Abstract
Although consensus is building that primary (PR) and secondary findings (SF) from genomic research should be offered to participants under some circumstances, data describing (1) actual choices of study participants and (2) factors associated with these choices are limited, hampering study planning. We conducted a cross-sectional analysis of choices made for return of PR and SF during informed consent by members of the first 247 families (790 individuals) enrolled in the Baylor-Hopkins Center for Mendelian Genomics, a genome sequencing study. Most (619; 78.3%) chose to receive SF and PR, 66 (8.4%) chose PR only, 65 (8.2%) wanted no results, and 40 (5.1%) chose SF only. Choosing SF was associated with an established clinical diagnosis in the proband (87.8 vs 79%, P=0.009) and European ancestry (EA) (87.7 vs 73%, P<0.008). Participants of non-European ancestry (NEA) were as likely as those of EA to choose SF when consented by a genetic counselor (GC) (82% NEA vs 88.3% EA, P=0.09) but significantly less likely when consented by a physician (67.4% NEA vs 85.4% EA, P=0.001). Controlling for proband diagnosis, individuals of NEA were 2.13-fold (95% CI: 1.11–4.08) more likely to choose SF when consented by a GC rather than a physician. Participants of NEA were 3-fold more likely than those of EA to decline all study results (14.7% NEA vs 5.4% EA, P<0.008). In this ethnically diverse population, whereas most participants desired PR and SF, more than 20% declined some or all results, highlighting the importance of research participant choice.
Introduction
With the expansion of exome and genome sequencing (ES/GS) for both research and clinical care, issues related to the return of ES/GS results, including secondary findings (SF) have risen to the forefront. The American College of Medical Genetics and Genomics (ACMG) defines SF as, ‘results that are not related to the indication for ordering the sequencing but that may nonetheless be of medical value or utility to the ordering physician and the patient'.1 A relatively broad policy consensus is emerging for return of results from sequencing done for clinical purposes. In 2013, the ACMG proposed a list of specific genes of medically actionable health significance to be analyzed for ‘secondary variants' in any sample on which ES/GS is performed by a clinical laboratory and recommended all pathogenic ‘secondary variants' in these genes should be offered to patients.1, 2 Broadly, these genes are those associated with hereditary cancer syndromes, inherited heart disease, connective tissue disorders predisposing to aneurysm, and malignant hyperthermia. The European Society of Human Genetics has also offered recommendations in this area. Their more general recommendations state that a healthcare professional should report ‘unsolicited genetic variants…indicative of serious health problems…that allow for treatment or prevention' in clinical genetic testing.3
In contrast, approaches to managing not only SF but also individual primary results (PR) of participants in research studies employing ES/GS remain unsettled.4, 5 In the ES/GS research context, individual PR refers to a causative genetic change found in an individual that is the basis of the condition being studied. There are limited recommendations6, 7, 8, 9, 10, 11, 12 and a lack of consensus as to best practices for managing individual genetic research results (PR and SF),13 hampering study planning.
Although studies have begun exploring research participants' preferences for return of individual ES/GS results, to date the literature is largely limited to studies reporting research participants' hypothetical preferences14 and future intentions15 to receive ES/GS results and smaller, qualitative studies exploring actual choices made.16, 17, 18 Large-cohort quantitative studies reporting actual choices have so far been limited to choices for SF in clinical settings19, 20 and choices made by a research cohort of familial cancer patients.21 The overarching theme across these studies is that research participants generally desire the return of both PR and SF. Nevertheless, this attitude is not uniform or stable.18 Furthermore, many of the studies had limited ethnic diversity among participants, which may limit their generalizability as studies suggest key differences associated with race and ethnic background in both attitudes towards genetic testing in general and the desire for genetic results.22, 23, 24
The combination of a limited literature and unsettled policy leaves researchers and Institutional Review Boards/Ethics Boards to plan studies and develop policy and practice for ES/GS research results with limited evidence or precedent to draw upon. This study is designed to help construct this evidence base by reporting actual choices for return of ES/GS research results made by the first cohort of participants enrolling in the Baylor-Hopkins Center for Mendelian Genomics (http://bhcmg.org/) via the Johns Hopkins site. The Baylor-Hopkins Center for Mendelian Genomics (henceforth referred to as the ‘Mendel Study') is funded by the National Human Genome Research Institute and the National Heart, Lung, and Blood Institute (#2UM1HG006542) and seeks to utilize ES/GS to find genetic causes of rare diseases likely to be Mendelian but of unknown molecular etiology. As part of the informed consent process, participants are given the option to receive PR related to the disease present in their family, as well as SF. In this manuscript we quantitatively report (1) Mendel Study participants' choices to receive PR and SF, (2) clinical, and demographic factors associated with these choices in this ethnically diverse population, and (3) whether choices were associated with genetic counselor vs physician consenter and parent/guardian vs participant consent.
Subjects and methods
Population and study enrollment
The study population included members of the first 247 families enrolled at the Johns Hopkins University site of the Mendel Study between November 2011 and March 2015. Adults and children with conditions suspected to have a primarily monogenic cause in whom available diagnostic testing (not including ES/GS) had not identified a causative mutation were eligible for the Mendel Study, as were their family members. As sequencing resources were limited, enrollment in the Mendel study was targeted to family members whose sample was most likely to be most informative for analysis based on clinical characteristics, pedigree, and suspected inheritance. Although Mendel Study participants were recruited from a variety of clinical specialties across our institution, all participants were consented to the study by a genetics provider (doctors, genetic counselors, or a nurse practitioner with genetics expertise). Each had received training to ensure consistency in the approach and content of the informed consent discussion.
The informed consent process, including choices for return of study results has been described previously.16, 25 Briefly, the consent process included a 20–40 min discussion during which the study purpose, ES/GS process, options for receiving PR and SF, process of clinical validation of study results, methods of sample collection, and sample and data storage were described. Informed consent of both affected individuals and family members was typically obtained in-person in association with a clinical visit. However, some participants were consented during a research visit or by telephone. At the time of consent, participants were given the option to receive PR (described in the consent form as ‘the genetic cause for the rare disorder or medical condition that led you or your family to take part in this research'), SF (described in the consent form as ‘a previously validated genetic cause for other serious medical conditions which you will likely develop as a result of a mutation we discover'), both, or neither. The consent form provided lines for participants to initial to opt in to receive each type of result. If a participant did not initial either line, he or she was recorded as choosing not to receive any results. The full consent form has been previously published.16
Data collection
Data for this study were obtained from two sources: (1) clinical, demographic, and family history data entered in the study database (PhenoDB)26 by the clinician enrolling the family in the Mendel Study and (2) data abstracted from study consent forms.
Clinical and demographic data were accessed from PhenoDB on 3/30/2015. Information in this database is input by the enrolling provider. Study variables included: sex, ancestry, clinical diagnosis if known or a phenotypic description if no diagnosis had been established, whether the participant was affected or unaffected, relationship to the proband, and suspected mode of inheritance based on the family pedigree. Ancestry categories in PhenoDB are: African, Asian, European, Native American, unknown, and ‘other' with the option to add additional details. Ancestry information was collected as part of pedigree construction. The existence of a clinical diagnosis was indicated by the entry of a Mendelian Inheritance in Man (MIM) number in that field in the database, whereas a description in the ‘unknown disorder' field indicated the lack of a clinical diagnosis.
Consent forms were manually reviewed by two members of the study team and choices to receive SF, PR, both or neither were recorded. Other data abstracted included (1) type of provider who signed the consent form documenting they had conducted the consent discussion (physician, genetic counselor, nurse practitioner) and (2) the relationship of the individual who provided consent to the study participant (an adult consenting for him/herself, a legally authorized representative (LAR) consenting for an adult, or a parent or LAR consenting for a minor child).
Statistical analysis
Categorical variables were reported as frequency (%) and compared between groups by the χ2 or Fisher's exact test. Bonferroni-corrected P-values were calculated by multiplying the individual, unadjusted P-values by 8 as 8 independent statistical tests were performed. The adjusted P-values were used to assess statistical significance and are reported in the manuscript. Univariate and multivariate logistic regression were used to identify independent predictors of choice to receive SF and PR. A P-value <0.05 was considered significant. SPSS (version 23; SPSS, Chicago, IL, USA) statistical software was used.
Results
Population
Of the 795 members of the first 247 families enrolled in the Mendel Study, 790 were eligible for inclusion in the current analysis. Five were excluded from further analysis after review of consent forms because their choices for return of study results were unclear.
As shown in Table 1, there was considerable variability in the phenotypes prompting study enrollment. Approximately half of participants (400/790; 50.6%) had a clinical diagnosis but no genetic basis for the disease had been established. The most common diagnoses were cardiovascular (22.5%), neurological/neurocutaneous (17.5%), or various syndromes associated with congenital malformations/multiple congenital anomalies (19%). For the other half of study participants (390/790; 49.4%), no clinical diagnosis had been established for the phenotype that prompted study enrollment. Congenital malformations/multiple congenital anomalies were disproportionately represented in this undiagnosed group (36.4%, P<0.001).
Table 1. Phenotype of proband that prompted study enrollment for all participants.
| Proband has a clinical diagnosis? | ||||||
|---|---|---|---|---|---|---|
| Yes | No | Total | ||||
| Category of condition | Number | Percent | Number | Percent | Number | Percent |
| Congenital malformation/multiple congenital anomalies | 76 | 19 | 142 | 36.41 | 218 | 27.59 |
| Cardiovascular | 90 | 22.50 | 38 | 9.74 | 128 | 16.20 |
| Neurological/neurocutaneous | 70 | 17.50 | 31 | 7.95 | 101 | 12.79 |
| Skeletal | 54 | 13.50 | 27 | 6.92 | 81 | 10.25 |
| Connective tissue | 24 | 6 | 14 | 3.59 | 38 | 4.81 |
| Neoplasm | 29 | 7.25 | 4 | 1.03 | 33 | 4.18 |
| Pulmonary | 0 | 0 | 32 | 8.21 | 32 | 4.05 |
| Muscular | 12 | 3 | 13 | 3.33 | 25 | 3.17 |
| Other | 10 | 2.50 | 12 | 3.08 | 22 | 2.78 |
| Dermatological | 16 | 4 | 5 | 1.28 | 21 | 2.66 |
| Gastrointestinal | 0 | 0 | 20 | 5.13 | 20 | 2.53 |
| Vision | 18 | 4.50 | 2 | 0.51 | 20 | 2.53 |
| Immune/autoimmune | 1 | 0.25 | 12 | 3.08 | 13 | 1.65 |
| No info given | 0 | 0 | 38 | 9.74 | 38 | 4.81 |
| Total | 400 | 100 | 390 | 100 | 790 | 100 |
Table 2 summarizes the demographic and clinical characteristics of study participants, consent characteristics, and choices made for return of results and presents the statistical association between clinical, demographic, and consent variables with choice to receive SF. Males and females were nearly equally represented, and the population was evenly split between affected and unaffected individuals. One-third of enrollees (30.3%) were probands, 58.7% were first-degree relatives, and the remainder were more distant relatives. The study included primarily individuals of European (white) ancestry, but nearly one-third had non-European ancestry. This included those with African (n=28), Asian (n=19), Native (North) American (n=2), Middle Eastern (n=29), and Latin American (n=98) ancestry, as well as a group for which ancestry was recorded as ‘other or unknown' (n=62). For analysis, ancestry was grouped as ‘European ancestry (EA)' and ‘non-European ancestry (NEA)' that included all other groups.
Table 2. Association of demographic, clinical, and consent variables with choices to receive genomic results at informed consent.
| Results chosen to receive | |||||||
|---|---|---|---|---|---|---|---|
| Total population | Refused secondary findings | Desired secondary findings | |||||
| Variable | # (%) | None | Primary result only | Secondary findings only | All results | P-valuea | Bonferroni-corrected P-valuea |
| Demographics | |||||||
| Sex | |||||||
| Female | 382 (48.4) | 33 (8.64) | 29 (7.59) | 14 (3.66) | 306 (80.1) | 0.848 | 1 |
| Male | 408 (51.6) | 32 (7.84) | 37 (9.07) | 26 (6.37) | 313 (76.7) | ||
| Ancestry | |||||||
| European | 552 (69.9) | 30 (5.43) | 38 (6.88) | 17 (3.08) | 467 (84.6) | <0.001 | <0.008 |
| Non-European | 238 (30.1) | 35 (14.7) | 28 (11.8) | 23 (9.66) | 152 (63.9) | ||
| Clinical characteristics | |||||||
| Relationship to proband | |||||||
| Proband | 239 (30.3) | 11 (4.60) | 35 (14.6) | 17 (7.11) | 176 (73.6) | 0.158 | 1 |
| First-degree relative | 464 (58.7) | 50 (10.8) | 26 (5.60) | 19 (4.09) | 369 (79.5) | ||
| ≥Second-degree relative | 87 (11) | 4 (4.60) | 5 (5.74) | 4 (4.60) | 74 (85.1) | ||
| Clinical status | |||||||
| Unaffected or unknown | 393 (49.7) | 46 (11.7) | 16 (4.07) | 16 (4.07) | 315 (80.2) | 0.567 | 1 |
| Affected | 397 (50.3) | 19 (4.79) | 50 (12.6) | 24 (6.05) | 304 (76.6) | ||
| Proband has a diagnosis | |||||||
| No | 390 (49.4) | 42 (10.8) | 40 (10.3) | 18 (4.62) | 290 (74.4) | 0.001 | 0.009 |
| Yes | 400 (50.6) | 23 (5.75) | 26 (6.50) | 22 (5.50) | 329 (82.3) | ||
| Inheritance pattern | |||||||
| Dominant | 294 (37.2) | 19 (6.46) | 17 (5.78) | 9 (3.06) | 249 (84.7) | <0.001 | <0.008 |
| Recessive or X-linked | 268 (33.9) | 21 (7.84) | 30 (11.2) | 10 (3.73) | 207 (77.2) | ||
| Isolated case/unknown | 228 (28.9) | 25 (11) | 19 (8.33) | 21 (9.21) | 163 (71.5) | ||
| Consent | |||||||
| Person providing consent | |||||||
| Self | 494 (62.5) | 47 (9.51) | 33 (6.68) | 23 (4.66) | 391 (79.1) | 0.767 | 1 |
| Parent or legal representative | 296 (37.5) | 18 (6.08) | 33 (11.1) | 17 (5.74) | 228 (77) | ||
| Consenter | |||||||
| Physician | 261 (33) | 31 (11.9) | 32 (12.3) | 20 (7.66) | 178 (68.2) | <0.001 | <0.008 |
| GC | 529 (67) | 34 (6.43) | 34 (6.43) | 20 (3.78) | 441 (83.4) | ||
| Total | 790 (100) | 65 (8.23) | 66 (8.35) | 40 (5.06) | 619 (78.4) | ||
P-values represent differences between those who desire secondary findings and those who do not; values in bold are significant at the 0.05 level.
Consent characteristics
Nearly two-thirds of participants (n=494, 62.5%) were adults able to provide consent for their own participation and hence made their own choices for which study results to receive. The remainder had choices made by a parent (n=287) or LAR (n=9). Two-thirds of consent discussions were conducted by either a genetic counselor (n=473) or the genetic nurse practitioner (n=56), with the remaining third conducted by a physician. For analysis, consenter was grouped into ‘doctor (MD)' and ‘genetic counselor (GC)' that included both genetic counselors and the genetic nurse practitioner.
Choices for return of results
Table 2 summarizes choices made for return of results and presents the association of clinical, demographic, and consent variables with SF choice made. The majority of participants chose to receive SF (n=659, 83.4%). Within this group, a small minority (n=40, 5.1%) declined PR and elected only SF. Among the 131 enrollees (16.6%) who declined SF, equal numbers chose to receive PR only (n=66, 8.35%) or to receive no sequencing results (n=65, 8.23%).
Predictors of SF choice
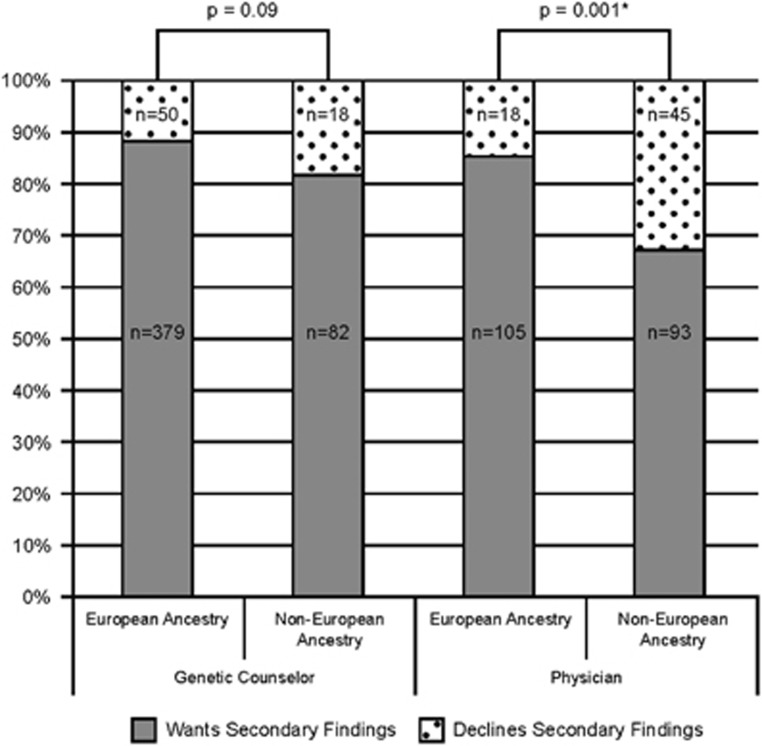
We next evaluated the association of demographic, clinical, and consent characteristics with choices to receive SF. As shown in Table 2, overall choice to receive SF was associated with having an established clinical diagnosis in the proband (87.8% with diagnosis vs 79% without, P=0.009), European ancestry (87.7 vs 73% NEA, P<0.008), and being consented by a GC (87.1 vs 75.9% MD, P<0.008), though the majority of all subpopulations elected to receive SF. There was no association of SF choice with sex, clinical status, inheritance pattern, or whether a participant vs a parent/LAR provided consent. As shown in Figure 1, there was an interaction between ancestry and consenting professional. Participants of non-European ancestry were as likely as those of EA to choose SF when a GC conducted informed consent (82% NEA vs 88.3% EA, P=0.09) but significantly less likely to choose SF when informed consent was conducted by a physician (67.4% NEA vs 85.4% EA, P=0.001).
Figure 1.
Association of choice to receive secondary findings with genetic counselor vs physician consenter. Participants of non-European ancestry were as likely as those of European ancestry to choose to receive secondary findings when a genetic counselor conducted informed consent (P=0.09) but significantly less likely to choose secondary findings when consented by a physician (P=0.001). *Statistical significance, P<0.05.
As ancestry was also associated with inheritance pattern, distribution of probands vs first degree vs more distant relatives, and proportion of participants providing consent for themselves rather than consent by a parent/LAR, we stratified the population by ancestry for further analysis. Table 3 shows the results of univariate and multivariate analysis of predictors of the choice to receive SF in the stratified population. As shown, controlling for having an established clinical diagnosis in the proband, participants of NEA were 2.13-fold (95% CI: 1.11–4.08) more likely to choose SF when a GC conducted informed consent. In contrast, the only significant predictor of choosing to receive SF among individuals of EA was being a distant relative, rather than a proband or first-degree family member (P=0.037).
Table 3. Predictors of choice to receive secondary findings based on results from univariate and multivariate logistic regression.
| European ancestry | Non-European ancestry | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Univariate | Multivariate | Univariate | Multivariate | |||||||||
| Variable | OR | 95% CI | P-value | OR | 95% CI | P-value | OR | 95% CI | P-value | OR | 95% CI | P-value |
| Demographics | ||||||||||||
| Female sex | 0.94 | (0.57–1.57) | 0.82 | — | — | — | 1.18 | (0.62–2.11) | 0.57 | — | — | — |
| Clinical characteristics | ||||||||||||
| Clinically affected | 0.90 | (0.54–1.50) | 0.69 | — | — | — | 0.76 | (0.43–1.36) | 0.35 | — | — | — |
| Relationship to probanda | — | — | 0.037 | — | — | 0.037 | — | — | 0.20 | — | — | — |
| First degree | 1.31 | (0.77–2.24) | 0.32 | 1.31 | (0.77–2.24) | 0.316 | 1.10 | (0.58–2.08) | 0.766 | — | — | — |
| ≥Second degree | 6.64 | (1.53–28.8) | 0.01 | 6.64 | (1.53–28.8) | 0.011 | 0.13 | (0.13–1.27) | 0.122 | — | — | — |
| Proband with clinical diagnosis | 1.18 | (0.71–1.97) | 0.52 | — | — | — | 4.23 | (2.26–7.92) | <0.001 | 4.16 | (2.21–7.83) | <0.001 |
| Inheritanceb | — | — | 0.30 | — | — | — | — | — | 0.33 | — | — | — |
| X-linked/autosomal recessive | 0.65 | (0.37–1.16) | 0.15 | — | — | — | 0.79 | (0.33–1.87) | 0.60 | — | — | — |
| Isolated or unknown | 0.98 | (0.50–1.91) | 0.95 | — | — | — | 0.56 | (0.24–1.31) | 0.18 | — | — | — |
| Consent | ||||||||||||
| Consenting for own enrollment | 1.14 | (0.67–1.94) | 0.62 | — | — | — | 0.78 | (0.44–1.43) | 0.45 | — | — | — |
| Consented by genetic counselor | 1.3 | (0.73–2.32) | 0.37 | — | — | — | 2.20 | (1.18–4.11) | 0.013 | 2.13 | (1.11–4.07) | 0.022 |
Bold values indicate statistical significance, P < 0.05.
Italic values denote the statistical significance of a categorical variable with multiple categories.
Proband is the reference category.
Autosomal dominant is the reference category.
Declining all study results
Although overall only 8.2% of study participants declined all study results, this choice was made by more than one in seven participants of NEA (14.7 vs 5.4% EA, P<0.008). We again stratified the population by ancestry to assess independent predictors of the decision to decline all study results. As shown in Table 4, in both the EA and NEA population, clinically affected participants were significantly less likely to decline all study results (EA P=0.038, NEA P=0.012). In addition, in the NEA population, declining all study results was associated with being a relative rather than the family proband (P=0.031), the proband not having an established diagnosis (P<0.008), and consenting for one's own enrollment (P=0.014) in univariate analysis. In multivariate analysis, only being clinically unaffected and the family proband lacking a clinical diagnosis remained significant independent predictors of the choice to decline all study results.
Table 4. Predictors of the choice to decline all study results based on results from univariate and multivariate logistic regression.
| European ancestry | Non-European ancestry | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Univariate | Multivariate | Univariate | Multivariate | |||||||||
| Variable | OR | 95% CI | P-value | OR | 95% CI | P-value | OR | 95% CI | P-value | OR | 95% CI | P-value |
| Demographics | ||||||||||||
| Female sex | 0.79 | (0.38–1.67) | 0.54 | — | — | — | 1.58 | (0.76–3.25) | 0.22 | — | — | — |
| Clinical characteristics | ||||||||||||
| Clinically affected | 0.44 | (0.20–0.96) | 0.038 | 0.44 | (0.20–0.96) | 0.038 | 0.36 | (0.16–0.80) | 0.012 | 0.21 | (0.14–0.73) | 0.007 |
| Relationship to probanda | — | — | 0.17 | — | — | — | — | — | 0.031 | — | — | — |
| First degree | 1.66 | (0.39–3.98) | 0.25 | — | — | — | 4.23 | (1.43–12.6) | 0.009 | — | — | — |
| ≥Second degree | 0.31 | (0.04–2.60) | 0.28 | — | — | — | 4.5 | (0.89–22.7) | 0.068 | — | — | — |
| Proband with clinical diagnosis | 1.03 | (0.49–2.15) | 0.93 | — | — | — | 0.21 | (0.09–0.47) | <0.001 | 0.19 | (0.081–0.44) | <0.001 |
| Inheritanceb | — | — | 0.36 | — | — | — | 0.28 | — | — | — | ||
| X-linked/autosomal recessive | 1.53 | (0.63–3.78) | 0.35 | — | — | — | 0.47 | (0.18–1.23) | 0.13 | — | — | — |
| Isolated or unknown | 1.91 | (0.77–4.70) | 0.16 | — | — | — | 0.75 | (0.30–1.90) | 0.55 | — | — | — |
| Consent | ||||||||||||
| Consenting for own enrollment | 1.21 | (0.55–2.71) | 0.63 | — | — | — | 2.75 | (1.23–6.16) | 0.014 | — | — | — |
| Consented by genetic counselor | 0.55 | (0.25–1.21) | 0.14 | — | — | — | 0.91 | (0.44–1.89) | 0.79 | — | — | — |
Abbreviations: 95 CI, 95% confidence interval; OR, odds ratio.
Proband is the reference category.
Autosomal dominant is the reference category.
Specifically declining primary results
A small group of participants (n=40, 5.1%) elected to receive SFs but refused PR. In comparison to the overall study population, these 40 participants were disproportionately male (n=26, 65%, P=0.083) and of NEA (n=23, 57.5% P<0.008). Membership in a family with/without a clinical diagnosis had no impact on likelihood of specifically declining primary results and surprisingly more than half of these individuals (n=24, 60%, P=0.206) were clinically affected. Physicians were disproportionately likely to have consented individuals who made this choice (n=20, 50%, P=0.019).
Discussion
This study is novel in that it quantitatively reports the actual choices for return of genomic research results made by members of families with suspected Mendelian disorders enrolling in an ES/GS study. Our large sample size and ethnically diverse population also allowed us sufficient power to (1) identify an important difference in desire for return of SF based on ancestry that may be modified by the professional conducting the consent discussion and (2) characterize an important subset of participants who declined to receive any study results.
Our findings that the majority of participants chose to receive both PR and SF are consistent with previous studies of preferences for return of ES/GS results including reports of choices made by patients undergoing clinical testing, qualitative analyses of participants enrolling in ES/GS research, and general population studies.15, 16, 17, 19, 20, 27, 28 The actual percentage of participants who chose or expressed a desire to receive SF has varied across studies, ranging from about 73 to 98%.15, 19, 21, 28, 29 Our finding that 83.4% of study participants chose to receive SF falls nicely within this range. However, it is lower than the percentages reported in two previous studies that quantified the desire for SF in the research setting. One study looked at 507 cancer research participants undergoing sequencing, and the other asked 311 ES/GS research participants about their intentions to receive SF prior to making the actual choice. These studies reported rates of desire for SF of 97.6 and 94.5%, respectively.15, 21 Importantly, both of these study populations were considerably less ethnically diverse than ours. A population-based survey of the desire for SF in the Australian population reported the lowest percentage of individuals desiring SF with only 73.1% of 800 individuals expressing the desire to receive SF in a hypothetical situation.28 Finally, two studies of actual choices in the clinical sequencing context, found desire for SF to be 76.1 and 93.5%.19, 20 It is important to note that in both of these studies based in clinical settings, participants had choices of various types of SF to receive, and these percentages represent only those individuals who agreed to receive all SFs.
Ancestry and choice of results
In the current study, participants of NEA were significantly more likely both to decline SF and to decline all results in general. The association of ancestry, ethnicity, or race with interest in ES/GS studies and their results has been explored previously, though not extensively. One such study found that African American participants were less likely than non-African American participants to indicate that they would participate in an ES/GS study and were similarly less likely to desire to receive any results from such a study.22 A focus-group study with underserved, culturally diverse populations found that Latino and Chinese participants had more favorable attitudes towards genetic testing than non-Hispanic whites, whereas African American participants had the least favorable attitudes.23 A 2004 telephone survey showed that although Latino and African American respondents were actually more likely than non-Hispanic whites to state that they would desire genetic testing for both treatable and untreatable conditions, these groups also believed that genetic testing would do more harm than good and held more negative beliefs about genetic testing.27 The authors of that study proposed that these negative beliefs together with fewer informational and financial resources may counteract the stated desire for testing. A study of African American, Latina, and Caucasian women in regards to genetic cancer testing showed that African American and Latina women had significantly greater medical mistrust than Caucasian women and that these groups of women were more likely than Caucasian women to agree that genetic testing is used “to show that their ethnic group is not as good as others…to interfere with the way God meant for people to be…and to interfere with the natural order of life.”24 Finally, one study showed that racial minorities had more negative emotional reactions and lower adherence intentions to genetic personalized medicine vignettes compared with conventional vignettes and as compared with the non-minorities.30 Although most of this literature is not in relation to ES/GS studies and each study includes only a subset of NEA groups, it does indicate that there are key differences in both attitudes towards genetic testing in general and the desire for genetic results among individuals of NEA as compared with the EA or non-Hispanic white individuals. Although we have not interviewed individuals in our cohort, our data suggest that there are, indeed, systematic differences in how individuals of NEA and EA view the desirability of ES/GS research results.
Individual conducting consent discussion
Our results suggest that the professional conducting the consent discussion may influence the choices made to receive SF by participants of NEA. Specifically, those of NEA consented by physicians were less likely to choose to receive SF than those consented by a GC or those of EA consented by either type of provider. We speculate that there are several potential causes of this difference. To begin, the genetic counseling profession emphasizes cultural competency, ethno-cultural sensitivity, and client-centered care as demonstrated in the practice-based competencies of the Accreditation Council for Genetic Counseling.31 Each of these aspects may allow NEA participants to feel more at ease and more likely to freely discuss their desires for results or concerns about them. This richer conversation could lead to greater clarification of values and beliefs and further understanding of the implications of SF. Possible support for this hypothesis may be seen in a qualitative study of GC and research coordinators providing consent for ES/GS studies, which demonstrated that GC were more likely than research coordinators to highlight challenging cases regarding decision-making, especially related to family dynamics or the value of sequencing for a particular participant.32 It is also possible that there are systematic differences in the attitudes of MDs and GCs toward the value of SF, which may lead to differences in what they say during the consent process. Of note, the difference between EA and NEA participants' choices by consenter was only significant for SF, not for PR, which could indicate that the role of a GC may be most important in this complex area.
The choice to decline all study results
Although the majority of participants chose to receive results, 21.6% declined to receive at least some type of genomic sequence result and a small portion of the study population declined all results. In our study, both the percentage who chose to decline some result and the percentage who declined all results was greater than in previously reported studies of intentions or actual decisions regarding ES/GS studies in the research setting.15, 21 Given the correlation between NEA and declining all results, this larger proportion that declined results in our sample may have been because of the greater diversity of ancestry in our population than in previously reported studies. It is interesting to consider why an individual would enroll in a study for which she/he actively declines all possible results. Altruism has been shown to be one motivating force among participants in ES/GS studies.33, 34 In the Mendel Study population, we have previously reported that participants are motivated to join to advance understanding of their condition and to benefit other affected families and patients in the future, in addition to any desire for personal or family benefit.35 A subset of Mendel Study participants were motivated to participate in part by pre-existing clinical relationships with study investigators. We suspect each of these reasons for enrolling might be associated with choice to enroll in the Mendel Study while also choosing to decline to receive study results.
Limitations
This study has several limitations, most of which stem from the study design. As a retrospective, cross-sectional analysis, there were limitations to the data available. For instance, ancestry information was limited to that entered by the research team at the time of enrollment. In particular, this study included both US and international participants, subsets of each of which could have been identified as NEA. However, data on which patients were international is not available, so we are unable to explore the role of nationality in choice of research results. In addition, participants were not randomized to GC or MD consenter which limits our ability to analyze the significance of the effect of provider on NEA choice of results. Finally, as consent setting (clinical visit, research visit, telephone call) was not recorded for each participant, we are unable to investigate its potential influence on PR and SF choice.
Summary and conclusions
This study has quantified the actual choices made for return of results of genomic sequencing research. In our clinically and ethnically diverse population of both affected and unaffected members of families with suspected Mendelian disease, we found that although most participants from all subpopulations desired genomic research results, an important percentage decline some or all results. Importantly, differences in choices are seen based on the ancestry of the individual and the type of healthcare professional conducting informed consent. These findings highlight areas for future research. Specifically, qualitative studies with various racial and ethnic groups exploring their thinking about genomic research may elucidate the role that ancestry and culture have in engagement with ES/GS research and help inform initiatives to enhance participation of individuals of NEA in genomic studies. In addition, a randomized-controlled trial assigning enrollees to GC or physician for consent may provide insight into the potential impact of provider on results choices, which would inform planning of future studies. Potential differences in the attitudes of MDs and GCs toward the value of SF could also be explored. Our findings also have relevant implications for policy and practice regarding ES/GS research studies. The high rate of desire of SF suggests that offers to return results from ES/GS research studies are likely to be favorably received. Although this does not mean that all studies have a duty to return results, many have suggested that a clear responsibility for offering SF exists when members of the research team also provide clinical care and when ES/GS research addresses a clinical question, as does the Mendel Study.6, 7, 10, 12 However, the fact that over one-fifth of the study population declined at least one type of result supports the current practice of this and other studies of allowing participants to choose which, if any, individual genetic research results to receive.4, 21 It is our hope that the results of this study will add to the growing information available regarding ES/GS research participants' preferences for results so as to inform future policy, research planning, and practice.
Acknowledgments
This work was partially supported by the Johns Hopkins McKusick-Nathans Institute of Genetic Medicine to complement activities of the Baylor-Hopkins Center for Mendelian Genomics that is funded by a grant from the NHGRI/NHLBI (2UM1HG006542). This research was supported in part by the National Human Genome Research Institute intramural research program (KF).
Footnotes
Amanda Bergner is currently employed by Ambry Genetics. Carolyn Applegate is a consultant for 23andMe.
References
- Green RC, Berg JS, Grody WW et al: ACMG recommendations for reporting of incidental findings in clinical exome and genome sequencing. Genet Med 2013; 15: 565–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ACMG: ACMG policy statement: updated recommendations regarding analysis and reporting of secondary findings in clinical genome-scale sequencing. Genet Med 2015; 17: 68–69. [DOI] [PubMed] [Google Scholar]
- van El CG, Cornel MC, Borry P et al: Whole-genome sequencing in health care. Eur J Hum Genet 2013; 21: 580–584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg JS, Amendola LM, Eng C et al: Processes and preliminary outputs for identification of actionable genes as incidental findings in genomic sequence data in the Clinical Sequencing Exploratory Research Consortium. Genet Med 2013; 15: 860–867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kullo IJ, Haddad R, Prows CA et al: Return of results in the genomic medicine projects of the eMERGE network. Front Genet 2014; 5: 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darnell AJ, Austin H, Bluemke DA et al: Commentary: a clinical service to support the return of secondary genomic findings in human research. Am J Hum Genet 2016; 98: 435–441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knoppers BM, Zawati MH, Sénécal K: Return of genetic testing results in the era of whole-genome sequencing. Nat Rev Genet 2015; 16: 553–559. [DOI] [PubMed] [Google Scholar]
- Knoppers BM, Avard D, Sénécal K, Zawati MH: Return of whole-genome sequencing results in paediatric research: a statement of the P3G international paediatrics platform. Eur J Hum Genet 2014; 22: 3–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabsitz RR, Mcguire A, Sharp RR et al: Ethical and practical guidelines for reporting genetic research results to study participants: updated guidelines from an NHLBI working group. Circ Cardiovasc Genet 2011; 3: 574–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarvik GP, Amendola LM, Berg JS et al: Return of genomic results to research participants: the floor, the ceiling, and the choices in between. Am J Hum Genet 2014; 94: 818–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiner C: Anticipate and communicate: Ethical management of incidental and secondary findings in the clinical, research, and direct-to-consumer contexts (December 2013 Report of the Presidential Commission for the Study of Bioethical Issues). Am J Epidemiol 2014; 180: 562–564. [DOI] [PubMed] [Google Scholar]
- Botkin JR, Belmont JW, Berg JS et al: Points to consider: ethical, legal, and psychosocial implications of genetic testing in children and adolescents. Am J Hum Genet 2015; 97: 6–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klitzman R, Appelbaum PS, Fyer A et al: Researchers' views on return of incidental genomic research results: qualitative and quantitative findings. Genet Med 2013; 15: 888–895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bollinger JM, Scott J, Dvoskin R, Kaufman D: Public preferences regarding the return of individual genetic research results: findings from a qualitative focus group study. Genet Med 2012; 14: 451–457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Facio FM, Eidem H, Fisher T et al: Intentions to receive individual results from whole-genome sequencing among participants in the ClinSeq study. Eur J Hum Genet 2012; 21: 261–265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergner AL, Bollinger J, Raraigh KS et al: Informed consent for exome sequencing research in families with genetic disease: the emerging issue of incidental findings. Am J Med Genet Part A 2014; 164: 2745–2752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sapp JC, Dong D, Stark C et al: Parental attitudes, values, and beliefs toward the return of results from exome sequencing in children. Clin Genet 2014; 85: 120–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright MF, Lewis KL, Fisher TC et al: Preferences for results delivery from exome sequencing/genome sequencing. Genet Med 2014; 16: 442–447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shahmirzadi L, Chao EC, Palmaer E, Parra MC, Tang S, Gonzalez KDF: Patient decisions for disclosure of secondary findings among the first 200 individuals undergoing clinical diagnostic exome sequencing. Genet Med 2014; 16: 395–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop C, Strong K, Dimmock D, Bishop CL: Choices of incidental findings of individuals undergoing genome wide sequencing, a single center's experience. Clin Genet 2016; 1–4. [DOI] [PubMed]
- Loud JT, Bremer RC, Mai PL et al: Research participant interest in primary, secondary, and incidental genomic findings. Genet Med 2016; 18: 1218–1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J-H, Crouch J, Jamal SM, Tabor HK, Bamshad MJ: Attitudes of African Americans toward return of results from exome and whole genome sequencing. Am J Med Genet A 2013; 161A: 1064–1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catz DS, Green NS, Tobin JN et al: Attitudes about genetics in underserved, culturally diverse populations. Commun Genet 2005; 8: 161–172. [DOI] [PubMed] [Google Scholar]
- Thompson HS, Valdimarsdottir HB, Jandorf L, Redd W: Perceived disadvantages and concerns about abuses of genetic testing for cancer risk: differences across African American, Latina and Caucasian women. Patient Educ Couns 2003; 51: 217–227. [DOI] [PubMed] [Google Scholar]
- Jurgens J, Ling H, Hetrick K et al: Assessment of incidental findings in 232 whole-exome sequences from the Baylor–Hopkins Center for Mendelian Genomics. Genet Med 2015; 17: 782–788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamosh A, Sobreira N, Hoover-Fong J et al: PhenoDB: a new web-based tool for the collection, storage, and analysis of phenotypic features. Hum Mutat 2013; 34: 566–571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Middleton A, Morley KI, Bragin E et al: Attitudes of nearly 7000 health professionals, genomic researchers and publics toward the return of incidental results from sequencing research. Eur J Hum Genet 2015; 24: 21–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleming J, Critchley C, Otlowski M, Stewart C, Kerridge I: Attitudes of the general public towards the disclosure of individual research results and incidental findings from biobank genomic research in Australia. Intern Med J 2015; 45: 1274–1279. [DOI] [PubMed] [Google Scholar]
- Vazquez LD, Kuhl EA, Shea JB et al: Age-specific differences in women with implantable cardioverter defibrillators: an international multi center study. Pacing Clin Electrophysiol 2008; 31: 1528–1534. [DOI] [PubMed] [Google Scholar]
- Butrick M, Roter D, Kaphingst K et al: Patient reactions to personalized medicine vignettes: an experimental design. Genet Med 2011; 13: 421–428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle DL, Awwad RI, Austin JC et al: 2013 Review and Update of the Genetic Counseling Practice Based Competencies by a Task Force of the Accreditation Council for Genetic Counseling. J Genet Counsel 2016; 25: 868–879. [DOI] [PubMed] [Google Scholar]
- Tomlinson AN, Skinner D, Perry DL, Scollon SR, Roche MI, Bernhardt BA: “Not Tied Up Neatly with a Bow”: professionals' challenging cases in informed consent for genomic sequencing. J Genet Couns 2016; 25: 62–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Facio FM, Brooks S, Loewenstein J, Green S, Biesecker LG, Biesecker BB: Motivators for participation in a whole-genome sequencing study: implications for translational genomics research. Eur J Hum Genet 2011; 19: 1213–1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanderson SC, Linderman MD, Suckiel SA et al: Motivations, concerns and preferences of personal genome sequencing research participants: baseline findings from the HealthSeq project. Eur J Hum Genet 2015; 24: 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergner A, Applegate C, Berrios C et al: Motivations for enrolling in genome-wide sequencing research: implications for returning results in families with inherited disease. ACMG Annu Clin Genet Meet 2014, poster 462.