertheless, the terrestrial influence is evident in post-storm nutrient pulses, in contrast with the generally low nutrients throughout much of the year (Johnson et al., 2013).
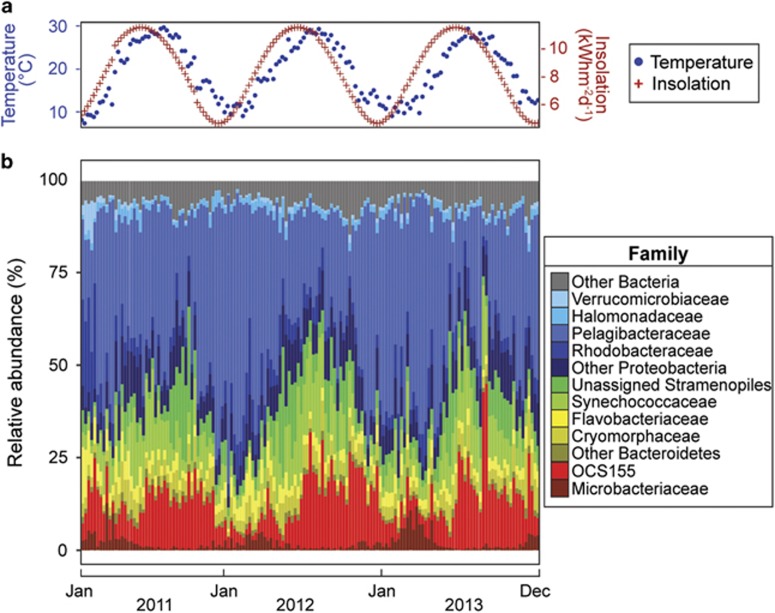
Figure 1.
Changes in environmental parameters and the microbial community at the Pivers Island Coastal Observatory (PICO). (a) Plot showing water temperature (°C) and predicted blue-sky isolation (kWh m−2 d−1) for weekly measurements over 3 years (January 2011–December 2013). (b) 16S rRNA gene sequence library composition based on RDP Classifier taxonomic identification of operational taxonomic units (OTUs; 97% sequence similarity). Taxonomy is displayed at the level of family. Families compromising less than 1% of total relative abundance over the entire data set and OTUs that were not identified at the family level are grouped at the phylum level, except for chloroplast sequences that are grouped at the order level. Taxonomic groups comprising an average of less than 1% relative abundance over the entire data set are labeled as ‘Other Bacteria'.
In this dynamic coastal environment, we explored microbial community composition by sequencing amplicon libraries of the 16S rRNA gene V3–V4 region and clustering these sequences into OTUs of at least 97% similarity (Edgar, 2013). As observed in other coastal locations, the bacterioplankton is dominated by Pelagibacteraceae (SAR11), Actinobacterial family OCS155 and Synechococaceae (Gilbert et al., 2012; Chow et al., 2013) and exhibits an annual cycle in the microbiome (Figure 1b). In winter, the relative abundance of Pelagibacteraceae (SAR11) increases, while the summer is enriched in the photosynthetic Synechococaceae and eukaryotic chloroplast sequences, as well as the Actinobacterial family OCS155. Thus, this site exhibits a typical coastal microbiome with seasonal patterns in both environmental parameters and the microbial community (Figure 1).
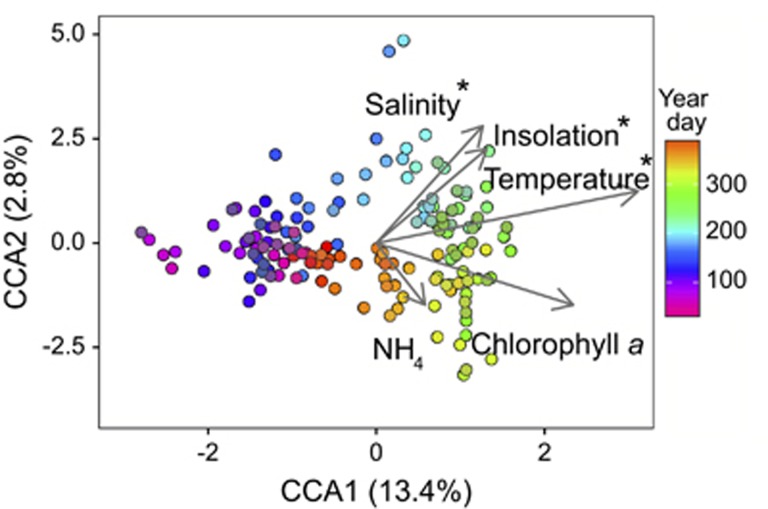
Although the microbial community follows a repeating annual trajectory, community change is not continuous: winter and summer communities are separated along the first CCA axis with a relatively rapid transition between these two states (Figure 2; Supplementary Figures S2 and S3b). These two seasonal microbial communities contrast with multiple distinct community states observed in other locations (Chow et al., 2013; Gifford et al., 2014, Alonso-Sáez et al., 2015; El-Swais et al., 2015). We may observe fewer community states in the PICO time series due to the absence of large spring and fall phytoplankton blooms and subsequent microbial community succession (Needham and Fuhrman, 2016; Teeling et al., 2016). However, like other time series, we do observe a strong annual pattern in the bacterial community (Fuhrman et al., 2006; Figure 2). To identify the environmental factors driving shifts between summer and winter communities, the CCA includes a constrained set of environmental variables determined using forward stepwise selection (P<0.01). Even though the first two CCA axes explain a small fraction of community variability (Figure 2), temperature, insolation and salinity are associated with community changes (P<0.001; Figure 2). This result mirrors that of previous surveys that found a large ‘seasonal driver,' which encompasses both light (day length) and water temperature (Gilbert et al., 2012; Chow et al., 2013, Sunagawa et al., 2015).
Figure 2.
Canonical correspondence analysis (CCA) biplot. CCA relating community composition to environmental variables for each sample from the PICO time series. The percent of variation in the microbial community explained by each axis is indicated in parentheses after the axis label. Environmental variables used in the CCA are represented by vectors, labels used in the figure are indicated by parentheses: water temperature (temperature), projected no-sky daily insolation (insolation), salinity, chlorophyll a and ammonium (NH4). Each circle represents the microbial community composition at a specific time point and the color gradient indicates the year day. Environmental variables marked with asterisks are statistically significant (P<0.001), as assessed by the marginal effects of terms.
Based on our observations, this seasonal driver results in rapid transitions between summer and winter communities roughly coinciding with fall and spring peaks in alpha diversity (Supplementary Figure S3a). This finding contrasts with winter diversity maxima reported in other locations (Gilbert et al., 2012; Ladau et al., 2013), yet winter diversity peaks may be an artifact of limited sampling coupled with greater winter community evenness or vertical mixing of microbes from different water depths (Caporaso et al., 2011; García et al., 2015). Although annual microbiome cycles reflect distinct summer and winter communities, the question remains: are these seasonal differences due to changes in abundance of ubiquitous taxa, or temporal partitioning of closely related taxa within families? To investigate this idea further, we looked at the dynamics of individual OTUs across seasons.
Seasonal signals in individual OTUs
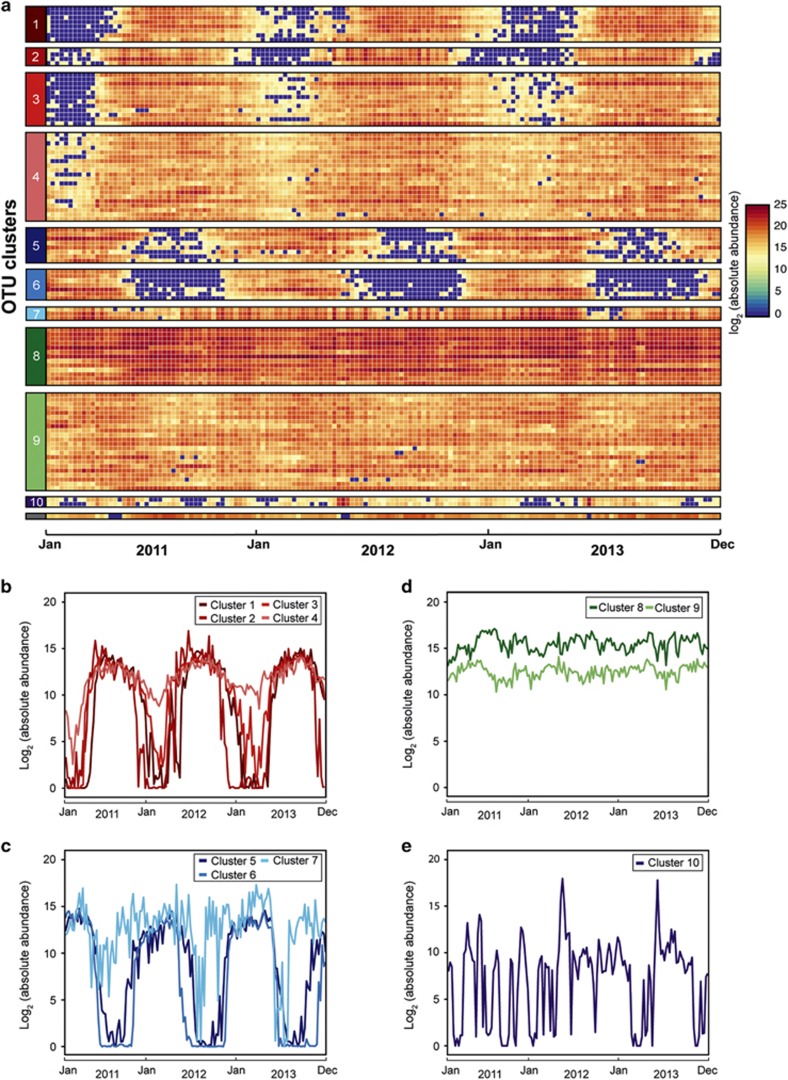
To understand how individual taxa rather than the whole community respond to environmental cues, we examined the temporal dynamics of OTUs' absolute abundances (rRNA operon-correct library relative abundance multiplied by total prokaryotic cell concentrations as determined using flow cytometry). Owing to the numerous biases inherent in amplicon library preparation including DNA extraction, amplification and sequencing (Hunt and Ward, 2015), we do not interpret absolute abundances as direct cell counts; however, this correction does normalize for the roughly five-fold seasonal change in the total prokaryotic cell concentration (Supplementary Figure S4). Visual inspection of an OTU heat map reveals a clear pattern across the microbial community: most taxa are strongly associated with either the summer or winter (Figure 3a). In contrast to recent research, which argues that many functionally redundant taxa can occupy a specific environmental niche (Louca et al., 2016), here we observe consistent taxa seasonally reoccurring over the 3 years of the study at this single site. To determine the extent of temporal niche partitioning, we used an unsupervised approach, using soft clustering to assign the 100 most abundant OTUs to 10 clusters. These clusters reflect differences in both temporal patterns and absolute abundance (Figure 3b–e). On the basis of visual similarities in temporal patterns, we then combined these clusters into four groups: ubiquitous, episodic, summer-associated and winter-associated (Figure 3). In this analysis, we observe OTUs present throughout the time series at relatively high absolute abundance (ubiquitous group: clusters 8 and 9; Figure 3d), including a number of OTUs belonging to the SAR11 clade. The episodic group (cluster 10) also exhibits a phylogenetic signal as both OTUs are identified as Stramenopile chloroplasts; these algal sequences display concordant dynamics that are not aligned along a seasonal axis but anecdotally appear to increase following storms (Hunt and Ward, 2015). Yet, seasonal association is the dominant pattern in this environment, as 62 of the 100 most abundant OTUs peak in either the winter or summer. This seasonality is much higher than observed in previous studies that classified between 10 and 30% of OTUs as seasonally associated (Chow et al., 2013; Alonso-Sáez et al., 2015). This strong seasonality could reflect methodological approaches: soft clustering, more frequent sampling and greater sequencing depth. Alternatively, PICO microbes may experience stronger environmental filtering due to large changes in environmental parameters. However, replicating this analysis using microbial communities from the Western English Channel again reveals strong seasonal associations (82 of the 100 most abundant OTUs), despite decreased sampling resolution, sequencing depth and the range of environmental variables (Supplementary Figure S5), suggesting that seasonality may be more readily detected using soft clustering compared with other approaches. Yet, in spite of strong seasonal associations at PICO, clusters exhibit slightly different patterns within seasonal groups; while some seasonally associated taxa are absent in the non-preferred season, others are detectable throughout the year. For example, cluster 4 is most abundant in the summer but is still observed at lower abundances in the winter (Figure 3b). Thus, although most OTUs exhibit strong seasonal preferences, these organisms may either be able to survive the wide range of environmental conditions that occur over the annual cycle or potentially exhibit sub-OTU seasonal partitioning.
Figure 3.
Seasonal patterns for the 100 most abundant OTUs over the 3-year PICO time series. (a) The heat map depicts the log2 (absolute abundance; cells ml−1) for each OTU, obtained by taking rRNA operon corrected relative library abundance and total prokaryotic cell counts to calculate a ‘cell count' for each OTU. OTUs are ordered based on patterns observed in soft clustering, shown as numbered boxes on the left side of the figure. The unnumbered gray OTU was not assigned to a cluster. (b–e) Show the centroids of the 10 soft clusters identified for the 100 most abundant OTUs over the entire PICO time series. Plots show the temporal dynamics of clusters that were visually assigned to (b) summer-associated, (c) winter-associated, (d) ubiquitous and (e) episodic groups.
For ubiquitous OTUs (Figure 3d), we investigated potential seasonal partitioning at the sub-OTU level to determine whether these taxa are likely to be generalists or instead partition resources at a finer scale. Fourteen of the OTUs in the ubiquitous group belong to the family Pelagibacteraceae, which has been previously shown to exhibit fine-scale ecological specialization (Eren et al., 2013); but sub-OTU habitat specialization may be common in the bacterioplankton (Yung et al., 2015). To test this hypothesis, we used minimum entropy decomposition (Eren et al., 2015) to examine potential sub-OTU temporal specialization in apparently ‘ubiquitous' OTUs using soft clustering (Supplementary Figure S6). Although the majority of sub-OTUs did not exhibit seasonal patterns, 41% were assigned to either summer- or winter-peaking clusters (Supplementary Table S2). These seasonally associated sub-OTUs are observed in a wide range of phylogenetic groups including SAR11, Synechococcaceae, Halomonadaceae, Flavobacteriaceae, OCS155 and Rhodobacteraceae, suggesting that sub-OTU seasonal patterns are a common feature in marine microbes (Johnson et al., 2006; Kashtan et al., 2014; Yung et al., 2015; Larkin et al., 2016). As seasonal pattern detection is limited by the phylogenetic resolution of the 16S rRNA gene, seasonal affiliation may be more widespread in the bacterioplankton than can be discerned using 16S rRNA gene libraries.
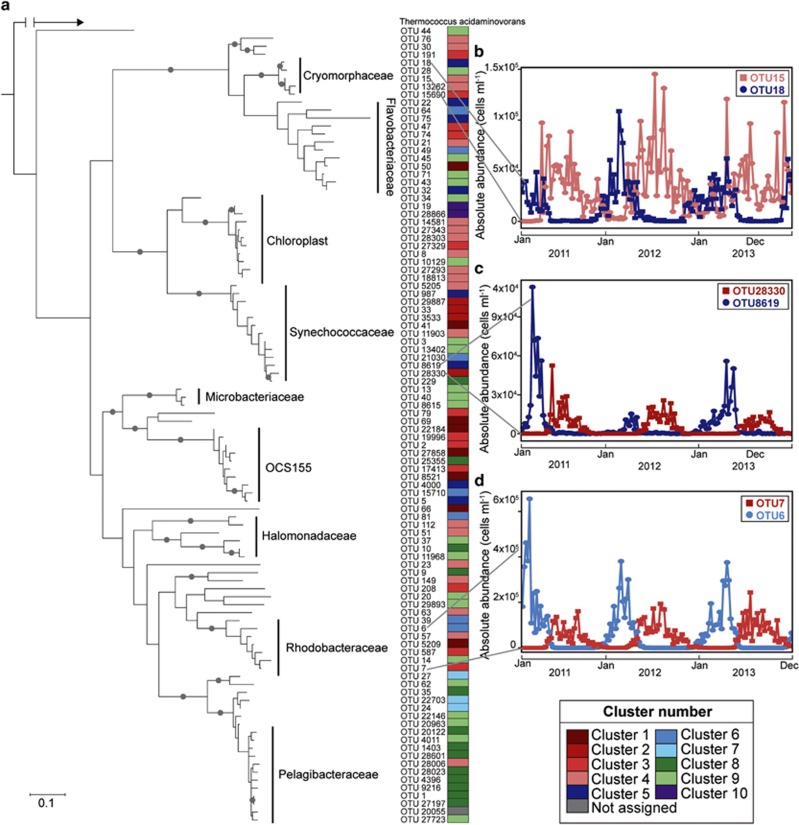
The persistence of many bacterial families throughout the year while individual OTUs or sub-OTUs are seasonally associated suggests that clades within a family might have similar ecological roles in summer and winter (Hunt et al., 2008; Yung et al., 2015) and is consistent with phylogenetic conservation of metabolic capabilities (Martiny et al., 2015). Further supporting this idea, with the exception of the ubiquitous SAR11 and the episodic algal OTUs, seasonal clusters are interspersed throughout a 16S rRNA gene phylogenetic tree (Figure 4a), where previously seasonal patterns were observed only in specific taxonomic groups (Alonso-Sáez et al., 2015). In our data, moreover, we see potential for closely related strains to temporally partition resources (Figure 4b–d). For example, two OTUs within the family Rhodobacteraceae appear to trade off: the dominant OTU within the genus Octadecabacter (OTU 6) and a Phaeobacter-like genus (OTU 7) alternate in dominance over seasonal cycles (Figure 4d). The Octadecabacter OTU peaks in the winter, whereas the Phaeobacter-like OTU exhibits highest absolute abundances in late summer, suggesting seasonal resource partitioning within bacterial families. As similar patterns were observed in a number of taxa (Figure 4b–d), temporal resource partitioning between closely related taxa appears to be a common phenomenon in the bacterioplankton. Although these taxa have different seasonal preferences, they may either fulfill equivalent biogeochemical functional roles or alternately could differ functionally on other ecological axes (Kashtan et al., 2014).
Figure 4.
Seasonal patterns of closely related taxa. (a) Partial 16S rRNA gene maximum-likelihood phylogenetic tree showing the 100 most abundant OTUs from the weekly PICO time series rooted with Thermococcus acidaminovorans. Bootstrap values greater than 80% are indicated by a gray circle at the node. Colored boxes on the right show the assigned clusters (from Figure 3). Larger clades are also labeled with the family assigned by RDP Classifier. (b) Absolute abundances of two OTUs within the genus Phaeocystidibacter (OTUs 15 and 18) in the family Cryomorphaceae over three years of weekly 16S rRNA gene libraries at the PICO time series. (c) Absolute abundances of two OTUs within the genus Synechococcus (OTUs 8619 and 28830). (d) Absolute abundances of two OTUs within the family Rhodobacteraceae, one identified as the dominant OTU within the genus Octadecabacter (OTU 6) and the other a Phaeobacter-like genus (OTU 7).
Seasonal changes in predicted function
With this seasonal switching between closely related taxa, we now ask whether summer and winter communities are largely functionally redundant or alternately whether seasonal environmental conditions select for distinct functions. To investigate potential biogeochemical implications of this seasonal OTU switching, we predict the functional capacity of the summer- and winter-associated microbial communities using existing genomes (Langille et al., 2013). Although this technique is limited by the ability of 16S rRNA gene sequences to resolve ecologically important units and the phylogenetic breadth and depth of marine microbial genomes, metagenomic prediction may nevertheless offer insight into the extent of both functional redundancy and differences in biogeochemical potential (but not rates) between the summer and winter communities (Hunt et al., 2013). Over the 3 years of observation, the dominant KEGG categories are relatively consistent (Supplementary Figure S7); however, summer- and winter-predicted metagenomes are statistically different (ANOSIM, P<0.001). Thus seasonal changes alter apparent biogeochemical potential in the coastal ocean in spite of apparent replacement by closely related strains. In fact, more than half of the KEGG categories adopt a seasonal pattern (analysis of variance, Tukey's test, P<0.0001), including biogeochemically important functions: energy metabolism (summer peak), membrane transport (winter peak), metabolism of cofactors and vitamins (winter peak) and carbohydrate metabolism (winter peak; Supplementary Figure S8). These seasonal predicted gene abundance patterns likely indicate changes in organic matter sources: summer increases in photosynthetic genes suggest production of labile carbon while the winter's more recalcitrant carbon may require additional transporters and carbohydrate degradation genes (Gifford et al., 2014; Sharma et al., 2014). Thus, although summer and winter PICO communities contain largely overlapping functions, seasonal changes in microbial communities alter predicted community metabolic potential.
Environmental drivers of microbiomes
In this study, we observe dramatic seasonal shifts in the microbial community, the abundance of specific OTUs as well as the metabolic potential. We propose two possible drivers for these repeated, seasonal environmental transitions: (i) microbial communities assemble based on the persistence of key environmental variables or (ii) that a keystone organism(s) changes in abundance or activity, likely due to an environmental factor, and triggers a community-wide change mediated by interactions (Hunt and Ward, 2015; Needham and Fuhrman, 2016). We focus on potential environmental drivers, as even an interaction-driven transition requires a seasonally sensitive keystone organism. Given the significance of both temperature and light for community composition in this and other locations (Figure 2; Supplementary Table S3), we predict that these variables are the primary drivers of the microbiome's seasonality (Supplementary Figure S2). Observational data is often poorly suited to separate the impacts of correlated environmental variables; however, cross-correlation analysis indicates seasonal OTUs exhibit stronger correlations and a shorter response lag with temperature compared to light (Supplementary Table S3). Further supporting the importance of temperature over light, the timing of cluster transitions varies on a yearly basis, for example, persistent cool weather in spring 2013 delayed seasonal warming and the appearance of summer-associated OTUs, indicating that temperature is likely to be the proximal driver of seasonal microbial community switching. While light is important for phytoplankton, which could serve as keystone microbes (Hunt and Ward, 2015, Needham and Fuhrman, 2016), temperature has the potential to directly impact the metabolic kinetics of all organisms (Brown et al., 2004). Thus, several lines of evidence point to temperature as the key driver of these community transitions, suggesting seasonal switching may be widespread in temperate regions of the world's oceans. Although temperature is consistently an important factor in determining microbial composition across a range of studies (Fuhrman et al., 2006, 2008; Johnson et al., 2006, Shade et al., 2010; Yung et al., 2015), this predominance of seasonal OTUs and repeated annual transitions at the OTU level have not been previously shown. Although a recent high-resolution (daily) study observed stronger correlations between specific phytoplankton and prokaryotic taxa than with environmental variables (Needham and Fuhrman, 2016), these short-term bloom successions are likely responding to different drivers (for example, DOM composition and nutrients) than those forcing the annual cycles we emphasize here. We previously speculated that OTU transitions might be less pronounced at locations with lower annual temperature ranges, compared with ~20 °C at the PICO study site. However, we observe similar summer- and winter-associated taxa at the Western Channel Observatory (Supplementary Figure S5), which has an annual temperature range of ~10 °C (Gilbert et al., 2012), suggesting that the persistence of a given environmental condition (Supplementary Figure S1b) may be more important than a specific temperature threshold.
Conclusions
Observations are one of the primary methods that scientists use to predict the ecologies of the largely uncultured microbial inhabitants of the world's oceans, yet the challenge is to link these observations to specific environmental drivers. Here we show that a dynamic microbial community inhabiting a temperate site exhibits winter and summer assemblages. On closer examination, community changes are mediated by switches between closely related taxa, suggesting that OTU-level differences in environmental preferences, most likely temperature-related, are responsible for seasonal cycles at the community level. Using 16S rRNA-based metagenome predictions reveals seasonal patterns in the predicted genome content. The question remains whether these predicted gene content changes are due to temperature preferences or the ability to degrade organic material where seasonal changes in composition is dependent on organic material, or perhaps co-selection of community members based on these two factors. The occurrence of temperature-mediated state changes has important implications for predicting microbial responses to future climate conditions: temperature changes result in dramatic responses in the microbiome that apparently influence biogeochemical potential.
Data deposition
Sequences reported in this paper have been submitted to the GenBank Sequence Read Archive under Bioproject #PRJNA309156. Oceanographic metadata are available at BCO-DMO under Project #2281.
Acknowledgments
We acknowledge the entire PICO team for help with environmental sampling. This work was supported by grants from the Gordon and Betty Moore Foundation (GBMF3768 to DEH) and the National Science Foundation (OCE1322950 to DEH; OCE1416665 to DEH and ZIJ; and a GRFP award to CSW).
Footnotes
Supplementary Information accompanies this paper on The ISME Journal website (http://www.nature.com/ismej)
The authors declare no conflict of interest.
Supplementary Material
References
- Alonso-Sáez L, Díaz-Pérez L, Morán XAG. (2015). The hidden seasonality of the rare biosphere in coastal marine bacterioplankton. Environ Microbiol 17: 3766–3780. [DOI] [PubMed] [Google Scholar]
- Baltar F, Palovaara J, Vila-Costa M, Salazar G, Calvo E, Pelejero C et al. (2015). Response of rare, common and abundant bacterioplankton to anthropogenic perturbations in a Mediterranean coastal site. FEMS Microbiol Ecol 91: fiv058. [DOI] [PubMed] [Google Scholar]
- Brown JH, Gillooly JF, Allen AP, Savage VM, West GB. (2004). Toward a metabolic theory of ecology. Ecology 85: 1771–1789. [Google Scholar]
- Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK et al. (2010). QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7: 335–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso JG, Paszkiewicz K, Field D, Knight R, Gilbert JA. (2011). The Western English Channel contains a persistent microbial seed bank. ISME J 6: 1089–1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chow C-ET, Sachdeva R, Cram JA, Steele JA, Needham DM, Patel A et al. (2013). Temporal variability and coherence of euphotic zone bacterial communities over a decade in the Southern California Bight. ISME J 7: 2259–2273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doney SC, Ruckelshaus M, Duffy JE, Barry JP, Chan F, English CA et al. (2012). Climate change impacts on marine ecosystems. Mar Sci 4: 11–37. [DOI] [PubMed] [Google Scholar]
- Dutkiewicz S, Scott JR, Follows M. (2013). Winners and losers: ecological and biogeochemical changes in a warming ocean. Glob Biogeochem Cycles 27: 463–477. [Google Scholar]
- Edgar RC. (2010). Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26: 2460–2461. [DOI] [PubMed] [Google Scholar]
- Edgar RC. (2013). UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat Methods 10: 996–998. [DOI] [PubMed] [Google Scholar]
- Edgar RC, Haas BJ, Clemente JC, Quince C, Knight R. (2011). UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 27: 2194–2200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Swais H, Dunn KA, Bielawski JP, Li WK, Walsh DA. (2015). Seasonal assemblages and short-lived blooms in coastal north-west Atlantic Ocean bacterioplankton. Environ Microbiol 17: 3642–3661. [DOI] [PubMed] [Google Scholar]
- Eren AM, Maignien L, Sul WJ, Murphy LG, Grim SL, Morrison HG et al. (2013). Oligotyping: differentiating between closely related microbial taxa using 16S rRNA gene data. Methods Ecol Evol 4: 1111–1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eren AM, Morrison HG, Lescault PJ, Reveillaud J, Vineis JH, Sogin ML. (2015). Minimum entropy decomposition: unsupervised oligotyping for sensitive partitioning of high-throughput marker gene sequences. ISME J 9: 968–979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuhrman JA, Hewson I, Schwalbach MS, Steele JA, Brown MV, Naeem S. (2006). Annually reoccurring bacterial communities are predictable from ocean conditions. Proc Natl Acad Sci USA 103: 13104–13109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuhrman JA, Steele JA, Hewson I, Schwalbach MS, Brown MV, Green JL et al. (2008). A latitudinal diversity gradient in planktonic marine bacteria. Proc Natl Acad Sci USA 105: 7774–7778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- García FC, Alonso-Sáez L, Morán XAG, López-Urrutia Á. (2015). Seasonality in molecular and cytometric diversity of marine bacterioplankton: the re-shuffling of bacterial taxa by vertical mixing. Environ Microbiol 17: 4133–4142. [DOI] [PubMed] [Google Scholar]
- Gifford SM, Sharma S, Moran MA. (2014). Linking activity and function to ecosystem dynamics in a coastal bacterioplankton community. Front Microbiol 5: 185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert JA, Steele JA, Caporaso JG, Steinbrück L, Reeder J, Temperton B et al. (2012). Defining seasonal marine microbial community dynamics. ISME J 6: 298–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guindon S, Dufayard J-F, Lefort V, Anisimova M, Hordijk W, Gascuel O. (2010). New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol 59: 307–321. [DOI] [PubMed] [Google Scholar]
- Hunt DE, David LA, Gevers D, Preheim SP, Alm EJ, Polz MF. (2008). Resource partitioning and sympatric differentiation among closely related bacterioplankton. Science 320: 1081–1085. [DOI] [PubMed] [Google Scholar]
- Hunt DE, Lin Y, Church MJ, Karl DM, Izzo LK, Tringe S et al. (2013). Relationship between abundance and specific activity of bacterioplankton in open ocean surface waters. Appl Environ Microbiol 79: 177–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunt DE, Ward CS. (2015). A network-based approach to disturbance transmission through microbial interactions. Front Microbiol 6: 1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson ZI, Wheeler BJ, Blinebry SK, Carlson CM, Ward CS, Hunt DE. (2013). Dramatic variability of the carbonate system at a temperate coastal ocean site (Beaufort, North Carolina, USA) is regulated by physical and biogeochemical processes on multiple timescales. PLoS ONE 8: e85117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson ZI, Zinser ER, Coe A, McNulty NP, Woodward EMS, Chisholm SW. (2006). Niche partitioning among Prochlorococcus ecotypes along ocean-scale environmental gradients. Science 311: 1737–1740. [DOI] [PubMed] [Google Scholar]
- Kashtan N, Roggensack SE, Rodrigue S, Thompson JW, Biller SJ, Coe A et al. (2014). Single-cell genomics reveals hundreds of coexisting subpopulations in wild Prochlorococcus. Science 344: 416–420. [DOI] [PubMed] [Google Scholar]
- Kozich JJ, Westcott SL, Baxter NT, Highlander SK, Schloss PD. (2013). Development of a dual-index sequencing strategy and curation pipeline for analyzing amplicon sequence data on the MiSeq Illumina sequencing platform. Appl Environ Microbiol 79: 5112–5120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar L, Futschik ME. (2007). Mfuzz: a software package for soft clustering of microarray data. Bioinformation 2: 5–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ladau J, Sharpton TJ, Finucane MM, Jospin G, Kembel SW, O'Dwyer J et al. (2013). Global marine bacterial diversity peaks at high latitudes in winter. ISME J 7: 1669–1677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langille MG, Zaneveld J, Caporaso JG, McDonald D, Knights D, Reyes JA et al. (2013). Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat Biotechnol 31: 814–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin AA, Blinebry SK, Howes C, Lin Y, Loftus SE, Schmaus CA et al. (2016). Niche partitioning and biogeography of high light adapted Prochlorococcus across taxonomic ranks in the North Pacific. ISME J 10: 1555–1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindh MV, Sjöstedt J, Andersson AF, Baltar F, Hugerth LW, Lundin D et al. (2015). Disentangling seasonal bacterioplankton population dynamics by high-frequency sampling. Environ Microbiol 17: 2459–2476. [DOI] [PubMed] [Google Scholar]
- Louca S, Parfrey LW, Doebeli M. (2016). Decoupling function and taxonomy in the global ocean microbiome. Science 353: 1272–1277. [DOI] [PubMed] [Google Scholar]
- Martiny JB, Jones SE, Lennon JT, Martiny AC. (2015). Microbiomes in light of traits: a phylogenetic perspective. Science 350: aac9323. [DOI] [PubMed] [Google Scholar]
- Massana R, Murray AE, Preston CM, DeLong EF. (1997). Vertical distribution and phylogenetic characterization of marine planktonic Archaea in the Santa Barbara Channel. Appl Environ Microbiol 63: 50–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Needham DM, Chow C-ET, Cram JA, Sachdeva R, Parada A, Fuhrman JA. (2013). Short-term observations of marine bacterial and viral communities: patterns, connections and resilience. ISME J 7: 1274–1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Needham DM, Fuhrman JA. (2016). Pronounced daily succession of phytoplankton, archaea and bacteria following a spring bloom. Nat Microbiol 1: 16005. [DOI] [PubMed] [Google Scholar]
- Nemergut DR, Schmidt SK, Fukami T, O'Neill SP, Bilinski TM, Stanish LF et al. (2013). Patterns and processes of microbial community assembly. Microbiol Mol Biol Rev 77: 342–356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oksanen J, Blanchet FG, Kindt R, Legendre P, Minchin PR, O'Hara R et al. (2013). Vegan: Community Ecology Package. R package version 2.
- Salter I, Galand PE, Fagervold SK, Lebaron P, Obernosterer I, Oliver MJ et al. (2015). Seasonal dynamics of active SAR11 ecotypes in the oligotrophic Northwest Mediterranean Sea. ISME J 9: 347–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shade A, Chiu CY, McMahon KD. (2010). Differential bacterial dynamics promote emergent community robustness to lake mixing: an epilimnion to hypolimnion transplant experiment. Environ Microbiol 12: 455–466. [DOI] [PubMed] [Google Scholar]
- Sharma AK, Becker JW, Ottesen EA, Bryant JA, Duhamel S, Karl DM et al. (2014). Distinct dissolved organic matter sources induce rapid transcriptional responses in coexisting populations of ProchlorococcusPelagibacter and the OM60 clade. Environ Microbiol 16: 2815–2830. [DOI] [PubMed] [Google Scholar]
- Steele JA, Countway PD, Xia L, Vigil PD, Beman JM, Kim DY et al. (2011). Marine bacterial, archaeal and protistan association networks reveal ecological linkages. ISME J 5: 1414–1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoddard SF, Smith BJ, Hein R, Roller BR, Schmidt TM. (2014). rrnDB: improved tools for interpreting rRNA gene abundance in bacteria and archaea and a new foundation for future development. Nucleic Acids Res 43: D593–D598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunagawa S, Coelho LP, Chaffron S, Kultima JR, Labadie K, Salazar G et al. (2015). Structure and function of the global ocean microbiome. Science 348: 1261359. [DOI] [PubMed] [Google Scholar]
- Teeling H, Fuchs BM, Bennke CM, Krüger K, Chafee M, Kappelmann L et al. (2016). Recurring patterns in bacterioplankton dynamics during coastal spring algae blooms. eLife 5: e11888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ter Braak CJ, Verdonschot PF. (1995). Canonical correspondence analysis and related multivariate methods in aquatic ecology. Aquatic Sci 57: 255–289. [Google Scholar]
- Thomas MK, Kremer CT, Klausmeier CA, Litchman E. (2012). A global pattern of thermal adaptation in marine phytoplankton. Science 338: 1085–1088. [DOI] [PubMed] [Google Scholar]
- Yawata Y, Cordero OX, Menolascina F, Hehemann J-H, Polz MF, Stocker R. (2014). Competition–dispersal tradeoff ecologically differentiates recently speciated marine bacterioplankton populations. Proc Natl Acad Sci USA 111: 5622–5627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yung C-M, Vereen MK, Herbert A, Davis KM, Yang J, Kantorowska A et al. (2015). Thermally adaptive tradeoffs in closely-related marine bacterial strains. Environ Microbiol 17: 2421–2429. [DOI] [PubMed] [Google Scholar]
- Yung C-M, Ward CS, Davis KM, Johnson ZI, Hunt DE. (2016). Insensitivity of diverse and temporally variable particle-associated microbial communities to bulk seawater environmental parameters. Appl Environ Microbiol 82: 3431–3437. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.