Abstract
Arbuscular mycorrhizal fungi (AMF) are crucial to plants and vice versa, but little is known about the factors linking the community structure of the two groups. We investigated the association between AMF and the plant community structure in the nearest neighborhood of Festuca brevipila in a semiarid grassland with steep environmental gradients, using high-throughput sequencing of the Glomeromycotina (former Glomeromycota). We focused on the Passenger, Driver and Habitat hypotheses: (i) plant communities drive AMF (passenger); (ii) AMF communities drive the plants (driver); (iii) the environment shapes both communities causing covariation. The null hypothesis is that the two assemblages are independent and this study offers a spatially explicit novel test of it in the field at multiple, small scales. The AMF community consisted of 71 operational taxonomic units, the plant community of 47 species. Spatial distance and spatial variation in the environment were the main determinants of the AMF community. The structure of the plant community around the focal plant was a poor predictor of AMF communities, also in terms of phylogenetic community structure. Some evidence supports the passenger hypothesis, but the relative roles of the factors structuring the two groups clearly differed, leading to an apparent decoupling of the two assemblages at the relatively small scale of this study. Community phylogenetic structure in AMF suggests an important role of within-assemblage interactions.
Introduction
Arbuscular mycorrhizal fungi (AMF) are one of the most important symbiont groups for plants, forming relationships with the majority of land plants and having a significant role in the acquisition of phosphorus (Smith and Read, 2008). Yet, despite some important progress in recent years, especially in relation to interactions with other soil biota or how AMF respond to management (Alguacil et al., 2014; Caravaca and Ruess, 2014; Leifheit et al., 2015; Knegt et al., 2016), there are many aspects of the assembly processes regulating the community ecology of these organisms that are poorly understood: a key challenge remains disentangling the relative contribution of dispersal limitation, environmental filtering and biotic interaction on AMF community structure (Vályi et al., 2016). The cryptic nature of the group and the complexity of the three-way interaction between plants, AMF and the environment complicate the study of the factors that regulate AMF community structure. Dispersal limitation remains one of the most complex aspects of AMF ecology (Zobel and Öpik, 2014): as for example reviewed in Vályi et al. (2016), AMF can disperse via local mycelium spread but also spores, hyphal fragments and colonized root fragments, and the importance of these mechanisms could be scale-dependent, although direct evidence is missing. Still, large AMF spores and hyphal fragments are mostly spread via zoochory, which implies limited dispersal capability and seems reflected by small-scale patterns in community structure (Mummey and Rillig, 2008; Dumbrell et al., 2010a; Horn et al., 2014). The effects of dispersal limitations are entangled with those of environmental gradients, biotic interactions within the AMF assemblage, and between AMF and plants (for example, Mummey and Rillig, 2008; Dumbrell et al., 2010a; Horn et al., 2014; Martinez-Garcia et al., 2015; García de León et al., 2016a, 2016b).
The study of AMF in grasslands is of particular importance as grassland ecosystems cover a significant proportion of the earth's surface, harbor the majority of herbaceous plant diversity (Shantz, 1954) and it is in grasslands that AMF reach their highest abundance and diversity (Treseder and Cross, 2006; Kivlin et al., 2011). Studies on plant biodiversity in grassland ecosystems at small scales have revealed connections between species richness of AMF and plants (Hiiesalu et al., 2014) and host plant effects on AMF community composition (Vályi et al., 2015). Still, effects can be very localized: AMF can form extended hyphal networks but spatial autocorrelation in their distribution is typically found at submeter scales (Mummey and Rillig, 2008), with a potential role for biotic interactions (Vályi et al., 2016). To date, only a few studies have taken this fact into account and applied a sufficiently fine-grained sampling design for a solid statistical analysis of the patterns generated by local processes (Dumbrell et al., 2010b; Horn et al., 2014).
AMF and plants form two sets of communities associated with each other but assembled through different processes that take place at different spatial and temporal scales (Zobel and Öpik, 2014). The plant set can drive the fungal set or vice versa (Figure 1), but which group is driving might depend on successional stage, which is linked to differences in dispersal processes between plants and AMF. Zobel and Öpik (2014) have used the concept of difference in dispersal between AMF and plants to revisit the Driver and Passenger hypotheses originally proposed by Hart et al. (2001). Zobel and Öpik (2014) also formulated the Habitat hypothesis to distinguish a situation where AMF and plant communities covary but are not directly causally linked, as opposed to the null hypothesis of no covariation (‘independence'). For example, during primary succession, plants typically arrive before AMF and then act as a potential filter to AMF: AMF are Passengers as they are following plants. However, dispersal limitation in an established AMF assemblage can cause the AMF assemblage to more strongly determine which plants will establish during secondary succession: the AMF assemblage becomes the Driver (Zobel and Öpik, 2014). Zobel and Öpik (2014) further predict that the Habitat hypothesis would be most common in regions with a stable community (for example, climax vegetation) where environmental variation within regions will cause a non-mechanistic covariation between AMF and plant communities. The general null hypothesis is that plants and AMF may vary independently of each other, which could possibly happen at very broad or global scales, where plants are more disperal limited than AMF seem to be (Kivlin et al., 2011; Öpik et al., 2013; Davison et al., 2015). Accordingly, Vályi et al. (2016) have recently proposed that the host effect is minimal at regional and global scales.
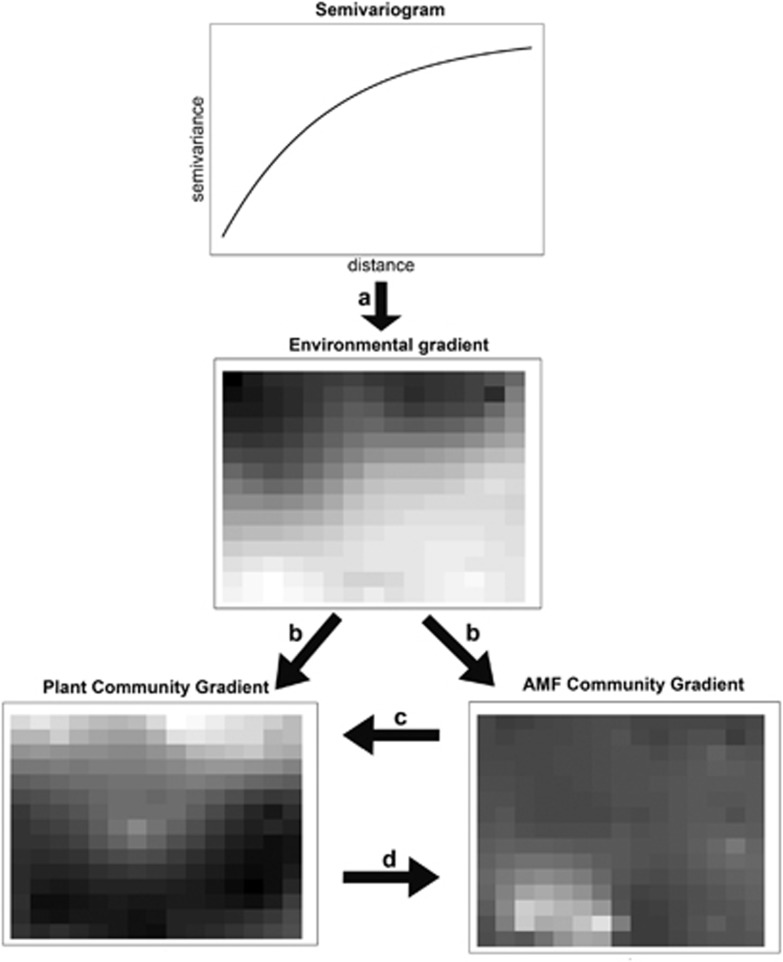
Figure 1.
Autocorrelation (Semivariogram) and trends in environmental variables create (arrow a) spatial structure and environmental gradients. Variation in the environment generates variation in plants and AMF (arrows b). AMF and plants can thus be structured by changes in habitat conditions, which can then simply lead to covariation between the two assemblages (Habitat hypothesis). Alternatively, AMF could either drive the plant assemblage (Driver hypothesis, arrow c) or be driven by the plant assemblage (Passenger hypothesis, arrow d). In all cases, the driving factors/assemblage (b–d) have a spatial structure that will be, at least partially, reflected by spatial structure in the driven assemblage. This spatial dependence calls for a spatially explicit approach to the testing of the three hypotheses. Spatial scale and successional stage have also been hypothesized to be the major factors in determining which among the Habitat, Driver and Passenger hypotheses apply to real systems. In addition to all these factors, AMF can also be structured by interactions within the assemblage, independently of plants, which has been hypothesized to happen at local scale and that could create very patchy distribution. All data are simulated.
There are studies that have touched upon components of these hypotheses. For example, AMF taxa are generally found to be able to colonize any AM (as opposed to non-AM) plant species (Klironomos, 2000), still there may be a bias towards easily cultivable species (Ohsowski et al., 2014) and ‘specificity' might be quantitative rather than qualitative (Vályi et al., 2015). Therefore, AM fungal communities and plant communities may still be directly causally correlated despite the perceived generalism of the AM symbiosis. A thorough account of the studies supporting the various hypotheses is given in Zobel and Öpik (2014) and we are aware of only two recent, observational studies that have addressed the subject (Martinez-Garcia et al., 2015; García de León et al., 2016a). However, a problematic aspect of observational field studies remains to tease apart cause and effect in the correlations between the two organism groups in the presence of spatial structure in the environment (Figure 1). To solve this problem, we applied a spatially explicit design to sample AMF and plant communities along a replicated steep but short (≈15 m) soil environmental gradient (Horn et al., 2014). We could therefore control for spatial patterns and environmental effects when testing for the effects of plants on AMF communities and vice versa. We used a standardized focal plant of high abundance to investigate environmental, plant and AMF community variation at sufficiently small scales. We also took into account the phylogenetic community structure of both plant and AMF assemblages to allow community relationships to occur at levels other than species/operational taxonomic unit (OTU) between and within the groups.
Our main aim was to collect for the first time multiple scales and high spatial resolution data to test the general null hypothesis that plant community structure, including phylogenetic structure, is independent of AMF community structure and vice versa. If the hypothesis were rejected, given the scales included in the study, we aimed to collect support for one or more of the three alternative hypotheses (Figure 1), with the overall goal of shedding light on the mutual relationships between plant and AMF communities.
Materials and methods
Study area and sample collection
Sampling was conducted in a nature protection area located in north-eastern Germany (Brandenburg, 52°27.778'N, 14°29.349'E), a Natura 2000 biodiversity hotspot that contains over 200 different plant species and combines floral elements of steppes and coastal habitats. Given the high diversity of plants (Ristow et al., 2011) and AMF (Horn et al., 2014), the area is very suitable for this study. We sampled by a hierarchical nesting of plots in April 2011: twelve 3 × 3 m2 plots were sampled at the four corners of three 15 × 15 m2 larger plots (henceforth called ‘macroplots') located on the slope of a hillside (Supplementary Figure S1). The distances between the macroplots ranged from 20 to 500 m (Supplementary Figure S2), leading to overall intersample distances from a few cm to 3 m (within a plot) and up to 500 m between macroplots. The uphill–downhill axes of the three macroplots were characterized by a steep textural gradient from sandy-loamy (uphill) to highly sandy (downhill) soils (Supplementary Figure S3). Soil parameters varied significantly and to a large extent (for example, almost 3 units of pH) along the texture gradient (Horn et al., 2015).
We assessed the local AM fungal community in the roots and surrounding soil of Festuca brevipila plants plus the neighboring plant species around these Festuca plants. F. brevipila is one of the most abundant species in sampled plots (Ristow et al., 2011; Horn et al., 2015). Soil cores (5 cm radius, 15 cm deep) were taken from five F. brevipila plants per plot, resulting in 60 (5 plants x 12 plots) sampling locations. Each sample position was random within the plot (minimum distance of 30 cm between any two samples in the same plot; Supplementary Figure S1). Plant presence/absence was assessed in the surrounding area in a radius of 15 cm around each soil core to target local interactions present in the rhizosphere of our focal plant (neighborhood plant community structure). This scale is consistent with the minimal observed spatial autocorrelation of AM fungi (30–100 cm; Mummey and Rillig, 2008).
Soil cores, including roots and plant material, were stored at −20 °C before analysis. Each soil core was thoroughly homogenized and subsampled for soil chemical analyses (Supplementary Information, part a). We measured water content, pH, carbon, nitrogen and phosphorus content of the soil, which are known to affect AMF community variation (Camenzind et al., 2014; Horn et al., 2014). Additionally, dehydrogenase activity was assessed as a proxy for microbial activity. Roots were washed in Millipore water before analysis.
DNA extraction, 454 pyrosequencing and OTU delineation
We extracted genomic DNA twice from each core, once from 150 mg of washed, fine-ground F. brevipila roots and once from 250 mg of soil material, which was sieved through a 2 mm mesh. We used the PowerSoil DNA Isolation Kit (MoBio Laboratories Inc., Carlsbad, CA, USA) following the procedure in the manufacturer's manual. We then created 454 pyrosequencing amplicon pools for the AMF using a nested PCR design, using the AMF-specific primer set SSUmAf and LSUmAr for the first and SSUmCf and LSUmBr for the second, nested PCR (Krüger et al., 2009). The amplified region spans genes for the SSU (small ribosomal subunit), the complete ITS (internal transcribed spacer) region and a part of the LSU (large ribosomal subunit). Subsequently, amplicons of ~600 bp in length were created from the AMF-specific PCR fragments using general fungal primers located in the LSU gene modified with 454 adapters and sample-specific barcode sequences (Supplementary Information, part b). The 454 sequencing was carried out on a Roche GS FLX+ system with titanium chemistry at the Göttingen Genomics Laboratory at the Georg-August University of Göttingen (Göttingen, Germany).
Sequences were denoized using the PyroNoise approach (Quince et al., 2009) implemented in Mothur (Schloss et al., 2009). The denoizing approach removes bad quality sequences, creates sequence clusters and removes chimera sequences. After denoizing and preclustering, sequences from roots and soil were clustered into OTUs using CROP (Clustering 16S rRNA for OTU Prediction; Hao et al., 2011), which uses a Bayesian clustering algorithm. This approach addresses species delineation uncertainty better than hierarchical clustering methods because of its flexible cutoff, thereby creating significantly less artifact OTUs than fixed cutoff clustering approaches (Hao et al., 2011). We checked the final OTU sequences against chimeras using the Mothur implementation of the uchime algorithm and the Krüger et al. (2012) SSU-ITS-LSU alignment, as well as the slayer algorithm against the sequences themselves. Default settings were used for both algorithms.
Owing to the nature of pyrosequencing, we found differences in read numbers for every sampling location, so we resampled the read numbers to equal amounts of 500 reads per sample using a bootstrap approach with 10 000 iterations per sample (Efron, 1979; Wehner et al., 2014). Samples with considerably lower read numbers than the estimated resampling threshold (<350 reads, equal to 70% of the resampling threshold) were discarded before resampling. Additionally, singletons were removed. All subsequent statistical analyses were carried out in R 3.1 (R Core Team, 2015).
Phylogenetic tree calculation
OTUs were annotated according to the results of a BLAST search against the NCBI nucleotide database (nt) before phylogenetic tree calculation. We calculated a phylogenetic tree for the AMF OTUs using RAxML (Stamatakis, 2006) to further refine the OTU definitions following our approach from a previous study (Horn et al., 2014). About 110 representative sequences of an SSU-ITS-LSU AMF reference alignment (Krüger et al., 2012) plus an outgroup sequence from the Chytridiomycota were added to our own sequences to determine the phylogenetic position of our OTUs. With the help of the phylogenetic tree, we removed sequences that clustered outside the Glomeromycotina and are therefore likely to be erroneous or non-AMF sequences.
Null model analysis and phylogenetic community structure
To account for non-random species associations potentially linked to biotic influences in AMF and plants, we performed null model analysis on plant and AMF species, respectively. Null models were created in EcoSim (Gotelli and Entsminger, 2012; for more detail see Supplementary Information, part c)
We included phylogenetic sorting of the respective communities as a potential driver of community structure (Horn et al., 2014). This approach tests the hypothesis that the relationship between AMF and plant communities is reflected at a phylogenetic level including, but not restricted to species/OTUs. We analyzed phylogenetic diversity within the AMF and plant communities separately. We chose the Daphne plant tree for our plant phylogenetic analysis (Durka and Michalski, 2012), which provides a complete set of phylogenetic distances for our plant data set. Phylogenetic distances between AMF OTUs were calculated using the Needleman–Wunsch implementation of Esprit (Sun et al., 2009). The distances between plant species were calculated as pairwise distances from the trimmed Daphne phylogenetic tree using the cophenetic.phylo function of the ape package (Paradis et al., 2004). Using the picante package (Kembel et al., 2010), we obtained two estimates of phylogenetic diversity: the standardized effect size of mean pairwise distance (SES-MPD), which calculates the net relatedness index from β-diversity with a null model; and intercommunity mean pairwise distance, that is phylogenetic distance between communities (Supplementary Information, part d). The mean values of the net relatedness index of all samples of AMF were then used as the α-diversity measure to judge the clustering (positive) or segregation (negative) of the overall AMF or plant community. Intercommunity mean pairwise distances were calculated as pairwise phylogenetic distances of the samples, based on pairwise genetic distances between OTUs and plant species. To include the intercommunity mean pairwise distance information in a subsequent variance partitioning analysis (Legendre and Legendre, 1998; Caruso et al., 2012), the distance matrices of plants and AMF were subjected to a principal coordinate analysis (PCoA), a generalization of ordinary principal component analysis (Legendre and Legendre, 1998) that is also the basis of distance-based redundancy analysis.
Models of correlations between plants and AMF
To test the null hypothesis of the study (that is, independence) robustly, we used three main multivariate and multiple regression analysis based on redundancy analysis (Horn et al., 2015 and Supplementary Information, part e). Specifically, we quantified how plant community variation was affected by variation in phylogenetic distance and community structure of AMF, and we also performed the vice versa analysis using plant phylogenetic community structure and plant community structure as a predictor of AM fungal community structure.
To visualize patterns of community structure, we used PCoA. For AMF, PCoA was applied to Hellinger-transformed data to prevent inflation in the weights of rare OTUs and work on an ecologically meaningful Euclidean space (Legendre and Legendre, 1998). For plants, PCoA was applied to the Jaccard distance matrix of the presence/absence data. We also used the kriging estimator (Ribeiro and Diggle, 2001) to display spatial structures in environmental variables and the PCoA axes. PCoA axes of the two assemblages were also plotted on a scatter plot to visualize correlation between the assemblages. We used Moran eigenvector mapping to account for spatial autocorrelation at multiple scales (Dray et al., 2006; Legendre et al., 2009; Supplementary Information, part e): the analysis produces a number of vectors that describe spatial patterns in species distribution at all the spatial scales resolvable by the sampling design. These vectors are sometimes referred to as ‘spatial factors' or ‘spatial effects', which implicitly describe spatial variation that may originate from a multitude of factors such as spatially structured environmental variation but also spatial variation not related to environmental variation, and/or unmeasured but spatially structured factors such as dispersal and biotic interactions. Spatial effects independent of environmental variables are often called ‘pure space' (for example, Legendre and Legendre, 1998).
We then used redundancy analysis and variance partitioning to test and quantify the effects of the community structure of one group on the other group by controlling for other covarying effects (space, environment, phylogeny).
Finally, to increase the statistical power of multivariate analysis (Warton et al., 2012) and so robustly test the null hypothesis, we also tested the generalized linear response of the relative abundance of AM fungal taxa to the plant community and vice versa using the manyglm function from the mvabund package (Wang et al., 2012; Warton et al., 2012). The test was performed on residuals after removing the contributions of environmental and spatial covariates.
All multivariate calculations were carried out in R, using the vegan (Oksanen et al., 2012), the spacemakeR (Dray, 2011) and geoR (Ribeiro and Diggle, 2001) packages.
Results
454 Pyrosequencing and OTU delineation
The clustered and denoized data set consisted of 325 putative AM fungal OTUs. During the resampling, we removed seven root and one soil sample based on minimal read numbers of 500 reads. Species accumulation curves showed a sufficient sampling depth (Supplementary Figures S4 and S5). After resampling and removal of singletons, 88 OTUs remained, of which 17 were removed since they clustered outside the Glomeromycotina subphylum (former Glomeromycota, see Spatafora et al., 2016, after Schüßler et al., 2001) as it is currently described. This resulted in a total of 71 OTUs used in all subsequent analyses. One representative sequence of each OTU is available from NCBI GenBank (https://www.ncbi.nlm.nih.gov/genbank/) under the accession numbers KX709382 to KX709452. The OTUs found in our tree span all known AMF families, indicating a fairly exhaustive coverage of the Glomeromycotina subphylum (Supplementary Figure S5). The root data set eventually consisted of 68 OTUs and the soil data set of 62 OTUs. Overall OTU richness per macroplot was comparable between these data sets, ranging from 30 to 43 in roots and from 28 to 43 in soil (Table 1). The dominant fungal groups in our soils and roots were Glomus spp. and Rhizophagus spp.
Table 1. AMF phylogeny and null model results from community abundance data.
| Sample size |
Phylogeny |
Null model |
|||
|---|---|---|---|---|---|
| OTUs | MPD | Effect size | P-value | ||
| All MPs root | 53 | 68 | 0.01 | 11.75 | <0.001 |
| MP1 root | 16 | 43 | −0.02 | 4.08 | 0.002 |
| MP2 root | 18 | 30 | −0.07 | 1.13 | 0.137 |
| MP3 root | 19 | 43 | 0.00 | −0.73 | 0.250 |
| All MPs soil | 59 | 62 | 0.01 | 19.42 | <0.001 |
| MP1 soil | 20 | 41 | 0.08 | 10.96 | <0.001 |
| MP2 soil | 19 | 28 | −0.14 | 10.66 | <0.001 |
| MP3 soil | 20 | 43 | 0.08 | 1.61 | 0.068 |
Abbreviations: AMF, arbuscular mycorrhizal fungi; MP, macroplot; OTU, operational taxonomic units.
Column names are: sample size, number of OTUs; MPD, the mean pairwise phylogenetic distance between individual communities (that is, samples). Positive effect sizes (C-score) and positive mean pairwise distances indicate segregated communities (species repel each other), whereas negative values represent an aggregated community (species attract each other). The rows ‘all MPs' show result across macroplots while the other rows show results for each macroplot (MP).
Community structure of AMF excluding plants
The AMF community was significantly segregated at the level of the entire data set. However, for the AMF communities in root samples, the effect was significant only for one of the macroplots and the whole data set (Table 1). For the soil community two out of three macroplots had significantly segregated assemblages and effect sizes were considerably higher in soil than in root data sets (Table 1).
There were no significant net relatedness index differences overall. Neither the root nor the soil sets of the phylogenetic data showed significantly segregated or aggregated communities on a per-macroplot or per-data-set basis.
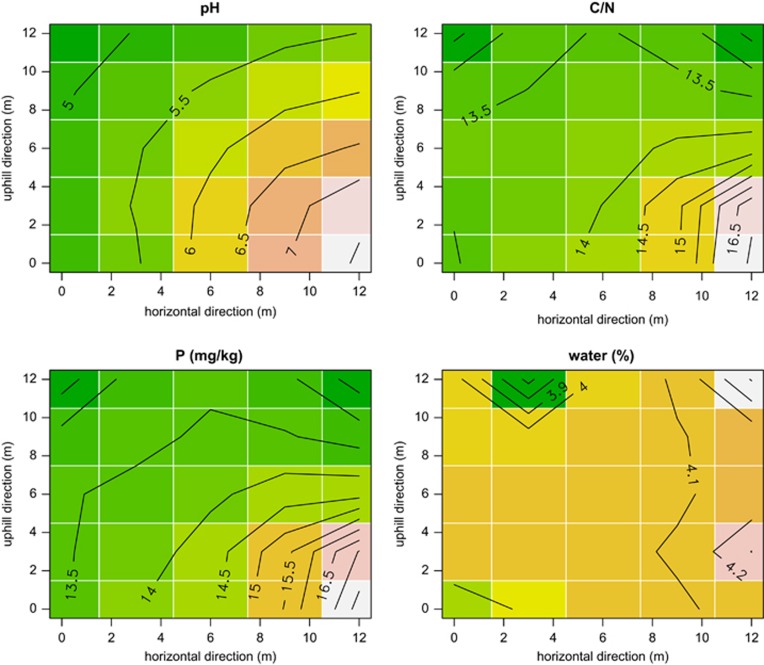
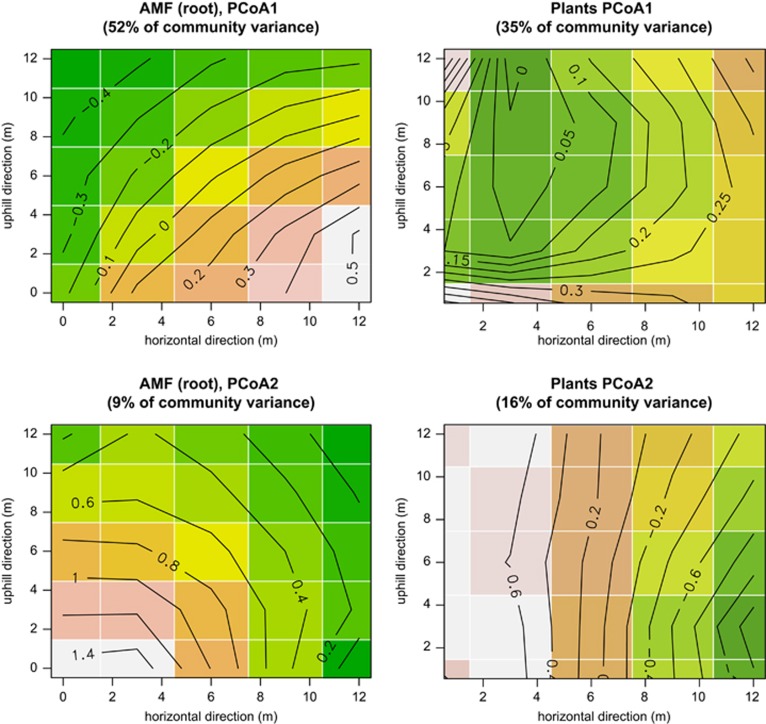
All measured environmental variables display a clear spatial gradient along the uphill direction (see four examples in Figure 2), although sometimes with an additional component of variation along the direction orthogonal to the uphill direction. At the macroplot scale, the spatial gradient in the first two axes of the PCoA of AMF (accounting for almost 2/3 of total variance) follow the environmental gradient more than the equivalent PCoA axis of plants do (Figure 3). When we excluded plants from the analysis and removed spatial effects, the effect of the measured environmental variables (pH, water content, C, N, C/N ratio, phosphorus, dehydrogenase activity) on AMF community structure was overall low. With an exception of the root data set from one macroplot, environmental data explained <10%. Pure space was a major predictor of the overall data set and within each macroplot, showing significant and large proportions (up to 31%) of explained variation (Supplementary Table S2). Phylogeny was the second largest explanatory component in the variance partitioning of the AMF without plants and up to 30% of variation could be explained by the phylogenetic distance of the AMF in our data set (Supplementary Table S2). Additionally, we found the spatial-phylogenetic effects accounted for a large fraction of the AMF variance.
Figure 2.
Kriging interpolation of four of the measured environmental variables as measured in one of the three macroplots (macroplot 1, see Supplementary Information). Plots were by construction aligned along a soil textural gradient on the slopes of a hillside (Supplementary Figure S1), with the gradient running along the uphill–downhill axis (y axis; Supplementary Figures S2 and S3). As we expected, the main gradient in major soil variables followed the uphill–downhill axis, although in the case of macroplot 1 water showed a patchy distribution.
Figure 3.
Kriging interpolation of the first two PCoA (see also Figure 4) axes of AMF and plants. Data are shown for macroplot 1, and are thus directly comparable with environmental variables presented in Figure 2. Spatial patterns in the structure of the two assemblages appear to be only poorly correlated. Similar patterns were observed in the other macroplots (not shown).
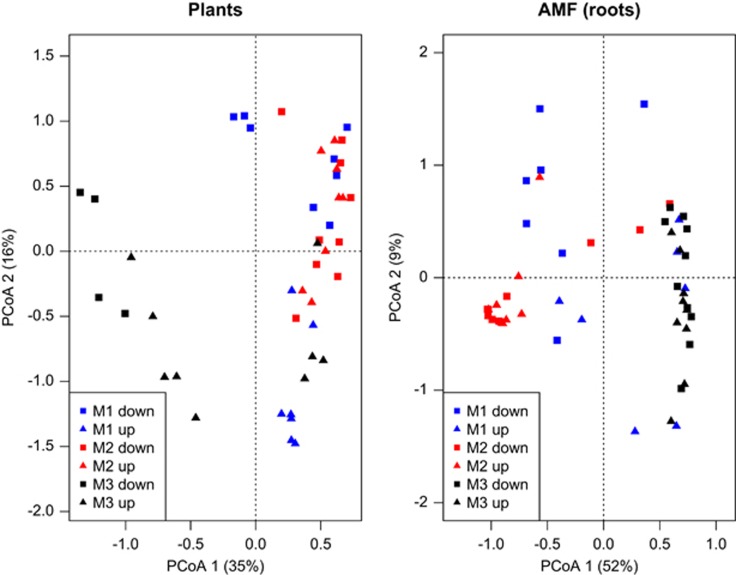
AMF–plant correlations
A PCoA ordination of all samples from all plots show that the plant assemblage seemed the most structured spatially: macroplot 3 clustered separately from macroplot 1 and 2 (see also Figure 4). The same clustering was not observed in AMF as clearly as in plants, neither in roots nor in soil. Scatter plots (Figure 5) of the first two PCoAs of AMF and plants revealed that gradients in the community structure of the two assemblages are correlated but with a confounding effect of spatial patterns at the broad scale separating the three macroplots (see for example Figures 5a and c). Still, after filtering out spatial autocorrelation, plant community structure accounted for a statistically significant amount of variation in the root AMF community, while plant phylogeny was not a significant predictor (Table 2). Instead, when we used the AMF community as a predictor of the plant community, the variation explained by the fungi was very low and not significant (Supplementary Table S3). Overall, these results reject the null hypothesis of the study, although the amount of variation uniquely attributable to the effect of plants on AMF is small (Table 2). GLM results were consistent with these results: plant community structure had significant effects on the AMF community in roots (P<0.001) and soil (P<0.001), but AMF communities did not show any significant effects when used as a predictor of plant community structure.
Figure 4.
PCoA ordination plots of plants and AMF. Individual samples are color labeled by macroplot (M1, blues; M2, red; M3, black) and symbol label in terms of uphill (up, triangle) or downhill (down, square) position of individual samples within the macroplot (see also Supplementary Figure S1). The plant assemblage appears to be more spatially structured in terms of the separation between M3 and M2+M1, with the latter two being geographically much closer to each other (Supplementary Figure S2). This clustering pattern is less evident in AMF.
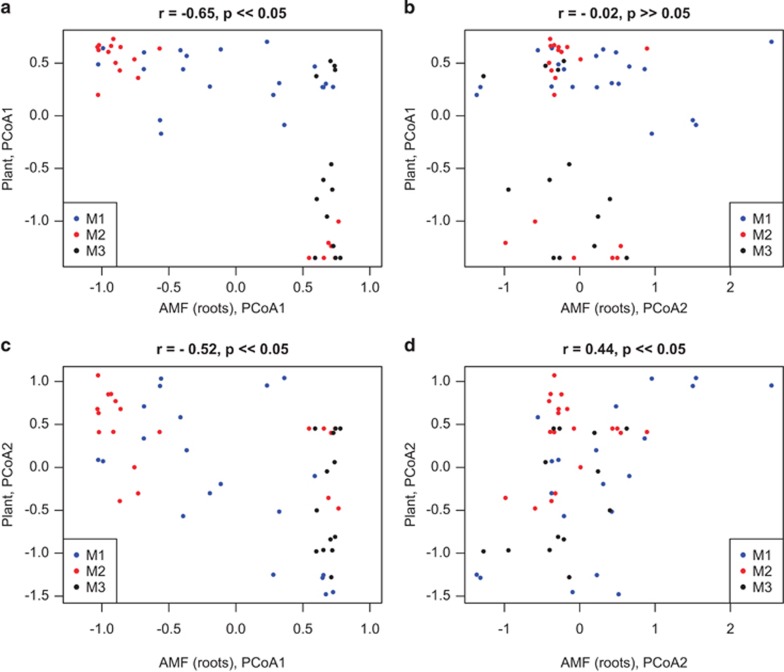
Figure 5.
Bivariate covariation of PCoA2 and 2 of both AMF (roots) and plants (see Figure 4) in all four possible combinations: (a) PCoA1 AMF vs PCoA1 plants; (b) PCoA2 AMF vs PCoA1 plants; (c) PCoA1 AMF vs PCoA2 plants; (d) PCoA2 AMF vs PCoA2 plants. Pearson's correlation coefficient (r) and relative P-value (P) is reported for each set of correlations. Individual samples are color labeled by macroplot (M1, blues; M2, red; M3, black). Some significant correlation is observed but seems driven by spatial structure between macroplots. For example, in a and c, M3 samples are clustered on the right-hand side, while in d, the observed positive correlation between the PCoA2 axes of plants and AMF is driven by variation internal to macroplot 1. These results suggest spatial dependence in the covariation between AMF and plants.
Table 2. Variance partitioning of the AMF community matrix with the plant community also included as a predictor of the AMF community.
| Source of variance |
Phylogeny |
Presence/absence |
||
|---|---|---|---|---|
| Root | Soil | Root | Soil | |
| Environment | 0NS | 0NS | 3*** | 0NS |
| Space | 30*** | 29*** | 19*** | 24*** |
| Plants | 0NS | 0NS | 4** | 0NS |
| Env+space | 4NT | 3NT | 11NT | 5NT |
| Space+plants | 0NT | 6NT | 11NT | 10NT |
| Env+plants | 0NT | 0NT | 0NT | 0NT |
| Env+space+plants | 3NT | 3NT | 0NT | 2NT |
| Unexplained | 63 | 59 | 52 | 54 |
Abbreviations: AMF, arbuscular mycorrhizal fungi; NS, not significant, NT, not testable.
The table is divided in two main blocks: phylogeny and presence/absence of plants. These blocks refer to how the effect of plants on AMF was evaluated. In the first two columns of results (phylogeny, root and soil), the effects of plants (row wise) is assessed by using plant phylogeny as a predictor of AMF. In the second two columns (presence/absence, root and soil), we used plant community structure as predictor of AMF. The other predictors were environment or env (soil properties) and space (geographic position). The plus sign in the Source of variance column stands for shared variation (it is not the sum of the variances explained by each predictor, for example, env+space is the spatially structured effect of the environment). Figures are percentage values of total variance. ***P<0.001; **P<0.01.
Discussion
Is the community structure of AMF independent of that of plants?
AMF and plants may affect each other's community dynamics depending on spatial and temporal scale, the latter especially in relation to succession (Zobel and Öpik, 2014). Evaluating which group is driving which other group is challenging because both groups may influence each other to some extent and possibly at different spatial and temporal scales (Martinez-Garcia et al., 2015; García de León et al., 2016a). Also, in a stable ecosystem (for example, climax) regional covariation between AMF and plants could arise as the effect of environmental gradients (Habitat hypothesis). Our results reflect this complexity of plant–AMF interactions in a species-rich grassland area at a range of small spatial scales but made clear some important points. First, AMF community variance is mostly accounted for by spatial factors and phylogenetic distance patterns in OTU composition. Second, plant communities were also strongly influenced by the soil environment, but AMF communities were not. Overall, AMF and plants showed different spatial structures and the relative roles of the tested factors clearly change between plant and AMF, which rules out the Habitat hypothesis. The strong influence of spatial factors on AMF communities aligns with the Driver hypothesis, but we did not find an effect of AMF on plants, thus refuting this hypothesis (Zobel and Öpik, 2014). Instead, when plant communities were used as a predictor of AMF, after taking into account all other effects (that is, environment, space), we found a significant effect of plants on AMF communities. We can thus reject the statistical null hypothesis that the groups are independent. Specifically, there is some support for AMF acting as Passengers. We have to note that reversing response and predictors (that is, AMF passenger or driver) in these multivariate statistical models is not trivial. For example, there is additional and not invertible information in the phylogenetic trees of each set of species.
Notwithstanding the aforementioned technicality and the statistical rejection of the null hypothesis, the complex set of correlations linking plants and AMF are relatively weak (whatever group plays the role of predictor or response), which implies that the interaction between plants and AMF are weak at the community level: plant community structure remains a modest predictor of AMF community structure compared with the other predictors employed in the analysis.
All these results are overall consistent with theoretical predictions put forward by Zobel and Öpik (2014): the scale of the study is relatively small, with a steep but short soil environmental gradient replicated a number of times at various distances (within plots and between plots), from tens of meters to a few hundred meters. At these scales, we can expect the absence of or weak dispersal limitation for plants but some dispersal limitation in AMF, and the texture gradient sampled along the hills may mimic a primary succession gradient in the plant assemblage (Horn et al., 2015). Under these conditions, the passenger ‘effect' should be at its strongest.
Which further mechanisms could underlie the observed patterns? More specifically, if AMF are passengers, why is the effect of plants apparently weak? It has been shown that plants may reward the best fungal partners with more carbohydrates (Bever et al., 2009; Kiers et al., 2011; Verbruggen et al., 2012) and that particular plant communities may cause the development of specific AMF communities (Hausmann and Hawkes, 2009). This is consistent with our observation that the neighborhood plant community of a dominant focal plant is a significant but not very strong predictor of the AMF community in its roots. Interestingly, we observed this effect only for the root assemblage and not for the soil assemblage and plant community phylogenetic structure seems to have no role in these effects.
The weakness of the observed effects of plant communities on AMF communities may be particular to the study system. For instance, the dominance of Glomus spp., Rhizophagus irregularis and other generalist taxa may cause effects to be less strong than in systems with higher evenness and/or specialist taxa. Another potential explanation is that other ecological interactions overwhelm the effect, as evidenced from the non-random phylogenetic community pattern of the AMF assemblage. Also, the grassland is dominated by several C3 grasses, which are not very dependent on mycorrhiza (Reinhart et al., 2012), and there is increasing evidence that these plants associate with generalist AMF taxa (Helgason et al., 2007; Öpik et al., 2009; Vályi et al., 2015).
Are AMF communities assembled through interspecific interactions?
As recently reviewed by Vályi et al. (2016), AMF communities are structured by a range of different processes, including environmental filtering, dispersal and biotic interactions (Lekberg et al., 2007; Peng et al., 2009; Dumbrell et al., 2010a, 2010b; Silva and Batalha, 2011). Biotic interaction at the interspecific level could have a major role in some cases. For example, negative interactions between AMF species competing for the same root space may result in the superior competitor persisting in the root (Hart et al., 2001; Thonar et al., 2014). In addition, greenhouse studies as well as field observational work have shown that net phylogenetic distance patterns can predict co-occurrence (Maherali and Klironomos, 2007; Horn et al., 2014) and AMF traits are phylogenetically conserved (Powell et al., 2009). For example, mechanisms such as facilitation or feedbacks between plants and AMF could be signaled by net phylogenetic distance patterns in community structure if closely related species received similar facilitation (Anacker et al., 2014). Here, the AMF assemblage was strongly segregated, while phylogenetic aggregation or segregation patterns were not significant, but with overall quite low mean pairwise distances between communities. This slightly contrasts with a previous analysis of AMF communities in the same sampling area as well as findings from other authors, which show local species pools to be phylogenetically clustered (Kivlin et al., 2011; Saks et al., 2014; Horn et al., 2014; Grilli et al., 2015). At the same time, when we excluded plants from the variance partitioning of AMF community matrix, up to 30% of AMF community variation could be explained by phylogenetic distance (Supplementary Table S2). Integrating all the available evidence (Kivlin et al., 2011; Saks et al., 2014; Horn et al., 2014; Grilli et al., 2015), including previous work from this site (Horn et al., 2014), AMF communities seem phylogenetically structured and very much spatially structured. Given the amount of variation accounted for by these effects and the fact that for plants environmental variation was the main structuring factor, we conclude that AMF communities in our sampling area assembled mostly independently of the plant community with a possibly important role of interactions within the AMF community. However, there is shared variation between environment, space and phylogenetically structured variation in AM fungal communities.
The processes behind shared variation (for example, spatially structured covariation between environmental and phylogenetic variation) cannot be explained solely on the basis of observational evidence. Experimental work will in the future be necessary to understand how this shared variation is generated. As already suggested by Zobel and Öpik (2014), in an ideal experiment either the plant or AMF community should be kept constant while varying the other community, and also in relation to changing environmental conditions (for example, soil properties such as pH) and different degrees of dispersal limitation. These experiments are challenging under field conditions, but we suggest that surveying AMF communities in plant assemblages under a range of primary and secondary succession stages (for example, García de León et al., 2016a) and manipulating vegetation to control the succession process will offer a valid starting point to move from patterns to the mechanisms. In that perspective, our study suggests to test for a potentially important role of biotic interactions within the AMF assemblage.
Acknowledgments
SH and TC acknowledge funding by the German science foundation (DFG Grant No. CA 987/1-1). TC was also supported by the project SENSE (Structure and Ecological Niche in the Soil Environment; EC FP7—631399—SENSE). We are grateful to four anonymous reviewers for their invaluable comments and suggestions, which have improved the quality of this work. Support during the 454 sequencing by the Göttingen Genomics Laboratory is gratefully acknowledged.
Footnotes
Supplementary Information accompanies this paper on The ISME Journal website (http://www.nature.com/ismej)
The authors declare no conflict of interest.
Supplementary Material
References
- Alguacil MM, Torrecillas E, Garcia-Orenes F, Roldan A. (2014). Changes in the composition and diversity of AMF communities mediated by management practices in a Mediterranean soil are related with increases in soil biological activity. Soil Biol Biochem 76: 34–44. [Google Scholar]
- Anacker BL, Klironomos JN, Maherali H, Reinhart KO, Strauss SY. (2014). Phylogenetic conservatism in plant-soil feedback and its implications for plant abundance. Ecol Lett 17: 1613–1621. [DOI] [PubMed] [Google Scholar]
- Bever JD, Richardson SC, Lawrence BM, Holmes J, Watson M. (2009). Preferential allocation to beneficial symbiont with spatial structure maintains mycorrhizal mutualism. Ecol Lett 12: 13–21. [DOI] [PubMed] [Google Scholar]
- Camenzind T, Hempel S, Homeier J, Horn S, Velescu A, Wilcke W et al. (2014). Nitrogen and phosphorus additions impact arbuscular mycorrhizal abundance and molecular diversity in a tropical montane forest. Glob Chang Biol 20: 3646–3659. [DOI] [PubMed] [Google Scholar]
- Caravaca F, Ruess L. (2014). Arbuscular mycorrhizal fungi and their associated microbial community modulated by Collembola grazers in host plant free substrate. Soil Biol Biochem 69: 25–33. [Google Scholar]
- Caruso T, Hempel S, Powell JR, Barto EK, Rillig MC. (2012). Compositional divergence and convergence in arbuscular mycorrhizal fungal communities. Ecology 93: 1115–1124. [DOI] [PubMed] [Google Scholar]
- Davison J, Moora M, Öpik M, Adholeya A, Ainsaar L, Bâ A et al. (2015). Global assessment of arbuscular mycorrhizal fungus diversity reveals very low endemism. Science 349: 970–973. [DOI] [PubMed] [Google Scholar]
- García de León D, Moora M, Öpik M, Neuenkamp L, Gerz M, Jairus T et al. (2016. a). Symbiont dynamics during ecosystem succession: co-occurring plant and arbuscular mycorrhizal fungal communities. FEMS Microbiol Ecol 92: fiw097. [DOI] [PubMed] [Google Scholar]
- García de León D, Moora M, Öpik M, Jairus T, Neuenkamp L, Vasar M et al. (2016. b). Dispersal of arbuscular mycorrhizal fungi and plants during succession. Acta Oecol 77: 128–135. [Google Scholar]
- Dray S. (2011). SpacemakeR: spatial modelling. R package version 0.0-5/r10. Available at: http://R-Forge.R-project.org/projects/sedar/ (accessed on 1 May 2015).
- Dray S, Legendre P, Peres-Neto PR. (2006). Spatial modelling: a comprehensive framework for principal coordinate analysis of neighbour matrices (PCNM). Ecol Model 196: 483–493. [Google Scholar]
- Dumbrell AJ, Nelson M, Helgason T, Dytham C, Fitter AH. (2010. a). Idiosyncrasy and overdominance in the structure of natural communities of arbuscular mycorrhizal fungi: is there a role for stochastic processes? J Ecol 98: 419–428. [Google Scholar]
- Dumbrell AJ, Nelson M, Helgason T, Dytham C, Fitter AH. (2010. b). Relative roles of niche and neutral processes in structuring a soil microbial community. ISME J 4: 337–345. [DOI] [PubMed] [Google Scholar]
- Durka W, Michalski SG. (2012). Daphne: a dated phylogeny of a large European flora for phylogenetically informed ecological analyses. Ecology 93: 2297–2297. [Google Scholar]
- Efron B. (1979). Bootstrap methods: another look at the jackknife. Ann Statist 7: 1–26. [Google Scholar]
- Gotelli NJ, Entsminger GL. (2012) EcoSim 7.72. Acquired Intelligence Inc. & Kesey-Bear: Jericho, VT, USA.
- Grilli G, Urcelay C, Galetto L, Davison J, Vasar M, Saks Ü et al. (2015). The composition of arbuscular mycorrhizal fungal communities in the roots of a ruderal forb is not related to the forest fragmentation process. Environ Microbiol 17: 2709–2720. [DOI] [PubMed] [Google Scholar]
- Hao X, Jiang R, Chen T. (2011). Clustering 16S rRNA for OTU prediction: a method of unsupervised Bayesian clustering. Bioinformatics 27: 611–618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hart MM, Reader RJ, Klironomos JN. (2001). Life-history strategies of arbuscular mycorrhizal fungi in relation to their successional dynamics. Mycologia 93: 1186–1194. [Google Scholar]
- Hausmann NT, Hawkes CV. (2009). Plant neighborhood control of arbuscular mycorrhizal community composition. New Phytol 183: 1188–1200. [DOI] [PubMed] [Google Scholar]
- Helgason T, Merryweather JW, Young JPW, Fitter AH. (2007). Specificity and resilience in the arbuscular mycorrhizal fungi of a natural woodland community. J Ecol 95: 623–630. [Google Scholar]
- Hiiesalu I, Pärtel M, Davison J, Gerhold P, Metsis M, Moora M et al. (2014). Species richness of arbuscular mycorrhizal fungi: associations with grassland plant richness and biomass. New Phytol 203: 233–244. [DOI] [PubMed] [Google Scholar]
- Horn S, Caruso T, Verbruggen E, Rillig MC, Hempel S. (2014). Arbuscular mycorrhizal fungal communities are phylogenetically clustered at small scales. ISME J 8: 2231–2242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horn S, Hempel S, Ristow M, Rillig MC, Kowarik I, Caruso T. (2015). Plant community assembly at small scales: Spatial vs. environmental factors in a European grassland. Acta Oecol 63: 56–62. [Google Scholar]
- Kembel SW, Cowan PD, Helmus MR, Cornwell WK, Morlon H, Ackerly DD et al. (2010). Picante: R tools for integrating phylogenies and ecology. Bioinformatics 26: 1463–1464. [DOI] [PubMed] [Google Scholar]
- Kiers ET, Duhamel M, Beesetty Y, Mensah JA, Franken O, Verbruggen E et al. (2011). Reciprocal rewards stabilize cooperation in the mycorrhizal symbiosis. Science 333: 880–882. [DOI] [PubMed] [Google Scholar]
- Kivlin SN, Hawkes CV, Treseder KK. (2011). Global diversity and distribution of arbuscular mycorrhizal fungi. Soil Biol Biochem 43: 2294–2303. [Google Scholar]
- Klironomos J. (2000). Host-specificity and functional diversity among arbuscular mycorrhizal fungi. Microbial Biosystems: New Frontiers. Proceedings of the 8th International Symposium on Microbial Ecology; Halifax, Nova Scotia, Canada. Atlantic Canada Society for Microbial Ecology.
- Knegt B, Jansa J, Franken O, Engelmoer DJP, Werner GDA, Bücking H et al. (2016). Host plant quality mediates competition between arbuscular mycorrhizal fungi. Fungal Ecol 20: 233–240. [Google Scholar]
- Krüger M, Krüger C, Walker C, Stockinger H, Schüßler A. (2012). Phylogenetic reference data for systematics and phylotaxonomy of arbuscular mycorrhizal fungi from phylum to species level. New Phytol 193: 970–984. [DOI] [PubMed] [Google Scholar]
- Krüger M, Stockinger H, Krüger C, Schüßler A. (2009). DNA-based species level detection of Glomeromycota: one PCR primer set for all arbuscular mycorrhizal fungi. New Phytol 183: 212–223. [DOI] [PubMed] [Google Scholar]
- Legendre P, Legendre L. (1998) Numerical Ecology. Elsevier Science: Amsterdam. [Google Scholar]
- Legendre P, Mi XC, Ren HB, Ma KP, Yu MJ, Sun IF et al. (2009). Partitioning beta diversity in a subtropical broad-leaved forest of China. Ecology 90: 663–674. [DOI] [PubMed] [Google Scholar]
- Leifheit EF, Verbruggen E, Rillig MC. (2015). Arbuscular mycorrhizal fungi reduce decomposition of woody plant litter while increasing soil aggregation. Soil Biol Biochem 81: 323–328. [Google Scholar]
- Lekberg Y, Koide RT, Rohr JR, Aldrich-Wolfe L, Morton JB. (2007). Role of niche restrictions and dispersal in the composition of arbuscular mycorrhizal fungal communities. J Ecol 95: 95–105. [Google Scholar]
- Maherali H, Klironomos JN. (2007). Influence of phylogeny on fungal community assembly and ecosystem functioning. Science 316: 1746–1748. [DOI] [PubMed] [Google Scholar]
- Martinez-Garcia LB, Richardson SJ, Tylianakis JM, Peltzer DA, Dickie IA. (2015). Host identity is a dominant driver of mycorrhizal fungal community composition during ecosystem development. New Phytol 205: 1565–1576. [DOI] [PubMed] [Google Scholar]
- Mummey DL, Rillig MC. (2008). Spatial characterization of arbuscular mycorrhizal fungal molecular diversity at the submetre scale in a temperate grassland. FEMS Microbiol Ecol 64: 260–270. [DOI] [PubMed] [Google Scholar]
- Ohsowski BM, Zaitsoff PD, Opik M, Hart MM. (2014). Where the wild things are: looking for uncultured Glomeromycota. New Phytol 204: 171–179. [DOI] [PubMed] [Google Scholar]
- Oksanen J, Guillaume Blanchet F, Kindt R, Legendre P, Minchin PR, O'Hara RB et al. (2012). vegan: Community ecology package. R package version 2.0-10. Available at: https://cran.r-project.org/web/packages/vegan/index.html (accessed on 20 January 2015).
- Öpik M, Metsis M, Daniell TJ, Zobel M, Moora M. (2009). Large-scale parallel 454 sequencing reveals host ecological group specificity of arbuscular mycorrhizal fungi in a boreonemoral forest. New Phytol 184: 424–437. [DOI] [PubMed] [Google Scholar]
- Öpik M, Zobel M, Cantero JJ, Davison J, Facelli JM, Hiiesalu I et al. (2013). Global sampling of plant roots expands the described molecular diversity of arbuscular mycorrhizal fungi. Mycorrhiza 23: 411–430. [DOI] [PubMed] [Google Scholar]
- Paradis E, Claude J, Strimmer K. (2004). APE: analyses of phylogenetics and evolution in R language. Bioinformatics 20: 289–290. [DOI] [PubMed] [Google Scholar]
- Peng Y, Chen G, Tian G, Yang X. (2009). Niches of plant populations in mangrove reserve of Qi'ao Island, Pearl River Estuary. Acta Ecol Sin 29: 357–361. [Google Scholar]
- Powell JR, Parrent JL, Hart MM, Klironomos JN, Rillig MC, Maherali H. (2009). Phylogenetic trait conservatism and the evolution of functional trade-offs in arbuscular mycorrhizal fungi. Proc R Soc Lond Ser B 276: 4237–4245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quince C, Lanzen A, Curtis TP, Davenport RJ, Hall N, Head IM et al. (2009). Accurate determination of microbial diversity from 454 pyrosequencing data. Nat Meth 6: 639–U627. [DOI] [PubMed] [Google Scholar]
- R Core Team. (2015) R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing: Vienna, Austria. [Google Scholar]
- Reinhart KO, Wilson GWT, Rinella MJ. (2012). Predicting plant responses to mycorrhizae: integrating evolutionary history and plant traits. Ecol Lett 15: 689–695. [DOI] [PubMed] [Google Scholar]
- Ribeiro Jr PJ, Diggle PJ. (2001). geoR: a package for geostatistical analysis. R-NEWS 1: 15–18. [Google Scholar]
- Ristow M, Rohner M-S, Heinken T. (2011). Die Oderhänge bei Mallnow und Lebus. Tuexenia Beih (Flora und Vegetation in Brandenburg) 4: 127–144. [Google Scholar]
- Saks Ü, Davison J, Öpik M, Vasar M, Moora M, Zobel M. (2014). Root-colonizing and soil-borne communities of arbuscular mycorrhizal fungi in a temperate forest understorey. Botany 92: 277–285. [Google Scholar]
- Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB et al. (2009). Introducing Mothur: Open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol 75: 7537–7541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schüßler A, Schwarzott D, Walker C. (2001). A new fungal phylum, the Glomeromycota: phylogeny and evolution. Mycol Res 105: 1413–1421. [Google Scholar]
- Shantz HL. (1954). The place of grasslands in the Earth's cover. Ecology 35: 3. [Google Scholar]
- Silva IA, Batalha MA. (2011). Plant functional types in Brazilian savannas: the niche partitioning between herbaceous and woody species. Perspect Plant Ecol Evol Syst 13: 201–206. [Google Scholar]
- Smith SE, Read DJ. (2008) Mycorrhizal symbiosis, 3rd edn. Academic Press: Burlington, MA, USA. [Google Scholar]
- Spatafora JW, Chang Y, Benny GL, Lazarus K, Smith ME, Berbee ML et al. (2016). A phylum-level phylogenetic classification of zygomycete fungi based on genome-scale data. Mycologia 108: 1028–1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis A. (2006). RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22: 2688–2690. [DOI] [PubMed] [Google Scholar]
- Sun YJ, Cai YP, Liu L, Yu FH, Farrell ML, McKendree W et al. (2009). ESPRIT: estimating species richness using large collections of 16S rRNA pyrosequences. Nucleic Acids Res 37: e76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thonar C, Frossard E, Smilauer P, Jansa J. (2014). Competition and facilitation in synthetic communities of arbuscular mycorrhizal fungi. Mol Ecol 23: 733–746. [DOI] [PubMed] [Google Scholar]
- Treseder KK, Cross A. (2006). Global distributions of arbuscular mycorrhizal fungi. Ecosystems 9: 305–316. [Google Scholar]
- Verbruggen E, El Mouden C, Jansa J, Akkermans G, Bucking H, West SA et al. (2012). Spatial structure and interspecific cooperation: theory and an empirical test using the mycorrhizal mutualism. Am Nat 179: E133–E146. [DOI] [PubMed] [Google Scholar]
- Vályi K, Mardhiah U, Rillig MC, Hempel S. (2016). Community assembly and coexistence in communities of arbuscular mycorrhizal fungi. ISME J 10: 2341–2351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vályi K, Rillig MC, Hempel S. (2015). Land-use intensity and host plant identity interactively shape communities of arbuscular mycorrhizal fungi in roots of grassland plants. New Phytol 205: 1577–1586. [DOI] [PubMed] [Google Scholar]
- Wang Y, Naumann U, Wright ST, Warton DI. (2012). mvabund—an R package for model-based analysis of multivariate abundance data. Methods Ecol Evol 3: 471–474. [Google Scholar]
- Warton DI, Wright ST, Wang Y. (2012). Distance-based multivariate analyses confound location and dispersion effects. Methods Ecol Evol 3: 89–101. [Google Scholar]
- Wehner J, Powell JR, Muller LAH, Caruso T, Veresoglou SD, Hempel S et al. (2014). Determinants of root-associated fungal communities within Asteraceae in a semi-arid grassland. J Ecol 102: 425–436. [Google Scholar]
- Zobel M, Öpik M. (2014). Plant and arbuscular mycorrhizal fungal (AMF) communities—which drives which? J Veg Sci 25: 1133–1140. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.