Key Points
Loss of Caspase-3 delays leukemogenesis in a mouse model for t(8;21) AML.
Loss of Caspase-3 triggers upregulation of ULK1 and induction of autophagy in leukemia-initiating cells.
Abstract
AML1-ETO (AE), a fusion oncoprotein generated by t(8;21), can trigger acute myeloid leukemia (AML) in collaboration with mutations including c-Kit, ASXL1/2, FLT3, N-RAS, and K-RAS. Caspase-3, a key executor among its family, plays multiple roles in cellular processes, including hematopoietic development and leukemia progression. Caspase-3 was revealed to directly cleave AE in vitro, suggesting that AE may accumulate in a Caspase-3–compromised background and thereby accelerate leukemogenesis. Therefore, we developed a Caspase-3 knockout genetic mouse model of AML and found that loss of Caspase-3 actually delayed AML1-ETO9a (AE9a)-driven leukemogenesis, indicating that Caspase-3 may play distinct roles in the initiation and/or progression of AML. We report here that loss of Caspase-3 triggers a conserved, adaptive mechanism, namely autophagy (or macroautophagy), which acts to limit AE9a-driven leukemia. Furthermore, we identify ULK1 as a novel substrate of Caspase-3 and show that upregulation of ULK1 drives autophagy initiation in leukemia cells and that inhibition of ULK1 can rescue the phenotype induced by Caspase-3 deletion in vitro and in vivo. Collectively, these data highlight Caspase-3 as an important regulator of autophagy in AML and demonstrate that the balance and selectivity between its substrates can dictate the pace of disease.
Introduction
The t(8;21), which leads to the expression of the AML1-ETO (AE) fusion transcription factor, represents the most frequent chromosomal translocation in acute myeloid leukemia (AML), occurring in ∼4% to 12% of adult and 12% to 30% of pediatric patients.1,2 The leukemogenicity of AE has been evaluated in multiple mouse models. AE-expressing transgenic mice do not develop leukemia in the absence of other secondary events, suggesting that cooperating events are required.3-6 Some mouse models of AE-driven AML have been developed, such as expression of AE in Cdkn1a-null hematopoietic stem cells (HSCs) or expression of AML1-ETO9a (AE9a), an alternatively spliced variant of AML1-ETO, in wild-type (WT) HSCs, which leads to fully penetrant AML after a prolonged latency.7,8 Our recent studies showed that both mouse models could accurately predict cooperating events in human t(8;21) AML.9
Caspase-3, an executioner caspase, plays multiple roles in cell processes, such as apoptosis, embryonic and hematopoietic development, and homeostasis.10-13 Caspase-3 has been found to be essential for normal brain development in some genetic mouse strains14; however, Caspase-3–deficient mice are viable and fertile in the C57BL/6 strain with no apparent defects in brain pathology.15,16 Caspase-3 has been shown to play important roles at multiple steps in embryonic stem cells and HSCs, affecting self-renewal and differentiation.17-19 In the hematopoietic system, loss of Caspase-3 leads to accelerated proliferation and impaired differentiation of bone marrow cells.19 Caspase-3 is also involved in the negative regulation of B-cell proliferation following antigen stimulation20 and activated Caspase-3 participates in T-cell proliferation in response to T-cell stimulation.21-23
It has been shown that uncleaved Caspase-3 levels are higher in the peripheral blood cells of AML patients compared with hematologically normal individuals, which suggests that the caspase pathway is dysregulated in AML.24 We and others have shown that AE is a direct substrate of Caspase-3 and the cleavage sites are TMPD188 and LLLD368.15,25,26 Moreover, a truncated AE protein (ΔAE), generated by cleavage of AE at Asp188, worked as a dominant-negative protein by interacting with AE and interfering with its oncogenic functions.27,28 Together, these data suggest that AE may accumulate in a Caspase-3 compromised background and thereby accelerate leukemogenesis. In this study, we sought to determine the role of Caspase-3 in leukemogenesis in vivo, by expressing AE9a in Caspase-3 knockout mouse model. We found that loss of Caspase-3 impaired self-renewal and delayed leukemogenesis by upregulating autophagy in a ULK1-dependent manner.
Materials and methods
Fetal liver transplantation
Fetal liver cells were isolated from embryonic day 14.5 (E14.5) embryos of WT and Caspase-3−/− mice and cultured in X-VIVO medium with 10 ng/mL interleukin-3, 10 ng/mL interleukin-6, and 100 ng/mL stem cell factor (Peprotech). Fetal liver cells were infected with retroviruses, which express AE9a in a MiGR1 vector (empty MiGR1 vector served as control). The efficiency of transduction was evaluated based on flow cytometry for green fluorescent protein (GFP) positivity. The C57Bl/6.SJL recipient mice were lethally irradiated with 950 cGy and transplanted with the transduced fetal liver cells by tail-vein injection. All mice were maintained and handled under viral antibody-free conditions in the University of Miami animal facility in accordance with the policies of the University of Miami Institutional Animal Care and Use Committee.
Homing
Fetal liver cells were isolated from E14.5 embryos of WT and Caspase-3−/− mice, then 5 × 106 cells were injected into lethally irradiated (950 cGy) C57Bl/6.SJL recipient mice. Bone marrow cells were harvested 18 hours following injection and donor-derived cells were identified by flow cytometry as CD45.2+Lin-Sca1+Mac1+ cells.
Secondary transplantation
Bone marrow and spleen cells were harvested from recipient mice that developed leukemia after receiving AE9a-WT or AE9a-Caspase-3−/− fetal liver cells. Bone marrow cells (1 × 105) were transplanted into sublethally irradiated recipient mice.
Limiting dilution transplantation
Transplantation of AE9a-WT or AE9a-Caspase-3−/− leukemia cells was performed as secondary transplantation. Limiting dilution experiments consisted of cohorts of mice that had received 106, 105, 104, 103, or 102 leukemia cells. The data were analyzed by ELDA software.29 The frequency of leukemic-initiating cells (LICs) in each population was calculated using L-Calc version 1.1 software (StemCell Technologies).
Serial replating assay
The AE9a- or AE- or MiGR1-transduced E14.5 fetal liver cells were sorted and 5000 cells were plated on methylcellulose media (MethoCult GF; StemCell Technologies). Seven days after plating the cells, clonogenic progenitors were determined and 1000 cells were replated weekly for 5 times.
Long-term culture–initiating cell assay
The AE9a- or AE-transduced E14.5 fetal liver cells were sorted and 5000 cells were cocultured with MS5 cells in minimal essential medium containing 12.5% fetal bovine serum, 12.5% horse serum, 1 μM hydrocortisone, and 1 nM dexamethasone. After 4 weeks of weekly semireplenishment of the medium, cells were harvested and plated on methylcellulose media for 10 days and the colonies were scored.
Electron microscope
MiGR1-, AE9a-, or AE-expressing fetal liver cells were sorted and fixed in indicating buffers. After dehydration with graded ethanol and propylene oxide, the cells were embedded in Epon. Thin sections were stained with uranyl acetate and lead citrate and observed on a transmission electron microscope (EM) (JEM-1400; JEOL).
Immunofluorescence
MiGR1-, AE9a-, or AE-expressing fetal liver cells were sorted and fixed by 4% paraformaldehyde in phosphate-buffered saline for 10 minutes at room temperature and permeabilized in 0.5% Triton X-100 in phosphate-buffered saline for 10 minutes. After blocking with 5% fetal bovine serum for 1 hour, cells were incubated with LC3 antibodies and then detected with Rhodamine (red)-conjugated secondary antibody for 2 hours followed by incubation with 4′,6-diamidino-2-phenylindole (DAPI) for 5 minutes. Finally, the cells were observed using an inverted system microscope (Olympus).
Overexpression of ULK1 and ULK1 mutants in 293T cells
pcDNA3-Flag-ULK1-WT, pcDNA3-Flag-ULK1-D102A, pcDNA3-Flag-ULK1-D356A, pcDNA3-Flag-ULK1-D485A, and pcDNA3-Flag-ULK1-D717A were transfected into 293T cells using Lipofectamine 2000. Forty-eight hours after transfection, cell lysates were harvested and used for western blot and immunoprecipitation assays.
Immunoprecipitation of Flag-tagged ULK1 and mutants
Cell lysates containing the Flag-tagged proteins were transferred to the microcentrifuge tube containing the anti-Flag M2 affinity gel and incubated for at least 3 hours at 4°C with gentle mixing. The affinity gel was collected by centrifugation (3000 rpm for 5 minutes). After washing 3 times with 10 gel volumes of wash buffer (100 mM Tris-HCl, 150 mM NaCl, pH 7.5–10× Tris-buffered saline, pH 7.5), the affinity gel was collected for western blot or in vitro cleavage assays.
In vitro cleavage of ULK1 by Caspase-3
The anti-Flag affinity gel containing Flag-ULK1 or Flag-ULK1 mutants was incubated with purified recombinant Caspase-3 (0.2 μg) in 20 mM N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid (HEPES; pH 7.2) containing 10 mM dithiothreitol and 10% (vol/vol) glycerol at 25°C for 120 minutes. The reaction was stopped by the addition of NuPAGE sample buffer and then subjected to western blot.
In vivo rescue of leukemogenesis by ULK1 shRNAs
Leukemia cells from the primary AE9a-expressing fetal liver transplantation models were infected with ULK1 short hairpin (sh) RNAs (scrambled shRNA served as control) and selected by puromycin for 24 hours. Cells were collected and the knockdown efficiency was tested by western blot. Cells (105) were injected into sublethally irradiated recipient mice. Four weeks after transplantation, complete blood counts (CBCs) and flow cytometry were performed to monitor the development of leukemia.
Colony formation assay for human CD34+ cells
Human CD34+ cells were first infected with lentiviruses, which express AE in a pCDH vector for 24 hours, and then infected with lentiviruses expressing Caspase-3 shRNAs (scrambled shRNA as control) for 48 hours. Cells were selected with puromycin for 48 hours and the GFP+ (AE-expressing) cells were isolated. Two thousand five hundred cells were cultured in Methocult GF-H4435 medium (StemCell Technologies) and the colony number was counted on day 10 using an inverted system microscope (Olympus).
Statistical analysis
All of the results were expressed as the mean ± standard error of the mean. Data were obtained from 3 independent experiments. Statistical analyses were performed using the Student t test. Survival functions were estimated using the Kaplan-Meier method and compared by the log-rank test. *P < .05 and **P < .01 are considered as statistically significant.
Results
Depletion of Caspase-3 delays leukemogenesis in an AE9a-driven leukemia model in vivo
Before utilizing Caspase-3 knockout mice for acute leukemia studies, we evaluated fetal hematopoiesis in WT and Caspase-3−/− mice using fetal liver cells collected from E14.5 embryos. The hematopoietic stem and progenitor cell (HSPC) frequencies and HSPC homing ability of the Caspase-3−/− mouse fetal liver cells were essentially normal (supplemental Figure 1, available on the Blood Web site).
We then expressed the AE9a oncogenic protein in fetal liver cells retrieved from Caspase-3–deficient mice and transplanted the transduced cells into lethally irradiated recipients, to determine their leukemogenic capacity (Figure 1A).
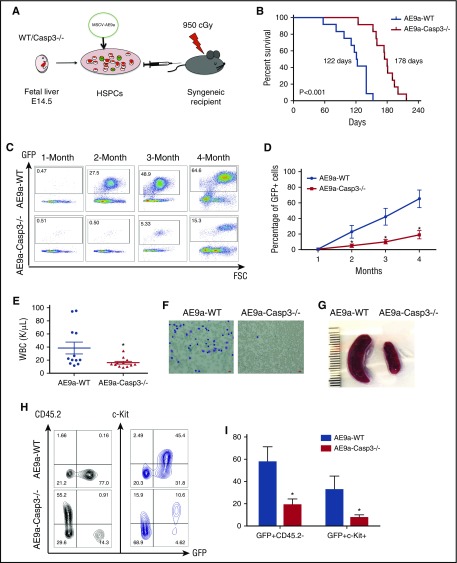
Figure 1.
Depletion of Caspase-3 delays AE9a-driven leukemogenesis in the fetal liver transplantation model. (A) The strategy of the AE9a fetal liver transplantation model. (B) Lethally irradiated recipient mice were injected with WT or Caspase-3 knockout (Casp3−/−) mouse E14.5 fetal liver cells transduced with AE9a. Loss of Caspase-3 prolongs the survival time of recipient mice. (178 days vs 122 days, n = 12, P < .001). (C-D) The AE9a-expressing cells proliferated less in the Caspase3−/− group compared with the WT group. Shown is the percentage of GFP+ (AE9a-expressing) cells in peripheral blood at different time points after transplantation. (E) The average WBC count in AE9a-Caspase-3−/− group was lower compared with the AE9a-WT group 2 months after transplantation. (F) The peripheral blood Giemsa staining shows less leukemia blast cells in the AE9a-Caspase-3−/− group compared with the AE9a-WT group 4 months after transplantation (scale bar, 10 μm). (G) The spleen size in the AE9a-Caspase-3−/− group was smaller compared with that of the AE9a-WT group 4 months after transplantation. (H-I) The frequencies of CD45.2-GFP+ cells and GFP+c-Kit+ cells in AE9a-Caspase-3−/− group were lower than in AE9a-WT group 4 months after transplantation. FSC, forward scatter; MSCV, murine stem cell virus.
The median survival for the recipients of AE9a-WT cells was 122 days compared with 178 days for the recipients of AE9a-Caspase-3−/− cells (Figure 1B). This prolonged survival was consistent with delayed leukemogenesis, as evidenced by the impaired expansion of AE9a-expressing cells in the peripheral blood of recipient mice after transplantation (Figure 1C-D). Two months after transplantation, the white blood cell (WBC) counts of the AE9a-Caspase-3−/− recipients (16.94 ± 2.063 × 109/L) were statistically lower than the AE9a-WT recipient group (38.65 ± 9.024 × 109/L) with a lower blast percentage in the AE9a-Caspase-3−/− group compared with the AE9a-WT group (Figure 1E). Unexpectedly, there were no significant differences between the 2 groups in terms of their red blood cell or platelet counts (supplemental Figure 2). Moreover, peripheral blood smears showed fewer leukemic blast cells in the AE9a-Caspase-3−/− group compared with the AE9a-WT group (Figure 1F). The CBC and blood smear data suggest that some of the AE9a-WT recipient mice developed leukemia as early as 2 months after transplantation. The spleen size of AE9a-Caspase-3−/− recipient mice was also significantly smaller than the AE9a-WT recipient mice 2 months after transplantation, suggesting less myeloid proliferation (Figure 1G). Four months after transplantation, the frequency of c-Kit+GFP+CD45.2 cells in the peripheral blood of the AE9a-Caspase-3−/− recipients was lower than the AE9a-WT recipient group (Figure 1H-I).
Histologic analysis of the peripheral blood, bone marrow, spleen, and liver, and flow cytometry analysis of bone marrow cells from both groups of moribund mice at end point, confirmed that the cause of death in these animals was AML (supplemental Figure 3).
Loss of Caspase-3 impairs LSC self-renewal and decreases LSC frequency
To determine the consequence of Caspase-3 deficiency on leukemia stem cell (LSC) self-renewal, we performed secondary transplantation and limiting dilution assays, transplanting either 105 bone marrow cells (containing 60% GFP+ cells, shown in Figure 2A-B; supplemental Figure 4A) or 106, 105, 104, 103, and 102 bone marrow cells collected from AE9a-WT and AE9a-Caspase-3−/− primary leukemia mice into sublethally irradiated mice (Figure 2C), respectively.
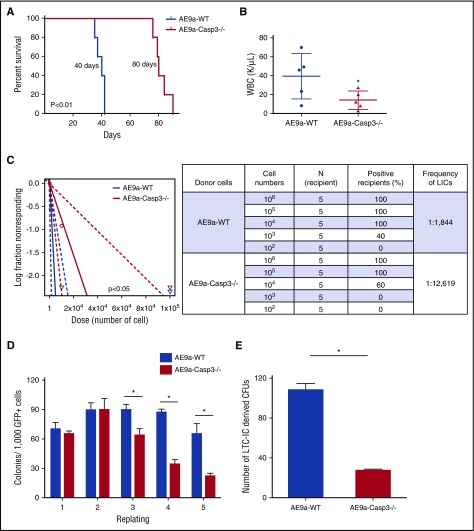
Figure 2.
Loss of Caspase-3 impairs leukemia maintenance and self-renewal of LSCs. (A) Survival of secondary transplantation model. Leukemia cells (1 × 105) from the AE9a-WT and AE9a-Caspase-3−/− group were injected into sublethally irradiated recipient mice. (B) WBC counts in the AE9a-WT and AE9a-Caspase3−/− groups in the secondary transplantation model 5 weeks after transplantation. (C) Loss of Caspase-3 significantly decreases the frequency of leukemia-initiating cells in the limiting dilution assay. The log-fraction plot (left panel) showing the results of the limiting dilution assay by using different dilutions of leukemia cells from AE9a-WT and AE9a-Caspase-3−/− groups in vivo (right panel). (D) Deletion of Caspase-3 decreases the self-renewal capacity of AE9a-expressing fetal liver cells in serial replating assays. Shown is the numbers of colonies generated from 1000 AE9a-expressing cells in each plating. (E) Deletion of Caspase-3 decreases the self-renewal capacity of AE9a-transduced fetal liver cells in LTC-IC assays. Shown is the number of colonies generated from 1000 AE9a-expressing cells. CFU, colony-forming unit.
The secondary transplanted AE9a-Caspase-3−/− group had a significantly longer survival time compared with the AE9a-WT group with a lower WBC count and less GFP+CD45.2−c-Kit+ cells in peripheral blood 5 weeks after transplantation (Figure 2A-B; supplemental Figure 4), consistent with the primary transplantation model (Figure 1). We found that loss of Caspase-3 significantly decreased the frequency of LSCs in AE9a-driven leukemia (1 in 1844 vs 1 in 12 619; P < .05) (Figure 2C).
We then performed serial replating assays and long-term culture–initiating cell (LTC-IC) assays using both AE9a and AE transduced fetal liver cells to examine the effect of Caspase-3 on self-renewal capacity in vitro. As shown in Figure 2D and supplemental Figure 6A, both the AE9a-Caspase-3−/− and AE-Caspase-3−/− cells had lower repopulating capacities (on the third, fourth, and fifth replating), compared with AE9a-WT or AE-WT cells. Moreover, the reduced number of colonies in the fifth replating could be partially rescued by re-expression of Caspase-3 back into AE9a-Caspase-3−/− fetal liver cells (supplemental Figure 6). In LTC-IC assays, AE9a-Caspase-3−/− and AE-Caspase-3−/− cells had fewer LTC-ICs than AE9a-WT and AE-WT cells, indicating that deletion of Caspase-3 reduced the frequency of LICs (Figure 2E; supplemental Figure 5B). Yet, loss of Caspase-3 did not alter the self-renewal capacity of MiGR1-transduced cells (supplemental Figure 7). In summary, loss of Caspase-3 impaired the self-renewal capacity and LSC frequency, thereby effectively delaying the initiation of AE9a-driven AML in the fetal liver transplantation models.
Loss of Caspase-3 promotes autophagy activity upon AE(9a) stress
To explore the mechanism of delayed leukemogenesis by Caspase-3 deletion, we measured cell death in the AE9a (Figure 3A-B) or AE (supplemental Figure 9A) transduced cells. We found that depletion of Caspase-3 significantly increased the percentage of 7-aminoactinomycin D (7AAD) single-positive cells as opposed to Annexin V+ cells, suggesting Caspase-3 depletion elicited cytotoxic effects in these cells through an apoptosis-independent manner.
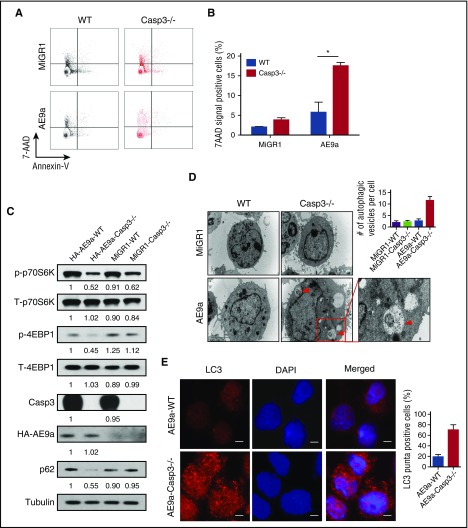
Figure 3.
Loss of Caspase-3 promotes cell death and induces autophagy in AE9a-expressing fetal liver cells. (A-B) Flow cytometric profiles (A) and statistical analysis of cell death (Annexin V−7AAD+) in MiGR1/AE9a-expressing fetal liver cells using Annexin V/7AAD assays. (C) Western blot of mTOR-autophagy signaling pathway proteins in MiGR1/AE9a-expressing fetal liver cell lysates. Compared with the AE9a-WT group, the protein levels of phosph-p70S6K and phosph-4EBP1 are lower compared with the AE9a-Caspase-3−/− group. (D) Immunofluorescent staining of LC3 (red) in AE-expressing fetal liver cells and quantification of percentage of LC3+ cells. More LC3 punctate can be observed in the AE-Caspase-3−/− group. (E) EM pictures of MiGR1/AE9a-expressing fetal liver cells and quantification of autophagic vesicles per cell. More autophagosomes and autolysosomes can be observed in the AE9a-Caspase-3−/− group (scale bar, 10 μm). Red triangles represent autophagic vesicles.
To identify the signaling pathways involved in this process, we used an apoptosis phosphoantibody array to interrogate the AE9a-expressing fetal liver cells and found that compared with the AE9a-WT fetal liver cells, mammalian target of rapamycin (mTOR)-signaling was markedly decreased in AE9a-Caspase3−/− fetal liver cells (data not shown). We confirmed the altered phosphorylation status of mTOR signaling components in the AE9a- and AE-expressing cells using several commercial antibodies (Figure 3C; supplemental Figure 9B).
As mTOR-signaling is a key regulator of autophagy, we evaluated the protein level of p62 (SQSTM1) which is an autophagy biomarker and found increased autophagic activity manifested by decreased p62 protein levels in AE(9a)-Caspase-3−/− cells compared with AE(9a)-WT cells (Figure 3C). LC3+ double-membrane degradation cargo is also considered to be the hallmark of autophagy activity. Therefore, we measured the conversion of LC3-I to LC3-II, LC3 puncta and autophagic vesicles. Increased LC3II was observed in AE9a-Caspase-3−/− cells upon inhibition of lysosomal turnover of LC3II by chloroquine compared with AE(9a)-WT cells, suggesting more conversion of LC3-I to LC3-II (supplemental Figure 8). We found loss of Caspase-3 in AE(9a)-expressing cells increased both the number of LC3 puncta and autophagic vesicles through immunofluorescence assays using LC3 antibodies (Figure 3D; supplemental Figure 9C) and EM (Figure 3E; supplemental Figure 9D). These data suggested that loss of Caspase-3 increased autophagy induction (flux) in AE(9a)-expressing cells.
Caspase-3 directly cleaves ULK1 on D485 in AE9a- and AE-expressing cells
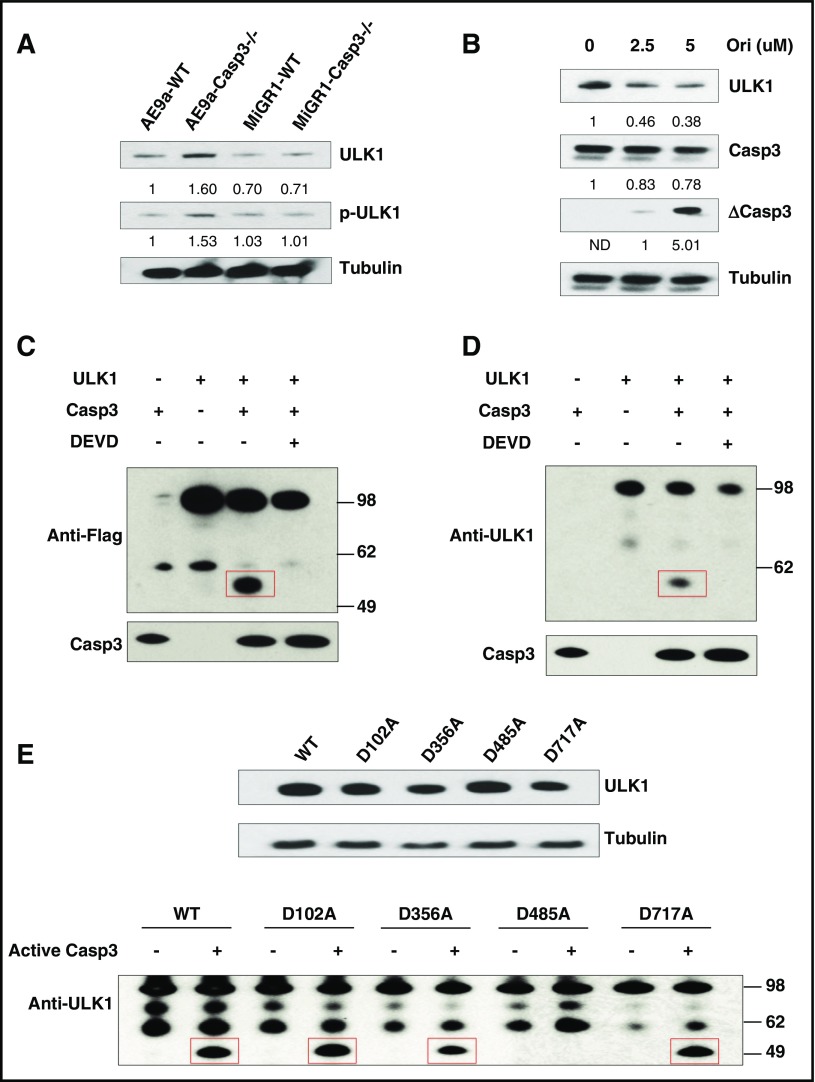
Caspase-3 exerts its protease function through direct binding and cleavage of its substrates. Therefore, we tested a panel of autophagy axis components, aiming to identify a biologically relevant Caspase-3 target in AE9a-expressing cells. We found that the levels of ULK1 (also known as ATG1, a direct target of mTOR), as well as phospho-ULK1, were significantly increased in the Caspase-3–deficient AE9a-expressing fetal liver cells compared with AE9a-WT or MiGR1-Caspase-3−/− cells (Figure 4A). The protein level of ULK1 decreased in AE9a-WT cells upon treatment with a Caspase activator, oridonin, validating ULK1 as a Caspase3 target (Figure 4B). We then performed an in vitro digestion assay to test whether ULK1 is a direct target of Caspase-3. Flag-tagged ULK1 was immunoprecipitated from transfected 293T cells and incubated with recombinant, active Caspase-3 in vitro; a ∼50-kDa fragment was detected on western blots using either an anti-epitope antibody or anti-ULK1 antibodies (Figure 4C-D). This fragment was not observed in the presence of a Caspase-3 inhibitor, Z-DEVD-fmk, suggesting the specificity of this digestion (Figure 4C-D).
Figure 4.
ULK1 is a substrate of Caspase-3. (A) Protein levels of ULK1 and phosphorylated ULK1 (p-ULK1) in AE9a-expressing fetal liver cells from WT and Caspase-3 KO mice. (B) Protein levels of ULK1 in AE9a-WT cells treated with oridonin (0, 2.5, and 5 μM) for 24 hours. (C-D) In vitro cleavage assays of ULK1 by recombinant Caspase-3. 293T cell were transfected with pcDNA3 and pcDNA3-Flag-ULK1. Flag-ULK1 protein was immunoprecipitated by Flag M2 beads. The precipitated Flag-ULK1 was incubated with or without recombinant active Caspase-3 (0.2 μg) in the presence and absence of 4 mM DEVD (Caspase-3 inhibitor) at 25°C for 2 hours. The samples were analyzed by western blots using anti-Flag (C) or anti-ULK1 (D) and Caspase-3 antibodies. (E) Caspase-3 cleaves ULK1 at D485. In vitro cleavage assays of ULK1-WT and ULK1-mutates by Caspase-3 were performed. 293T cell were transfected with pcDNA3-Flag-ULK1-WT and pcDNA3-Flag-ULK1 mutants were overexpressed in 293T cells (top panel). Flag-ULK1 proteins were immunoprecipitated by Flag-M2 affinity gel. The precipitated Flag-ULK1-WT and Flag-ULK1 mutants were incubated with or without recombinant active Caspase-3 (0.2 μg) at 25°C for 2 hours. The cleaved samples were analyzed by western blots using anti-ULK1 antibodies and anti-Tubulin antibody (bottom panel).
Caspase-3 recognizes a tetrapeptide motif Asp-x-x-Asp (DXXD) on its substrates and hydrolyzes peptide bonds after aspartic acid residues. Therefore, to identify the cleavage site(s) of ULK1 by Caspase-3, we generated ULK1 mutants (D102A, D356A, D485A, and D717A) which compromised its potential Caspase-3 recognition sites. Only the D485A mutation abolished its in vitro cleavage by Caspase-3 (Figure 4E). This correlated with the size of the ∼50-kDa ULK1 cleavage product, suggesting that activated Caspase-3 cleaves ULK1 at D485.
The impaired self-renewal capacity and leukemogenicity of AE9a-Caspase-3−/− cells can be restored by ULK1 inhibition
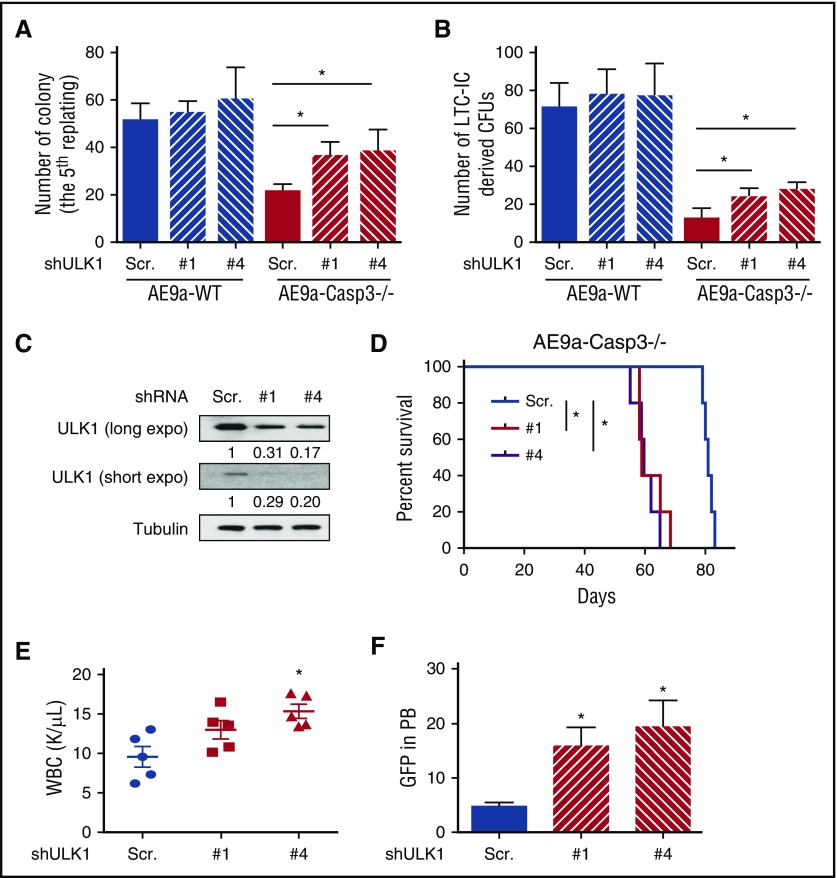
To determine whether the cleavage of ULK1 by Caspase-3 regulates the self-renewal capacity and leukemogenicity of AE9a-expressing fetal liver cells, we performed in vitro and in vivo rescue assays with ULK1 inhibition. As shown in Figure 5A-B, the decreased colony numbers in the fifth replating and the decreased numbers of LTC-ICs in the AE9a-Caspase-3−/− group were both attenuated by depleting ULK1 using 2 distinct shRNAs. Similar results were also seen using a small-molecule inhibitor of ULK1 (SBI-0206965)30 (supplemental Figure 10).
Figure 5.
The phenotype of AE9a-Caspase-3−/− or AE-Caspase-3−/− cells induced by Caspase-3 deficiency can be rescued by ULK1 inhibition. (A) Average colony numbers in the fifth replating resulting from plating of 1000 AE9a-expressing fetal liver cells with ULK1 knockdown by shRNAs. (B) Average number of LTC-IC–driven CFUs in LTC-IC assays resulting from 1000 AE9a-expressing fetal liver cells with ULK1 knockdown by shRNAs. (C) The knockdown efficiency of ULK1 in AE9a-Caspase-3−/− leukemia cells before transplantation. (D) The survival of recipient mice transplanted with AE9a-Caspase-3−/− leukemia cells expressing shULK1s were shorter than AE9a-Caspase-3−/− leukemia cells expressing scrambled shRNA in a secondary transplantation model. (E-F) Four weeks after transplantation, the recipient mice transplanted with AE9a-Caspase-3−/− leukemia cells expressing shULK1s had higher WBC counts and more GFP+ (AE9a-expressing) cells. PB, peripheral blood.
We then sought to knockdown ULK1 in primary leukemia cells derived from recipient mice and evaluate their growth potential in secondary transplantation assays. We found that the recipient mice who received the shULK1-expressing AE9a-Caspase-3−/− cells had a shorter life span than the mice who received scrambled shRNA-transduced cells (Figure 5C-D; supplemental Figure 11A-B). These mice also had higher WBC and more AE9a-expressing cells (GFP+) in their peripheral blood 4 weeks after transplantation, demonstrating that the delayed leukemogenicity of AE9a-Caspase3−/− leukemia cells was restored by ULK1 knockdown in vivo (Figure 5D-E; supplemental Figure 11C-D).
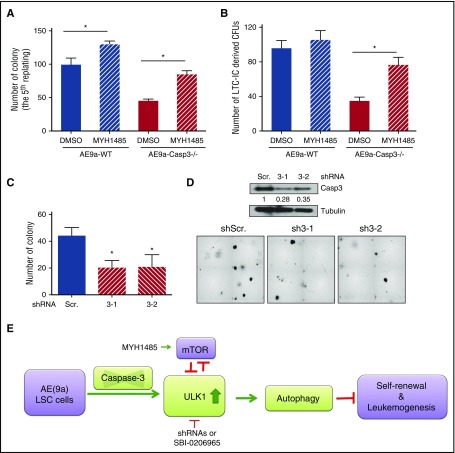
As shown in supplemental Figure 12, knockdown of ULK1 also increased the phosphorylation levels of p70S6K and 4EBP1, suggesting that the ULK1 protein level negatively regulates mTOR signaling. Therefore, we tried to rescue the self-renewal capacity of AE9a- or AE-expressing cells in the presence of the mTOR activator, MYH1485. We found that MYH1485 also partially restored the serial replating capacity and LTC-IC numbers in the AE9a-Caspase3−/− or AE-Caspase-3−/− group (Figure 6A-B; supplemental Figure 13).
Figure 6.
Caspase-3 regulates colony formation of AE-expressing LSCs via autophagy modulation. (A) Average numbers of colonies in the fifth replating resulting from plating 1000 AE9a-expressing fetal liver cells with treatment of MHY1485, an mTOR activator. (B) Average numbers of LTC-IC–driven CFUs in LTC-IC assays resulting from 1000 AE9a-expressing fetal liver cells with treatment of MHY1485, an mTOR activator. (C) The knockdown efficiency of ULK1 in AE-expressing human CD34+ cells. (D) Average number of colonies resulting from 2500 AE-expressing human CD34+ cells with Caspase-3 knockdown by shRNAs (scrambled shRNA served as control) (top panel) and the morphology of the colonies (bottom panel). (E) Working model of the effect of depletion of Caspase-3 on AE(9a)-driven leukemogenesis. DMSO, dimethyl sulfoxide.
Knockdown of Caspase-3 reduces the colony formation of AE-expressing human CD34+ cells
Finally, we tested the effect of Caspase-3 depletion on AE-expressing human CD34+ cells. As shown in Figure 6C-D, the colony-forming capacity was decreased by knockdown of Caspase-3 in AE-expressing human CD34+ cells.
In summary, the deficiency of Caspase-3 in AE(9a)-expressing LSCs increased the ULK1 level; this activates autophagy, inhibits mTOR signaling, decreases self-renewal capacity, and ultimately delays the initiation and progression of AE(9a)-driven leukemia (Figure 6E).
Discussion
In this study, we demonstrate that Caspase-3 is not only an apoptosis executioner but also a regulator of autophagy in AML cells. Loss of Caspase-3 delays leukemogenesis, with impaired leukemia initiation and maintenance in an AE9a-expressing fetal liver transplantation model; depletion or loss of Caspase-3 triggers decreased self-renewal capability with increased autophagy activity. These data suggest that the induction of autophagy activation by Caspase-3 deletion may critically affect leukemogenesis.
Autophagy is an intercellular degradation process, which has a variety of physiological and pathophysiological roles, including intracellular protein and organelle clearance, cell death, and tumor suppression.31-35 Autophagy was reported to predominantly drive a cytodestructive cascade in human AML that promotes oncogenic fusion protein clearance and leads to cell differentiation and cell death.36 The autophagy-driven degradation of these fusion oncoproteins (eg, PML-RARA and BCR-ABL) regulates leukemia cell differentiation and cell death, and can contribute to treatment response.37,38 In human AML, blasts have reduced expression of autophagy-related genes, decreased autophagic flux, and the accumulation of unhealthy mitochondria, indicating that low autophagy activity provides an advantage for the growth of transformed cells. Taken together, the dysregulation of autophagy may be an important step in leukemia initiation and progression.
Moreover, induction of autophagy may be beneficial to human AML treatment. It has been shown that autophagy plays essential roles in the effectiveness of a number of chemotherapy drugs for t(8;21) AML patients. Autophagy was shown to contribute to both cytarabine and idarubicin’s efficacy in AML cells.39,40 Autophagy is also required for mitoxantrone-induced immunogenic signaling in tumors.41 Moreover, arsenic trioxide,42,43 vitamin D3,44 vitamin K2,45 eupalinin A (sesquiterpene lactone),46 BAY11-7082 (NF-κB inhibitor),47 morphinone (a morphin derivative),48 APO866 (NAD biosynthesis inhibitor),49 and platonin50 have all been reported to induce leukemia cell death via activation of autophagy.51 Therefore, autophagy may play a general role in the treatment of leukemia.
Although our previous work showed that AE could be degraded in a Caspase-3–dependent manner in leukemia cells by treatment with Eriocalyxin B or Oridonin,25,28 we found comparable levels of AE and AE9a protein when we expressed AE or AE9a in fetal liver cells from Caspase-3−/− mice vs WT mice. Moreover, the D188A, D368A, or double D188AD368A mutations in AE or AE9a did not affect the self-renewal capability of AE- or AE9a-expressing WT fetal liver cells, suggesting that the WT level of Caspase-3 did not affect the expression of AE or AE9a in fetal liver cells. Furthermore, loss of Caspase-3 in AE- or AE9a-expressing fetal liver cells, did not change in the level of AE or AE9a protein, indicating that AE or AE9a is not a substrate of autophagy, consistent with reports by other groups.52
Because mTOR is a negative regulator of autophagy, we checked the mTOR signaling pathway including the phosphorylation status of P70S6K and 4EBP1, 2 mTOR downstream targets. We found decreased P-P70S6K and P-4EBP1 in the AE9a-Caspase-3−/− group compared with the AE9a-WT group, suggesting a downregulation of mTOR signaling. Moreover, the impaired self-renewal induced by loss of Caspase-3 in vitro can be partially rescued by MHY1485, an mTOR activator. In AML, mTOR activation is seen in nearly all AML cases, as is phosphorylation of downstream targets such as p70S6, S6RP, and 4EBP1.53-55 Clinical data suggest that mTOR inhibitors have potential therapeutic roles in a number of hematologic malignancies, including acute lymphoblastic leukemia, chronic myelogenous leukemia, mantle cell lymphoma, anaplastic large-cell lymphoma, and lymphoproliferative disorders.56 All of these studies reveal that targeting mTOR-centered autophagy may have broad therapeutic benefit.
Finding that autophagy induction and mTOR signaling pathway downregulation are both triggered by loss of Caspase-3, we proceeded to identify a novel and direct substrate of Caspase-3, ULK1, which is also a substrate of mTOR. ULK1, a serine/threonine UNC-51-like kinase, is the homology of Atg1 in mammalian cells57-60 which controls the initiation of autophagy. Meanwhile, ULK1 can also act as negative regulator of mTOR by phosphorylating its substrates, Raptor and S6K1,61 and hindering substrate binding. Thus, ULK1 inhibits cell proliferation by blocking the kinase activity of mTOR.62 We found loss of Caspase-3 increased both the protein level and phosphorylation of ULK1 in AE9a- or AE-expressing fetal liver cells; and the upregulation of ULK1 triggered the induction of autophagy and inhibition of mTOR activation. We further identified and confirmed D485 as the cleavage site of ULK1 by Caspase-3 by in vitro cleavage assays and site-direct mutagenesis. Besides autophagy regulation mediated by ULK1, other effects, or other substrates, could certainly be at play in this phenotype. We examined a variety of other ATGs that have been reported to be substrates of Caspase-3, including Atg16L63 and Atg4,64,65 and found no changes in their level of expression, suggesting that these Caspase-3 substrates did not contribute to the observed phenotypes in our model.
Furthermore, inhibition of ULK1 by shRNAs not only rescued the self-renewal deficiency of AE9a-Caspase-3−/− fetal liver cells in vitro but also rescued the delayed leukemogenesis of AE9a-Caspase-3−/− leukemia cells in vivo, indicating that ULK1 may serve as a novel therapeutic target for AML. SBI-0206965, a small-molecule inhibitor of ULK1, also rescued the self-renewal defect of AE9a-Caspase-3−/− fetal liver cells. Although SBI-0206965 can act synergistically with mTOR inhibition in lung cancer cell lines and this compound is still under preclinical evaluation. Determining where to best test this drug remains challenging, as the role of autophagy in tumor initiation, progression, and resistance to treatment is still largely unknown and context dependent.
In our model, the accumulation of ULK1 caused by the absence of Caspase-3 impaired self-renewal and leukemogenicity of AE9a-expressing LSCs, suggesting that the interaction between Caspase-3 and ULK1 dictates the pace of AE-driven leukemogenesis. Our study suggests that autophagy activation is critical for leukemogenesis and that ULK1, the autophagy activator, is a promising new therapeutic target for AML.
Acknowledgments
The authors thank the other members of the laboratory of S.D.N., and Jianping Li from the laboratory of F.-C.Y., for their assistance and thoughtful input. The authors thank the Sylvester Comprehensive Cancer Center (SCCC)–Flow Cytometry Core Facility and SCCC-Immunohistochemistry Core Facilities shared services carrying out this work.
This work was supported by funding from the American Cancer Society (L.W.), Gabrielle’s Angel Foundation (L.W.), the Leukemia Research Foundation (L.W.), the Stanley J. Glaser Foundation (L.W.), a grant from the National Institutes of Health National Cancer Institute R01CA166835 (S.D.N.), and the National Natural Science Foundation of China (grant nos. 81470316, 81470334, 81670122, 81622003, 81670094, and 81371677). This work was also supported by the Bureau of Major R&D Programs, Chinese Academy of Sciences (XDA12010310); the Bureau of Frontier Sciences and Education, Chinese Academy of Sciences (QYZDB-SSW-SMC027); the Chinese National Key Basic Research Project (2013CB966801); the National Key Research and Development Plan of China (2016YFC0902202); and the Shanghai Municipal Education Commission–Gaofeng Clinical Medicine Grant Support (20152506).
Footnotes
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Authorship
Contribution: N.M., Y.T., G.C., X.-J.S., M.S., D.H., and B.C. performed the experiments and analyzed data; D.L.K. and K.A. analyzed data; F.L., S.M.G., C.M., M.X., F.-C.Y., X.-J.S., S.C., and Z.C. reviewed the manuscript; N.M. designed the studies and wrote the manuscript; L.W. and S.D.N. designed, supervised the study, analyzed data, and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Stephen D. Nimer, University of Miami Miller School of Medicine, Miami, FL 33136; e-mail: snimer@med.miami.edu; or Lan Wang, Shanghai Institutes for Biological Sciences, 320 Yueyang Rd, Shanghai 200031, China; e-mail: lwang@sibs.ac.cn.
References
- 1.Müller AM, Duque J, Shizuru JA, Lübbert M. Complementing mutations in core binding factor leukemias: from mouse models to clinical applications. Oncogene. 2008;27(44):5759-5773. [DOI] [PubMed] [Google Scholar]
- 2.Nimer SD, Moore MA. Effects of the leukemia-associated AML1-ETO protein on hematopoietic stem and progenitor cells. Oncogene. 2004;23(24):4249-4254. [DOI] [PubMed] [Google Scholar]
- 3.Hatlen MA, Wang L, Nimer SD. AML1-ETO driven acute leukemia: insights into pathogenesis and potential therapeutic approaches. Front Med. 2012;6(3):248-262. [DOI] [PubMed] [Google Scholar]
- 4.Rhoades KL, Hetherington CJ, Harakawa N, et al. . Analysis of the role of AML1-ETO in leukemogenesis, using an inducible transgenic mouse model. Blood. 2000;96(6):2108-2115. [PubMed] [Google Scholar]
- 5.Higuchi M, O’Brien D, Kumaravelu P, Lenny N, Yeoh EJ, Downing JR. Expression of a conditional AML1-ETO oncogene bypasses embryonic lethality and establishes a murine model of human t(8;21) acute myeloid leukemia. Cancer Cell. 2002;1(1):63-74. [DOI] [PubMed] [Google Scholar]
- 6.Yuan Y, Zhou L, Miyamoto T, et al. . AML1-ETO expression is directly involved in the development of acute myeloid leukemia in the presence of additional mutations. Proc Natl Acad Sci USA. 2001;98(18):10398-10403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yan M, Kanbe E, Peterson LF, et al. . A previously unidentified alternatively spliced isoform of t(8;21) transcript promotes leukemogenesis. Nat Med. 2006;12(8):945-949. [DOI] [PubMed] [Google Scholar]
- 8.Peterson LF, Yan M, Zhang DE. The p21Waf1 pathway is involved in blocking leukemogenesis by the t(8;21) fusion protein AML1-ETO. Blood. 2007;109(10):4392-4398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hatlen MA, Arora K, Vacic V, et al. . Integrative genetic analysis of mouse and human AML identifies cooperating disease alleles. J Exp Med. 2016;213(1):25-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McIlwain DR, Berger T, Mak TW. Caspase functions in cell death and disease. Cold Spring Harb Perspect Biol. 2013;5(4):a008656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Porter AG, Jänicke RU. Emerging roles of caspase-3 in apoptosis. Cell Death Differ. 1999;6(2):99-104. [DOI] [PubMed] [Google Scholar]
- 12.Zeiss CJ, Neal J, Johnson EA. Caspase-3 in postnatal retinal development and degeneration. Invest Ophthalmol Vis Sci. 2004;45(3):964-970. [DOI] [PubMed] [Google Scholar]
- 13.Lo SC, Wang Y, Weber M, Larson JL, Scearce-Levie K, Sheng M. Caspase-3 deficiency results in disrupted synaptic homeostasis and impaired attention control. J Neurosci. 2015;35(5):2118-2132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Leonard JR, Klocke BJ, D’Sa C, Flavell RA, Roth KA. Strain-dependent neurodevelopmental abnormalities in caspase-3-deficient mice. J Neuropathol Exp Neurol. 2002;61(8):673-677. [DOI] [PubMed] [Google Scholar]
- 15.Lu Y, Chen GQ. Effector caspases and leukemia. Int J Cell Biol. 2011;2011:738301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kuida K, Zheng TS, Na S, et al. . Decreased apoptosis in the brain and premature lethality in CPP32-deficient mice. Nature. 1996;384(6607):368-372. [DOI] [PubMed] [Google Scholar]
- 17.Abdul-Ghani M, Megeney LA. Rehabilitation of a contract killer: caspase-3 directs stem cell differentiation. Cell Stem Cell. 2008;2(6):515-516. [DOI] [PubMed] [Google Scholar]
- 18.Fujita J, Crane AM, Souza MK, et al. . Caspase activity mediates the differentiation of embryonic stem cells. Cell Stem Cell. 2008;2(6):595-601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Janzen V, Fleming HE, Riedt T, et al. . Hematopoietic stem cell responsiveness to exogenous signals is limited by caspase-3. Cell Stem Cell. 2008;2(6):584-594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Woo M, Hakem R, Furlonger C, et al. . Caspase-3 regulates cell cycle in B cells: a consequence of substrate specificity. Nat Immunol. 2003;4(10):1016-1022. [DOI] [PubMed] [Google Scholar]
- 21.Alam A, Cohen LY, Aouad S, Sékaly RP. Early activation of caspases during T lymphocyte stimulation results in selective substrate cleavage in nonapoptotic cells. J Exp Med. 1999;190(12):1879-1890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kennedy NJ, Kataoka T, Tschopp J, Budd RC. Caspase activation is required for T cell proliferation. J Exp Med. 1999;190(12):1891-1896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Miossec C, Dutilleul V, Fassy F, Diu-Hercend A. Evidence for CPP32 activation in the absence of apoptosis during T lymphocyte stimulation. J Biol Chem. 1997;272(21):13459-13462. [DOI] [PubMed] [Google Scholar]
- 24.Estrov Z, Thall PF, Talpaz M, et al. . Caspase 2 and caspase 3 protein levels as predictors of survival in acute myelogenous leukemia. Blood. 1998;92(9):3090-3097. [PubMed] [Google Scholar]
- 25.Wang L, Zhao WL, Yan JS, et al. . Eriocalyxin B induces apoptosis of t(8;21) leukemia cells through NF-kappaB and MAPK signaling pathways and triggers degradation of AML1-ETO oncoprotein in a caspase-3-dependent manner. Cell Death Differ. 2007;14(2):306-317. [DOI] [PubMed] [Google Scholar]
- 26.Lu Y, Peng ZG, Yuan TT, Yin QQ, Xia L, Chen GQ. Multi-sites cleavage of leukemogenic AML1-ETO fusion protein by caspase-3 and its contribution to increased apoptotic sensitivity. Leukemia. 2008;22(2):378-386. [DOI] [PubMed] [Google Scholar]
- 27.Zhen T, Wu CF, Liu P, et al. . Targeting of AML1-ETO in t(8;21) leukemia by oridonin generates a tumor suppressor-like protein. Sci Transl Med. 2012;4(127):127ra38. [DOI] [PubMed] [Google Scholar]
- 28.Zhou GB, Kang H, Wang L, et al. . Oridonin, a diterpenoid extracted from medicinal herbs, targets AML1-ETO fusion protein and shows potent antitumor activity with low adverse effects on t(8;21) leukemia in vitro and in vivo. Blood. 2007;109(8):3441-3450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hu Y, Smyth GK. ELDA: extreme limiting dilution analysis for comparing depleted and enriched populations in stem cell and other assays. J Immunol Methods. 2009;347(1-2):70-78. [DOI] [PubMed] [Google Scholar]
- 30.Egan DF, Chun MG, Vamos M, et al. . Small molecule inhibition of the autophagy kinase ULK1 and identification of ULK1 substrates. Mol Cell. 2015;59(2):285-297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yang ZJ, Chee CE, Huang S, Sinicrope FA. The role of autophagy in cancer: therapeutic implications. Mol Cancer Ther. 2011;10(9):1533-1541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Klionsky DJ, Abdalla FC, Abeliovich H, et al. . Guidelines for the use and interpretation of assays for monitoring autophagy. Autophagy. 2012;8(4):445-544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kenific CM, Debnath J. Cellular and metabolic functions for autophagy in cancer cells. Trends Cell Biol. 2015;25(1):37-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Klionsky DJ, Abdelmohsen K, Abe A, et al. . Guidelines for the use and interpretation of assays for monitoring autophagy (3rd edition). Autophagy. 2016;12(1):1-222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mizushima N. Autophagy: process and function. Genes Dev. 2007;21(22):2861-2873. [DOI] [PubMed] [Google Scholar]
- 36.Zhang SP, Niu YN, Yuan N, et al. . Role of autophagy in acute myeloid leukemia therapy. Chin J Cancer. 2013;32(3):130-135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang Z, Cao L, Kang R, et al. . Autophagy regulates myeloid cell differentiation by p62/SQSTM1-mediated degradation of PML-RARα oncoprotein. Autophagy. 2011;7(4):401-411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Torgersen ML, Simonsen A. Autophagy: friend or foe in the treatment of fusion protein-associated leukemias? Autophagy. 2013;9(12):2175-2177. [DOI] [PubMed] [Google Scholar]
- 39.Chen L, Guo P, Jia P, Tong J, Hu J, Li J. Autophagy is an important event for low dose cytarabine treatment in acute myeloid leukemia U937 cell line [abstract]. Blood. 2014;124(21). Abstract 5209. [Google Scholar]
- 40.Ristic B, Bosnjak M, Arsikin K, et al. . Idarubicin induces mTOR-dependent cytotoxic autophagy in leukemic cells. Exp Cell Res. 2014;326(1):90-102. [DOI] [PubMed] [Google Scholar]
- 41.Michaud M, Xie X, Bravo-San Pedro JM, Zitvogel L, White E, Kroemer G. An autophagy-dependent anticancer immune response determines the efficacy of melanoma chemotherapy. OncoImmunology. 2014;3(7):e944047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Duffy A, Le J, Sausville E, Emadi A. Autophagy modulation: a target for cancer treatment development. Cancer Chemother Pharmacol. 2015;75(3):439-447. [DOI] [PubMed] [Google Scholar]
- 43.Zheng X, Seshire A, Rüster B, et al. . Arsenic but not all-trans retinoic acid overcomes the aberrant stem cell capacity of PML/RARalpha-positive leukemic stem cells. Haematologica. 2007;92(3):323-331. [DOI] [PubMed] [Google Scholar]
- 44.Wang J, Lian H, Zhao Y, Kauss MA, Spindel S. Vitamin D3 induces autophagy of human myeloid leukemia cells. J Biol Chem. 2008;283(37):25596-25605. [DOI] [PubMed] [Google Scholar]
- 45.Danilenko M, Wang X, Studzinski GP. Carnosic acid and promotion of monocytic differentiation of HL60-G cells initiated by other agents. J Natl Cancer Inst. 2001;93(16):1224-1233. [DOI] [PubMed] [Google Scholar]
- 46.Itoh T, Ohguchi K, Nozawa Y, Akao Y. Intracellular glutathione regulates sesquiterpene lactone-induced conversion of autophagy to apoptosis in human leukemia HL60 cells. Anticancer Res. 2009;29(4):1449-1457. [PubMed] [Google Scholar]
- 47.Fabre C, Carvalho G, Tasdemir E, et al. . NF-kappaB inhibition sensitizes to starvation-induced cell death in high-risk myelodysplastic syndrome and acute myeloid leukemia. Oncogene. 2007;26(28):4071-4083. [DOI] [PubMed] [Google Scholar]
- 48.Takeuchi R, Hoshijima H, Nagasaka H, et al. . Induction of non-apoptotic cell death by morphinone in human promyelocytic leukemia HL-60 cells. Anticancer Res. 2006;26(5A):3343-3348. [PubMed] [Google Scholar]
- 49.Nahimana A, Attinger A, Aubry D, et al. . The NAD biosynthesis inhibitor APO866 has potent antitumor activity against hematologic malignancies. Blood. 2009;113(14):3276-3286. [DOI] [PubMed] [Google Scholar]
- 50.Chen YJ, Huang WP, Yang YC, et al. . Platonin induces autophagy-associated cell death in human leukemia cells. Autophagy. 2009;5(2):173-183. [DOI] [PubMed] [Google Scholar]
- 51.Watson AS, Riffelmacher T, Stranks A, et al. . Autophagy limits proliferation and glycolytic metabolism in acute myeloid leukemia. Cell Death Discov. 2015:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Torgersen ML, Engedal N, Bøe SO, Hokland P, Simonsen A. Targeting autophagy potentiates the apoptotic effect of histone deacetylase inhibitors in t(8;21) AML cells. Blood. 2013;122(14):2467-2476. [DOI] [PubMed] [Google Scholar]
- 53.Fransecky L, Mochmann LH, Baldus CD. Outlook on PI3K/AKT/mTOR inhibition in acute leukemia. Mol Cell Ther. 2015;3:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Récher C, Dos Santos C, Demur C, Payrastre B. mTOR, a new therapeutic target in acute myeloid leukemia. Cell Cycle. 2005;4(11):1540-1549. [DOI] [PubMed] [Google Scholar]
- 55.Chow S, Minden MD, Hedley DW. Constitutive phosphorylation of the S6 ribosomal protein via mTOR and ERK signaling in the peripheral blasts of acute leukemia patients. Exp Hematol. 2006;34(9):1183-1191. [DOI] [PubMed] [Google Scholar]
- 56.Teachey DT, Grupp SA, Brown VI. Mammalian target of rapamycin inhibitors and their potential role in therapy in leukaemia and other haematological malignancies. Br J Haematol. 2009;145(5):569-580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Matsuura A, Tsukada M, Wada Y, Ohsumi Y. Apg1p, a novel protein kinase required for the autophagic process in Saccharomyces cerevisiae. Gene. 1997;192(2):245-250. [DOI] [PubMed] [Google Scholar]
- 58.Mizushima N. The role of the Atg1/ULK1 complex in autophagy regulation. Curr Opin Cell Biol. 2010;22(2):132-139. [DOI] [PubMed] [Google Scholar]
- 59.Gammoh N, Florey O, Overholtzer M, Jiang X. Interaction between FIP200 and ATG16L1 distinguishes ULK1 complex-dependent and -independent autophagy. Nat Struct Mol Biol. 2013;20(2):144-149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mizushima N, Yoshimori T, Ohsumi Y. The role of Atg proteins in autophagosome formation. Annu Rev Cell Dev Biol. 2011;27:107-132. [DOI] [PubMed] [Google Scholar]
- 61.Dunlop EA, Hunt DK, Acosta-Jaquez HA, Fingar DC, Tee AR. ULK1 inhibits mTORC1 signaling, promotes multisite Raptor phosphorylation and hinders substrate binding. Autophagy. 2011;7(7):737-747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lee SB, Kim S, Lee J, et al. . ATG1, an autophagy regulator, inhibits cell growth by negatively regulating S6 kinase. EMBO Rep. 2007;8(4):360-365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Murthy A, Li Y, Peng I, et al. . A Crohn’s disease variant in Atg16l1 enhances its degradation by caspase 3. Nature. 2014;506(7489):456-462. [DOI] [PubMed] [Google Scholar]
- 64.Betin VM, Lane JD. Caspase cleavage of Atg4D stimulates GABARAP-L1 processing and triggers mitochondrial targeting and apoptosis. J Cell Sci. 2009;122(Pt 14):2554-2566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhu Y, Zhao L, Liu L, et al. . Beclin 1 cleavage by caspase-3 inactivates autophagy and promotes apoptosis. Protein Cell. 2010;1(5):468-477. [DOI] [PMC free article] [PubMed] [Google Scholar]