Abstract
The Basque population inhabits the Franco-Cantabrian region in southwest Europe where Palaeolithic human groups took refuge during the Last Glacial Maximum. Basques have been an isolated population, largely considered as one of the most ancient European populations and it is possible that they maintained some pre-Neolithic genetic characteristics. This work shows the results of mitochondrial DNA analysis of seven ancient human remains from the Cave of Santimamiñe in the Basque Country dated from Mesolithic to the Late Roman period. In addition, we compared these data with those obtained from a modern sample of Basque population, 158 individuals that nowadays inhabits next to the cave. The results support the hypothesis that Iberians might have been less affected by the Neolithic mitochondrial lineages carried from the Near East than populations of Central Europe and revealed the unexpected presence of prehistoric maternal lineages such as U5a2a and U3a in the Basque region. Comparison between ancient and current population samples upholds the hypothesis of continuity of the maternal lineages in the area of the Franco-Cantabrian region.
Introduction
During the last years genetic markers of uniparental lineages, such as Y-chromosome and mitochondrial DNA (mtDNA), arose as suitable tools for tracking population migrations.1 Specifically, mtDNA is often used for the challenging genetic analysis of prehistoric remains.1, 2 DNA analysis of ancient remains has evidenced the arrival of some human groups from the Near East to the European continent during the Neolithic period.3, 4 Nonetheless, the effect of these humans on the pre-existing European genetic pool and how their culture expanded remains unclear.5, 6 Based on genetic data of current populations, the Franco-Cantabrian region (south-western Europe) appears as a keystone in the post-glacial settlement of the European continent.1, 7, 8, 9 Even though human remains of pre-Neolithic period are very scarce, archaeological data from this region, such as abundant cultural records of Palaeolithic hunter-gatherer human groups living during the Last Glacial Maximum, suggest that this was the most densely populated European region along the Upper Palaeolithic.10
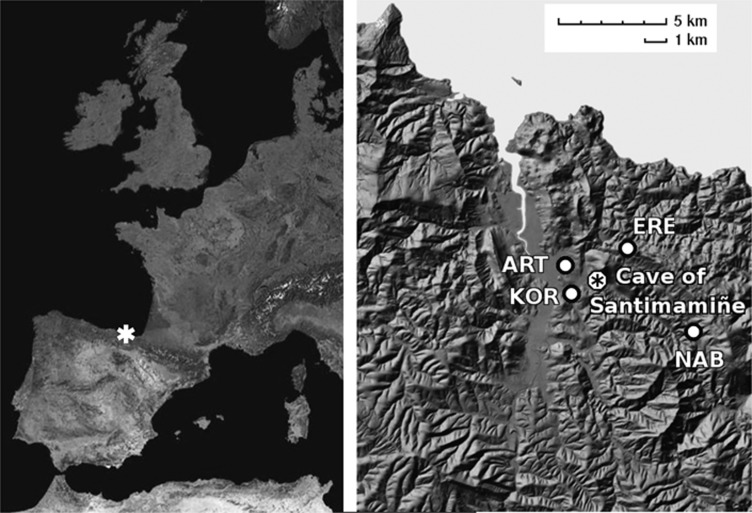
The Cave of Santimamiñe is located at the oriental side of the Oka river basin and very close to its estuary, <10 km from the current shore of Cantabrian sea, within the area of settlement of the Franco-Cantabrian refuge (Figure 1). It is a key archaeological site, well known by its valuable Palaeolithic cave art and selected as a World Heritage Site by UNESCO. Evidences of continued human occupation from the Lower Magdalenian (ca. 15 500 calibrated years before Common Era, cal BCE) to the Bronze Age (ca. 1500 cal BCE), including some sporadic occupation along Late Roman period, have been found in this cave.11 Moreover, several human remains from different historical periods have been found in the post-Palaeolithic stratigraphic levels of the Cave of Santimamiñe, the oldest one belonging to Mesolithic period.11, 12
Figure 1.
Location of the Cave of Santimamiñe in the North of the Iberian Peninsula. The Basque villages around the cave, where samples of the current autochthonous population were taken, are marked by white dots. ART, Gautegiz-Arteaga; ERE, Ereño; KOR, Kortezubi; NAB, Nabarniz.
The analysis of the DNA of human remains could contribute to unravel genetic composition of the Palaeolithic population and the human evolution of this region. In contrast with several studies focused on the analysis of the current autochthonous population, there are only a few reports on pre-Neolithic samples from the Franco-Cantabrian region.13, 14 These studies are grounded in the supposedly genetic continuity between the Palaeolithic hunter-gatherers and the autochthonous Basque population.15, 16
Herein we report a study on ancient maternal lineages of the Franco-Cantabrian region and their persistence in modern populations. It is based on the combination of ancient and modern mitochondrial DNA data. We analysed and compared mtDNA from ancient remains found in the Cave of Santimamiñe and from modern autochthonous Basque individuals (from four villages surrounding the Cave, into the region of Busturialdea, Biscay, Spain).
Materials and methods
Ancient human remains selected for this study were found in the Cave of Santimamiñe throughout numerous archaeological dig campaigns (1918–1926, 1960–1962).11 We selected seven teeth attached to the jawbone without visible signs of damage (such as crown abrasion, cracks or cavities). Isotopic analyses were performed by Beta Analytic (Miami, FL, USA). Age dates provided in this study were 2σ calibrated (95% probability). Every step involving ancient samples was carried out in laboratories exclusively dedicated to ancient DNA analysis. DNA extraction was performed by adsorption to silica gel membrane, following a protocol modified from Marshall et al.17 Hypervariable segments HVS-I and HVS-II of the mtDNA control region were amplified by PCR using five overlapping fragments of ∼175 base pairs (bp) each one (Supplementary Table S1). Samples were subjected to different additional analyses to confirm the results and to probe their authenticity, such as cloning amplified sequences, analysing haplogroup diagnostic polymorphisms located in the mtDNA coding region by different techniques, and the independent analysis of some samples by an external laboratory of Strasbourg (France).
Saliva samples were collected with mouthwash from 158 autochthonous Basques living in four villages surrounding the area of the Cave of Santimamiñe (Figure 1), a region known as Busturialdea (Biscay, Spain), following procedures in accordance with the ethical standards of the Declaration of Helsinki. DNA was extracted from saliva samples following a standard phenol–chloroform protocol.18 The entire control region of mtDNA was amplified and sequenced as described by Cardoso et al.19 In some samples coding region polymorphism determinant of H1j1 haplogroup was analysed following the same procedure, using primers shown in Supplementary Table S1. All the control region sequences of the population of Busturialdea are available online at GenBank under accession numbers KR697593—KR697750. Likewise, haplotypes have been deposited into EMPOP (http://empop.online)20 under accession number EMP00668.
Mitochondrial sequences of ancient and modern samples were compared to the revised Cambridge Reference Sequence,21 (rCRS, RefSeq NC_012920.1). Haplogroup assignment was performed according to PhyloTree Build 17 (http://www.phylotree.org).22 Multivariate analysis of ancient European populations through Principal Component Analysis (PCA) was performed using PAST v3.01.23 Population statistics were calculated with Arlequin v3.5.24 Software SplitsTree v425 was used to draw the neighbour-joining tree which represented the relation between the haplotypes of modern and ancient samples, and the median-joining network was performed with software Network v5 (http://www.fluxus-engineering.com),26 which also calculated the time elapsed between ancient and modern lineages.
A more detailed explanation of the methods employed in this study can be found on Appendix (Supplementary Appendix).
Results and discussion
Isotopic analysis of ancient samples from Santimamiñe
Seven human remains from the Cave of Santimamiñe were studied. In order to establish their antiquity and obtain information about their diet these remains were radiocarbon dated and subjected to the analysis of stable isotopes of carbon 13C/12C (δ13C) and nitrogen 15N/14N (δ15N; Table 1, Supplementary Dataset S1 and Supplementary Appendix).
Table 1. Isotopic data of the samples from the Cave of Santimamiñe analysed in this study, the cultural period to which they presumably belong and their corresponding mtDNA haplotypes and haplogroups.
| Stable isotopes | 14C Dating | Haplotypea | |||||
|---|---|---|---|---|---|---|---|
| Sampleb | δ13C (‰) | δ15N (‰) | 2σ Calibration | Period | HVS-I (16 159–16 406) | HVS-II (12-408) | Haplogroup |
| S.15I | −16.1 | 10.6 | 240–390 cal CE | Late Roman | rCRS | 214G, 263G, 315.1C | H |
| S.16G | −19.4 | 8.9 | 1450–1380 cal BCE, 1340–1320 cal BCE | Bronze Age | 16343G, 16390A | 73G, 150T, 263G, 309.1C, 315.1C | U3a |
| S.I.147 | −19.9 | 8.8 | 1600–1570 cal BCE, 1560–1550 cal BCE, 1540–1440 cal BCE | Bronze Age | 16299G, 16311C | 263G, 309.1C, 309.2C, 315.1C | R0 |
| S.7J | −19.4 | 8.6 | 1740–1610 cal BCE | Bronze Age | 16192T, 16270T | 73G, 146C, 150T, 263G, 309.1C, 315.1C | U5b |
| S.11N | −19.8 | 9.2 | 1740–1610 cal BCE | Bronze Age | rCRS | 263G, 309.1C, 315.1C | H1 |
| S.17G | −18.8 | 10.4 | 2200–2010 cal BCE, 2000–1980 cal BCE | Chalcolithic | 16185T, 16294T, 16296T, 16304C, 16311C, 16366T | Not analysed | T2b |
| S.12N | −16.3 | 10.4 | 5210–4950 cal BCE | Mesolithic | (16114A)c, 16192T, 16256T, 16270T, 16294T | 54A, 73G, 263G, 309.1C, 315.1C | U5a2a |
Abbreviations: BCE, years before Common Era; mtDNA, mitochondrial DNA; rCRS, revised Cambridge Reference Sequence.
The polymorphisms showed are changes referred to rCRS.21
Sample names noted in this table, as well as those used in all the text, are the shortened names used in our laboratory. In Supplementary Table S1 the relation between each laboratory sample name and its archaeological label is shown.
The polymorphism in brackets was revealed through the authentication analyses performed in the laboratory of Strasbourg (France), since it is out of the region analysed in our laboratory.
The analysis of the 14C radioactive isotope dated the antiquity of these remains in a wide period of time, ranging from 5210 cal BCE to 390 calibrated years of Common Era (cal CE). Data from four of these individuals were comprised in a short time period, between 1740 and 1320 cal BCE.
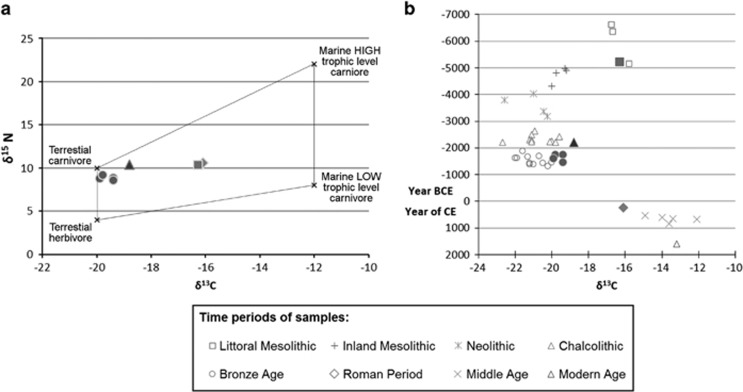
The oldest sample found in the Cave of Santimamiñe (S.12N) was dated in the transition between the 5th and 6th millennia cal BCE, which is a critical time period due to the controversy that exists around the arrival of Neolithic period to the Franco-Cantabrian region.27, 28 In this case, a more thorough analysis of the stable isotopes of carbon and nitrogen, combined with 14C, made possible to infer the cultural period to which individual S.12N belongs (Figure 2). Values of δ13C and δ15N of sample S.12N were consistent with a marine diet mainly based on invertebrates (Figure 2a), which was characteristic of Mesolithic groups living close to the littoral (Figure 2b). Thus, although the presence of Neolithic groups along Franco-Cantabrian region at the same time could not be discarded, the oldest individual from the Cave of Santimamiñe showed a Mesolithic diet, probably indicating the persistence of this subsistence way of life in the area at least until 5210–4950 cal BCE.
Figure 2.
Ratios of carbon and nitrogen stable isotopes of the ancient remains from the Cave of Santimamiñe. (a) Distribution of the samples into the four (theoretical) extreme human dietary types as Richards and Hedges46 based on their stable isotope content. (b) Evolution of δ13C content in human remains from the Cantabrian region over time (age of the samples is based on 14C dating, without calibration) modified from Arias47 to include samples from our study.
On the contrary, the values of δ13C and δ15N of the remaining samples of Santimamiñe matched with the type of diets that would be supposed to the cultural periods in which each sample was dated by 14C (Figure 2).
Authenticity of ancient DNA results
Beside the laboratory strict conditions and precautions followed during the manipulation and analysis of these samples, the genetic results obtained from the ancient remains of Santimamiñe were authenticated through diverse analyses (Supplementary Dataset S1). Samples with sufficient tooth powder (57.1%) were subjected to two independent DNA extractions, and the concordance of the control region sequences obtained was confirmed. Cloning of the amplified products and duplicated analysis by an external laboratory was performed over almost a third of the samples as proposed by Cooper and Poinar.29 Finally, the analysis of coding region SNPs, through different methodologies (direct sequencing, minisequencing or MALDI-TOF mass spectrometry analysis), was carried out to corroborate the haplogroup obtained by control region analysis on 71.4% of the samples.
Altogether, the results of all samples, except for S.17G, were confirmed by one or more additional analyses. Moreover, results of cloning (Supplementary Dataset S2) and analyses performed on the external laboratory were consistent with our results, and showed no trace of contamination. In the same way, none of the haplotypes obtained from the human remains of Santimamiñe was coincident with the haplotypes of the researchers, except for the sample S.11N, which shared mitochondrial lineage with one researcher. However, bearing in mind that it is the most common haplotype among the current European population and that the other authentication analyses corroborated the result of this ancient sample, contamination along the analysis process was discarded.
Mitochondrial diversity in the ancient remains from the Cave of Santimamiñe
First one human jawbone was selected for a pilot analysis (S.17G) with the aim of determining the inhibition effect on the DNA extracted from Santimamiñe remains and their suitability for genetic analyses. As positive results were obtained, six additional human remains were analysed.
The quantification by qPCR of the DNAs extracted showed very low amounts of nuclear DNA (Supplementary Dataset S1), even though, it was possible to amplify mtDNA in all the samples.
Sequencing of 645 bp of mtDNA control region was carried out with five overlapping fragments: two fragments for HVS-I and three fragments for HVS-II. The unique exception was sample S.17G, where only HVS-I was sequenced, due to the scarce DNA recovered from this remain and the previous tests carried out in the pilot analysis. The analysis of HVS-II usually does not go beyond 73 A/G position in ancient DNA studies, but in our study, the analysis of the whole segment provided valuable information about each sample. The analysis of these sequences resulted in seven different mitochondrial haplotypes, showing that all the remains included in this study belonged to different maternally unrelated individuals (Table 1).
The diversity of haplotypes is also shown at the haplogroup level. A complete analysis of HVS-II allowed detecting some private polymorphisms of the haplotypes that, not only made possible to differentiate the maternal lineage of all the samples, but, in some cases, it was also helpful to make a more accurate phylogenetic classification. Most of the lineages belonged to R0 and U macrohaplogroups, with three individuals each, being T2b the only linage of JT macrohaplogroup found in the cave.
Overall, genomic and isotopic data highlight that the most ancient haplogroup found in the Cave of Santimamiñe is U5a2a, belonging to the individual S.12N. This finding pointed to the presence of this haplogroup in the northern fringe of the current Basque Country at least 7000 years ago.
Phylogenetic history
Lineages H1, T2b and U5b were observed in Santimamiñe between 2200 and 1610 cal BCE. All of them have been found widely distributed along the European prehistory, with several human remains scattered through this continent.2, 30, 31, 32 These three subhaplogroups have the highest frequencies in the extant European populations. Strikingly, with the exception of T2b, whose frequency peak is located in the North of Italy,33 subhaplogroups H1 and U5b reach their highest frequencies in the North of the Iberian Peninsula, specifically in the region where the Franco-Cantabrian refuge was settled during the Upper Palaeolithic.8, 34, 35
Two subhaplogroups found in Santimamiñe, U5a2a and U3a, are scarce in prehistoric remains and virtually absent in current European populations. However, these haplogroups of the samples S.12N and S.16G are of great interest because they are the most ancient evidence of the presence of subhaplogroups U5a2a and U3a found to date in Western Europe (Supplementary Appendix).
Therefore, these findings show that subhaplogroup U3a, which probably arose in the Near East and entered the European continent with Neolithic migrations,36 had already reached the western area of the European continent in the Bronze Age, about 3300 years ago. On the other hand, U5a2a was present in a Mesolithic population from Western Europe 7000 years ago, around its first appearance in Central Europe,3 even though nowadays U5a2a is only found at very low frequencies in central and northern Europe.37, 38, 39
Prehistoric European genetic context
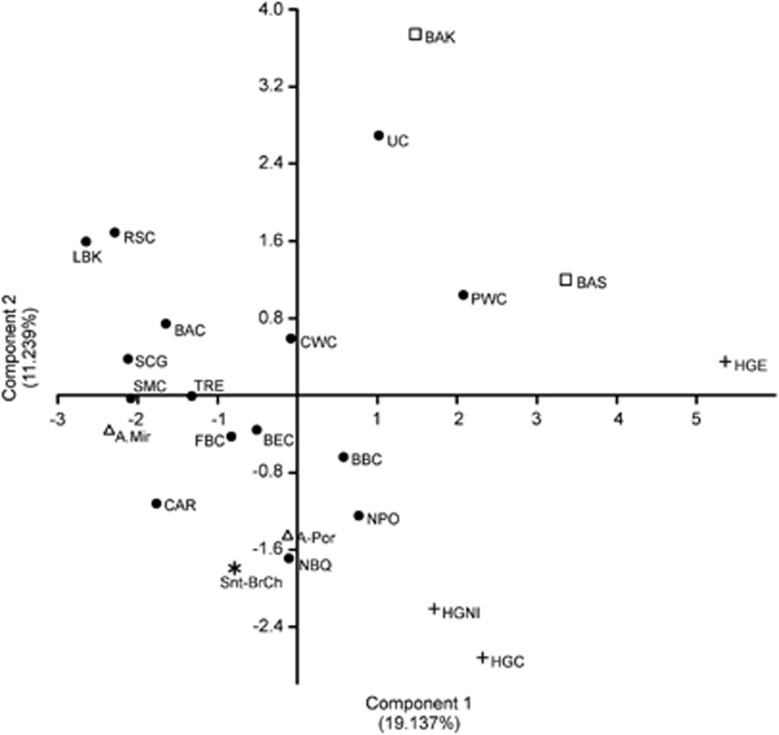
The contextualisation of the human remains of the Cave of Santimamiñe into the European prehistory is not a simple issue due to the different antiquity of these samples. Therefore, four individuals of Santimamiñe, which dated into a relatively narrow period of time, ranging from Chalcolithic to Bronze Age, were grouped together (Snt-BrCh) to study their phylogenetic link with other prehistoric European remains (Supplementary Dataset S3). On the other hand, Mesolithic sample from Santimamiñe, S.12N, was joined to previously studied pre-Neolithic/hunter-gatherer remains from the northern Iberian Peninsula (HGNI).13, 14 A PCA was performed with haplogroup frequencies of European populations from different prehistoric periods (Figure 3, Supplementary Dataset S3).
Figure 3.
Graphic representation of the PCA of European prehistoric populations based on their haplogroup frequencies. Each population is represented by different symbols. The symbol refers to the cultural period being cross for hunter-gatherers and pre-Neolithic groups; circle for Neolithic populations; triangle for Chalcolithic period and square for Bronze Age. The group of samples from Santimamiñe of Chalcolithic and Bronze Age is represented by an asterisk. Data are shown in Supplementary Dataset S3.
The graphic shows that pre-Neolithic individuals of the Northern Iberian Peninsula (HGNI), including S.12N, spread similarly to hunter-gatherers of Central Europe (HGC). The group formed by Chalcolithic and Bronze Age remains of Santimamiñe appeared closer to other Iberian populations, and specially, to those from the North of the Iberian Peninsula, such as the Neolithic group of Basque Country and Navarre (NBQ) and the Chalcolithic group of El Portalón in Atapuerca (A-Por). However, remaining population groups of Central and Eastern Europe showed scarce genetic relationship with Chalcolithic and Bronze Age individuals from Santimamiñe. Indeed, every group from the Iberian Peninsula spreads similarly, close to the negative axis of the second component, except for El Mirador from Atapuerca (A-Mir), which shows quite different haplogroup frequencies, and is placed near to other Central European population groups.
Overall, pre-Neolithic group of North Iberia showed a close relation with most of the chronologically later groups of the Iberian Peninsula, while, on the contrary, on Central Europe hunter-gatherers appeared more distant from Neolithic and following Central European populations. Biplot PCA (Supplementary Figure S1) showed that this proximity between populations of different periods of Northern and Western Iberia was due to a higher persistence of U and U5b haplogroups in this region even after the arrival of Neolithic culture. This could confirm that Iberian populations, especially those distant from Mediterranean coastal regions, were less affected by the Neolithic mitochondrial lineages carried from Near East than population groups of Central Europe, as it was previously pointed by Brand et al.5
Current population of Busturialdea
The phylogeographic nature of mtDNA is very useful to trace the movements of human groups based on the frequency distribution of haplogroups in current populations and to infer their origin. However, to find the place of origin of more precise subhaplogroups is not easy. Nevertheless, there are some exceptions on lineages that have remained in isolated populations or in populations from regions that have not been influenced by great migration waves through the history, and thus, they have preserved their characteristic lineages over time. Autochthonous Basque population of Franco-Cantabrian region is one of these exceptional populations. It preserves autochthonous subhaplogroups rarely found out of this region, some of which have been described in the last years.15, 16, 34, 40
With the aim of studying the presence of the ancient mitochondrial lineages from past times in the present, the control region of mtDNA was analysed in a sample of 158 autochthonous Basque individuals belonging to the population inhabiting the vicinity of the Cave of Santimamiñe, the region of Busturialdea (Figure 1,Supplementary Appendix). MtDNA haplotypes (Supplementary Dataset S4), haplogroup frequencies (Supplementary Table S2) and descriptive statistics (Supplementary Table S3) of this population can be accessed in the Supplementary Material.
In this population the most remarkable lineages found were those of subhaplogroups H1j1 and U5b1f1a. The two most frequent haplotypes belong to these subhaplogroups, which have been suggested as autochthonous lineages of the Franco-Cantabrian region. Subhaplogroup H1j1, which only presents the polymorphism 16129A in the control region, was described by Behar et al.15 as an autochthonous haplogroup of the Franco-Cantabrian region.15, 16 This haplotype has been found with a remarkable frequency among numerous populations of the North of the Iberian Peninsula, especially in the Basque area, or in populations of Basque ancestry.15, 35, 41, 42 In this population sample of Busturialdea 18 individuals (11.39%) were classified into H1j1 subhaplogroup by the analysis of the coding region polymorphism T4733C.
Subhaplogroup U5b1f1a, with a frequency of 5.06% in Busturialdea, is also remarkable. It represents 10% of the mitochondrial diversity of the current populations of the Franco-Cantabrian region, reaching a maximum frequency of ∼24% in the populations next to the Pyrenees and decreasing to the West.16 Due to its frequency distribution pattern and its absence in populations out of the Franco-Cantabrian region, U5b1f1a was proposed as an autochthonous lineage of the Basque population, which could have originated during the Younger Dryas (12 800–11 500 years ago), with a splitting age of 11 985 years.16
Comparison between ancient individuals of Santimamiñe and current population of Busturialdea
To determine the presence of any kinship between past and present populations from Busturialdea, haplotypes of ancient samples from the Cave of Santimamiñe and current population of the area were studied all together by means of a neighbour-joining tree (Supplementary Figure S3). Even if they do not share the same haplotypes, the tree confirmed that these populations are not completely unlinked since their lineages gather similarly, belonging to common phylogenetic branches. Indeed, the comparison at the haplogroup level between samples of our study showed that most of the subhaplogroups found in the population that nowadays inhabits the vicinity of Santimamiñe were already present in the populations that passed by the cave in prehistoric times, to be named, H1, T2b and U5b.
The difficulty of finding the same haplotypes of the ancient human remains in current populations has been addressed in previous studies of ancient DNA.30 This fact could be due to the extinction of the ancient lineages throughout history, but also, to the high mutation rate that characterises mtDNA, and specially the control region.43 This high ability to change could modify the ancient haplotype with new polymorphisms or through back mutation of the previous ones, doing improbable to find an exact match between ancient and modern lineages. According to the mutation rate of regions HVS-I and HVS-II calculated by Rieux et al.,44 which is calibrated with aDNA samples, the time elapsed would suppose the appearance of up to 1.7 mutations on the lineages of Santimamiñe until today.
By this reason, an alignment analysis was performed with the haplotypes from the Cave of Santimamiñe that belong to the same phylogenetic branch but had incomplete match with lineages of current population of Busturialdea, such as R0 (without HV0), U5a, U5b and T2 (Supplementary Dataset S5). Almost 80% of the samples from Busturialdea included in this alignment presented between one and three differences against those of Santimamiñe (Supplementary Figure S2). According to the mutation rate previously mentioned, direct descendants could present up to two differences, which is fulfilled in more than 50% of them.
Moreover, a median-joining network was performed (Supplementary Figure S4a) including samples from the Cave of Santimamiñe, the modern population of Busturialdea and pre-Neolithic individuals from the Northern Iberian Peninsula.13, 14 The samples selected for this analysis were those belonging to the phylogenetic branch of haplogroup U5, since it is the most represented on pre-Neolithic period and some of its subhaplogroups are highly frequent in current Basque population, what has been seen as a sign of genetic continuity in this region.16, 41, 45 The estimated time between the selected different ancestors and their respective descendents is close to the time elapsed between these dated individuals (Supplementary Figure S4), supporting the possible genetic link among ancient and current populations (Supplementary Appendix).
Thus, even if ancient remains analysed in this study cannot be considered as a population in itself and the phylogenetic link between ancient and current population has been probably diluted over time, the findings of this study might uphold the hypothesis of continuity.
In conclusion, the uncovering of lineages that inhabited this region in the prehistory, and especially before the arrival of Neolithic, shed light over its ancient populations and brings us closer to the disentanglement of the population that sheltered in the Franco-Cantabrian region. This study revealed the presence of a new mitochondrial haplotype on the pre-Neolithic northern Iberian Peninsula. In addition, DNA analysis of Chalcolithic and Bronze Age samples of Santimamiñe contributed to place the temporal context of the Near Eastern mitochondrial haplogroups that arrived to the Northern Iberian Peninsula with the Neolithic wave, and their effect over previous populations.
On the other hand, the results obtained from the comparison between the ancient samples from Santimamiñe and current autochthonous Basque population from Busturialdea are in line with the existence of continuity of the maternal lineages in the area of the Franco-Cantabrian region.
Acknowledgments
We are deeply indebted to all the people from the villages surrounding the Cave of Santimamiñe who voluntarily participated in this study, as well as to the Museum of Archaeology of Biscay, who has kindly provided the ancient samples. We are grateful to PhD Lourdes Basabe-Desmonts for reviewing this manuscript, to PhD Maite Álvarez for her human and technical assistance provided on the DNA Bank Service (SGIker) of the University of the Basque Country (UPV/EHU, European funding: ERDF and ESF). This study was funded by the Basque Government (Grupo Consolidado IT-424-07 and IT-833-13), the Provincial Council of Biscay and the University of the Basque Country UPV/EHU (ELDUNANOTEK UFI11/32). Leire Palencia-Madrid was granted by the Basque Government (Dpto. de Educación, Universidades e Investigación) during the execution of this study.
Footnotes
Supplementary Information accompanies this paper on European Journal of Human Genetics website (http://www.nature.com/ejhg)
The authors declare no conflict of interest.
Supplementary Material
References
- Soares P, Achilli A, Semino O et al: The archaeogenetics of Europe. Curr Biol 2010; 20: R174–R183. [DOI] [PubMed] [Google Scholar]
- Lacan M, Keyser C, Crubezy E, Ludes B: Ancestry of modern Europeans: contributions of ancient DNA. Cell Mol Life Sci 2013; 70: 2473–2487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bramanti B, Thomas MG, Haak W et al: Genetic discontinuity between local hunter-gatherers and central Europe's first farmers. Science 2009; 326: 137–140. [DOI] [PubMed] [Google Scholar]
- Fu Q, Rudan P, Pääbo S, Krause J: Complete mitochondrial genomes reveal neolithic expansion into Europe. PLoS One 2012; 7: e32473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandt G, Szecsenyi-Nagy A, Roth C, Alt KW, Haak W: Human paleogenetics of Europe—the known knowns and the known unknowns. J Hum Evol 2015; 79: 73–92. [DOI] [PubMed] [Google Scholar]
- Posth C, Renaud G, Mittnik A et al: Pleistocene mitochondrial genomes suggest a single major dispersal of non-africans and a late glacial population turnover in Europe. Curr Biol 2016; 26: 827–833. [DOI] [PubMed] [Google Scholar]
- Torroni A, Bandelt HJ, Macaulay V et al: A signal, from human mtDNA, of postglacial recolonization in Europe. Am J Hum Genet 2001; 69: 844–852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Achilli A, Rengo C, Magri C et al: The molecular dissection of mtDNA haplogroup H confirms that the Franco-Cantabrian glacial refuge was a major source for the European gene pool. Am J Hum Genet 2004; 75: 910–918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira L, Richards M, Goios A et al: High-resolution mtDNA evidence for the late-glacial resettlement of Europe from an Iberian refugium. Genome Res 2005; 15: 19–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamble C, Davies W, Pettitt P, Hazelwood L, Richards M: The archaeological and genetic foundations of the European population during the Late Glacial: implications for ‘agricultural thinking'. Camb Archaeol J 2005; 15: 193–223. [Google Scholar]
- López Quintana JC, Guenaga Lizasu A: Revisión estratigráfica del depósito arqueológico de la Cueva de Santimamiñe (Kortezubi, Bizkaia): campaña de 2004 a 2006. KOBIE 2011; 1: 7–70. [Google Scholar]
- Aranzadi Unamuno Td, Barandiarán JM Exploraciones en la caverna de Santimamiñe (Basondo: Cortézubi): 3a. Memoria, yacimientos azilienses y paleolíticos; Exploraciones en la caverna de Lumentxa (Lequeitio). Bilbao, 1935..
- Hervella M, Izagirre N, Alonso S et al: Ancient DNA from hunter-gatherer and farmer groups from Northern Spain supports a random dispersion model for the Neolithic expansion into Europe. PLoS One 2012; 7: e34417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Quinto F, Schroeder H, Ramirez O et al: Genomic affinities of two 7000-year-old Iberian hunter-gatherers. Curr Biol 2012; 22: 1494–1499. [DOI] [PubMed] [Google Scholar]
- Behar DM, Harmant C, Manry J et al: The Basque paradigm: genetic evidence of a maternal continuity in the Franco-Cantabrian region since pre-Neolithic times. Am J Hum Genet 2012; 90: 486–493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardoso S, Valverde L, Alfonso-Sanchez MA et al: The expanded mtDNA phylogeny of the Franco-Cantabrian region upholds the pre-neolithic genetic substrate of Basques. PLoS One 2013; 8: e67835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall PL, Stoljarova M, Schmedes SE, King JL, Budowle B: A high volume extraction and purification method for recovering DNA from human bone. Forensic Sci Int Genet 2014; 12: 155–160. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T: Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory: New York, NY, USA, 1989. [Google Scholar]
- Cardoso S, Zarrabeitia MT, Valverde L, Odriozola A, Alfonso-Sanchez MA, de Pancorbo MM: Variability of the entire mitochondrial DNA control region in a human isolate from the Pas Valley (northern Spain). J Forensic Sci 2010; 55: 1196–1201. [DOI] [PubMed] [Google Scholar]
- Parson W, Dür A: EMPOP—a forensic mtDNA database. Forensic Sci Int Genet 2007; 1: 88–92. [DOI] [PubMed] [Google Scholar]
- Andrews RM, Kubacka I, Chinnery PF, Lightowlers RN, Turnbull DM, Howell N: Reanalysis and revision of the Cambridge reference sequence for human mitochondrial DNA. Nat Genet 1999; 23: 147. [DOI] [PubMed] [Google Scholar]
- van Oven M, Kayser M: Updated comprehensive phylogenetic tree of global human mitochondrial DNA variation. Hum Mutat 2009; 30: E386–E394. [DOI] [PubMed] [Google Scholar]
- Hammer Ø, Harper D, Ryan P: PAST-Palaeontological statistics software package for education and data analysis. Palaeontol Electron 2001; 4: 9. [Google Scholar]
- Excoffier L, Lischer HE: Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Mol Ecol Resour 2010; 10: 564–567. [DOI] [PubMed] [Google Scholar]
- Huson DH, Bryant D: Application of phylogenetic networks in evolutionary studies. Mol Biol Evol 2006; 23: 254–267. [DOI] [PubMed] [Google Scholar]
- Bandelt HJ, Forster P, Röhl A: Median-joining networks for inferring intraspecific phylogenies. Mol Biol Evol 1999; 16: 37–48. [DOI] [PubMed] [Google Scholar]
- González Morales MR, Straus LG, Diez Castillo A, Ruiz Cobo J: La costa y el interior en la Época Postglacial: las Transiciones Epipaleolítico-Mesolítico-Neolítico en la Región Vasco-Cantábrica. Munibe Antropol Arkeol 2004; 56: 61–78. [Google Scholar]
- Alday Ruiz A: El final del Mesolítico y los inicios del Neolítico en la Peínsula Ibérica: cronología y fases. Munibe Antropol Arkeol 2009; 60: 157–173. [Google Scholar]
- Cooper A, Poinar HN: Ancient DNA: do it right or not at all. Science 2000; 289: 1139. [DOI] [PubMed] [Google Scholar]
- Brotherton P, Haak W, Templeton J et al: Neolithic mitochondrial haplogroup H genomes and the genetic origins of Europeans. Nat Commun 2013; 4: 1764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandt G, Haak W, Adler CJ et al: Ancient DNA reveals key stages in the formation of central European mitochondrial genetic diversity. Science 2013; 342: 257–261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunther T, Valdiosera C, Malmstrom H et al: Ancient genomes link early farmers from Atapuerca in Spain to modern-day Basques. Proc Natl Acad Sci USA 2015; 112: 11917–11922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pala M, Olivieri A, Achilli A et al: Mitochondrial DNA signals of late glacial recolonization of Europe from near eastern refugia. Am J Hum Genet 2012; 90: 915–924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarez-Iglesias V, Mosquera-Miguel A, Cerezo M et al: New population and phylogenetic features of the internal variation within mitochondrial DNA macro-haplogroup R0. PLoS One 2009; 4: e5112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardoso S, Alfonso-Sanchez MA, Valverde L et al: The maternal legacy of Basques in northern navarre: new insights into the mitochondrial DNA diversity of the Franco-Cantabrian area. Am J Phys Anthropol 2011; 145: 480–488. [DOI] [PubMed] [Google Scholar]
- Richards M, Macaulay V, Hickey E et al: Tracing European founder lineages in the Near Eastern mtDNA pool. Am J Hum Genet 2000; 67: 1251–1276. [PMC free article] [PubMed] [Google Scholar]
- Hedman M, Brandstätter A, Pimenoff V et al: Finnish mitochondrial DNA HVS-I and HVS-II population data. Forensic Sci Int 2007; 172: 171–178. [DOI] [PubMed] [Google Scholar]
- Tillmar AO, Coble MD, Wallerström T, Holmlund G: Homogeneity in mitochondrial DNA control region sequences in Swedish subpopulations. Int J Legal Med 2010; 124: 91–98. [DOI] [PubMed] [Google Scholar]
- Lembring M, van Oven M, Montelius M, Allen M: Mitochondrial DNA analysis of Swedish population samples. Int J Legal Med 2013; 127: 1097–1099. [DOI] [PubMed] [Google Scholar]
- Gómez-Carballa A, Olivieri A, Behar DM, Achilli A, Torroni A, Salas A: Genetic continuity in the Franco-Cantabrian region: new clues from autochthonous mitogenomes. PLoS One 2012; 7: e32851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alfonso-Sanchez MA, Cardoso S, Martinez-Bouzas C et al: Mitochondrial DNA haplogroup diversity in Basques: a reassessment based on HVI and HVII polymorphisms. Am J Hum Biol 2008; 20: 154–164. [DOI] [PubMed] [Google Scholar]
- Cardoso S, Villanueva-Millán MJ, Valverde L et al: Mitochondrial DNA control region variation in an autochthonous Basque population sample from the Basque Country. Forensic Sci Int Genet 2012; 6: e106–e108. [DOI] [PubMed] [Google Scholar]
- Vigilant L, Pennington R, Harpending H, Kocher TD, Wilson AC: Mitochondrial DNA sequences in single hairs from a southern African population. Proc Natl Acad Sci USA 1989; 86: 9350–9354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieux A, Eriksson A, Li M et al: Improved calibration of the human mitochondrial clock using ancient genomes. Mol Biol Evol 2014; 31: 2780–2792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malyarchuk B, Derenko M, Grzybowski T et al: The peopling of Europe from the mitochondrial haplogroup U5 perspective. PLoS One 2010; 5: e10285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards MP, Hedges REM: Stable isotope evidence for similarities in the types of marine foods used by late mesolithic humans at sites along the atlantic coast of Europe. J Archaeol Sci 1999; 26: 717–722. [Google Scholar]
- Arias Cabal P: Determinaciones de isótopos estables en restos humanos de la región Cantábrica: aportación al estudio de la dieta de las poblaciones del Mesolítico y el Neolítico. Munibe Antropol Arkeol 2005; 57: 359–374. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.