 (4,7di-OMe)Sα2,3Gal or (4,7di-OMe)Sα2,6Gal is used for the highly specific and rapid detection of influenza viruses.
(4,7di-OMe)Sα2,3Gal or (4,7di-OMe)Sα2,6Gal is used for the highly specific and rapid detection of influenza viruses.
Abstract
Rapid and precise detection of influenza viruses in a point of care setting is critical for applying appropriate countermeasures. Current methods such as nucleic acid or antibody based techniques are expensive or suffer from low sensitivity, respectively. We have developed an assay that uses glucose test strips and a handheld potentiostat to detect the influenza virus with high specificity. Influenza surface glycoprotein neuraminidase (NA), but not bacterial NA, cleaved galactose bearing substrates, 4,7di-OMe N-acetylneuraminic acid attached to the 3 or 6 position of galactose, to release galactose. In contrast, viral and bacterial NA cleaved the natural substrate, N-acetylneuraminic acid attached to the 3 or 6 position of galactose. The released galactose was detected amperometrically using a handheld potentiostat and dehydrogenase bearing glucose test strips. The specificity for influenza was confirmed using influenza strains and different respiratory pathogens that include Streptococcus pneumoniae and Haemophilus influenzae; bacteria do not cleave these molecules. The assay was also used to detect co-infections caused by influenza and bacterial NA. Viral drug susceptibility and testing with human clinical samples was successful in 15 minutes, indicating that this assay could be used to rapidly detect influenza viruses at primary care or resource poor settings using ubiquitous glucose meters.
Introduction
The Centers for Disease Control and Prevention, Atlanta, GA, USA reports that influenza and pneumonia are the 8th leading cause of death in the US.1,2 According to the World Health Organization, influenza infects approximately 0.4–0.6 billion children and 0.2–5.0 billion adults worldwide. In addition to seasonal influenza, emerging pandemic strains can infect a large number of people rapidly. For example, the H1N1 2009 “swine” strain pandemic reported in March 2009 in Mexico spread rapidly to over 74 countries in less than two months.3 While influenza vaccines are recommended especially for the most vulnerable groups, only ∼70% of people over the age of 65 and 50% of children in the age group 6–18 received vaccines in the US for the 2015–2016 season. This makes other countermeasures that include rapid diagnostics highly significant. The typical detection methods for influenza viruses are viral isolation and growth using standard cell culture, nucleic acid identification and antibody based immunoassays. Cell culture and nucleic acid based identification are expensive, require trained personnel and are not particularly suited for resource poor settings.4 Antibody based immunochromatography tests are user friendly, but suffer from low clinical sensitivity, especially at low pathogen counts. These strip tests are typically used to “rule out” the infection,5 and are followed up with further testing.6 An ideal biosensor should meet the ASSURED (Affordable, Sensitive, Selective, User friendly, Rapid and Robust, Equipment free and Deliverable to end-users) criteria7–9 that are particularly important for fast spreading respiratory infections. If diagnostics are made affordable and user-friendly, monitoring influenza disease progression and effectiveness of the therapy can also be achieved.
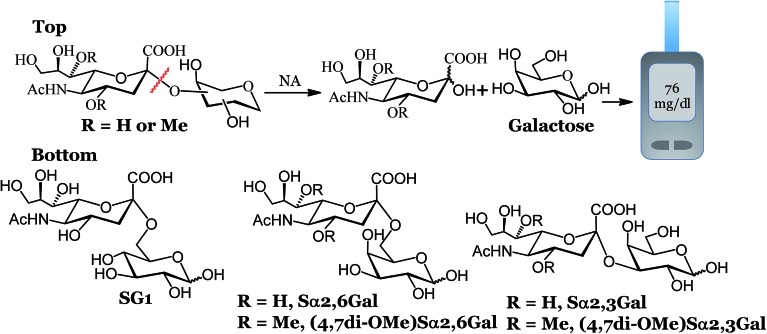
We recently developed an electrochemical assay to detect influenza viruses using a glucose bearing substrate.10 The neuraminidase (NA) enzyme, present in high abundance on the surface of influenza viruses,11 cleaves this substrate to release glucose, which can be measured amperometrically using a glucose meter. Glucose meters also meet the ASSURED criteria and are used by millions worldwide and are increasingly being used to detect a variety of non-glucose targets.12–17 In our previous report, we used SG1 (Fig. 1) to detect 19 unique influenza strains in 1 hour. However, SG1 is cleaved by NA from any source, human, bacterial or viral, which makes differentiation between influenza virus and other pathogens problematic unless an additional step of using antiviral drugs is included. Here, we designed compounds that detect influenza NA and the virus with high precision within 15 minutes. We used a portable potentiostat and commercial Accu-Chek test strips that use dehydrogenase enzymes that detect galactose or glucose. We demonstrated antiviral susceptibility and detection of clinically relevant amounts of influenza in spiked human nasal swabs, indicating that this assay can be used to detect influenza in a clinical or home setting.
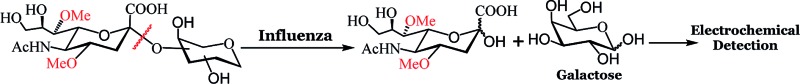
Fig. 1. Workflow and structure of the glycans used for the electrochemical detection. Top: electrochemical detection of influenza and S. pneumoniae. Bottom: structure of the glycans used for the electrochemical assay.
Results and discussion
To develop highly specific substrates for influenza viral NA, we introduced a larger methoxy group at the 4 and 7 positions of N-acetyl neuraminic acid (sialic acid). We reasoned that the binding pocket of influenza NA can accommodate a larger group at the 4 and 7 positions of sialic acid. Indeed, X-ray structures (Fig. S1, ESI†) and functional assays from Zanamivir and Oseltamivir18–22 have confirmed that bacterial or human NA possess a smaller binding pocket and cannot accommodate larger groups. Several groups have exploited the unique nature of the virus active site to develop highly specific NA inhibitors that have amine/guanidine groups at the 4 position of sialic acid.19,23,24 The amine or guanidine at the 4 position of sialic acid interacts with Glu119 and Glu227 present in the binding pocket of influenza NA. The same unique feature of the active site of influenza NA has been used to develop specific substrates for influenza virus.25–27 Recently, Bennett and coworkers demonstrated that NA from virus or bacteria can accommodate a variety of groups at the 4 position of sialic acid, each molecule possessing a range of cleavage efficiency.28 As a first step towards the development of highly specific substrates for influenza, we chose to introduce a methoxy group at the 4 and 7 positions of sialic acid and coupled it to the 3 or 6 hydroxyl group of d-galactose to establish proof of concept that these modifications lead to exclusive selectivity. We chose d-galactose instead of d-glucose for three reasons. (i) The Accu-Chek strips detect galactose and glucose since it uses a dehydrogenase enzyme.29,30 In contrast, glucose meters that use glucose oxidase detect only glucose and not galactose. (ii) Glucose may be present in the nasal or throat swabs of some patients; diabetic patients have low levels of glucose in their nasal cavity due to paracellular leakage, active co-transport, and facilitated diffusion31 that may interfere with the readings. (iii) Sialic acid attached to galactose and not glucose, is the natural substrate for NAs and therefore, could lead to a better fit at the active site.
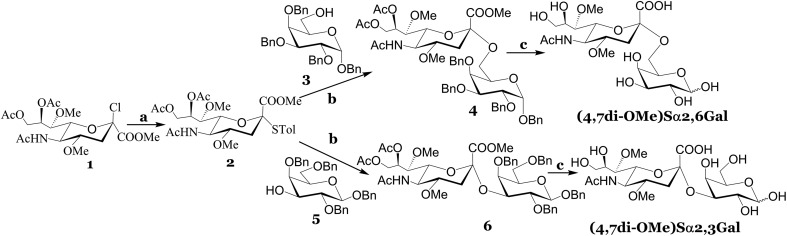
The synthesis of (4,7di-OMe)Sα2,3Gal and (4,7di-OMe)Sα2,6Gal is depicted in Scheme 1. Our strategy was to introduce the methoxy groups to the 4 and 7 positions of sialic acid before coupling to a suitably protected d-galactose acceptor. To this end, we synthesized 1, which was obtained from sialic acid using previously reported procedures.25,26 The chloride derivative, 1 was reacted with 4-methyl thiophenol to afford 2 in decent yield. The reaction of 2 with the suitably protected d-galactose acceptors, 3 or 5 and NIS as promoter gave the alpha isomers, 4 or 6 in tolerable yields. A negligible amount (<5%) of the beta isomer was observed for both reactions, however, this isomer was separated using standard column chromatography. The alpha linkage was confirmed using NMR spectroscopy; the H3eq of the sialic acid component for the alpha isomer in the coupled product 4 resonates at 2.8 ppm in the 1H NMR and the C2 carbon of the sialic acid resonates at 99.0 ppm in the 13C NMR. Global deprotection using a standard one-pot three step procedure resulted in the desired compounds in near quantitative yield. Naturally occurring sialosides, Sα2,3Gal and Sα2,6Gal, a portable potentiostat and Accu-Chek test strips to detect galactose were purchased. We found that the minimum detectable concentration of galactose over background signal was 0.3 mM from the standard curve for different concentrations of galactose (Fig. S2, ESI†). Therefore, we used a substrate concentration of 0.3 mM in all of our studies.
Scheme 1. Reagents and conditions: (a) 4-methyl thiophenol, DCM, DIEPA, rt, 12 h; (b) TfOH, NIS, DCM, –50 °C, 2 h, 40% for compound 4 and 37% for compound 6; (c) (i) NaOMe, MeOH, rt, 1 h; (ii) Pd(OH)2/C/H2, EtOH, rt, 12 h; (iii) 0.05 N NaOH in H2O, rt, 2 h, 94% for (4,7di-OMe)Sα2,6Gal and 88% for (4,7di-OMe)Sα2,3Gal over the three steps.
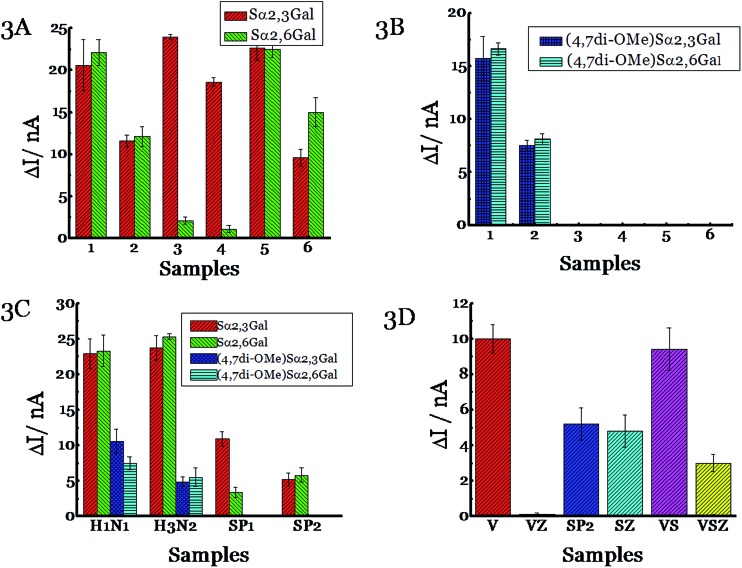
Next, we tested the ability of NAs to cleave the substrates before testing with pathogens. Influenza NAs are arranged into two groups based on their sequences with N1, N4, N5, N8 and N2, N3, N6, N7, N9 being part of groups 1 and 2, respectively.32,33 We chose N1 and N2 as representatives of the two major groups. N1 from H5N1 (A/Anhui/1/2005) was incubated with Sα2,3Gal or Sα2,6Gal substrate and the current was measured using a potentiostat (Fig. 2). A discernable signal was obtained in 1 h corresponding to 60% of substrate cleavage, and the difference between the signal and background is shown in Fig. 3A. A signal can also be obtained in 15 minutes (Fig. S3, ESI†). A similar result was obtained when N2 from influenza A, H3N2/Babol/36/2005, was used, although the intensity of the signal was slightly lower, presumably because the activity of the N2 strain is not as high as the N1 strain. When recombinant bacterial NAs (from Streptococcus pneumoniae, Salmonella typhimurium, Clostridium perfringens and Arthrobacter ureafaciens) were exposed to Sα2,3Gal or Sα2,6Gal, we found all four NAs cleave Sα2,3Gal with high efficiency; however, two of the NAs (Streptococcus pneumoniae and Salmonella typhimurium) cleave Sα2,6Gal at a negligible rate as they do not recognize Sα2,6Gal glycans well. Some of these NAs have been derived from pathogens present in the digestive tract; therefore, these glycans could potentially be used to detect those pathogens if NAs from these pathogens are excreted in appreciable amounts in the urine or stool. Next, we tested the ability of these six NAs, with (4,7di-OMe)Sα2,3Gal or (4,7di-OMe)Sα2,6Gal. We found that N1 and N2 from the two different groups of influenza cleave these new derivatives specifically and NAs from bacteria do not cleave the glycans, demonstrating excellent specificity.
Fig. 2. Image of the electrochemical system used in these studies. The strips are connected to a potentiostat, which is connected to a smartphone. Left: Accu-Chek Aviva strips. Middle: portable potentiostat. Right: smartphone.
Fig. 3. Detection of NA using (A) natural substrates Sα2,3Gal and Sα2,6Gal or (B) new substrates (4,7di-OMe)Sα2,3Gal and (4,7di-OMe)Sα2,6Gal. 50 U of different enzymes [(1) H5N1 NA (A/Anhui/1/2005), (2) H3N2 NA (A/Babol/36/2005), (3) NA from Streptococcus pneumoniae specific for α2-3 linkages, (4) NA from Salmonella typhimurium specific for α2-3 linkages, (5) NA from Clostridium perfringens specific for α2-3, α2-6 and α2-8 linkages and (6) NA from Arthrobacter ureafaciens specific for α2-3, α2-6, α2-8 and α2-9 linkages] was incubated with Sα2,3Gal or Sα2,6Gal at 37 °C for 1 h. (C) Detection of influenza and S. pneumoniae. Different substrate [(Sα2,3Gal:Sα2,6Gal; (4,7di-OMe)Sα2,3Gal or (4,7di-OMe)Sα2,6Gal)] was incubated with different pathogens [H1N1 (A/Brisbane/59/2007); H3N2 (A/Victoria/361/2011); SP1 (serotype 1, ATCC 6301) or SP2 (serotype 1, ATCC6305)] at 37 °C for 1 h. (D) Monitoring antiviral efficacy. 10 ng of Zanamivir was premixed with different pathogens [(H3N2 A/Victoria/361/2011, VZ), SP2 (serotype 1, ATCC 6305, SZ), or H3N2 A/Victoria/361/2011 and SP2 (VSZ)] for 30 min before the addition of Sα2,3Gal. Also shown is data obtained without the addition of Zanamivir to the pathogens [(H3N2 A/Victoria/361/2011, V), SP2 (serotype 1, ATCC 6305, SP2), or A/Victoria/361/2011 and SP2 (VS)]. The current was measured using Accu-Chek Aviva strips (Roche Diagnostics) with the 10 s average current recorded 5 s following substrate introduction at a working potential of –0.15 V. The y axis, ΔI, represents the difference in current before and after the addition of the substrate. All experiments were performed in triplicate independently on different days to demonstrate scientific rigor.
The cleavage rate of the new derivatives is slower than Sα2,3Gal and Sα2,6Gal; this is to be expected as the new derivatives are not the natural substrate (Fig. 3B). Next, we tested the ability of virus and bacteria, not just the recombinant NAs because recombinant NAs are not the same as the natural NAs. While enzymes may work well under controlled conditions, the assay may not work with intact viruses and bacteria. Viral NAs are embedded on the cell surface as a tetramer, whereas recombinant NAs from the manufacturer are present as a mixture of monomers, dimers and tetramers. Also, bacteria produce multiple NAs. For example, S. pneumoniae produces three types of NA, one of which is a trans-sialidase.34 To this end, we cultured influenza viruses and S. pneumoniae and characterized them using plaque assays. Since S. pneumoniae has been reported to use NA as a virulence factor, we grew S. pneumoniae in the presence of sialic acid as it has been demonstrated that the bacteria produce more NAs when sialic acid is added to the growth media.35 It was gratifying to observe the viral strains, H1N1 (A/Brisbane/59/2007) and H3N2 (A/Victoria/361/2011) cleave all four compounds extremely well; in contrast, the two strains of S. pneumoniae (serotype 1, ATCC 6301 and serotype 1, ATCC 6305) cleave only the natural substrates, (Fig. 3C) and not (4,7di-OMe)Sα2,3Gal or (4,7di-OMe)Sα2,6Gal. We also demonstrated that Haemophilus influenzae, another respiratory pathogen, does not cleave (4,7di-OMe)Sα2,3Gal (Fig. S4, ESI†). Thus, the new derivatives can be used to detect influenza virus specifically. The assay can also detect influenza B virus in addition to influenza A, but not influenza C (Fig. S4, ESI†), as influenza C virus does not possess sialic acid cleavage activity.36,37
The next set of experiments was designed to monitor antiviral efficacy. We premixed a H3N2 strain (A/Victoria/361/2011) with the anti-viral drug, Zanamivir i.e. Relenza, before adding the natural substrate, Sα2,3Gal. Zanamivir was used because strains develop resistance to Oseltamivir more frequently than Zanamivir;38 in 2009, the majority of circulating H1N1 strains were resistant to Oseltamivir.39 Galactose was not released because Zanamivir blocks the action of viral NA. When we performed the same experiment with S. pneumoniae (serotype 1, ATCC 6305) instead of influenza virus, we found that galactose was released because Zanamivir does not block the action of bacteria (Fig. 3D). Next, we mixed the same virus and bacteria to simulate co-infections, followed by addition of Zanamivir and subsequent incubation of Sα2,3Gal. We found that galactose was released because the antiviral does not block the action of bacteria, however, the signal is lower indicating that the antiviral drug targeted influenza and not the bacterium. Taken together, this assay can be used to detect influenza viruses and monitor treatment efficacy. An added feature of this assay is that it can also be potentially used to detect co-infections caused by bacteria that overexpress NA as a virulence factor by using the natural and the modified substrate. For example, S. pneumoniae produces increased amounts of bacterial NA as a virulence factor during the infection process.40 It has been shown that the cleavage of the terminal sialic acid on host cells exposes the underlying glycans, GalNAc β1-4Gal, which serve as receptors for opportunistic pathogens to increase adhesion41 and the advancement of S. pneumoniae into the middle ear leading to otitis media in a chinchilla model.42,43 Increase/decrease in the activity of influenza NA and bacterial NA can be measured as a function of time to follow the infection process using ubiquitous glucose meters. This becomes important from a clinical standpoint, as the treatment regimen for influenza virus and bacterial infections are different; physicians typically treat bacterial infections with antibiotics and influenza virus with antivirals.
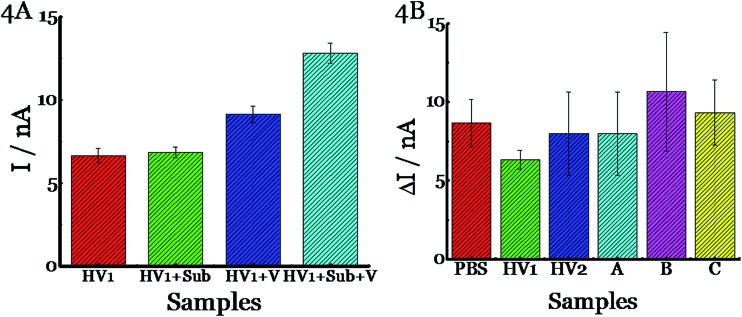
The next set of experiments were designed to study the matrix effects. Since there is a plethora of natural flora in the nasal cavity, we were concerned that the human clinical samples may either have galactose in them and/or endogenous enzymes may cleave the substrate resulting in false positives. To this end, we obtained nasal swabs from human volunteers who did not show any signs of influenza and clinical samples from patients who exhibited influenza like symptoms, but did not have influenza as determined by qRT-PCR. First, we tested nasal swabs in buffer and found minimal background (HV1, Fig. 4A). Spiking the sample with substrate (HV1+Sub) or H1N1 A/Brisbane/59/2007 (HV1+V) resulted in no appreciable signal. The HV1+Sub result indicates that there is nothing in the sample that cleaves the substrate and the HV1+V result indicates there is no endogenous compound that can be cleaved by the virus to release galactose. A signal was observed only when the sample with spiked with both virus and substrate (HV1+Sub+V, Fig. 4A). We repeated these experiments with multiple samples that include buffer (PBS), two healthy human volunteer samples and three clinical samples from patients who did not have influenza (Fig. 4B). The spiked virus was detected in all samples, indicating that this assay could be used in a clinical setting. Most importantly, measuring the signal before and after the introduction of the substrate eliminates any background signal due to endogenous galactose or glucose or other interferences. The limit of detection of this assay for H1N1 A/Brisbane/59/2007 and H3N2 A/Victoria/361/2011 was determined to be 102 pfu per sample and S. pneumoniae (serotype 1, ATCC 6305) was 102 cfu per sample in 15 minutes (Fig. S5–S7, ESI†). This is clinically significant as patients entering the clinic typically carry 103–108 viral particles in their nose or throat.44,45 With longer incubation times, the limit of detection is 102 pfu per sample as this is an enzyme based system (Fig. S5, ESI†).
Fig. 4. Detection of spiked H1N1 influenza virus (A/Brisbane/59/2007, 2.5 × 105 pfu per sample) in human samples using (4,7di-OMe)Sα2,3Gal. (A) A nasal swab from human volunteer 1 was incubated with PBS only (HV1), substrate (HV1+Sub), virus (HV1+V), or substrate + virus (HV1+Sub+V) at 37 °C for 15 min. The y axis, I, represents the current. (B) (4,7di-OMe)Sα2,3Gal was incubated with different matrices such as PBS, HV1, HV2, A, B, or C at 37 °C for 15 min. PBS is buffer, and HV1 and HV2 are nasal swabs from human volunteers. A, B and C are samples from patients who did not have influenza as determined by rRT-PCR. The current was measured using Accu-Chek Aviva strips (Roche Diagnostics) with the 10 s average current recorded 5 s following substrate introduction at a working potential of –0.15 V. The y axis, ΔI, represents the difference in current before and after the addition of the substrate. All experiments were performed in triplicate independently on different days to demonstrate scientific rigor.
Conclusions
We have developed a novel assay that can detect clinically relevant influenza viruses with high specificity within 15 minutes, which makes this technique highly relevant for a point of care setting. While we used a portable potentiostat and commercially available test strips as part of our protocol, glucose meters could also be used.16 The advantages of this assay are that it meets the ASSURED criteria and uses existing technology for a new purpose i.e. a glucose meter to detect and differentiate these pathogens. Antiviral susceptibility can also be measured rapidly. An added feature of this assay is that disease progression and response to therapy can be tracked using the natural substrate and the new derivatives to monitor increased/decreased activity of viral and bacterial NAs. For example, other pathogens present in the respiratory tract can cause secondary infections after the initial attack by influenza has diminished. S. pneumoniae, a pathogen that already exists in the respiratory tract can cause secondary infections particularly in patients with weak (e.g. children and senior citizens) or compromised (e.g. patients with human immunodeficiency virus, tuberculosis or organ transplants) immune systems.46 Multiple studies have demonstrated that the initial influenza infection leads to subsequent bacterial infection, typically caused by S. pneumoniae; this combination “one-two punch” is lethal resulting in intensive care hospitalizations and death, especially in conjunction with pandemic influenza strains.47–53 We anticipate that this concept will be applied to the detection of other infectious agents or other human disorders.
Experimental section
Materials
α2-3 NA S (S. pneumoniae), α2-3 NA (Salmonella typhimurium), α2-3,6,8 NA (Clostridium perfringens) and α2-3,6,8,9 NA A (Arthrobacter ureafaciens) were purchased from New England Biolabs, Ipswich, MA. H5N1 (A/Anhui/1/2005) and H3N2 (A/Babol/36/2005) neuraminidase were purchased from Sino Biological Inc., Beijing, China. S. pneumoniae ATCC 6301 and S. pneumoniae ATCC 6305 were purchased from American Type Culture Collection (ATCC). Sα2,6Gal and Sα2,3Gal were purchased from Carbosynth Limited, Berkshire, UK. Accu-Chek Aviva strips were purchased from Amazon, Seattle, WA. Influenza strains were obtained from BEI resources, NIAID. All experiments with human clinical samples were approved by Georgia State University Institutional Review Board.
Bacterial growth
Briefly, S. pneumoniae ATCC 6301 and S. pneumoniae ATCC 6305 were inoculated in Brain Heart Infusion (BHI) broth augmented with 0.16 g L–1 of sialic acid to enhance NA production by the bacteria.35 The bacterial concentration was determined to be 106 CFU mL–1 using filtration and the colony count method. Bacteria was lysed using the method described by Giacomini et al. 54 To 100 μL of bacterial samples, 0.01% SDS and 20 μL of chloroform were added. The samples were vigorously vortexed for 30 s and incubated at 28 °C for 5 min to allow bacterial lysis.
Detection of different substrates
50 U of the neuraminidase was mixed with 1 μL of GlycoBuffer (10X, pH 5.5) and 1.5 μL of substrate (2 mM). DI H2O was added to bring the volume to 10 μL. The mixture was incubated at 37 °C for 1 h. 1.5 μL of this solution was used to measure the current using Accu-Chek Aviva strips for an average of 10 s recorded 5 s following substrate introduction at a working potential of –0.15 V.
Detection of influenza virus and S. pneumoniae
50 μL of A/Brisbane/59/2007 (1.25 × 107 pfu mL–1), A/Victoria/361/2011 (3.5 × 106 pfu mL–1), S. pneumoniae (serotype 1, ATCC 6301, 106 CFU mL–1), or S. pneumoniae (serotype 1, ATCC 6305, 106 CFU mL–1) was mixed with 8.8 μL of substrate (2 mM). The resulting mixture was incubated at 37 °C for 1 h. 1.5 μL of this solution was used to measure the current using Accu-Chek Aviva strips for an average of 10 s recorded 5 s following substrate introduction at a working potential of –0.15 V as described previously.55
Antiviral efficacy study
10 ng of Zanamivir was premixed with 50 μL of H3N2 A/Victoria/361/2011 (3.5 × 106 pfu mL–1), S. pneumoniae (serotype 1, ATCC 6305, 106 CFU mL–1), or a mixture of virus and bacteria for 30 min at rt. Next, the solution was incubated with Sα2,3Gal for 1 h at 37 °C. 1.5 μL of this solution was used to measure the current using Accu-Chek Aviva strips for an average of 10 s recorded 5 s following substrate introduction at a working potential of –0.15 V.
Detection of clinical samples
25 μL of PBS, HV1, HV2, A, B, or C was mixed with 25 μL of virus (H1N1 A/Brisbane/59/2007, 1.25 × 107 pfu mL–1), followed by the addition of 8.8 μL of 2 mM solution of (4,7di-OMe)Sα2,3Gal in PBS. The resulting mixture was incubated at 37 °C for 15 min. 1.5 μL of this solution was used to measure the current using Accu-Chek Aviva strips for an average of 10 s recorded 5 s following substrate introduction at a working potential of –0.15 V.
Acknowledgments
We are grateful to NIH-NIAID (R01-AI089450) for funding and BEI resources, and NIAID for the influenza reagents.
Footnotes
References
- Heron M., National Vital Statistics Reports: from the Centers for Disease Control and Prevention, National Center for Health Statistics, National Vital Statistics System, 2016, vol. 65, pp. 1–95. [Google Scholar]
- Short K. R., Habets M. N., Hermans P. W. M., Diavatopoulos D. A. Future Microbiol. 2012;7:609–624. doi: 10.2217/fmb.12.29. [DOI] [PubMed] [Google Scholar]
- New influenza A (H1N1) virus: WHO guidance on public health measures, Wkly Epidemiol Rec, 2009, vol. 84, pp. 261–264. [PubMed] [Google Scholar]
- Wang S., Lifson M. A., Inci F., Liang L. G., Sheng Y. F., Demirci U. Expert Rev. Mol. Diagn. 2016;16:449–459. doi: 10.1586/14737159.2016.1142877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horita N., Miyazawa N., Kojima R., Kimura N., Inoue M., Ishigatsubo Y., Kaneko T. Respirology. 2013;18:1177–1183. doi: 10.1111/resp.12163. [DOI] [PubMed] [Google Scholar]
- Balish A., Warnes C. M., Wu K., Barnes N., Emery S., Berman L., Shu B., Lindstrom S., Xu X., Uyeki T., Shaw M., Klimov A. and Villanueva J., MMWR Wkly Rep, 2009, vol. 58, pp. 826–829. [Google Scholar]
- Caliendo A. M., Gilbert D. N., Ginocchio C. C., Hanson K. E., May L., Quinn T. C., Tenover F. C., Alland D., Blaschke A. J., Bonomo R. A., Carroll K. C., Ferraro M. J., Hirschhorn L. R., Joseph W. P., Karchmer T., Machintyre A. T., Reller L. B., Jackson A. F. Clin. Infect. Dis. 2013;57(3):S139-170. doi: 10.1093/cid/cit578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- UNICEF/UNDP/World Bank/WHOSpecial Programme for Research and Training in Tropical Diseases. Accessible quality-assured diagnostics: annual report 2009, World Health Organization, 2010.
- Peeling R. W., Holmes K. K., Mabey D. and Ronald A., Sexually Transmitted Infections, 2006, vol. 82, suppl 5, pp. v1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Dhawane A. N., Sweeney J., He Y., Vasireddi M., Iyer S. S. Angew. Chem., Int. Ed. Engl. 2015;54:5929–5932. doi: 10.1002/anie.201412164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris A., Cardone G., Winkler D. C., Heymann J. B., Brecher M., White J. M., Steven A. C. Proc. Natl. Acad. Sci. U. S. A. 2006;103:19123–19127. doi: 10.1073/pnas.0607614103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chavali R., Gunda N. S. K., Naicker S., Mitra S. K. Anal. Methods. 2014;6:6223–6227. [Google Scholar]
- Mohapatra H., Phillips S. T. Chem. Commun. 2013;49:6134–6136. doi: 10.1039/c3cc43702g. [DOI] [PubMed] [Google Scholar]
- Zhang J., Xiang Y., Novak D. E., Hoganson G. E., Zhu J., Lu Y. Chem.–Asian J. 2015;10:2221–2227. doi: 10.1002/asia.201500642. [DOI] [PubMed] [Google Scholar]
- Wang Q., Wang H., Yang X., Wang K., Liu R., Li Q., Ou J. Analyst. 2015;140:1161–1165. doi: 10.1039/c4an02033b. [DOI] [PubMed] [Google Scholar]
- Lan T., Zhang J., Lu Y. Biotechnol. Adv. 2016;34:331–341. doi: 10.1016/j.biotechadv.2016.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurale B. P., Dhawane A. N., Cui X., Das A., Zhang X., Iyer S. S. Anal. Chem. 2016;88:4248–4253. doi: 10.1021/acs.analchem.5b03943. [DOI] [PubMed] [Google Scholar]
- von Itzstein M., Wu W.-Y., Kok G. B., Pegg M. S., Dyason J. C., Jin B., Phan T. V., Smythe M. L., White H. F., Oliver S. W., Colman P. M., Varghese J. N., Ryan D. M., Woods J. M., Bethell R. C., Hotham V. J., Cameron J. M., Penn C. R. Nature. 1993;363:418–423. doi: 10.1038/363418a0. [DOI] [PubMed] [Google Scholar]
- von Itzstein M. Nat. Rev. Drug Discovery. 2007;6:967–974. doi: 10.1038/nrd2400. [DOI] [PubMed] [Google Scholar]
- Varghese J. N., McKimm-Breschkin J. L., Caldwell J. B., Kortt A. A., Colman P. M. Proteins. 1992;14:327–332. doi: 10.1002/prot.340140302. [DOI] [PubMed] [Google Scholar]
- Varghese J. N., Laver W. G., Colman P. M. Nature. 1983;303:35–40. doi: 10.1038/303035a0. [DOI] [PubMed] [Google Scholar]
- Gut H., Xu G., Taylor G. L., Walsh M. A. J. Mol. Biol. 2011;409:496–503. doi: 10.1016/j.jmb.2011.04.016. [DOI] [PubMed] [Google Scholar]
- Kim J. H., Resende R., Wennekes T., Chen H. M., Bance N., Buchini S., Watts A. G., Pilling P., Streltsov V. A., Petric M., Liggins R., Barrett S., McKimm-Breschkin J. L., Niikura M., Withers S. G. Science. 2013;340:71–75. doi: 10.1126/science.1232552. [DOI] [PubMed] [Google Scholar]
- Yang Y., He Y., Li X., Dinh H., Iyer S. S. Bioorg. Med. Chem. Lett. 2014;24:636–643. doi: 10.1016/j.bmcl.2013.11.077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liav A., Hansjergen J. A., Achyuthan K. E., Shimasaki C. D. Carbohydr. Res. 1999;317:198–203. doi: 10.1016/s0008-6215(99)00058-0. [DOI] [PubMed] [Google Scholar]
- Shimasaki C. D., Achyuthan K. E., Hansjergen J. A., Appleman J. R. Philos. Trans. R. Soc., B. 2001;356:1925–1931. doi: 10.1098/rstb.2001.10006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Achyuthan K. E., Pence L. M., Appleman J. R., Shimasaki C. D. Luminescence. 2003;18:131–139. doi: 10.1002/bio.714. [DOI] [PubMed] [Google Scholar]
- Shidmoossavee F. S., Watson J. N., Bennet A. J. J. Am. Chem. Soc. 2013;135:13254–13257. doi: 10.1021/ja405916q. [DOI] [PubMed] [Google Scholar]
- Dimeski G., Jones B. W., Tilley V., Greenslade M. N., Russell A. W. Ann. Clin. Biochem. 2010;47:358–365. doi: 10.1258/acb.2010.009291. [DOI] [PubMed] [Google Scholar]
- Ceriotti F., Kaczmarek E., Guerra E., Mastrantonio F., Lucarelli F., Valgimigli F., Mosca A. J. Diabetes Sci. Technol. 2015;9:268–277. doi: 10.1177/1932296814563351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pezzulo A. A., Gutierrez J., Salm K. S., McConnell K. S., Karp P., Stoltz D. A., Zabner J. FASEB J. 2009;23:570.3. [Google Scholar]
- Russell R. J., Haire L. F., Stevens D. J., Collins P. J., Lin Y. P., Blackburn G. M., Hay A. J., Gamblin S. J., Skehel J. J. Nature. 2006;443:45–49. doi: 10.1038/nature05114. [DOI] [PubMed] [Google Scholar]
- Air G. M. Influenza Other Respir. Viruses. 2012;6:245–256. doi: 10.1111/j.1750-2659.2011.00304.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu G., Kiefel M. J., Wilson J. C., Andrew P. W., Oggioni M. R., Taylor G. L. J. Am. Chem. Soc. 2011;133:1718–1721. doi: 10.1021/ja110733q. [DOI] [PubMed] [Google Scholar]
- Gualdi L., Hayre J. K., Gerlini A., Bidossi A., Colomba L., Trappetti C., Pozzi G., Docquier J. D., Andrew P., Ricci S., Oggioni M. R. BMC Microbiol. 2012;12:200. doi: 10.1186/1471-2180-12-200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nerome K., Ishida M., Nakayama M. Arch. Virol. 1976;50:241–244. doi: 10.1007/BF01320578. [DOI] [PubMed] [Google Scholar]
- Vlasak R., Krystal M., Nacht M., Palese P. Virology. 1987;160:419–425. doi: 10.1016/0042-6822(87)90013-4. [DOI] [PubMed] [Google Scholar]
- Tamura D., Sugaya N., Ozawa M., Takano R., Ichikawa M., Yamazaki M., Kawakami C., Shimizu H., Uehara R., Kiso M., Kawakami E., Mitamura K., Kawaoka Y. Clin. Infect. Dis. 2011;52:432–437. doi: 10.1093/cid/ciq183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moscona A. N. Engl. J. Med. 2009;360:953–956. doi: 10.1056/NEJMp0900648. [DOI] [PubMed] [Google Scholar]
- Mitchell A. M., Mitchell T. J. Clin. Microbiol. Infect. 2010;16:411–418. doi: 10.1111/j.1469-0691.2010.03183.x. [DOI] [PubMed] [Google Scholar]
- Krivan H. C., Roberts D. D., Ginsburg V. Proc. Natl. Acad. Sci. U. S. A. 1988;85:6157–6161. doi: 10.1073/pnas.85.16.6157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linder T. E., Daniels R. L., Lim D. J., DeMaria T. F. Microb. Pathog. 1994;16:435–441. doi: 10.1006/mpat.1994.1043. [DOI] [PubMed] [Google Scholar]
- Tong H. H., Blue L. E., James M. A., DeMaria T. F. Infect. Immun. 2000;68:921–924. doi: 10.1128/iai.68.2.921-924.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee N., Chan P. K., Hui D. S., Rainer T. H., Wong E., Choi K. W., Lui G. C., Wong B. C., Wong R. Y., Lam W. Y., Chu I. M., Lai R. W., Cockram C. S., Sung J. J. J. Infect. Dis. 2009;200:492–500. doi: 10.1086/600383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suess T., Remschmidt C., Schink S. B., Schweiger B., Heider A., Milde J., Nitsche A., Schroeder K., Doellinger J., Braun C., Haas W., Krause G., Buchholz U. PLoS One. 2012;7:e51653. doi: 10.1371/journal.pone.0051653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Brien K. L., Wolfson L. J., Watt J. P., Henkle E., Deloria-Knoll M., McCall N., Lee E., Mulholland K., Levine O. S., Cherian T., Dis H. P. G. B. Lancet. 2009;374:893–902. doi: 10.1016/S0140-6736(09)61204-6. [DOI] [PubMed] [Google Scholar]
- Palacios G., Hornig M., Cisterna D., Savji N., Bussetti A. V., Kapoor V., Hui J., Tokarz R., Briese T., Baumeister E., Lipkin W. I. PLoS One. 2009;4:e8540. doi: 10.1371/journal.pone.0008540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brundage J. F., Shanks G. D. Emerging Infect. Dis. 2008;14:1193–1199. doi: 10.3201/eid1408.071313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Brien K. L., Walters M. I., Sellman J., Quinlisk P., Regnery H., Schwartz B., Dowell S. F. Clin. Infect. Dis. 2000;30:784–789. doi: 10.1086/313772. [DOI] [PubMed] [Google Scholar]
- Morens D. M., Taubenberger J. K., Fauci A. S. J. Infect. Dis. 2008;198:962–970. doi: 10.1086/591708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCullers J. A., Bartmess K. C. J. Infect. Dis. 2003;187:1000–1009. doi: 10.1086/368163. [DOI] [PubMed] [Google Scholar]
- Kash J. C., Walters K. A., Davis A. S., Sandouk A., Schwartzman L. M., Jagger B. W., Chertow D. S., Li Q., Kuestner R. E., Ozinsky A., Taubenberger J. K. mBio. 2011;2:e00172-11. doi: 10.1128/mBio.00172-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y., Tu W., Lam K. T., Chow K. H., Ho P. L., Guan Y., Peiris J. S., Lau Y. L. J. Virol. 2015;89:2013–2023. doi: 10.1128/JVI.02455-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giacomini A., Corich V., Ollero F. J., Squartini A., Nuti M. P. FEMS Microbiol. Lett. 1992;100:87–90. doi: 10.1111/j.1574-6968.1992.tb14024.x. [DOI] [PubMed] [Google Scholar]
- Cha K. H., Jensen G. C., Balijepalli A. S., Cohan B. E., Meyerhoff M. E. Anal. Chem. 2014;86:1902–1908. doi: 10.1021/ac4040168. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.