Abstract
Background
Gene bodies of vertebrates and flowering plants are occupied by the histone variant H3.3 and DNA methylation. The origin and significance of these profiles remain largely unknown. DNA methylation and H3.3 enrichment profiles over gene bodies are correlated and both have a similar dependence on gene transcription levels. This suggests a mechanistic link between H3.3 and gene body methylation.
Results
We engineered an H3.3 knockdown in Arabidopsis thaliana and observed transcription reduction that predominantly affects genes responsive to environmental cues. When H3.3 levels are reduced, gene bodies show a loss of DNA methylation correlated with transcription levels. To study the origin of changes in DNA methylation profiles when H3.3 levels are reduced, we examined genome-wide distributions of several histone H3 marks, H2A.Z, and linker histone H1. We report that in the absence of H3.3, H1 distribution increases in gene bodies in a transcription-dependent manner.
Conclusions
We propose that H3.3 prevents recruitment of H1, inhibiting H1’s promotion of chromatin folding that restricts access to DNA methyltransferases responsible for gene body methylation. Thus, gene body methylation is likely shaped by H3.3 dynamics in conjunction with transcriptional activity.
Electronic supplementary material
The online version of this article (doi:10.1186/s13059-017-1221-3) contains supplementary material, which is available to authorized users.
Keywords: H3.3, Histone variants, H2A.Z, Linker histone H1, DNA methylation, Chromatin
Background
Two major types of histone H3 variants evolved in multicellular eukaryotes, H3.1 and H3.3, distinguished by a few amino acid residues as well as their expression patterns and modes of deposition by distinct chaperones [1–3]. While H3.1 expression is coupled to DNA replication, H3.3 expression occurs throughout the cell cycle [4]. H3.3 is a crucial chromatin component required for development in Drosophila [5, 6], mouse [7], and Xenopus [8]. Notably H3.3 and HIRA are required for reprogramming events during development in animals [9–13] and plants [14, 15].
H3.3 is associated with actively expressed genes in both animals and plants [4, 16–19]. More specifically, genome-wide analysis of chromatin immunoprecipitation (ChIP) in several model organisms, including plants, showed that H3.3 is predominantly enriched near transcription end sites (TES) of genes and positively associated with transcription [18–21], suggesting a direct mechanistic link between H3.3 enrichment and transcription. This distinctive pattern of H3.3 over genes overlaps with the enrichment of RNA polymerase II (RNAPII) [19, 21]. However, H3.3 knockdown has a limited impact on transcription in Drosophila [5] and mouse embryonic stem cells (mESCs) [22]. Thus, the functional relationship between H3.3 enrichment and transcriptional activity remains unresolved.
Transcriptional activity has also been related to DNA methylation on gene bodies in mammals, Arabidopsis, and other plants [23–25]. In mammals, gene body methylation is maintained by the recruitment of DNA methyltransferase by H3K36me3 [26]. In mammals, H3.3K36me3 is positively correlated with transcriptional activity, elongation, and splicing [27], thus providing a potential mechanism to explain the link between transcription and gene body methylation. However, the prevalence and overall shape of the profile of gene body methylation observed in mammals is not present in other animal groups. In the plant lineage, gene body methylation is largely absent in green algae and bryophytes but is present in most vascular plants [28–31]. In Arabidopsis, the similarity between the profiles of gene body methylation and enrichment of H3.3 suggests a link but the mechanism involved remains unknown. To investigate this question, we engineered Arabidopsis lines deficient in H3.3 and report decreased gene body methylation in these lines. We further identify that H3K36 methylation and other transcription-related H3 modifications do not play a role in gene body methylation. Instead, we show that H3.3 prevents the deposition of the linker histone H1 on gene bodies, and relaxes chromatin in correlation with transcriptional activity. We propose that this action of H3.3 promotes access to DNA methyltransferase and explains the origin of the transcription-dependent profile of gene body methylation in Arabidopsis.
Results
H3.3 impacts plant development
In Arabidopsis H3.3 is encoded by three HISTONE 3 RELATED (HTR) genes, HTR4 (At4g40030), HTR5 (At4g40040), and HTR8 (At5g10980), which are highly expressed throughout development [14, 32]. To obtain a knockout line devoid of H3.3 genes we combined T-DNA insertion lines to generate the double mutant combinations htr4/htr8 and htr5/htr8, which were phenotypically normal despite the absence of the respective full length transcripts (Additional file 1: Table S1; Additional file 2: Figure S1a, b). To obtain a complete H3.3 knockout mutant, we designed a CRISPR/Cas9-based approach to functionally delete both HTR4 and HTR5 (Additional file 1: Tables S1 and S2; Additional file 2: Figure S1c). htr4;htr5 double homozygous mutants were crossed to htr8; however, we were not able to isolate triple homozygous plants (Additional file 2: Figure S1d). Reciprocal crosses suggested that the knockout of H3.3 impaired male gametogenesis and caused embryo lethality (Additional file 2: Figure S1d). We concluded that it would be impossible to obtain somatic tissues completely devoid of H3.3 using this strategy.
As an alternative, to effectively reduce levels of H3.3 transcripts in vegetative tissues we combined the alleles htr4 and htr8 with artificial microRNAs (amiRNAs) targeting HTR5 (Additional file 1: Table S2; Additional file 2: Figure S2a). We constructed two amiRNAs (amiR-HTR5-I/II) targeting different regions of the HTR5 transcript and introduced them into plants segregating from htr4/+;htr8/htr8 plants. In contrast to double homozygous htr4/htr4;htr8/htr8 plants that looked similar to wild type (WT; Additional file 2: Figure S2b), htr4/htr4;htr8/htr8 plants that carried either amiR-HTR5-I or amiR-HTR5-II (collectively referred to as h3.3kd lines) showed serration of leaf margins, reduced growth, and partial sterility (Fig. 1a; Additional file 1: Table S2; Additional file 2: Figure S2b, c). Transcriptome analyses by RNA-seq revealed that HTR5 transcript levels were reduced in h3.3kd plants (Additional file 2: Figure S2d). As a control, the introduction of an amiR-HTR5-II resistant version (rH3.3) into h3.3kd led to the partial rescue of the phenotypic defects observed in h3.3kd plants (Additional file 1: Table S2; Additional file 2: Figure S2b, c), confirming that H3.3 knockdown was responsible for the morphological defects observed in h3.3kd plants. We noted that serrated leaf margins are prominent in mutants for the H3.3 chaperone complex [15]. Transcriptome analyses in h3.3kd plants revealed that the reduction of transcript levels of the three H3.3 genes caused increased levels of three out of five H3.1 genes, while the expression levels of genes putatively involved in H3 deposition were not significantly misregulated (Additional file 1: Table S3; Additional file 2: Figure S2d). Because we did not observe any phenotypes in plants overexpressing H3.1-GFP [14], it appears unlikely that phenotypes observed in h3.3kd plants resulted from the increased expression of H3.1 variants. Overall, the loss of H3.3 dosage relative to the total pool of H3 led to pleiotropic phenotypic defects, while a complete loss of H3.3 caused lethality. Thus, H3.3 is an essential, non-redundant component of plant chromatin.
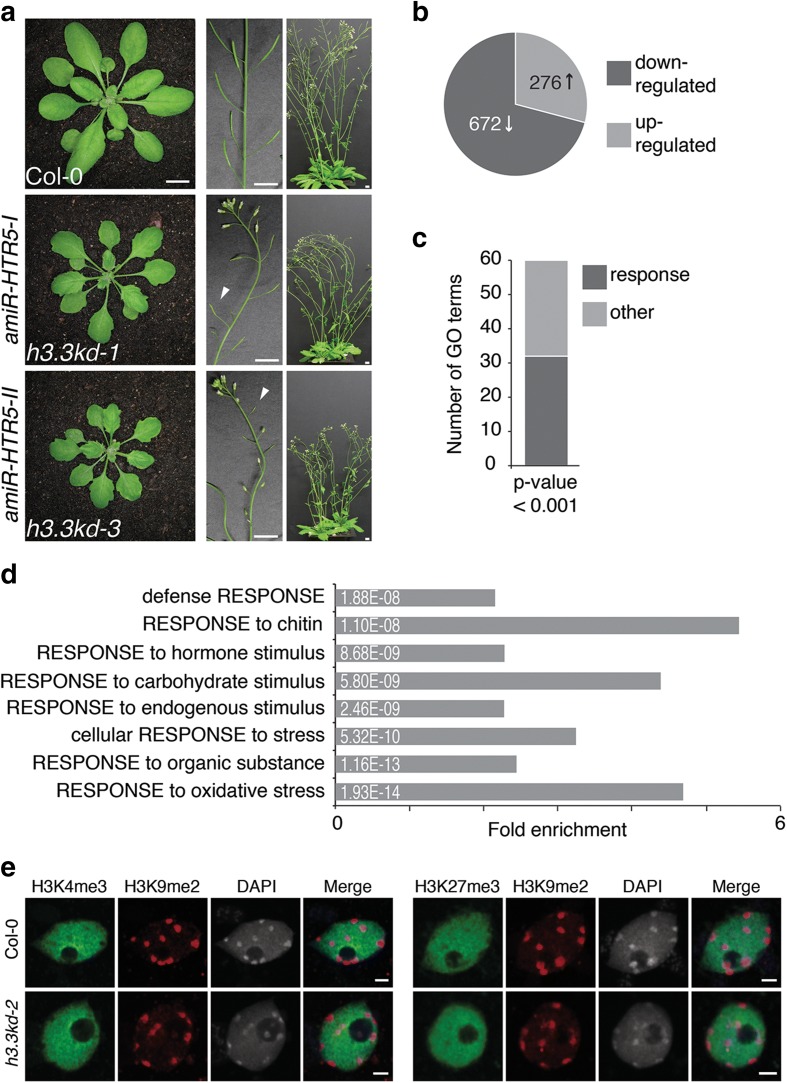
Fig. 1.
Knockdown of H3.3 causes various phenotypic defects and misregulation of response genes. a The impact of H3.3 knockdown on plant growth and development includes serrated leaf shape and smaller rosette size (left panel), partial sterility (middle panel) and reduced height (right panel) of flowering transgenic h3.3kd plants compared to wild type (Col-0). WT control plants (top row) are shown with two independent transgenic lines, both double homozygous for htr4 and htr8 (htr4/htr8) alleles with either pHTR5-amiR-HTR5-I (middle row; h3.3kd-1) or pHTR5-amiR-HTR5-II (bottom row; h3.3kd-3) artificial miRNAs. b The total number of significantly up- and downregulated genes in RNA-seq data from h3.3kd-3 compared to WT plants. c Summary of the Gene Ontology (GO) analysis of misregulated genes in h3.3kd-3 compared to WT. The bar graph represents the number of response related GO terms compared to others with p values less than 0.001. d Enrichment and p values for selected GO terms. The complete list can be found in Additional file 5. e Chromatin localization of H3K4me3, H3K27me3, and H3K9me2 in WT Col-0 and h3.3kd-2 plants as detected by immunofluorescence in nuclei isolated from mature leaves. DAPI staining shown in grey
Impact of h3.3kd on transcription
H3.3 knockdown caused a variety of developmental defects in plants, suggesting transcription misregulation. We assessed the impact of H3.3 knockdown on transcription using RNA-seq analysis. To minimize secondary effects of H3.3 knockdown on transcription from differences in development, we used WT and h3.3kd plants at the seedling stage, where phenotypic defects of h3.3kd are less severe. Over 900 genes were significantly misexpressed in h3.3kd (Fig. 1b; Additional files 3 and 4), with the majority being downregulated. However, the transcriptional changes in h3.3kd were not tightly correlated with enrichment of H3.3 in WT over promoters and/or gene bodies (Additional file 1: Table S4). Hence, although there is a clear correlation between transcriptional activity and H3.3 enrichment over bodies of active genes [18, 19] and promoters [17], H3.3 might not be directly required for transcription.
Gene expression is associated with specific chromatin modifications of histone H3. To investigate the global chromatin architecture in h3.3kd we performed immunofluorescence staining. Euchromatin marked by H3K4me3 and H3K27me3 and heterochromatin marked by H3K9me2 showed similar patterns in nuclei from WT and h3.3kd leaves (Fig. 1e). Thus, global chromatin organization remained intact in h3.3kd. H3K4me3 and H3K36me3 accompany transcriptionally active genes [33]. We compared the profiles of these modifications in WT and h3.3kd across gene bodies and observed very little impact on the H3K4me3 profile, consistent with the fact that H3.3 is not enriched at the 5′ end of genes (Additional file 2: Figure S3a). However, we found that H3K36me3 profiles were affected by the loss of H3.3 (Additional file 2: Figure S3b). Promoters of genes downregulated in h3.3kd versus WT showed reduced levels of H3K36me3 (Additional file 2: Figures S3d). In contrast, upregulation of genes in h3.3kd versus WT correlated with elevated H3K36me3 levels at the 5′ end of genes over gene bodies (Additional file 2: Figure S3f). In contrast to H3K36me3, the levels and profiles of H3K4me3 did not accompany changes in transcriptional activity (Additional file 2: Figure S3c, e).
The loss of H3.3 affected transcription of a relatively limited number of genes. Gene ontology (GO) analysis of downregulated genes revealed a large variety of response processes, including environmental and endogenous stimuli (Fig. 1c, d; Additional file 5). In contrast to stably expressed housekeeping genes, responsive genes are typically differentially regulated during development or in response to stress or other stimuli [34]. A comprehensive analysis of a large set of Arabidopsis expression data led to the identification of a set of hypervariable genes that most dramatically change expression levels between different tissues or in response to stimuli and housekeeping genes with nearly constant expression [34]. Of the 123 identified hypervariable genes, 44 were downregulated in h3.3kd, while only one of the 379 housekeeping genes was affected (p = 8.58 e-35, hypergeometric probability; see “Methods” for details). We thus conclude that H3.3 is not required for gene expression in a global manner. Yet, the loss of H3.3 directly or indirectly affects the expression of subsets of particularly dynamic, responsive, and hypervariable genes.
Loss of DNA methylation in h3.3kd
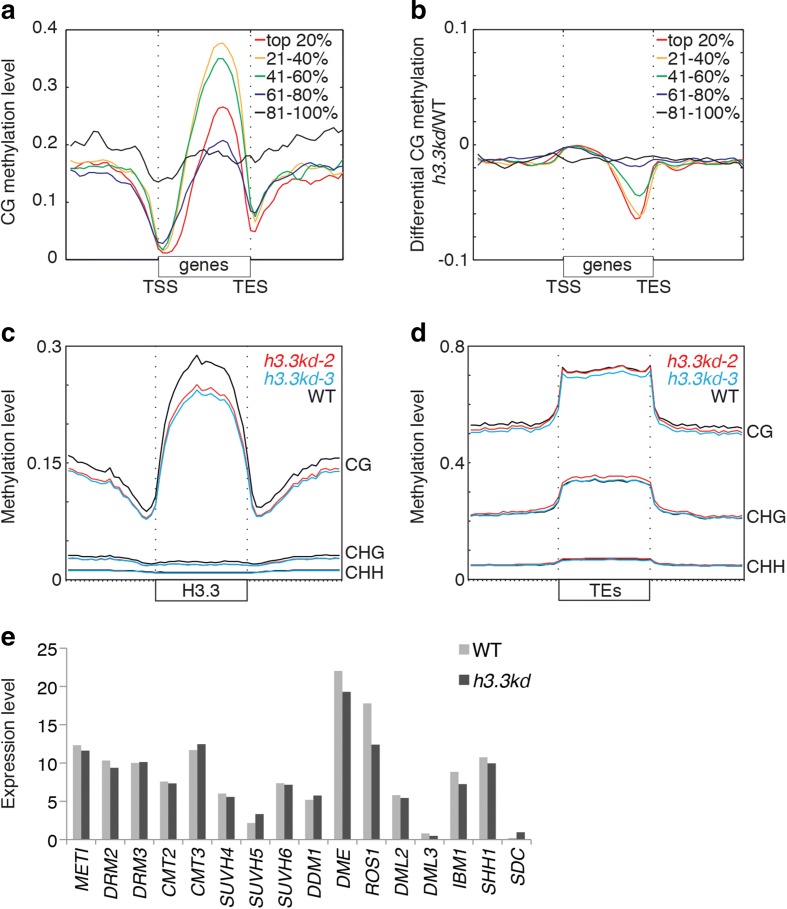
Many chromatin marks other than H3.3 correlate with gene expression, including DNA methylation. Gene body methylation consists of DNA methylation in CG contexts, that is, is enriched towards the 3′ end of active genes [35–37], partially overlapping with the predominant domain of H3.3 enrichment [18, 19]. The similarities between these two patterns prompted us to investigate the impact of H3.3 depletion on the deposition of DNA methylation over gene bodies using genome-wide sequencing of bisulfite-converted DNA (BS-seq) from mature leaves of WT and h3.3kd plants. In agreement with previous reports [35–38], methylation levels over gene bodies increased towards the TES and were highest over expressed genes, but not the most highly expressed genes in WT plants (Fig. 2a). In h3.3kd plants we noticed a distinct loss of CG methylation over gene bodies. We identified 16,711 hypomethylated regions (hypo-CG-DMRs) in h3.3kd; 89% of hypo-CG-DMRs overlapped with gene bodies and 70% of hypo-CG-DMRs overlapped with H3.3 enriched regions (defined in [18]). Loss of CG methylation over gene bodies was more severe with increasing gene expression level in the WT (Fig. 2b). More generally, DNA methylation levels, normally highly increased over H3.3-enriched regions in WT plants, were reduced in h3.3kd (Fig. 2c). In contrast, only 3.1% of TEs in the genome overlapped with hypo-CG-DMRs (Additional file 6), and DNA methylation profiles over transposable elements (TEs) remained largely unaffected in all sequence contexts (Fig. 2d), indicating that the impact of h3.3kd on DNA methylation was specific to gene bodies and other H3.3 enriched regions. In conclusion, H3.3 appeared to be required for the deposition or maintenance of DNA methylation over gene bodies.
Fig. 2.
Depletion of DNA methylation over gene bodies and H3.3-enriched regions in h3.3kd. a–d Genome-wide BS-seq results showing enrichment profiles of DNA methylation in WT, h3.3kd-2, and h3.3kd-3. a CG DNA methylation patterns over genes in WT plants. Genes were aligned from transcription start site (TSS) to transcription end site (TES) and grouped into quintiles according to their level of expression. b Relative CG methylation levels over gene bodies in h3.3kd compared to WT. Note pronounced loss of methylation at the 3′ ends of highly and moderately expressed genes. c, d DNA methylation levels in all contexts (CG, CHG, and CHH) over H3.3-enriched regions (c) and TEs (d) in WT and h3.3kd. e Expression of DNA methylation-related factors in WT compared to h3.3kd-3 (RNA-seq)
The linker histone H1 links H3.3 and DNA methylation
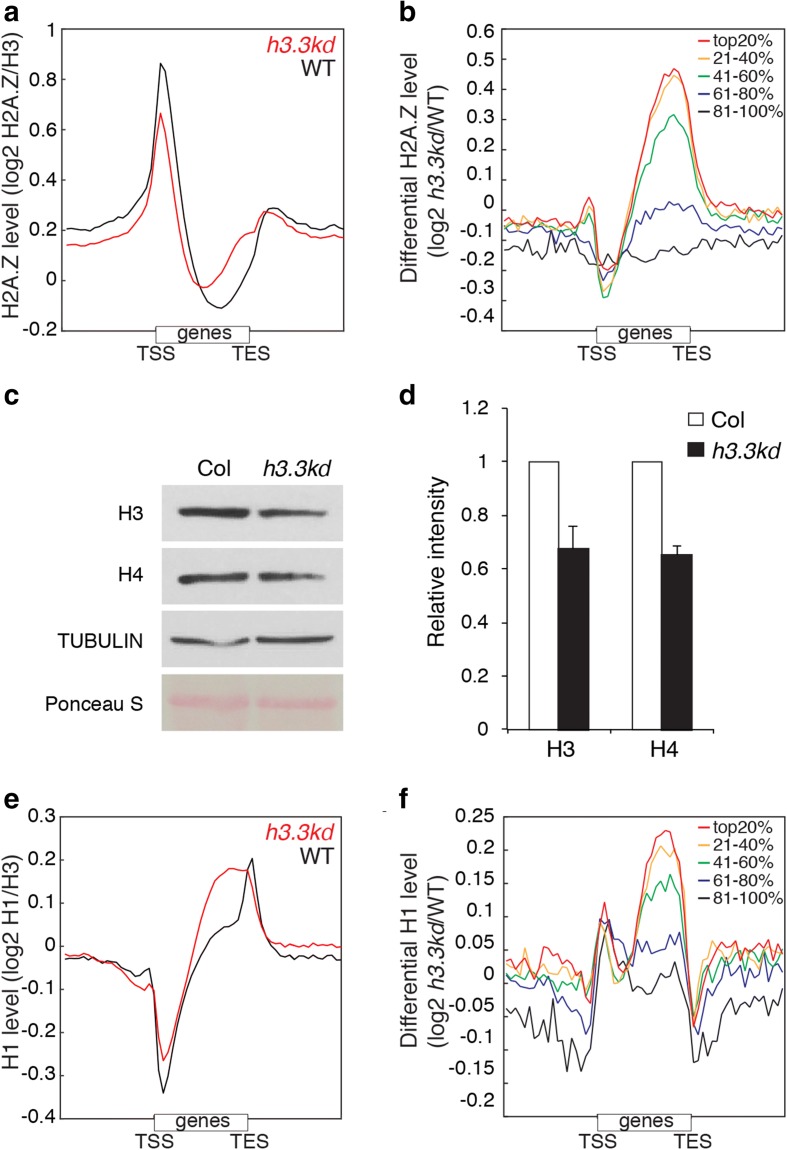
We searched for a mechanism to explain the loss of DNA methylation over gene bodies in H3.3 knockdown lines. Gene body methylation antagonizes the deposition of the histone variant H2A.Z [39]. We profiled H2A.Z in h3.3kd and found that H2A.Z increased over gene bodies in h3.3kd compared to WT (Fig. 3a), particularly towards 3′ gene ends where we observed a decrease of DNA methylation (Fig. 2a). This modified profile of H2A.Z in the h3.3kd correlated positively with gene expression and was most pronounced over highly expressed genes (Fig. 3b), which also exhibit the strongest levels in H3.3 [18, 19] and DNA methylation. It was reported that H2A.Z does not antagonize DNA methylation [40], leading to the conclusion that H3.3 promotes DNA methylation, which in concert with H3.3 prevents deposition of H2A.Z towards the 3′ end of gene bodies.
Fig. 3.
Loss of H3.3 impacts chromatin profiles of H2A.Z and H1. a, b ChIP-seq profiles depicting the enrichment of H2A.Z in WT and h3.3kd over all genes (a) and the differential profiles of h3.3kd versus WT over genes separated according to their level of expression (b). c Western blot on total H3 and H4 in Col-0 and h3.3kd plants. One of three replicates with similar results is shown. d Quantification of H3 and H4 protein abundance in Col-0 and h3.3kd plants from western blot analysis in three replicates. Bars represent standard deviation, n = 3. e, f ChIP-seq profiles depicting the enrichment of H1 in WT and h3.3kd over all genes (e) and the differential profiles of h3.3kd versus WT over genes separated according to expression level (f)
The loss of H3.3 had no effect on the expression of DNA demethylases, DNA methyltransferases, and associated factors (Fig. 2e). Thus, the loss of DNA methylation on gene bodies in h3.3kd is not likely due to decreased activity of DNA methyltransferases or increased activity of DNA demethylases. The profile of H3K36me3 in WT and h3.3kd are not correlated with gene body methylation (Additional file 2: Figure S3), thus supporting that mechanisms involved in gene body methylation in Arabidopsis do not rely on this mark, in contrast with mammals. The loss of H3.3 expression caused overexpression of genes encoding H3.1, H2A, H2B, and H4 (Additional file 1: Table S3). The predicted increased levels of core histones could compensate for the deficit of nucleosomes. We thus performed western blotting and observed that total H3 and H4 protein levels were reduced in h3.3kd versus WT (Fig. 3c, d). We thus concluded that the depletion of H3.3 results in lower density of nucleosomes over chromatin in general, but likely more pronounced over active genes, where H3.3 is highly enriched in WT plants.
In eukaryotes, the linker histone H1 binds to exposed linker DNA between nucleosomes and promotes chromatin folding [41, 42]. In vitro experiments suggest that H1 deposition into the chromatin is anti-correlated with nucleosome density [43]. In Arabidopsis it was proposed that H1 prevents access of DNA methyltransferases to pericentromeric heterochromatin, thus explaining the increase of DNA methylation over these regions in loss of function mutants for H1 [44]. We observed an increased expression of the three genes encoding H1 in h3.3kd versus WT (Additional file 1: Table S3), suggesting that H1 density could increase over gene bodies to compensate for the reduction of nucleosome density caused by the loss of H3.3. This hypothesis was supported by a marked invasion of gene bodies by H1 in h3.3kd in comparison with WT (Fig. 3e). The change of profile of H1 was correlated positively with gene expression (Fig. 3f) and anti-correlated with the loss of gene body methylation in h3.3kd (Fig. 2). These results suggest that H3.3 counters the deposition of H1 over gene bodies. The lack of H3.3 is responsible for the relative enrichment in H1 that opposes DNA methylation over gene bodies in a transcription-dependent manner.
Discussion
Our study shows that H3.3 is required for transcription of a subset of genes with a marked effect on genes involved in responses to environmental or developmental cues. These genes are subjected to transcriptional reprogramming and harbor a chromatin environment distinct from genes constitutively expressed and involved in basic cell functions. Hence, in Arabidopsis, H3.3 loss-of-function affects genes regulating predominantly development and response to the environment but not housekeeping genes. Similarly, loss of H3.3 affects development in Xenopus, Drosophila, and mouse [6–8], while in transcriptionally stable human embryonic stem cells the impact of H3.3 loss-of-function on gene expression is limited and abnormalities occur only upon their differentiation [22]. H3.3 opposes the deposition of H2A.Z at the 3′ end of gene bodies. H2A.Z also affects expression of hypervariable genes. Nucleosomes containing both H2A.Z and H3.3 are unstable [45] and we propose that these two variants promote the turnover of nucleosomes and likely provide a specific chromatin environment, enabling faster adaptation of the chromatin composition to the requirements of the transcriptional machinery.
Recent analyses have shown that bryophytes and the flowering plant Eutrema salsugineum are devoid of gene body methylation, thus questioning its requirement in land plants [28]. Yet, gene body methylation has been implicated in several transcription-related processes, like maintaining the constitutive expression of housekeeping genes, preventing erratic transcription, or enhancing the accuracy of splicing [46]. Our analysis reveals an unexpected link between H3.3 enrichment and gene body DNA methylation.
H3K36me3 has been shown in mammals to facilitate docking of de novo methyltransferase to chromatin [26]. In Arabidopsis, however, the profile of H3K36me3 over gene bodies is opposite to that of DNA methylation and H3K36me3 levels over gene bodies increased in h3.3kd, specifically at 5′ gene ends, opposite to the domain where DNA methylation is most affected. These observations are incompatible with the idea that H3K36me3 could recruit DNA methyltransferases over gene bodies. This conclusion is also coherent with the absence of a significant change of gene body methylation in the mutant sdg8 with reduced H3K36me3 [47], suggesting that the mechanisms that recruit DNA methyltransferases to gene bodies are distinct between flowering plants and mammals. It is possible that a specific modification present only or primarily on H3.3 participates in recruiting DNA methyltransferases. The maintenance DNA methyltransferase MET1 contains a bromo adjacent homology domain (BAH) of unknown binding specificity, which might recognize an H3.3-specific mark enriched over gene bodies and H3.3 regions. Yet the identity of this mark remains unknown. Another alternative is the possibility that a mark present only on H3.1 prevents the recruitment of DNA methyltransferases. In h3.3kd, ectopic enrichment of H3.1 would then lead to a decrease of gene body methylation.
Conclusions
We envisage that H3.3 enrichment regulates the recruitment of DNA methyltransferases. The specific profile of H3.3 enrichment over genes is required to retain an adequate density of nucleosomes to maintain suitable chromatin structure for transcription, while providing sufficient accessibility to DNA methyltransferases that methylate gene bodies. When the supply of H3.3 decreases, the linker histone H1 invades gene bodies, while the profile of H1 in promoters does not change significantly. Notably, the anti-correlation of H3.3 and H1 appears to be a conserved mechanism to maintain an open chromatin formation. H3.3 knockdown in Drosophila and mouse leads to increased H1 levels [48, 49]. The ability of H1 to promote folding [50] predictably reduces chromatin accessibility, thus preventing access to DNA methyltransferases and reducing gene body methylation. Low gene body methylation levels might then allow ectopic recruitment of H2A.Z containing nucleosomes across the gene bodies. We propose that H3.3 is crucial to safeguard ideal chromatin structure suitable for transcription by maintaining an optimal nucleosome density and preventing H1 deposition over gene bodies. Transcriptional activity promotes H3.3 deposition and DNA methyltransferases maintain the characteristic gene body methylation profile that is positively correlated with gene expression. Gene body methylation prevents deposition of unstable H2A.Z/H3.3 nucleosomes [45, 51], which are generally found at nucleosome-free regions like at promoters. This negative feed-back loop between H3.3, H1, DNA methylation, and H2A.Z might sustain steady transcriptional activity across active genes. This mechanism involves proteins with functions and properties conserved in multicellular eukaryotes, suggesting that it might also play a role in regulating gene body methylation in mammals.
Methods
Plant material and growth conditions
All wild-type (WT), mutant, and transgenic lines were in the Columbia-0 (Col-0) ecotype. T-DNA insertion lines were analyzed by PCR-based genotyping and the absence of full-length transcript was confirmed by RT-PCR using primers located in the 5′ and 3′ UTRs. H3.3 T-DNA insertion lines are listed in Additional file 1: Table S1 and primer sequences in Additional file 1: Table S5. For double mutant combinations, lines htr4-1 (N582765), htr4-2 (N807939), or htr5-3 (N846395) were combined with htr8-2 (N641101). Plants were grown directly on soil in growth rooms with short day (SD) condition for a period of 4–5 weeks and then shifted to long day (LD) condition. Pictures were taken at different growth stages of soil-grown plants. For the methylation analysis, mature leaves of soil-grown plants were harvested after the shift to LD. For RNA- and ChIP-seq analysis, ethanol-sterilized seeds were grown on 1× MS (Murashige and Skoog) medium with glucose at LD conditions in a Percival incubator for 10 days. All seeds were incubated at 4 °C in the dark for 3–5 days prior to germination.
Cloning and transgenic lines
Genetic backgrounds of transgenic lines used in this study are listed in Additional file 1: Table S2. A CRISPR/Cas9 vector pKIR1.0 [52] containing tandem sgRNA cassettes for HTR4 and HTR5 was used to generate htr4;htr5 double mutants. Target sequences for HTR4 and HTR5 were 5′-gCCTCCGGTGGACTTACGAG-3′ and 5′-gCAGCTCGTAAGTCTACTGG-3′, respectively. Cas9-free T2 seeds were selected by absence of seed fluorescence, and T2 plants were screened by direct sequencing of HTR4 and HTR5 gene loci to obtain htr4;htr5 double homozygous mutants.
Cloning was done using the Gateway® Cloning Technology (Invitrogen) or PCR-based site-directed mutagenesis. The artificial miRNAs (amiRNAs) amiR-HTR5-I and amiR-HTR5-II were designed and cloned according to the original protocol [53], with minor modifications. Sequences of the amiRNAs can be found in Additional file 2: Figure S2. Primers are listed in Additional file 1: Table S5. Briefly, primers were combined in three independent PCR reactions using M13-fwd and M13-rev primers instead of primers A and B of the original protocol (http://wmd3.weigelworld.org). PCR products were purified (QIAquick PCR Purification Kit, Qiagen) and combined in a fusion PCR using primers designed to add attB1/attB2 tails. Resulting amiRNAs were recombined using the Gateway® BP ClonaseTM II enzyme into pDONRTM221 and used for subsequent Multisite Gateway® recombination with the LR ClonaseTM II Plus enzyme (all Invitrogen) into a modified, Multisite Gateway® compatible pAlligator-MGW binary plasmid, under control of the HTR5 promoter. The HTR5 promoter [14] was amplified adding attB4/attB1r sites and recombined into pDONRTM P4-P1R (Invitrogen). For the third cassette in the Multisite Gateway® system a short nucleotide sequence was designed and recombined with attB2r/attB3 sites into pDONRTM P2R-P3 (Invitrogen), as “empty plasmid”. Resulting binary plasmids pHTR5-amiR-HTR5-I (pHW358) and pHTR5-amiR-HTR5-II (pHW359) were transformed into plants descending from a htr4-2/+;htr8-2 parent, and therefore segregating for the htr4-2 T-DNA insertion, using a simplified floral dip method. Primary transgenic plants were selected by green fluorescence in dry seeds, grown, and phenotypically characterized. For each transgene a subset of lines with single T-DNA insertion was followed into subsequent generations.
The HTR5 gene was amplified and cloned into pDONRTM221 as described above. Two independent steps of site-directed mutagenesis were performed on this plasmid to introduce silent mutations, rendering the resulting rHTR5 transcript resistant against amiRNA-HTR5-II targeting and to introduce a STOP codon. Multisite Gateway® recombination resulted in the final binary plasmid pHTR5-rHTR5 (pHW375) in pSRR4R3-19ST, which were transformed into htr4-2;htr8-2 double homozygous plants. Primary transgenic lines were selected by red fluorescence in dry seeds, phenotypically characterized, and, for a subset of lines, followed into subsequent generations. F1 seeds resulting from crosses to h3.3kd-3 were confirmed by green and red fluorescence in dry seeds. Phenotypic analysis of rosette size, leaf serration, and silique length was carried out in F1 plants. Plant genotypes are listed in Additional file 1: Table S2.
Immunofluorescence
Immunofluorescence experiments were performed as described previously [54] on isolated nuclei from mature leaves of h3.3kd-1 (not shown) and h3.3kd-2 plants (Fig. 2a), with similar results. We used antibodies from Abcam (H3K9me2, ab1220; H3K4me3, ab8580). The H3K27me3 antibody was a generous gift from Thomas Jenuewein, Max Planck Institute of Immunobiology and Epigenetics in Freiburg, Germany.
RNA-seq and microarray analysis
RNA was extracted from pooled 10-day-old seedlings grown on 1× MS medium with glucose as described above, using the RNeasy Mini Kit (Qiagen). Concentration and purity were determined with Nanodrop measurement and, dependent on the values, total RNA extracts were subjected to additional ethanol precipitation or used directly for subsequent steps. For RNA-seq, total RNA was sent to AITbiotech (Singapore) for library generation, sequencing on the Illumina platform, and subsequent data analysis. Briefly, after mRNA enrichment and fragmentation to ~200-bp fragments, first strand cDNA synthesis with random hexamer-primers and second strand cDNA synthesis, cDNA was purified with a QIAquick PCR purification kit (Qiagen). After end repair and A-tailing, sequencing adaptors were ligated to the fragments; fragments were purified on agarose gel, PCR amplified, and sequenced on a HiSeqTM2000 (Illumina). TopHat, cufflinks, cuffmerge, and cuffdiff were used for analysis, TAIR9 was used for alignment, and expression values are FPKM values. Gene Ontology analysis was performed using DAVID (http://david.abcc.ncifcrf.gov/) [55, 56]. A single dataset was obtained and results were confirmed based on additional analyses of microarrays (Additional file 2: Figure S2e). We performed technical duplicates using 10-day-old plate-grown seedlings of h3.3kd (htr4;htr8;amiRHTR5-II) and h3.3kd/+ (htr4;htr8;amiRHTR5-II/+). For microarray analysis, total RNA extracted from 10-day-old seedlings by an RNeasy Mini Kit (Qiagen) was hybridized to an ATH1(Affymetrix) array according to the manufacturer’s instructions. Biotin was used to label extracts. Fragmentation, hybridization, washing, and scanning were performed at BSF (Singapore). All datasets were normalized by log2GC-RMA (Robust Multichip Average) methods.
The specific enrichment in hypervariable genes affected by the loss of H3.3 was statistically significant [57]. We applied the hypergeometric test as follows: population size, 21340 (expressed genes in Col-0, FPKM >0.5 in the RNA-Seq); number of successes in population, 123 (all hypervariable genes); sample size, 669 (downregulated genes excluding HTR4, 5, and 8); number of successes in sample, 44 (overlap downregulated genes and hypervariable genes). Result is p(X ≥ 44) = 8.e-35.
ChIP-seq analysis
ChIP was also performed on 10-day-old plate-grown seedlings and according to a previously described procedure [19]. The following commercially available antibodies were used: H3 (Abcam, ab1791), H3K4me3 (Abcam, ab8580), H3K36me3 (Abcam, ab9050), and H1 (Agrisera, AS11 1801). The H2A.Z antibody was generated in our laboratory and has been described elsewhere [54]. ChIP-seq libraries were generated according to the manufacturer’s instructions and sequenced on a HiSeq2000 (Illumina). Reads were mapped to the TAIR10 genome by allowing up to two mismatches and only retaining reads that map uniquely to the genome using Bowtie [58]. Reads mapping to the same coordinates were removed, and data were normalized by total number of uniquely mapping reads. For the metaplots, flanking regions were scaled to the same length as the gene body (middle region). Duplicates were obtained for all datasets.
Methylation analysis
Mature leaves from Col-0 WT and h3.3kd-2 and h3.3kd-3 were collected and ground in liquid nitrogen to a fine powder. DNA was extracted using the DNeasy Plant Mini Kit (Qiagen) according to the manufacturer’s instructions and Bi-seq and data analysis were carried out as previously described [47]. Briefly, identical reads were collapsed into one read, and reads were mapped to the TAIR10 genome using BS seeker [59] and methylation levels were calculated as #C/(#C + #T). For the metaplots, flanking regions were scaled to the same length as the gene body (middle region). Differentially methylated regions (DMRs) in h3.3kd were defined with a method similar to a previously published DMR-finding approach [47]. The genome was binned into 100-bp tiles, where tiles with less than 20 C + T calls in any sample were omitted, and tiles with Benjamini–Hochberg-corrected FDR <0.01 (Fisher’s exact test) and a 20% absolute methylation reduction were selected. Only tiles that met these criteria in two biological replicates of h3.3kd data were retained and finally tiles within 200 bp of each other were merged.
Additional files
Supplemental tables. Table S1 Summary of H3.3 T-DNA insertion lines. Table S2 Plant genetic backgrounds. Table S3 Expression of selected chromatin factors in WT and h3.3kd. Table S4 Limited overlap between H3.3 enrichment in WT and transcriptional changes in h3.3kd. Table S5 Primer sequences. (DOCX 36 kb)
Supplemental figures. Figure S1 Generation of h3.3KO. Figure S2 Generation of h3.3kd. Figure S3 Impact of h3.3kd on active chromatin modifications. (DOCX 1094 kb)
List of genes significantly downregulated in h3.3kd-3 compared to WT. (XLS 171 kb)
List of genes significantly upregulated in h3.3kd-3 compared to WT. (XLS 116 kb)
GO term categories of significantly downregulated genes in h3.3kd-3 compared to WT. (XLS 86 kb)
List of hypomethylated TEs in h3.3kd-3. (XLS 190 kb)
Acknowledgements
We thank the UCLA BSCRC BioSequencing Core facility; Meredith Calvert for help with imaging and Mahnaz Akhavan for Illumina sequencing; ABRC and NASC for providing seeds; Hiroki Tsutsui and Tetsuya Higashiyama for the vector used to obtain H3.3 knockout lines; the Vienna Biocenter Core Facilities (VBCF) for plant cultures and next-generation sequencing.
Funding
HW, RY, XN, LJ, and FB were funded by Temasek LifeSciences Laboratory and Gregor Mendel Institute. FB was also supported by FWF grant P 28320-B21. DJ was supported by EMBO fellowship ALTF 1129-2013. HS was supported by a Dissertation Year Fellowship from UCLA and is an HHMI Fellow of the Damon Runyon Cancer Research Foundation. Work in the Jacobsen lab was supported by NIH grant GM60398. SEJ is an investigator of the Howard Hughes Medical Institute. HT was supported by the Japan Society for the Promotion of Science (Overseas Research Fellowship No. 601). YT and TK were supported by grants from the Japanese Ministry of Education, Culture, Sports, Science and Technology (19207002 and 19060014).
Availability of data and materials
Raw sequence read data for all the samples used are deposited at the Gene Expression Omnibus with accession numbers GSE96873 and GPL198 and can be found at https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE95175; https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE96834; https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GPL198.
Authors’ contributions
HW and RY performed most of the experiments with the help of LJ, BJ, SH, and XN. YT performed bisulfite sequencing, HT and DJ isolated and analyzed the H3.3 knockout line, DJ performed the western blotting. HS analyzed high-throughput sequencing data. FB and HW wrote the manuscript assisted by HS, RY, YT, TK, and SJ. FB conceived and financed the study with the help of TK and SJ. All authors read and approved the final manuscript.
Competing interests
The authors declare that they have no competing interests relevant to the work and data reported here.
Ethics approval
The authors declare that the study did not raise ethical issues in the context of the current legislation.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Footnotes
Electronic supplementary material
The online version of this article (doi:10.1186/s13059-017-1221-3) contains supplementary material, which is available to authorized users.
Contributor Information
Steven E. Jacobsen, Email: Jacobsen@ucla.edu
Frédéric Berger, Email: frederic.berger@gmi.oeaw.ac.at.
References
- 1.Talbert PB, Henikoff S. Histone variants on the move: substrates for chromatin dynamics. Nat Rev Mol Cell Biol. 2017;18:115–26. doi: 10.1038/nrm.2016.148. [DOI] [PubMed] [Google Scholar]
- 2.Filipescu D, Müller S, Almouzni G. Histone H3 variants and their chaperones during development and disease: contributing to epigenetic control. Annu Rev Cell Dev Biol. 2014;30:615–46. doi: 10.1146/annurev-cellbio-100913-013311. [DOI] [PubMed] [Google Scholar]
- 3.Talbert PB, Ahmad K, Almouzni G, Ausió J, Berger F, Bhalla PL, et al. A unified phylogeny-based nomenclature for histone variants. Epigenetics Chromatin. 2012;5:7. doi: 10.1186/1756-8935-5-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ahmad K, Henikoff S. The histone variant H3.3 marks active chromatin by replication-independent nucleosome assembly. Mol Cell. 2002;9:1191–200. doi: 10.1016/S1097-2765(02)00542-7. [DOI] [PubMed] [Google Scholar]
- 5.Hodl M, Basler K. Transcription in the absence of histone H3.3. Curr Biol. 2009;19:1221–6. doi: 10.1016/j.cub.2009.05.048. [DOI] [PubMed] [Google Scholar]
- 6.Sakai A, Schwartz BE, Goldstein S, Ahmad K. Transcriptional and developmental functions of the H3.3 histone variant in Drosophila. Curr Biol. 2009;19:1816–20. doi: 10.1016/j.cub.2009.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Couldrey C, Carlton MB, Nolan PM, Colledge WH, Evans MJ. A retroviral gene trap insertion into the histone 3.3A gene causes partial neonatal lethality, stunted growth, neuromuscular deficits and male sub-fertility in transgenic mice. Hum Mol Genet. 1999;8:2489–95. doi: 10.1093/hmg/8.13.2489. [DOI] [PubMed] [Google Scholar]
- 8.Szenker E, Lacoste N, Almouzni G. A developmental requirement for HIRA-dependent H3.3 deposition revealed at gastrulation in Xenopus. Cell Rep. 2012;1:730–40. doi: 10.1016/j.celrep.2012.05.006. [DOI] [PubMed] [Google Scholar]
- 9.Akiyama T, Suzuki O, Matsuda J, Aoki F. Dynamic replacement of histone H3 variants reprograms epigenetic marks in early mouse embryos. PLoS Genet. 2011;7:e1002279. doi: 10.1371/journal.pgen.1002279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jullien J, Astrand C, Szenker E, Garrett N, Almouzni G, Gurdon JB. HIRA dependent H3.3 deposition is required for transcriptional reprogramming following nuclear transfer to Xenopus oocytes. Epigenetics Chromatin. 2012;5:17. doi: 10.1186/1756-8935-5-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Loppin B, Bonnefoy E, Anselme C, Laurencon A, Karr TL, Couble P. The histone H3.3 chaperone HIRA is essential for chromatin assembly in the male pronucleus. Nature. 2005;437:1386–90. doi: 10.1038/nature04059. [DOI] [PubMed] [Google Scholar]
- 12.Orsi GA, Algazeery A, Meyer RE, Capri M, Sapey-Triomphe LM, Horard B, et al. Drosophila yemanuclein and HIRA cooperate for de novo assembly of H3.3-containing nucleosomes in the male pronucleus. PLoS Genet. 2013;9:e1003285. doi: 10.1371/journal.pgen.1003285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Santenard A, Torres-Padilla M-E. Epigenetic reprogramming in mammalian reproduction: Contribution from histone variants. Epigenetics. 2009;4:80–4. doi: 10.4161/epi.4.2.7838. [DOI] [PubMed] [Google Scholar]
- 14.Ingouff M, Rademacher S, Holec S, Šoljić L, Xin N, Readshaw A, et al. Zygotic resetting of the HISTONE 3 variant repertoire participates in epigenetic reprogramming in Arabidopsis. Curr Biol. 2010;20:2137–43. doi: 10.1016/j.cub.2010.11.012. [DOI] [PubMed] [Google Scholar]
- 15.Nie X, Wang H, Li J, Holec S, Berger F. The HIRA complex that deposits the histone H3.3 is conserved in Arabidopsis and facilitates transcriptional dynamics. Biol Open. 2014;3:794–802. doi: 10.1242/bio.20148680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McKittrick E, Gafken PR, Ahmad K, Henikoff S. Histone H3.3 is enriched in covalent modifications associated with active chromatin. Proc Natl Acad Sci U S A. 2004;101:1525–30. doi: 10.1073/pnas.0308092100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shu H, Nakamura M, Siretskiy A, Borghi L, Moraes I, Wildhaber T, et al. Arabidopsis replacement histone variant H3.3 occupies promoters of regulated genes. Genome Biol. 2014;15:R62. doi: 10.1186/gb-2014-15-4-r62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stroud H, Otero S, Desvoyes B, Ramírez-Parra E, Jacobsen SE, Gutierrez C. Genome-wide analysis of histone H3.1 and H3.3 variants in Arabidopsis thaliana. Proc Natl Acad Sci U S A. 2012;109:5370–5. doi: 10.1073/pnas.1203145109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wollmann H, Holec S, Alden K, Clarke ND, Jacques PE, Berger F. Dynamic deposition of histone variant H3.3 accompanies developmental remodeling of the Arabidopsis transcriptome. PLoS Genet. 2012;8:e1002658. doi: 10.1371/journal.pgen.1002658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Goldberg AD, Banaszynski LA, Noh K-M, Lewis PW, Elsaesser SJ, Stadler S, et al. Distinct factors control histone variant H3. 3 localization at specific genomic regions. Cell. 2010;140:678–91. doi: 10.1016/j.cell.2010.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ray-Gallet D, Woolfe A, Vassias I, Pellentz C, Lacoste N, Puri A, et al. Dynamics of histone h3 deposition in vivo reveal a nucleosome gap-filling mechanism for H3.3 to maintain chromatin integrity. Mol Cell. 2011;44:928–41. doi: 10.1016/j.molcel.2011.12.006. [DOI] [PubMed] [Google Scholar]
- 22.Banaszynski LA, Wen D, Dewell S, Whitcomb SJ, Lin M, Diaz N, et al. Hira-dependent histone H3.3 deposition facilitates prc2 recruitment at developmental loci in ES cells. Cell. 2013;155:107–20. doi: 10.1016/j.cell.2013.08.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zemach A, McDaniel IE, Silva P, Zilberman D. Genome-wide evolutionary analysis of eukaryotic DNA methylation. Science. 2010;328:916–9. doi: 10.1126/science.1186366. [DOI] [PubMed] [Google Scholar]
- 24.Zilberman D, Gehring M, Tran RK, Ballinger T, Henikoff S. Genome-wide analysis of Arabidopsis thaliana DNA methylation uncovers an interdependence between methylation and transcription. Nat Genet. 2007;39:61–9. doi: 10.1038/ng1929. [DOI] [PubMed] [Google Scholar]
- 25.To TK, Saze H, Kakutani T. DNA Methylation within transcribed regions. Plant Physiol. 2015;168:1219–25. doi: 10.1104/pp.15.00543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Baubec T, Colombo DF, Wirbelauer C, Schmidt J, Burger L, Krebs AR, et al. Genomic profiling of DNA methyltransferases reveals a role for DNMT3B in genic methylation. Nature. 2015;520:243–7. doi: 10.1038/nature14176. [DOI] [PubMed] [Google Scholar]
- 27.Wagner EJ, Carpenter PB. Understanding the language of Lys36 methylation at histone H3. Nat Rev Mol Cell Biol. 2012;13:115–26. doi: 10.1038/nrm3274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bewick AJ, Ji L, Niederhuth CE, Willing E-M, Hofmeister BT, Shi X, et al. On the origin and evolutionary consequences of gene body DNA methylation. Proc Natl Acad Sci U S A. 2016;113:9111–6. doi: 10.1073/pnas.1604666113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Takuno S, Ran J-H, Gaut BS. Evolutionary patterns of genic DNA methylation vary across land plants. Nat Plants. 2016;2:15222. doi: 10.1038/nplants.2015.222. [DOI] [PubMed] [Google Scholar]
- 30.Vidalis A, Živković D, Wardenaar R, Roquis D, Tellier A, Johannes F. Methylome evolution in plants. Genome Biol. 2016;17:264. doi: 10.1186/s13059-016-1127-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ausin I, Feng S, Yu C, Liu W, Kuo HY, Jacobsen EL, et al. DNA methylome of the 20-gigabase Norway spruce genome. Proc Natl Acad Sci U S A. 2016;113:E8106–13. doi: 10.1073/pnas.1618019113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Okada T, Endo M, Singh MB, Bhalla PL. Analysis of the histone H3 gene family in Arabidopsis and identification of the male-gamete-specific variant AtMGH3. Plant J. 2005;44:557–68. doi: 10.1111/j.1365-313X.2005.02554.x. [DOI] [PubMed] [Google Scholar]
- 33.Roudier F, Ahmed I, Bérard C, Sarazin A, Mary-Huard T, Cortijo S, et al. Integrative epigenomic mapping defines four main chromatin states in Arabidopsis. EMBO J. 2011;30:1928–38. doi: 10.1038/emboj.2011.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Aceituno FF, Moseyko N, Rhee SY, Gutierrez RA. The rules of gene expression in plants: organ identity and gene body methylation are key factors for regulation of gene expression in Arabidopsis thaliana. BMC Genomics. 2008;9:438. doi: 10.1186/1471-2164-9-438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cokus SJ, Feng S, Zhang X, Chen Z, Merriman B, Haudenschild CD, et al. Shotgun bisulphite sequencing of the Arabidopsis genome reveals DNA methylation patterning. Nature. 2008;452:215–9. doi: 10.1038/nature06745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lister R, O’Malley RC, Tonti-Filippini J, Gregory BD, Berry CC, Millar AH, et al. Highly integrated single-base resolution maps of the epigenome in Arabidopsis. Cell. 2008;133:523–36. doi: 10.1016/j.cell.2008.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang X, Yazaki J, Sundaresan A, Cokus S, Chan SW, Chen H, et al. Genome-wide high-resolution mapping and functional analysis of DNA methylation in Arabidopsis. Cell. 2006;126:1189–201. doi: 10.1016/j.cell.2006.08.003. [DOI] [PubMed] [Google Scholar]
- 38.Tran RK, Henikoff JG, Zilberman D, Ditt RF, Jacobsen SE, Henikoff S. DNA methylation profiling identifies CG methylation clusters in Arabidopsis genes. Curr Biol. 2005;15:154–9. doi: 10.1016/j.cub.2005.01.008. [DOI] [PubMed] [Google Scholar]
- 39.Zilberman D, Coleman-Derr D, Ballinger T, Henikoff S. Histone H2A.Z and DNA methylation are mutually antagonistic chromatin marks. Nature. 2008;456:125–9. doi: 10.1038/nature07324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Coleman-Derr D, Zilberman D. Deposition of histone varinat H2A.Z within gene bodies regulates responisve genes. PLoS Genet. 2012;8(10):e1002988. doi: 10.1371/journal.pgen.1002988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Woodcock CL, Skoultchi AI, Fan Y. Role of linker histone in chromatin structure and function: H1 stoichiometry and nucleosome repeat length. Chromosom Res. 2006;14:17–25. doi: 10.1007/s10577-005-1024-3. [DOI] [PubMed] [Google Scholar]
- 42.Hergeth SP, Schneider R. The H1 linker histones: multifunctional proteins beyond the nucleosomal core particle. EMBO Rep. 2015;16:1439–53. doi: 10.15252/embr.201540749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Routh A, Sandin S, Rhodes D. Nucleosome repeat length and linker histone stoichiometry determine chromatin fiber structure. Proc Natl Acad Sci U S A. 2008;105:8872–7. doi: 10.1073/pnas.0802336105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zemach A, Kim MY, Hsieh P-H, Coleman-Derr D, Eshed-Williams L, Thao K, et al. The Arabidopsis nucleosome remodeler DDM1 allows DNA methyltransferases to access H1-containing heterochromatin. Cell. 2013;153:193–205. doi: 10.1016/j.cell.2013.02.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jin C, Felsenfeld G. Nucleosome stability mediated by histone variants H3.3 and H2A.Z. Genes Dev. 2007;21:1519–29. doi: 10.1101/gad.1547707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Coleman-Derr D, Zilberman D. DNA methylation, H2A.Z, and the regulation of constitutive expression. Cold Spring Harb Symp Quant Biol. 2012;77:147–54. doi: 10.1101/sqb.2012.77.014944. [DOI] [PubMed] [Google Scholar]
- 47.Stroud H, Greenberg MVC, Feng S, Bernatavichute YV, Jacobsen SE. Comprehensive analysis of silencing mutants reveals complex regulation of the Arabidopsis methylome. Cell. 2013;152:352–64. doi: 10.1016/j.cell.2012.10.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Braunschweig U, Hogan GJ, Pagie L, van Steensel B. Histone H1 binding is inhibited by histone variant H3.3. EMBO J. 2009;28:3635–45. doi: 10.1038/emboj.2009.301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lin C-J, Conti M, Ramalho-Santos M. Histone variant H3.3 maintains a decondensed chromatin state essential for mouse preimplantation development. Development. 2013;140:3624–34. doi: 10.1242/dev.095513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Robinson PJJ, Rhodes D. Structure of the “30 nm” chromatin fibre: a key role for the linker histone. Curr Opin Struct Biol. 2006;16:336–43. doi: 10.1016/j.sbi.2006.05.007. [DOI] [PubMed] [Google Scholar]
- 51.Henikoff S. Labile H3.3 + H2A.Z nucleosomes mark “nucleosome-free regions”. Nat Genet. 2009;41:865–6. doi: 10.1038/ng0809-865. [DOI] [PubMed] [Google Scholar]
- 52.Tsutsui H, Higashiyama T. pKAMA-ITACHI vectors for highly efficient CRISPR/Cas9-mediated gene knockout in Arabidopsis thaliana. Plant Cell Physiol. 2017;58:46–56. doi: 10.1093/pcp/pcw191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schwab R, Ossowski S, Riester M, Warthmann N, Weigel D. Highly specific gene silencing by artificial microRNAs in Arabidopsis. Plant Cell. 2006;5:1121–33. doi: 10.1105/tpc.105.039834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yelagandula R, Stroud H, Holec S, Zhou K, Feng S, Zhong X, et al. The histone variant H2A.W defines heterochromatin and promotes chromatin condensation in Arabidopsis. Cell. 2014;158:98–109. doi: 10.1016/j.cell.2014.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.da Huang W, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.da Huang W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 57.Wu Z, Irizarry RA. Preprocessing of oligonucleotide array data. Nat Biotechnol. 2004;22:656–8. doi: 10.1038/nbt0604-656b. [DOI] [PubMed] [Google Scholar]
- 58.Langmead B, Schatz MC, Lin J, Pop M, Salzberg SL. Searching for SNPs with cloud computing. Genome Biol. 2009;10:R134. doi: 10.1186/gb-2009-10-11-r134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chen P-Y, Cokus SJ, Pellegrini M. BS Seeker: precise mapping for bisulfite sequencing. BMC Bioinformatics. 2010;11:203. doi: 10.1186/1471-2105-11-203. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental tables. Table S1 Summary of H3.3 T-DNA insertion lines. Table S2 Plant genetic backgrounds. Table S3 Expression of selected chromatin factors in WT and h3.3kd. Table S4 Limited overlap between H3.3 enrichment in WT and transcriptional changes in h3.3kd. Table S5 Primer sequences. (DOCX 36 kb)
Supplemental figures. Figure S1 Generation of h3.3KO. Figure S2 Generation of h3.3kd. Figure S3 Impact of h3.3kd on active chromatin modifications. (DOCX 1094 kb)
List of genes significantly downregulated in h3.3kd-3 compared to WT. (XLS 171 kb)
List of genes significantly upregulated in h3.3kd-3 compared to WT. (XLS 116 kb)
GO term categories of significantly downregulated genes in h3.3kd-3 compared to WT. (XLS 86 kb)
List of hypomethylated TEs in h3.3kd-3. (XLS 190 kb)
Data Availability Statement
Raw sequence read data for all the samples used are deposited at the Gene Expression Omnibus with accession numbers GSE96873 and GPL198 and can be found at https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE95175; https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE96834; https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GPL198.